Abstract
Proteins can be engineered for synthetic biology through circular permutation, a sequence rearrangement where native protein termini become linked and new termini are created elsewhere through backbone fission. However, it remains challenging to anticipate a protein’s functional tolerance to circular permutation. Here, we describe new transposons for creating libraries of randomly circularly permuted proteins that minimize peptide additions at their termini, and we use transposase mutagenesis to study the tolerance of a thermophilic adenylate kinase (AK) to circular permutation. We find that libraries expressing permuted AK with either short or long peptides amended to their N-terminus yield distinct sets of active variants and present evidence that this trend arises because permuted protein expression varies across libraries. Mapping all sites that tolerate backbone cleavage onto AK structure reveals that the largest contiguous regions of sequence that lack cleavage sites are proximal to the phosphotransfer site. A comparison of our results with a range of structure-derived parameters further showed that retention of function correlates to the strongest extent with the distance to the phosphotransfer site, amino acid variability in an AK family sequence alignment, and residue-level deviations in superimposed AK structures. Our work illustrates how permuted protein libraries can be created with minimal peptide additions using transposase mutagenesis, and they reveal a challenge of maintaining consistent expression across permuted variants in a library that minimizes peptide additions. Furthermore, these findings provide a basis for interpreting responses of thermophilic phosphotransferases to circular permutation by calibrating how different structure-derived parameters relate to retention of function in a cellular selection.
Introduction
Genomic rearrangements that alter protein length (gene duplication, fission, and fusion) underlie the evolution of circularly permuted proteins in nature1-4, sets of related tertiary structures that are encoded by different arrangements of primary structure. Unfortunately, bioinformatics approaches used to discover circularly permuted proteins have not provided a complete understanding of tolerance to permutation because they utilize sequences and structures after natural selection has occurred5-7. Combinatorial experiments represent an alternative strategy to study how circular permutation influences protein function. Selections of libraries encoding circularly permuted variants of natural proteins have revealed sequence elements that are critical for folding, stability, and activity8,9, and they have shown that protein activity can be improved through circular permutation10-13. Combinatorial experiments have also been used to create protein switches for synthetic biology by randomly inserting circularly permuted variants of natural proteins into other protein domains14,15. While these studies have demonstrated the value of circular permutation for protein design and synthetic biology, they have not yet revealed how to reliably enrich libraries in functional permuted proteins16,17.
Within a cell, a protein’s tolerance to circular permutation can be disrupted through multiple mechanisms. Permuted protein translation and total enzyme activity could at times be attenuated as permuted genes present an unfavorable genetic context to a ribosomal binding site (RBS). Studies examining the effect of genetic context on RBS strength have shown that translation initiation can vary by orders of magnitude when protein-coding sequences present varying genetic contexts to an RBS18. Changes in protein contact order arising from permutation can also decrease protein stability19,20, alter folding21,22, and inhibit the formation of native residue-residue contacts required for activity23,24. Furthermore, the new termini created by permutation can generate structural incompatibilities that disrupt activity. Some library synthesis methods attach peptides to the new termini25, creating bulk that must be accommodated in the structure, while other methods delete residues26. The extent to which each of these mechanisms shapes protein tolerance to random circular permutation within a cellular selection remains challenging to anticipate. This challenge arises because the minimal activity needed to support cellular growth can be disrupted by a decrease in the translation initiation rate, an increase in the free energy of folding, a local structural perturbation that disrupts substrate binding or catalysis, a change in a dynamic motion that is critical to catalysis, or an increase in the protein degradation rate.
The effects of random circular permutation on protein function have been widely studied using proteins from mesophilic organisms. These studies have revealed that polypeptides involved in early folding events and stability display a low tolerance to backbone fission arising from permutation8,9. Structure-derived parameters extracted from these studies have been used to develop an algorithm (CP site predictor, CPred), which scores the likelihood that proteins fold and function upon circular permutation27,28. To date, the quality of CPred predictions has not been tested with a thermophilic protein, and it remains unclear how well CPred anticipates the results of a combinatorial experiment involving proteins with extreme stability. Thermophilic proteins are expected to display a lower susceptibility to the destabilizing effects of permutation compared with mesophilic proteins when assayed under similar conditions29-32, suggesting that models trained on marginally-stable proteins may be suboptimal for anticipating which of the possible permuted variants will retain function in protein homologs with a range of thermostabilities. While active circularly permuted variants of mesophilic proteins are expected to be similarly active within the context of thermophilic orthologs under similar cellular conditions, only a fraction of the inactive circularly permuted variants of mesophilic proteins are expected to be inactive within the context of more thermostable homologs.
To better understand how a thermophilic protein’s functional tolerance to random circular permutation relates to different aspects of protein structure (e.g., accessible surface area, contact order, flexibility, sequence variability and structural conservation), we created libraries of circularly permuted Thermotoga neapolitana adenylate kinase (TnAK) with peptides of varying length amended to their termini and selected these libraries for functional variants at 40°C. TnAK was chosen for these experiments because it displays extreme thermostability (Tm ≥ 99°C) but retains high activity at temperatures (40°C) where Escherichia coli selections can be employed to analyze functional tolerance to permutation33,34. In addition, AK structure has been extensively studied so permutation tolerance measurements with this protein can be compared to a range of structure-derived metrics. Previous studies have revealed that AK contains multiple domains, including a rigid core that binds substrates and mobile AMP binding and lid domains whose motions are critical to catalysis35,36.
By comparing the functional variants selected from our libraries with all possible variants, we show that a tradeoff exists between minimizing peptide additions at the N-terminus of circularly permuted proteins and maintaining consistent expression across variants in a library. In addition, we provide evidence that the tolerance of a thermophilic phosphotransferase to circular permutation correlates strongest with three parameters, including: (i) the distance between the backbone fission and AK phosphotransfer site, (ii) AK family sequence variability at residues near the fission site, and (iii) root mean square deviation (RMSD) of residues near the fission site in superimposed AK structures. Among the structure-derived metrics analyzed, these three structure-derived metrics correlated with functional conservation to the greatest extent, while no significant correlation was observed using CPred27,28.
Results and Discussion
AK tolerance to circular permutation
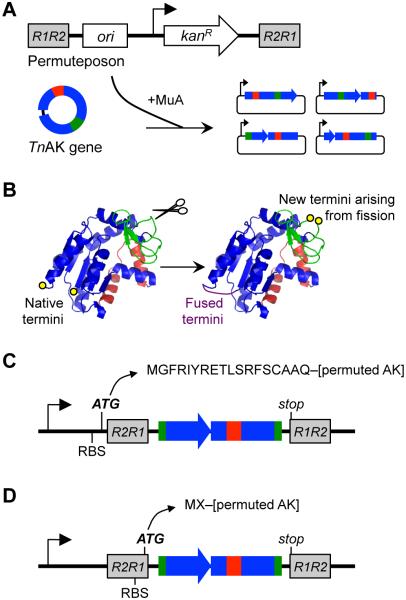
We previously reported a method that simplifies the construction of circularly permuted protein libraries called PERMutation Using Transposase Engineering, PERMUTE25. With PERMUTE, a transposon containing all of the attributes of a vector (referred to as a permuteposon) is randomly inserted into a gene of interest to create a library of vectors that express different permuted variants of a protein (Figure 1A). In these variants, the N- and C-termini in the original protein become covalently linked through an Ala-Ala-Ala peptide, and new termini are created elsewhere through backbone fission (Figure 1B). The initial permuteposon developed for PERMUTE (designated P1) amends an eighteen amino acid tag to the N-terminus of permuted proteins (Figure 1C). This long peptide is added because the RBS used to initiate translation is separated from the permuted gene by the DNA sequence at the end of the permuteposon that is recognized by MuA transposase. To decrease the size of the peptide that PERMUTE adds to the N-terminus of permuted proteins, we synthesized two new permuteposons (P2 and P3) with RBSs closer to the permuteposon ends (Figure S1). When used in PERMUTE, P2 and P3 add two extra amino acids to the N-terminus of permuted proteins (Figure 1D), a methionine followed by a residue whose identity depends on the permuted variant being expressed. P2 contains previously described mutations within the transposase recognition sequence that introduce an RBS37. P3 also contains mutations within this recognition sequence, but these encode a novel RBS.
Figure 1.
AK circular permutation using laboratory evolution. (A) Permuteposon insertion into the TnAK gene by MuA transposase yields vectors that express (B) circularly permuted TnAK. (C) P1 initiates translation before the R2R1 transposase recognition sequence and amends an eighteen-residue peptide to the N-termini of permuted AK. (D) P2 and P3 amend only two residues because they contain an RBS within the transposase recognition sequence.
P1 was previously used to create vectors that express circularly permuted TnAK25. Selection of this P1 library for active variants identified 15 permuted TnAK that retain activity even when fused to a long peptide at their N-terminus. To determine if smaller peptide additions at the N-terminus affect the number and identity of permuted TnAK discovered in a combinatorial experiment, we used P2 and P3 to create TnAK libraries and selected these libraries for active variants using Escherichia coli CV233, a strain with a temperature-sensitive AK that displays growth defects ≥40°C that can be complemented by TnAK (Figure S2). We also selected our P1 library for additional functional TnAK. Non-exhaustive DNA sequencing identified 68 unique vectors with in frame TnAK that complemented E. coli CV2 growth at 42°C (Figure S3), including 21 from the P1, 24 from the P2, and 23 from the P3 libraries. In total, these vectors express ~25% of the possible permuted TnAK variants. All three of the libraries yielded active permuted TnAK arising from backbone fission within the AMP binding, core, and lid domains. Only 7% of the permuted TnAK were discovered in both the P1 (long peptide) and the P2 or P3 (short peptide) libraries (Figure S4A). Greater overlap (30%) was observed between the variants selected from the P2 and P3 libraries (Figure S4B), which express identical variants using different RBSs.
Permuted protein expression variability
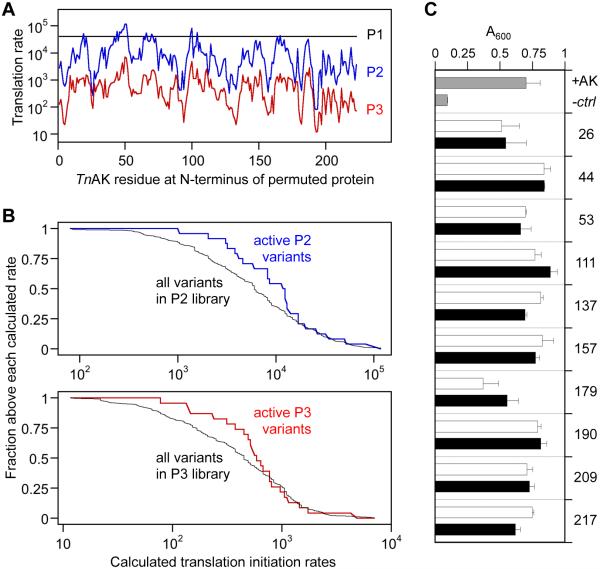
We hypothesized that protein expression variability contributed to the non-degenerate sets of active permuted TnAK selected from our libraries, since different RBSs are used to initiate translation in each library. Genetic context can modulate the strength of translation initiation from a single RBS18, and vectors in our P2 and P3 libraries express permuted proteins whose sequences vary immediately downstream of the RBS. To estimate how much protein expression varies within each library, we used a thermodynamic model to calculate the relative translation initiation rates of every possible permuted TnAK18. Figure 2A shows that the permuteposon that amends the longest peptide addition to the N-terminus of permuted proteins (P1) yields a high and consistent rate of translation initiation across the different permuted TnAK. In contrast, the permuteposons that amend small peptides, P2 and P3, yield rates that vary up to 506- and 270-fold, respectively. These findings suggest that there is a tradeoff between minimizing the size of the peptide added to the N-terminus of permuted proteins and minimizing expression variability across variants in a library.
Figure 2.
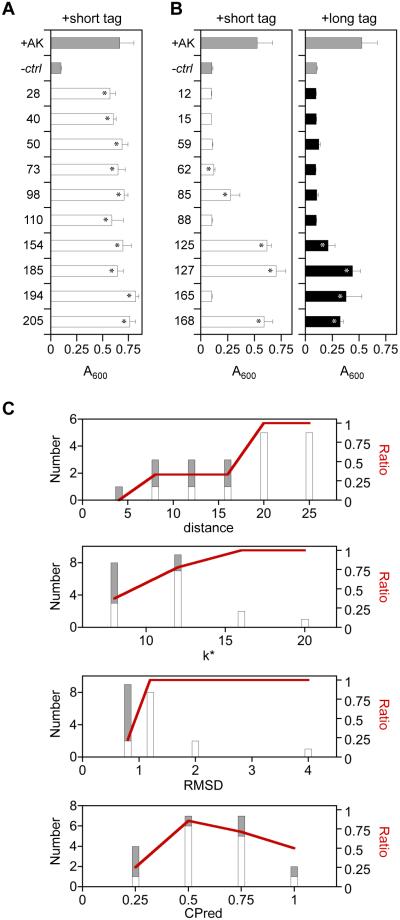
Relationship between translation initiation and protein function. (A) Translation initiation rates for all circularly permuted TnAK in vectors created using the P1 (black), P2 (blue), and P3 (red) permuteposons. These values represent the calculated translation initiation rate from the intended start codon. (B) The fraction of active P2 and P3 variants above each rate was compared with the fraction of total possible variants having translation initiation rates above each calculated value. (C) The absorbance at 600 nm of E. coli CV2 after 15 hours of growth when transformed with pET expression vectors that contain genes encoding native TnAK (+AK), permuted TnAK with short (white bars) and long (black bars) N-terminal peptide tags, or no protein (-ctrl). All variants displayed significant growth compared to the negative control (t-test; p < 0.05), and only variant 179 with a short tag displayed growth that was significantly lower than TnAK (p < 0.01). Error bars represent ±1σ for n = 4.
To determine whether the active variants discovered in our selections were influenced by protein expression, we compared the calculated translation initiation rates of the active permuted TnAK and every theoretically possible variant in the P2 and P3 libraries. We quantified the fraction of active variants and the fraction of all variants with values above each calculated translation initiation rate (Figure 2B). For example, 24 out of 24 active variants (1.0) in the P2 library had rates >1,000, while only 194 out of 220 possible variants (0.88) had translation rates >1,000. Calculations that compared this enrichment above higher translation rates revealed that the fraction of active variants exceeded the fraction of total variants across 97% of the calculated rates in the P2 library. Enrichment was also observed in the P3 library. However, it was only observed across 70% of the translation rates. Application of a one-tailed Mann-Whitney-Wilcoxon test yielded a 98% probability that the median expression for active permuted TnAK in the P2 library is greater than the value for all theoretically possible variants. A lower probability (91%) was obtained with the P3 library. Because alternative translation initiation sites could contribute to protein expression, we performed calculations that considered permuted TnAK synthesized from the start codon and alternative in frame start sites (Figure S5). With this analysis, both libraries yielded a >98% probability that the median expression level of active variants is greater than the median for all variants. Thus, permuted TnAK that maximize calculated translation initiation have a higher likelihood of exhibiting activity than those chosen at random from our P2 and P3 libraries.
The finding that active P2 and P3 variants display higher than average calculated translation initiation suggested that many of our permuted TnAK might be active in the context of both short and long peptide additions beyond those discovered in our selections. For example, active permuted TnAK discovered in the P1 library (long peptide) might not have been discovered in the P2 and P3 libraries (small peptide) because these libraries do not express these equivalent permuted proteins at sufficiently high levels to complement E. coli CV2. To evaluate the effects of peptide additions on permuted TnAK activity and to minimize any expression challenges when comparing the effects of the peptide tags, we created pairs of vectors that use a strong constitutive promoter to express identical permuted TnAK as fusions to short and long peptides and examined their ability to complement E. coli CV2. All of the circularly permuted TnAK complemented CV2 growth at 42°C in the presence of short and long tags (Figure 2C), and 95% of the vectors complemented growth to the same extent as native TnAK. These findings suggest that the size of a peptide added to the N-terminus of permuted TnAK does not correlate with retention of phosphotransfer activity.
Mapping permuted sequences onto AK structure
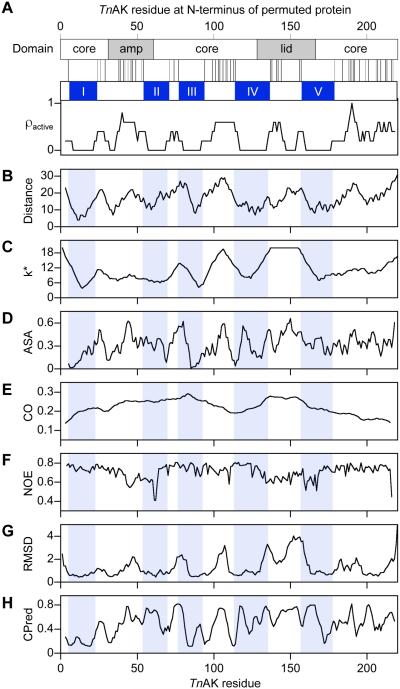
A previous study examining the in vitro properties of every possible circularly permuted variant of dihydrofolate reductase led to the proposal that contiguous stretches of primary structure that are intolerant to backbone fission (arising from permutation) are critical for folding from the unfolded to native state9. To determine how AK structure is related to permutation tolerance, we performed a similar analysis with TnAK. We combined the data from all three of our libraries for this analysis because our expression studies suggested that tolerance to permutation is independent of the extra residues amended to protein termini. A comparison of all sites where TnAK tolerates new termini upon circular permutation (Figure 3A) reveals five contiguous peptides longer than 15 residues within the TnAK primary structure that appear intolerant to backbone cleavage (labeled peptides I-V). These peptides map onto the rigid core domain and the mobile AMP binding and lid domains, including regions that represent the domain boundaries38.
Figure 3.
Relationship between AK structure and the pattern of permutation tolerance. (A) A comparison of AK domain structure with the peptide bonds that can be broken by permutation without disrupting TnAK function reveals five contiguous peptides that lack fission sites, I-V. The dispersion of tolerated backbone fragmentation sites (ρactive) was calculated using a sliding window of five residues. For all residues within TnAK, profiles were generated that show: (B) the distance of each αC to the γ-Pi of Ap5A, (C) k*, the number of unique amino acids in a sequence alignment of 100 AK orthologs, (D) the accessible surface area, (E) the relative contact orders of permuted variants that begin with that residue, (F) the NOE values, (G) the positional structural deviation calculated from 45 pairwise superpositions of Ap5A-bound AK orthologs, and (H) the CPred scores.
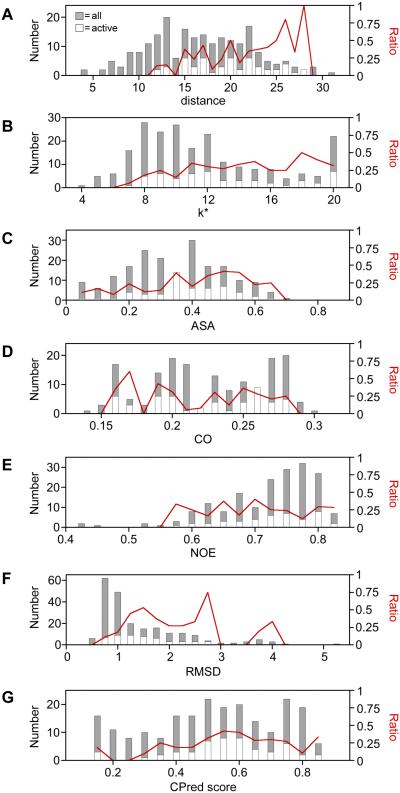
Mapping peptides I-V onto AK structure (Figure S6) shows that they are proximal to P1,P5-Di(adenosine-5')pentaphosphate (Ap5A), a bisubstrate analog for AMP and ATP39. To quantify how residues within peptides I-V relate to the site of catalysis, we calculated the distance between the αC within peptides I-V and the γ phosphate (γ-Pi) in Ap5A, a proxy for the site of catalysis, and we compared these values with the distances to all other αC (Figure 3B). Among the active variants discovered in our selections, we observed distances that ranged from 12 to 28 Å (Figure 4A). In contrast, our libraries are predicted to contain αC whose distances from the γ-Pi range from 4 to 31Å. The ratio of active to total variants at each distance was found to increase as distance increases. A statistically significant correlation was observed between the retention of function and distance using a Spearman's rank correlation (RSR = 0.315; two-tailed t test, p < 0.00001).
Figure 4.
The dispersion of variants across the different values of each metric represented in our libraries, including (A) distance [RSR = 0.315, p = 0.000002], (B) k* [RSR = 0.207, p = 0.002], (C) accessible surface area [RSR = 0.157, p = 0.02], (D) contact order [RSR = −0.061, p = 0.4], (E) NOE [RSR = 0.012, p = 0.9], (F) RMSD [RSR = 0.218, p = 0.001], and (G) CPred score [RSR = 0.115, p = 0.09]. For each structure-derived metric, all of the theoretically possible permuted TnAK in our libraries (gray bars) is compared with the subset of variants that complement bacterial growth (open bars). The fraction of active variants (red line) is shown across the different values of each metric. Spearman's rank correlation coefficients (RSR) were calculated using structure-derived metric values for all possible variants in the libraries and scoring variants as active or not detected as active. P values are from a two-tailed t-test.
Previous studies observed a weak correlation between the number of unique amino acids tolerated at each native position in a protein family and the distance to the site of catalysis40,41. This observation suggested that there might be a correlation between regions of sequence conservation in AK orthologs and the five peptides (I-V) that display a low tolerance to backbone fission. To test this idea, we generated a multiple sequence alignment using one hundred AK ortholog sequences, calculated the mutational tolerance (k*) at each position, i.e., the number of unique amino acids observed at each position, and compared k* values within peptides I-V with all other sites in the primary structure (Figure S7A). Residues within peptides I-V display an average k* value (8.9) that is lower than the value calculated for all native sites (11.4). This enrichment of low k* within peptides I-V becomes more pronounced when a sliding window is used to calculate k*. Averaging the positional k* data over a 13 residue sliding window generates a profile where the permuted variants with the highest k* (9% of the possible variants) have backbone fission uniformly outside of peptides I-V (Figure 3C). When profiles are generated using smaller sliding windows, the highest k* positions (k* = 20) are not fully resolved from peptides I-V (Figure S7B-F). Among the active variants discovered in our selections, we observed k* values that ranged from 6.9 to 20 (Figure 4B), while our libraries were designed to encode variants with k* values that ranged from 3.8 to 20. The ratio of active to total variants at each k* value revealed that the proportion of active variants discovered increases significantly as k* increases (RSR = 0.206; two-tailed t test, p < 0.002).
To further explore how AK tolerance to permutation relates to structure, we compared our pattern of mutational tolerance with four additional structural properties, including accessible surface area (ASA), contact order (CO), conformation variability measured using nuclear magnetic resonance (NMR) spectroscopy, and RMSD at each native position in superimposed AK crystal structures. To test whether active TnAK variants are enriched at surface exposed sites that can best accommodate this bulk, we compared the dispersion of ASA for all possible backbone fission sites with the ASA in peptides I-V (Figure 3D) and the active variants (Figure 4C). This analysis revealed a significant enrichment of functional variants as ASA increases, but the significance of this correlation (RSR = 0.115; two-tailed t test, p = 0.02) was weaker than the correlations for both distance and k*. We next investigated whether there was a correlation between tolerance to permutation and the order in which contacting residues within TnAK are synthesized by the ribosome (Figure 3E), a property that can influence folding rates42. The highest fraction of functional variants was observed at low contact order values (Figure 4D), although the fraction was only ~2-fold higher than that observed at the highest contact orders, and there was no significant correlation observed between CO and retention of function (RSR = −0.06; two-tailed t test, p = 0.4).
AK phosphotransferases undergo dynamic conformational fluctuations that are thought to be critical to function35,36. Residue-specific measurements of these dynamics by NMR spectroscopy have provided evidence that the opening of the mobile domains is rate limiting for catalysis43. In addition, measurements of 15N-1H nuclear Overhauser enhancement (NOE) have shown that residues in the AMP binding and lid domains are more flexible than those in the core domain44. Circular permutation is expected to alter flexibility at the new termini created through backbone fission, which could be preferentially tolerated in regions of low or high flexibility. To test this idea, we compared the dispersion of 15N-1H NOE values for all possible residues with the values for residues in peptides I-V (Figure 3F). This comparison revealed that the two regions with the greatest flexibility (lowest NOE) overlap with peptides I-V. The dispersion of NOE values for active variants was also compared with all possible variants (Figure 4E). Active variants were not discovered at the lowest NOE values (0.4 to 0.55), which represent the regions of highest flexibility, but instead were similarly dispersed across the higher NOE values (0.55 to 0.8), which are most prevalent in the library. No significant correlation was observed between retention of function and NOE values (RSR = 0.012; two-tailed t test, p = 0.9).
To evaluate whether there is a relationship between permutation tolerance and crystallographic data, the RMSD from superpositions of AK crystal structures was compared with permutation tolerance. Ten Ap5A-bound structures were used for these calculations to ensure that comparisons involved the same closed AK conformation. The ten AK used for these calculations display a broad range of pairwise identities, 29 to 74% (Figure S8). Figure 3G shows the RMSD at each native site calculated using all ten structures, which represents the average positional values calculated using 45 pairwise structure superpositions. This profile contains seven contiguous peptides with RMSD ≤1 Å. A comparison of the number of residues within peptides I-V above each RMSD value with all other sites shows that there is an enrichment of low RMSD sites within the peptides that lacked backbone cleavage sites. Among the active variants discovered in our selections, we observed RMSD values that ranged from 0.52 to 3.92 Å (Figure 4F), while our libraries were designed to encode variants with RMSD values that ranged from 0.45 to 5.04 Å. The ratio of active to total variants at each RMSD value revealed that the proportion of active variants discovered increases as RMSD increases from 0.5 to 2.75 Å. For variants having an RMSD >2.75 Å, which includes approximately 7% of the total possible permuted proteins, the ratio of active variants decreased. Analysis of the correlation between all possible RMSD values and retention of function revealed a significant correlation (RSR = 0.218; two-tailed t test, p = 0.001).
To determine how AK sequence identity affects the RMSD profile, we calculated the positional RMSD using AK with defined levels of sequence divergence, including AK displaying pairwise identities that range from 20-29%, 30-39%, 40-49%, 50-59%, 60-69%, and 70-79%. The topologies of each RMSD profile were similar (Figure S9). All of the RMSD profiles contained contiguous peptides with RMSD ≤1 Å that overlapped with peptides I-V.
Previous measurements of permuted protein tolerance have been used to develop an algorithm (CPred) that anticipates permuted protein folding and function28. To determine how CPred predictions relate to the results from our experiments, we calculated the CPred score for all possible permuted variants and compared the scores for variants arising from backbone fission within peptides I-V with all possible variants (Figure 3H). Among the six minima in the profile with the lowest CPred scores, predicting a low likelihood of functioning, only three overlapped with peptides I-V. We also calculated the fraction of active variants discovered at each CPred score (Figure 4G). No significant correlation was observed between the retention of function and CPred score (RSR = 0.115; two-tailed t test, p = 0.09). The highest proportion of active variants were discovered at intermediate CPred scores, rather than the highest values, and the bin with the highest CPred scores only displayed a ~2-fold higher enrichment of active variants compared with the bin with the lowest CPred scores.
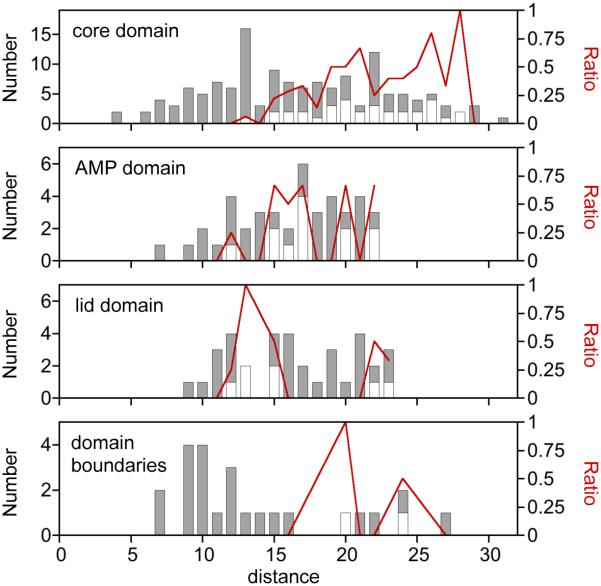
Functional analysis of permuted TnAK libraries provided evidence that distance between the αC at permuted protein termini and the γ-Pi of Ap5A represent the strongest correlation with retention of protein function. To determine if this trend varies across AK domains, we compared the distance values for all variants within each domain with the distance values for active variants. This analysis revealed that the rigid core domain displays the strongest distance dependence enrichment of functional variants (Figure 5). The core also contains the largest fraction of total residues that are proximal to the substrate (≤10 Å) and the largest fraction of residues that are distal (>22 Å) from the substrate. The AMP binding domain displayed a similar distance-dependent enrichment of active variants as the core domain between 10 and 20 Å. However, the lid domain had a higher enrichment of active variants at lower distances (10 and 15 Å) compared with the enrichment observed in the core and AMP binding domains. Since domain motions are thought to be critical to AK function43, we also examined the extent to which TnAK tolerates new termini within the residues at the domain boundaries. These regions display a large dispersion of possible distances and are only enriched in active variants at the largest distances.
Figure 5.
Comparison of αC to γ-Pi distances within each AK domain. The dispersion of variants across the different distance values was calculated for the core, lid, and AMP binding domains. A similar analysis was performed with the domain boundary regions, defined as the six-residue window at the junction of each domain. For each domain, all of the theoretically possible permuted TnAK in our libraries (gray bars) is compared with the subset of variants that complement bacterial growth (open bars). The fraction of active variants (red line) is shown across the different values of each metric.
Two of our structure-derived metrics represent a measure of evolutionary conservation (k* and RMSD), suggesting that these may correlate with one another. To determine how these biophysical parameters relate to one another and distance, we examined the pairwise relationships between each parameter in all possible permuted TnAK (Figure S10). We find that RMSD and k* display the strongest linear correlation (R = 0.75), while weaker correlations are observed between distance and k* (R = 0.49) and RMSD and distance (R = 0.58).
Evaluating trends using rationally-designed variants
The functional variants selected from our libraries are thought to represent a subset of the possible permuted TnAK that retain activity, since only one hundred complementing variants were sequenced from each library. Additional permuted TnAK are expected to function in vivo, and these active variants are predicted to display a dispersion of distance, k*, and RMSD values that are similar to those values observed in our selection experiments. Thus, some variants arising from backbone cleavage within peptides I-V are expected to retain function in cases where those variants display high distance, k* or RMSD values. To test these ideas, we created expression vectors for twenty circularly permuted TnAK that were not discovered in our selections and examined their ability to complement E. coli CV2 when expressed as fusions to the short peptide tags created in the P2 and P3 libraries. These vectors used a strong constitutive promoter to express permuted TnAK to maximize our sensitivity for detecting retention of function. Ten vectors expressed permuted TnAK that arose from backbone fission within peptides I-V, and ten vectors expressed variants that arose from fission outside of peptides I-V. All ten of the permuted variants that were created by introducing backbone fission sites outside of peptides I-V complemented E. coli CV2 growth to the same extent as native TnAK (Figure 6A). In contrast, five of the variants created by introducing backbone fission sites within peptides I-V were unable to complement bacterial growth (Figure 6B), two partially complemented growth, and three exhibited full complementation. Because long tags result in more consistent translation initiation across different permuted proteins (Figure 2A), we also examined the complementation of these latter ten variants when expressed as fusions to long peptide tags created in the P1 library. Among the long tag variants, six were unable to complement E. coli CV2, two displayed partial complementation, and two displayed full complementation. Among the variants that were analyzed with the two different tags (Figures 2C and 6B), the majority (85%) display a phenotype that is independent of the tag amended.
Figure 6.
Activities of rationally-designed permuted TnAK. E. coli CV2 growth after 15 hours at 42°C is reported for cells transformed with pET vectors that constitutively express permuted TnAK variants that arise from fission at (A) backbone locations outside of peptides I-V, and (B) within peptides I-V. Permuted TnAK were expressed with the long and short peptides amended in the P1 (closed bars) and P2/P3 (open bars) libraries. Error bars represent ±1σ for n ≥ 4. Asterisks indicate significant growth compared with the negative control (t-test; p < 0.05). (C) Relationship between retention of function in rationally-designed variants and distance, k*, RMSD, and CPred score. All of the rationally-designed variants (gray bars) are compared with the subset of permuted TnAK that fully complemented bacterial growth (open bars). The fraction of active variants (red line) is shown across the different values of each metric.
A comparison of bacterial complementation by rationally-designed TnAK variants and CPred values provides additional evidence that this algorithm is limited in its ability to anticipate TnAK tolerance to permutation (Figure 6C). With CPred, active variants displayed the greatest enrichment at intermediate CPred scores (0.25 to 0.75). The highest range of CPred values (0.75 to 1.0) had ~2-fold more active variants compared with the lowest range of values (0 to 0.25), similar to the trends obtained from our library selections. As observed in our selection experiments, inactive variants were enriched to a greater extent at low distance, k*, and RMSD values. Among the variants with a distance ≤15 Å (n = 10), five variants were inactive, 2 variants weakly complemented growth, and 3 variants fully complemented growth. All ten variants with distance >15 Å fully complemented growth, consistent with the dispersion of active variants observed in our library selections. Among the variants with RMSD values ≤0.78 Å (n = 9), only 2 variants fully complemented bacterial growth and 2 weakly complemented growth, while all 11 variants with RMSD >0.8 Å fully complemented bacterial growth. Among the variants with k* values < 9 (n = 8), only 3 variants fully complemented bacterial growth, between 9 and 12 (n = 9) 7 variants fully complemented growth, and above 12 (n = 3), all variants fully complemented growth. These findings illustrate how the pattern of mutational tolerance obtained from a library selection can be used to calibrate predictions about the retention of function in permuted proteins.
Biophysical implications
The structure-based metrics evaluated herein correlate to varying extents with the active variants selected from our libraries. Our results show that TnAK tolerance to circular permutation correlates the strongest with the distance between the phosphotransfer site (γ-Pi) and the protein termini created by permutation. We attribute this trend to the disruptive nature of backbone fission within regions of the structure whose arrangement is most critical to substrate binding and catalysis.
Because a highly thermostable protein was used in this study34, and function was evaluated at a temperature that is >50°C lower than the melting temperature of the protein, many of the permuted TnAK are predicted to fold into a parent-like topology29-31. However, because peptides were introduced into TnAK at permuted protein termini, which are predicted to alter local structure at the site of backbone fragmentation, permutation is expected to alter structure and conformational flexibility at residues proximal to the site of backbone fragmentation. Three additional metrics displayed significant correlations with retention of function, including variability in an AK family sequence alignment (k*), variability in superimposed AK structures (RMSD), and accessible surface area of the residue at the new termini. Among these metrics, k* and RMSD yielded the smallest p values. In contrast, CPred, CO and NOE did not reveal significant correlations with retention of function.
The lack of a correlation with CPred was surprising, given that this algorithm was specifically designed to anticipate protein tolerance to circular permutation27. CPred was developed by fitting 46 structure-derived metrics to permutation tolerance data in five proteins, including dihydrofolate reductase9, disulfide oxidoreductase8, green fluorescent protein45-47, myoglobin48, and phosphoribosylanthranilate isomerase49. In this data set, viable circularly permuted proteins were defined by conservation of structure rather than function27. The reason why CPred fails to capture our trends is not known. However, our results suggest that catalytic activity, expression, and thermostability may be important parameters to consider in parallel when building algorithms. Our trends may also differ from past studies with other families of enzymes because we generate distinct sequence diversity from those studies. Early permutation studies used library generation methods that can yield scars of varying size at the termini of permuted proteins 8,26, which could influence the extent to which permutation disrupts protein function.
A fundamental design challenge exposed by our studies is the maintenance of consistent protein expression across vectors in a circularly permuted library. Unlike other classes of mutations, permuted proteins present a different genetic context to the RBS being used to initiate translation when they lack modifications at their termini. Using a thermodynamic model for translation initiation18, we show that our selections yielded active variants whose median expression is significantly greater than the median calculated for all possible variants in our P2 and P3 libraries. These findings suggest that a tradeoff exists between maintaining consistent translation initiation rates across all variants in a permuted protein library and minimizing the size of the peptide amended to the beginning of permuted protein. In libraries that minimize peptide additions to the termini, the sensitivity of a screen (or selection) will largely determine the fraction of active permuted variants that can be discovered using a cellular assay due to the variability in protein expression. Thus, deep-sequencing approaches used to comprehensively map protein tolerance to other classes of mutations40,50 may at times be limited in their utility with permuted protein libraries that display such variable expression. In bacteria, this expression challenge could be overcome by expressing permuted protein libraries using strong constitutive promoters. Alternatively, permuted proteins can be expressed with peptides fused to their N-terminus that maintain the RBS in a consistent genetic context, as illustrated with our P1 library. Our results suggest that peptide addition has little effect on the permutation tolerance of TnAK, which has extremely high stability34. Among the variants whose function was assayed in the presence of different peptides tags, a majority displayed similar complementation with both peptides. Biochemical studies that examine the effects of tag sequence and length on protein activity (and stability) will be required to determine why a small fraction of these variants retain function with only one of the peptide tags. In case of marginally-stable AKs, large peptide additions may be more disruptive to folding and stability, so further improved library methods are needed that minimize peptide additions and maintain consistent expression from an RBS.
Early structural studies provided evidence that AKs exhibit dynamic motions that are critical to function. In substrate-bound AK structure, the AMP binding and lid domains are closed over the active site36. In contrast, substrate-free AK displays an open conformation that is more favorable for substrate binding35. Site-specific measurements of protein dynamics have revealed that the lid opening rates are rate limiting for catalysis43, implicating the large-scale motions predicted from crystallography as critical for catalysis51. Further support for the importance of AK dynamics has come from mutational studies. Measurements analyzing the biophysical effects of surface-exposed glycine mutations within the lid domain revealed that these sequence changes perturb local conformational dynamics and substrate binding without disrupting the ground state crystal structure52. Because backbone fission arising from circular permutation increases local conformational fluctuations11,53,54 like glycine mutations, and TnAK displays extreme stability34, this finding suggests that some of our inactive permuted TnAK might be inactive yet still retain an AK-like structure. However, it is unclear if increased motions within the lid domain are always detrimental to function. An AK recombination study showed that increased motions within this mobile domain may actually improve activity. An AK chimera created by grafting the AMP binding and lid domains from a mesophilic AK onto a thermophilic core domain displayed increased activity compared with both parent proteins over all temperatures analyzed55. Since mesophilic and thermophilic AK differ in their conformational dynamics when assayed at the same temperature38,43, this observation suggests that increased conformational flexibility arising from permutation may at times enhance catalytic activity. The extent to which circular permutation tunes TnAK activity and substrate binding by enhancing conformational motions will require intensive biochemical studies that compare the ground state structures, protein dynamics, substrate binding and thermostability of the different active permuted variants discovered herein.
The new permuteposons described herein, which have hybrid ribosomal and transposase binding sites, will be useful for future studies that seek to create libraries of randomly permuted proteins that lack large peptide additions at their protein termini.
These permuteposons can be used to generate many libraries in parallel to compare how orthologs with a range of sequences and stabilities differ in their permutation tolerance. One advantage of this library construction approach is its ability to generate proteins with constrained sequence diversity that avoids random deletions and insertions upon permutation, which can arise with other methods8,26. When used with thermophilic proteins, these permuteposons should also be useful for identifying the subset of variants that are most likely to retain structure within a protein family. This approach can be used to quickly identify permuted proteins for use in the design of molecular switches through domain insertion, in which different permuted variants of a protein are inserted into a second protein domain to create fused polypeptides with new allosteric functions14,15. By avoiding inactive variants during molecular switch library construction, alternative sequence space parameters (e.g., linker length and composition) can be more thoroughly sampled in screens and selections. Although design strategies have been proposed for optimal peptide linkers56, this aspect of molecular switch design remains an open challenge.
Materials And Methods
Permuteposon design
The P2 and P3 permuteposons were built by PCR amplifying P1 with primers that alter the transposase recognition sequences. The P2 permuteposon was constructed with the splitposon transposase recognition sequence that was developed for constructing split protein libraries and used to discover split variants of T7 RNA polymerase37. The P3 permuteposon was built by making two point mutations within the RBS of the P2 permuteposon. While these mutations are in the region of the transposon necessary for recognition and transposition, they match the terminal recognition sequence found in the native Mu transposon57. Like P1, P2 introduces a single stop codon after permuted genes when used to construct PERMUTE libraries, while P3 has stop codons in three different frames.
Constructing PERMUTE libraries
P2 and P3 (100 ng) were incubated with 300 ng pMM1 and MuA (1 unit) for 16 hours at 37°C as described25. After inactivation at 75°C, DNA was transformed into MegaX DH10B E. coli and grown on LB agar containing kanamycin (25 μg/mL) and chloramphenicol (15 μg/mL) at 43°C. Lawns of cells were obtained and harvested by scraping, pMM1-P2 (or pMM1-P3) hybrids were purified and digested with NotI, P2-AK genes (and P3-AK genes) were isolated using gel electrophoresis, and DNA was circularized through ligation to yield the final libraries. The protocol for building the P2 and P3 libraries was similar to that previously described25, which is estimated to sample ≥70% of the possible permuted AK variants. The percentage sampled was estimated using the library size and number of cfu obtained from the PERMUTE transformation step that yielded the lowest number of colonies58.
Bacterial complementation
Selections were performed by transforming the P1, P2, and P3 libraries into E. coli CV2 using electroporation, plating cells on LB-agar and incubating plates at 40°C for 48 hours. Complementing vectors were isolated from each library (~100) and sequenced. The activities of in frame clones selected from libraries were verified by rescreening for complementation at 42°C (see Supplemental Information for sequences). This was achieved by transforming sequenced vectors into E. coli CV2, obtaining single colonies on LB-agar plates containing kanamycin (25 μg/mL) at 30°C, using multiple colonies to inoculate LB cultures containing kanamycin (25 μg/mL) within a 96-well deep well plate, growing these cultures at 30°C for 28 hours, transferring 1 μL of each overnight culture to fresh LB kanamycin (25 μg/mL) in a Costar 3595 Flat Bottom 96-well plate using a 96-pin replicator, and monitoring cell growth at 42°C using a TECAN infinite M1000 plate reader by measuring the absorbance at 600 nm every 10 minutes for 15 hours. The total phosphotransferase activity required to complement E. coli CV2 growth was estimated by comparing the AK activity in the extracts of cells that were complemented by AK to differing extents59 with the activity of TnAK at the temperature of our selection34. This comparison suggests that a minimum of ~400 TnAK molecules are required for complementation.
Translation initiation calculations
A thermodynamic model for translation initiation18 was used to calculate relative translation initiation rates of every possible permuted TnAK gene within the genetic context of permuteposons P1, P2, and P3. The rates at the intended start codon were calculated by using 60 base pair windows of sequence that encompasses each intended start codon.
Vectors for expressing permuted variants
Permuted TnAK discovered in a single library were cloned into pET26b expression vectors that produce each variant with N-terminal peptide modifications that occur in the P1 and P2/P3 libraries. The T7 promoter in pET26b has high basal activity when transformed into E. coli CV2, which has a DE3 lysogen, allowing it to function as a constitutive promoter without requiring the addition of an inducer. pET26b-derived plasmids were also created that express twenty permuted TnAK that were not discovered in the P1, P2, or P3 libraries.
Structural calculations
Pairwise structural superpositions of Ap5A-bound AK orthologs 36,51,60-65 were generated using Visual Molecular Dynamics 1.9.266. The positional RMSD values obtained from each calculation were related to TnAK by generating pairwise sequence alignments of TnAK with each AK using ClustalW267. At native positions where the superposition yielded a zero value because the reference structure contained an inserted residue compared with the second structure used for the calculations, RMSD was assigned a large value (5 Å) to account for high tolerance of that native position to amino acid insertions. For each permuted variant, ASA was calculated for the residue at the N-terminus using ASAView68, and residue level values are reported for a sliding window of five residues. The relative contact order of each permuted variant was calculated as described69. Calculations of ASA, contact order, and distance were performed using the crystal structure of Bacillus stearothermophilus AK, a thermophilic ortholog of TnAK60. NOE values were derived from measurements with Escherichia coli AK44. The Bacillus stearothermophilus AK structure60 was also used as an input for the online CPred server28. ASA, contact order, CPred and distance values were related to TnAK using a sequence alignment generated using ClustalW.
Mutational tolerance calculations
A multiple sequence alignment (MSA) of 100 AK sequences was generated using MUSCLE70. Mutational tolerance (k*) was calculated as the number of unique amino acids at each TnAK native site. Any sites containing a gap in AK sequences within the MSA were given a k* value of 20. The UniProt numbers for each AK include: B9K8A7, P27142, P16304, P69441, D9RPQ5, X5B753, Q9HXV4, F9ZKP0, Q98N36, Q3SIR8, P9WKF5, Q601J4, O83604, J7RC67, P10772, Q9PQP0, P0DH56, O69172, P58117, P53398, C5CC42, C5CKV9, C4L8U2, C4K2F8, C1KZF9, C6BV58, A5I7I5, C4Z2V1, Q9KTB7, B8E732, B3CT26, B9M6F7, B9MI80, C0ZIK1, A1QZK3, B9KFZ2, A9W4R9, C1D6J0, B8ELE2, F8I5Q5, E0RS39, H1XTN4, F2LXS1, F0SR16, F0SY28, D6THC3, G7V8W1, F7YY03, E3CUM4, E3DPV1, D5V3W5, D3DH81, E1SWA7, C9LTF0, E4TEN8, E3H906, H2CAP7, H1Y541, E8UBV9, I2EYI5, I4B7F3, F4KNH2, F4L5F9, G0IVS5, F2NMQ6, F2IKB1, F0NXZ5, F3ZT36, F9Z919, K9Z2P9, L8N2D5, K9XY97, K9W5D1, H1ZCM5, F0SDA0, E6S961, D7BCZ4, D7CVF8, D7BK89, E4RVW9, F2N8M7, G8THM5, F7YDS0, E4U9G4, E4TT93, I8RFP1, Q47XA8, F6ET29, E4TB16, Q5NQ43, D0MI16, G2SAB0, F6DB56, S4YD16, G0JRR2, C6CQR4, P56104, D5WS67, D3QBA0.
Statistical analysis
A two-tailed t test was used to examine whether the complementation of our permuted TnAK differed from a vector lacking or containing TnAK. Variants were scored as fully complementing if the mean of the permuted variant was not significantly different from cells expressing native TnAK, while variants were scored as partially complementing if the supported growth that was significantly greater than cells lacking TnAK and significantly less than that observed with cells expressing TnAK. A one-tailed Mann-Whitney-Wilcoxon was used to evaluate the probability that the median expression for active permuted TnAK in our libraries is different from the value for all theoretically possible variants. The relationship between each structure-derived metric and retention of function was quantified using the Spearman's rank correlation coefficient.
Supplementary Material
Acknowledgements
We are grateful for financial support from the National Science Foundation (1150138) and Robert A. Welch Foundation (C-1614). AMJ and EET were partially funded by a training fellowship from the Keck Center of the Gulf Coast Consortia, on the Houston Area Molecular Biophysics Program, National Institute of General Medical Sciences (NIGMS) T32GM008280. AMJ and JTA were supported by the National Science Foundation Graduate Research Fellowship Program (NSF GFRP) under grant number (R3E821). THSS was supported by the National Defense Science & Engineering Graduate Fellowship Program and by a Fannie and John Hertz Foundation Fellowship.
Footnotes
Supporting Information Available
Figure S1. Permuteposons used to create libraries.
Figure S2. Assessing AK function using bacterial complementation.
Figure S3. Complementation strength of variants selected from each library.
Figure S4. Comparison of permuted TnAK discovered in each library.
Figure S5. Translation initiation rates calculated using intended and alternative start codons.
Figure S6. Mapping peptides I-V onto AK structure.
Figure S7. Mutational tolerance calculated using a multiple sequence alignment.
Figure S8. Sequence variability for AK orthologs used in RMSD calculations.
Figure S9. Positional RMSD calculated using AK structures of varying identity.
Figure S10. Comparison of distance, k*, and RMSD for each possible variant.
Supplementary Sequences. Selected and rationally-designed permuted TnAK. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- (2).Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat. Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- (3).Weiner J, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- (4).Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays. 2006;28:973–978. doi: 10.1002/bies.20474. [DOI] [PubMed] [Google Scholar]
- (5).Jung J, Lee B. Circularly permuted proteins in the protein structure database. Protein Sci. 2001;10:1881–1886. doi: 10.1110/ps.05801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lo W-C, Lyu P-C. CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships. Genome Biol. 2008;9:R11. doi: 10.1186/gb-2008-9-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bliven SE, Bourne PE, Prlić A. Detection of circular permutations within protein structures using CE-CP. Bioinformatics. 2015;31:1316–1318. doi: 10.1093/bioinformatics/btu823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J. Mol. Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- (9).Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nat. Struct. Biol. 2000;7:580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- (10).Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- (11).Reitinger S, Yu Y, Wicki J, Ludwiczek M, D'Angelo I, Baturin S, Okon M, Strynadka NCJ, Lutz S, Withers SG, McIntosh LP. Circular permutation of Bacillus circulans xylanase: a kinetic and structural study. Biochemistry. 2010;49:2464–2474. doi: 10.1021/bi100036f. [DOI] [PubMed] [Google Scholar]
- (12).Guntas G, Kanwar M, Ostermeier M. Circular permutation in the Ω-loop of TEM-1 β-lactamase results in improved activity and altered substrate specificity. PLoS ONE. 2012;7:e35998. doi: 10.1371/journal.pone.0035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Daugherty AB, Govindarajan S, Lutz S. Improved biocatalysts from a synthetic circular permutation library of the flavin-dependent oxidoreductase old yellow enzyme. J. Am. Chem. Soc. 2013;135:14425–14432. doi: 10.1021/ja4074886. [DOI] [PubMed] [Google Scholar]
- (14).Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yu Y, Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol. 2011;29:18–25. doi: 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- (17).Liberles DA, Teichmann SA, Bahar I, Bastolla U, Bloom J, Bornberg-Bauer E, Colwell LJ, de Koning APJ, Dokholyan NV, Echave J, Elofsson A, Gerloff DL, Goldstein RA, Grahnen JA, Holder MT, Lakner C, Lartillot N, Lovell SC, Naylor G, Perica T, Pollock DD, Pupko T, Regan L, Roger A, Rubinstein N, Shakhnovich E, Sjölander K, Sunyaev S, Teufel AI, Thorne JL, Thornton JW, Weinreich DM, Whelan S. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012;21:769–785. doi: 10.1002/pro.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang T, Bertelsen E, Benvegnu D, Alber T. Circular permutation of T4 lysozyme. Biochemistry. 1993;32:12311–12318. doi: 10.1021/bi00097a006. [DOI] [PubMed] [Google Scholar]
- (20).Protasova NYu, Kireeva ML, Murzina NV, Murzin AG, Uversky VN, Gryaznova OI, Gudkov AT. Circularly permuted dihydrofolate reductase of E. coli has functional activity and a destabilized tertiary structure. Protein Eng. 1994;7:1373–1377. doi: 10.1093/protein/7.11.1373. [DOI] [PubMed] [Google Scholar]
- (21).Lindberg M, Tångrot J, Oliveberg M. Complete change of the protein folding transition state upon circular permutation. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- (22).Nobrega RP, Arora K, Kathuria SV, Graceffa R, Barrea RA, Guo L, Chakravarthy S, Bilsel O, Irving TC, Brooks CL, Matthews CR. Modulation of frustration in folding by sequence permutation. Proc Natl Acad Sci USA. 2014;111:10562–10567. doi: 10.1073/pnas.1324230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Creation of a zymogen. Nat. Struct. Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Butler JS, Mitrea DM, Mitrousis G, Cingolani G, Loh SN. Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase. Biochemistry. 2009;48:3497–3507. doi: 10.1021/bi900039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mehta MM, Liu S, Silberg JJ. A transposase strategy for creating libraries of circularly permuted proteins. Nucleic Acids Res. 2012;40:e71–e71. doi: 10.1093/nar/gks060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Graf R, Schachman HK. Random circular permutation of genes and expressed polypeptide chains: application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lo W-C, Dai T, Liu Y-Y, Wang L-F, Hwang J-K, Lyu P-C. Deciphering the preference and predicting the viability of circular permutations in proteins. PLoS ONE. 2012;7:e31791. doi: 10.1371/journal.pone.0031791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lo W-C, Wang L-F, Liu Y-Y, Dai T, Hwang J-K, Lyu P-C. CPred: a web server for predicting viable circular permutations in proteins. Nucleic Acids Res. 2012;40:W232–7. doi: 10.1093/nar/gks529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. U.S.A. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- (31).Besenmatter W, Kast P, Hilvert D. Relative tolerance of mesostable and thermostable protein homologs to extensive mutation. Proteins. 2007;66:500–506. doi: 10.1002/prot.21227. [DOI] [PubMed] [Google Scholar]
- (32).Segall-Shapiro TH, Nguyen PQ, Santos Dos ED, Subedi S, Judd J, Suh J, Silberg JJ. Mesophilic and hyperthermophilic adenylate kinases differ in their tolerance to random fragmentation. J. Mol. Biol. 2011;406:135–148. doi: 10.1016/j.jmb.2010.11.057. [DOI] [PubMed] [Google Scholar]
- (33).Haase GH, Brune M, Reinstein J, Pai EF, Pingoud A, Wittinghofer A. Adenylate kinases from thermosensitive Escherichia coli strains. J. Mol. Biol. 1989;207:151–162. doi: 10.1016/0022-2836(89)90446-4. [DOI] [PubMed] [Google Scholar]
- (34).Vieille C, Krishnamurthy H, Hyun H-H, Savchenko A, Yan H, Zeikus JG. Thermotoga neapolitana adenylate kinase is highly active at 30 degrees C. Biochem. J. 2003;372:577–585. doi: 10.1042/BJ20021377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Müller CW, Schlauderer GJ, Reinstein J, Schulz GE. Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure. 1996;4:147–156. doi: 10.1016/s0969-2126(96)00018-4. [DOI] [PubMed] [Google Scholar]
- (36).Müller CW, Schulz GE. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. J. Mol. Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- (37).Segall-Shapiro TH, Meyer AJ, Ellington AD, Sontag ED, Voigt CA. A “resource allocator” for transcription based on a highly fragmented T7 RNA polymerase. Molecular Systems Biology. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Krishnamurthy H, Munro K, Yan H, Vieille C. Dynamics in Thermotoga neapolitana adenylate kinase: 15N relaxation and hydrogen-deuterium exchange studies of a hyperthermophilic enzyme highly active at 30 degrees C. Biochemistry. 2009;48:2723–2739. doi: 10.1021/bi802001w. [DOI] [PubMed] [Google Scholar]
- (39).Lienhard GE, Secemski II. P 1 ,P 5 -Di(adenosine-5')pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J. Biol. Chem. 1973;248:1121–1123. [PubMed] [Google Scholar]
- (40).Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene's fitness landscape. Mol. Biol. Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Abriata LA, Palzkill T, Dal Peraro M. How structural and physicochemical determinants shape sequence constraints in a functional enzyme. PLoS ONE. 2015;10:e0118684. doi: 10.1371/journal.pone.0118684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Plaxco KW, Larson S, Ruczinski I, Riddle DS, Thayer EC, Buchwitz B, Davidson AR, Baker D. Evolutionary conservation in protein folding kinetics. J. Mol. Biol. 2000;298:303–312. doi: 10.1006/jmbi.1999.3663. [DOI] [PubMed] [Google Scholar]
- (43).Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- (44).Tugarinov V, Shapiro YE, Liang Z, Freed JH, Meirovitch E. A novel view of domain flexibility in E. coli adenylate kinase based on structural mode-coupling (15)N NMR relaxation. J. Mol. Biol. 2002;315:155–170. doi: 10.1006/jmbi.2001.5231. [DOI] [PubMed] [Google Scholar]
- (45).Topell S, Hennecke J, Glockshuber R. Circularly permuted variants of the green fluorescent protein. FEBS Lett. 1999;457:283–289. doi: 10.1016/s0014-5793(99)01044-3. [DOI] [PubMed] [Google Scholar]
- (46).Hsu S-TD, Blaser G, Jackson SE. The folding, stability and conformational dynamics of beta-barrel fluorescent proteins. Chem Soc Rev. 2009;38:2951–2965. doi: 10.1039/b908170b. [DOI] [PubMed] [Google Scholar]
- (47).Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- (48).Ribeiro EA, Ramos CHI. Circular permutation and deletion studies of myoglobin indicate that the correct position of its N-terminus is required for native stability and solubility but not for native-like heme binding and folding. Biochemistry. 2005;44:4699–4709. doi: 10.1021/bi047908c. [DOI] [PubMed] [Google Scholar]
- (49).Akanuma S, Yamagishi A. Identification and characterization of key substructures involved in the early folding events of a (beta/alpha)8-barrel protein as studied by experimental and computational methods. J. Mol. Biol. 2005;353:1161–1170. doi: 10.1016/j.jmb.2005.08.070. [DOI] [PubMed] [Google Scholar]
- (50).Firnberg E, Ostermeier M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS ONE. 2012;7:e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- (52).Schrank TP, Bolen DW, Hilser VJ. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc Natl Acad Sci USA. 2009;106:16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hahn M, Piotukh K, Borriss R, Heinemann U. Native-like in vivo folding of a circularly permuted jellyroll protein shown by crystal structure analysis. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10417–10421. doi: 10.1073/pnas.91.22.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Pieper U, Hayakawa K, Li Z, Herzberg O. Circularly permuted beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1997;36:8767–8774. doi: 10.1021/bi9705117. [DOI] [PubMed] [Google Scholar]
- (55).Bae E, Phillips GN. Roles of static and dynamic domains in stability and catalysis of adenylate kinase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2132–2137. doi: 10.1073/pnas.0507527103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Choi JH, Laurent AH, Hilser VJ, Ostermeier M. Design of protein switches based on an ensemble model of allostery. Nat Commun. 2015;6:6968. doi: 10.1038/ncomms7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Goldhaber-Gordon I, Early MH, Gray MK, Baker TA. Sequence and positional requirements for DNA sites in a mu transpososome. J. Biol. Chem. 2002;277:7703–7712. doi: 10.1074/jbc.M110342200. [DOI] [PubMed] [Google Scholar]
- (58).Bosley AD, Ostermeier M. Mathematical expressions useful in the construction, description and evaluation of protein libraries. Biomol. Eng. 2005;22:57–61. doi: 10.1016/j.bioeng.2004.11.002. [DOI] [PubMed] [Google Scholar]
- (59).Kawai M, Kidou S, Kato A, Uchimiya H. Molecular characterization of cDNA encoding for adenylate kinase of rice (Oryza sativa L.) Plant J. 1992;2:845–854. doi: 10.1046/j.1365-313x.1992.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]
- (60).Berry MB, Phillips GN. Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+ Proteins. 1998;32:276–288. doi: 10.1002/(sici)1097-0134(19980815)32:3<276::aid-prot3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- (61).Bae E, Phillips GN. Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J. Biol. Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- (62).Thach TT, Luong TT, Lee S, Rhee D-K. Adenylate kinase from Streptococcus pneumoniae is essential for growth through its catalytic activity. FEBS Open Bio. 2014;4:672–682. doi: 10.1016/j.fob.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Davlieva M, Shamoo Y. Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus marinus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009;65:751–756. doi: 10.1107/S1744309109024348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Abele U, Schulz GE. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995;4:1262–1271. doi: 10.1002/pro.5560040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wild K, Grafmüller R, Wagner E, Schulz GE. Structure, catalysis and supramolecular assembly of adenylate kinase from maize. Eur. J. Biochem. 1997;250:326–331. doi: 10.1111/j.1432-1033.1997.0326a.x. [DOI] [PubMed] [Google Scholar]
- (66).Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (67).Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- (68).Ahmad S, Gromiha M, Fawareh H, Sarai A. ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- (70).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.