Abstract
Bacteriophytochrome photoreceptors (BphP) are knotted proteins that have been developed as near-infrared fluorescent protein (iRFP) reporters of gene expression. To explore how rearrangements in the peptides that interlace into the knot within the BphP photosensory core affect folding, we subjected iRFP to random circular permutation using an improved transposase mutagenesis strategy and screened for variants that fluoresce. We identified twenty seven circularly permuted iRFP that display biliverdin-dependent fluorescence in Escherichia coli. The variants with the brightest whole cell fluorescence initiated translation at residues near the domain linker and knot tails, although fluorescent variants were discovered that initiated translation within the PAS and GAF domains. Circularly permuted iRFP retained sufficient cofactor affinity to fluoresce in tissue culture without the addition of biliverdin, and one variant displayed enhanced fluorescence when expressed in bacteria and tissue culture. This variant displayed a similar quantum yield as iRFP, but exhibited increased resistance to chemical denaturation, suggesting that the observed signal increase arose from more efficient protein maturation. These results show how the contact order of a knotted BphP can be altered without disrupting chromophore binding and fluorescence, an important step towards the creation of near-infrared biosensors with expanded chemical-sensing functions for in vivo imaging.
Keywords: biliverdin, circular permutation, knot, near-infrared fluorescent protein, phytochrome, protein engineering
INTRODUCTION
The first description of a knotted protein was controversial because of the entropic penalty that is associated with interlacing a polypeptide chain.1,2 Since that time, approximately one thousand knotted structures have been deposited in the protein data bank,3 representing approximately 1% of the total structures deposited, suggesting that this structural feature is present in diverse macromolecules that support cellular reactions. A recent experimental study provided evidence that small knotted proteins (≤160 residues) spontaneously fold within the context of a cell free translation system without forming misfolded species,4 although knot formation in these proteins was found to occur post-translationally and was rate limiting for folding. At times, knots may also provide extra mechanical stability to proteins.5 However, the lower free energy relative to the unfolding transition is thought to come at the expense of folding rates, since the introduction of a knot into a polypeptide decreased the folding rate by over an order of magnitude.6 While this protein design study demonstrated that knotted proteins can arise from an unknotted topology by changing protein contact order (CO), our understanding of knotted protein tolerance to other sequence changes that alter CO remains limited.
In nature, changes in CO arise as genes experience duplication, fusion, and fission.7 Support for this idea has come from the discovery of circularly permuted proteins that share the same topology but differ in the location of their NH2 and COOH termini.8 Circular permutation has also been used in the lab for protein design,9 where it has yielded proteins with improved activity,10,11 altered ligand binding,12 and enhanced stability.13 Additionally, circularly permuted variants of natural proteins have been leveraged to construct biosensors and molecular switches through domain insertion,14,15 as well as artificial zymogens.16,17 An artificial zymogen has been successfully designed using a knotted protein. This zymogen was built by connecting the termini of a knotted fluorescent protein using a small protease-cleavable linker that inhibits folding and cofactor-binding that are both required for maturation into a protein that displays fluoescence.17 When exposed to a caspase that cleaves the linker, the resulting protein fragments matured into a fluorescent protein, consistent with that observed in fragmentation studies.18 While this study showed how to inactivate a knotted protein using circular permutation,17 there have been no systematic studies examining the structural tolerance of a knotted protein to random circular permutation.
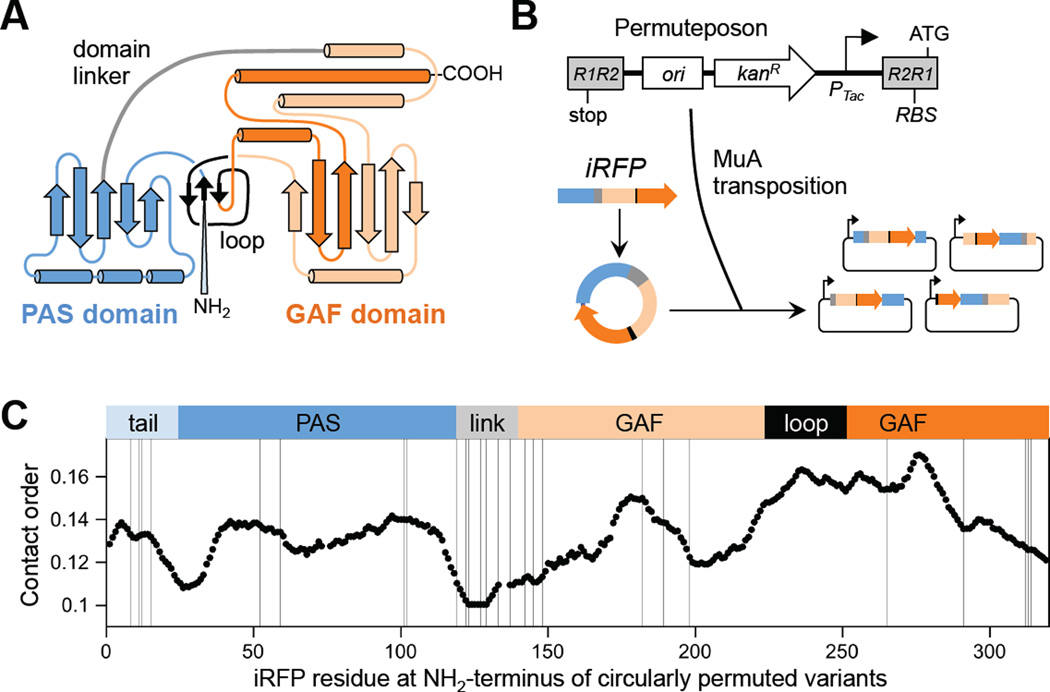
Bacteriophytochrome photoreceptors containing a knot are well suited for studies of permutation tolerance because structural conservation can be easily measured in mutants by analyzing fluorescence. BphP use light-dependent conformational changes in their knotted photosensory core to alter the activity of their signaling domains.19 Structural studies have shown that this photosensory core is made up of a pair of domains, the Per/ARNT/Sim (PAS) and cGMP phosphodiesterase/adenylylcyclase/FhlA (GAF) domains (Figure 1a), which interlace into the knot and bind linear tetrapyrrole cofactors such as biliverdin.20,21 When this cofactor experiences photochemical isomerization, it interconverts the PAS-GAF photosensory core between two different conformational states that differ in their activation of adjacent signaling domains.22 Because the PAS-GAF core interacts with far red light that is well suited for deep tissue imaging, photosensory cores have been mutated and screened to create variants with enhanced near-infrared fluorescence, such as IFP and iRFP.23–25 However, these and other BphP have not been subjected to random circular permutation, and it remains unclear which of the possible circularly permuted BphP are best suited for constructing near-infrared biosensors or light-dependent switches using domain insertion.
Figure 1.
iRFP tolerance to circular permutation. (A) In iRFP, a knot is formed when the NH2 terminus of the PAS domain passes through a loop created by the GAF domain. (B) Random insertion of a permuteposon into a circular iRFP gene using the transposase MuA yields a library of cp-iRFP vectors. (C) Relationship between domain structure, contact order, and the location of the NH2 termini created by permutation. The fission sites in fluorescent cp-iRFP are mapped (with lines) onto BphP (PBD = 1ZTU) to relate their location to the NH2 tail (cyan), PAS domain (blue), domain linker (grey), loop from the GAF domain (black) and remainder of the GAF domain (orange). Each line represents the first residue in the cp-iRFP.
EXPERIMENTAL PROCEDURES
Permuteposon design
Permuteposon P4 was built by mutating a previously described permuteposon to introduce a RBS into the R2R1 transposase recognition sequence.26 In addition, a tac promoter containing a lacI operator was inserted after the kanamycin-resistance cassette (KanR) but before the R2R1 sequence. In contrast to the previously described permuteposon, which constitutively expresses permuted proteins with an 18 amino acid peptide scar at their NH2-terminus,26 P4 allows for inducible expression of permuted proteins and only adds a two amino acid scar to protein termini.
Library construction
DNA encoding the iRFP gene was PCR amplified from pBAD/His-B-iRFP24 using primers that introduce NotI restriction sites and 37 base pairs between the first and last codon of iRFP and the adjacent NotI site. These extra base pairs and the NotI restriction sites will ultimately encode a peptide linker having fifteen residues (GGSGGSAAAGGSGGS) that links the original NH2 and COOH-termini of iRFP in each permuted variant. This long linker was used in our design because the iRFP residues that become connected through permutation are separated by ~35Å.20 The product of this reaction was digested with NotI, size selected using agarose gel electrophoresis, purified using Zymoclean Gel DNA Recovery Kit (Zymo Research), and circularized by incubating with T4 DNA ligase for 16 hours at 16°C. The P4 permutepsoon (100 ng) was inserted into the circularized iRFP (60 ng) by incubating these DNA in a 20 µL reaction containing 1 µL (0.22 µg/µL) of HyperMu MuA transposase (ThermoFisher Scientific) and buffer for 16 hours at 37°C. After terminating the reaction at 70°C for 10 min, DNA was purified using DNA Clean & Concentrator (Zymo Research), electroporated into MegaX DH10B electrocompetent E. coli (Invitrogen), and grown on Luria Broth (LB)-agar plates containing 25 µg/mL kanamycin at 37°C. Lawns of cells (>50,000 cfu) were obtained after overnight growth, cells were harvested by scraping, and total plasmid DNA was purified using a Qiagen DNA Miniprep kit. The ensemble of DNA was linearized using NotI, and iRFP-transposon hybrids (2.9 kb) were separated from larger DNA, which are thought to represent iRFP containing multiple inserted permuteposons. The iRFP-transposon hybrids were circularized by ligation for 16 hours at 16°C to create the final library.
Screening
E. coli XL1 Blue transformed with the final library were grown on LB-agar plates containing 50 µg/mL kanamycin at 37°C. Single colonies from these plates were used to inoculate 200 µL cultures in 96-well deep well plates containing 25 µg/mL kanamycin. Deep well plates were grown at 37°C while shaking at 250 rpm. After 18 hours of growth, stationary phase cultures were diluted by adding 600 µL LB containing 25 µg/mL kanamycin. After growing for 1 hour at 37°C, 200 µL LB containing isopropyl β-D-1-thiogalactopyranoside (IPTG) and BV (Frontier Scientific) were added to final concentrations of 0.5 mM and 80 µM, respectively. Deep well plates were grown for additional 4 hours in the dark at either 23 or 37°C. A fraction (200 µL) of each culture was transferred to clear polystyrene 96-well V-bottom plates (Rainin), cells were pelleted by centrifuging at 3,000×g for 5 minutes, supernatant containing excess BV was removed, and whole cell absorbance (600 nm) and fluorescence (λex = 690 nm; λem = 700–725 nm) was measured using a Tecan M1000 plate reader. Measurements were performed using 5 nm bandwidths in top mode with 1 nm step size. For each variant, the emission intensities measured from 705 to 720 nm were averaged, and this average value was normalized to absorbance in each well to obtain the relative near-infrared fluorescence signals of each variant. As negative controls, each plate contained four cultures of cells transformed with a vector lacking the iRFP gene. Cultures that yielded absorbance-normalized fluorescence signals that were ≥5σ higher than the signal obtained from cells lacking the iRFP gene were sequenced (n = 81) and given names that correspond to the iRFP residue encoded by the first codon in the permuted open reading frame.
Vector construction
All of the permuted iRFP were PCR amplified and cloned into pBAD/His-B (Invitrogen) vector using standard cloning to create plasmids that use the araBAD promoter to express each permuted iRFP with a NH2-terminal (His)6 tag. The (His)6 tag was fused to each variant through a 27 residue polypeptide having the sequence GMASMTGGQQMGRDLYDDDDKDPSSRS. To create mammalian vectors that express permuted iRFP, select variants were PCR amplified using primers that introduce BglII and SalI restriction sites and an HA tag to C-terminus. PCR amplified variants were cloned into pEGFP-C1 vector (Clontech) using standard cloning. All vectors were sequence verified.
Whole cell fluorescence measurements
Circularly permuted iRFP yielding significant fluorescence over background were transformed into E. coli XL1 Blue using heat shock, and grown on LB-agar plates containing 50 µg/mL kanamycin at 37°C. After overnight growth, three or more colonies derived from each vector were used to inoculate 3 mL LB cultures in 15 mL Falcon tubes containing 50 µg/mL kanamycin. After 16 hour growth at 37°C and 250 rpm, 1 mL of cells were harvested by centrifugation, resuspended in 1 mL LB medium, and used to inoculate a fresh 4 mL LB culture containing 0.5 mM IPTG, 80 µM BV, and 50 µg/mL kanamycin. Cells were grown for 5 hours at the indicated temperatures while shaking at 250 rpm in the dark, washed with 25% glycerol (1 mL), and resuspended in 25% glycerol (1 mL) to a similar density. Aliquots (200 µL) of each resuspended sample were transferred to four wells in transparent flat-bottom 96-well plates (Corning), and whole cell absorbance (600 nm) and fluorescence (λex = 690 nm, λem = 700–800 nm) was acquired using a Tecan M1000 plate reader. Emission data was normalized to absorbance in each well, and data reported represent the average of measurements performed on samples derived from three or more colonies. A vector that expresses full-length iRFP (pBAD/His-B-iRFP) and circularized P4 lacking the iRFP gene were used as positive and negative controls, respectively.
BV dependence
Arabinose-inducible vectors encoding circular permuted iRFP were transformed into E. coli XL1 Blue, and individual colonies were used to inoculate LB cultures containing 100 µg/mL ampicillin. After 16 hour incubation at 37°C and 250 rpm, cells (2 mL) were harvested by centrifugation and used to inoculate fresh 8 mL LB cultures containing 1 mM arabinose, 100 µg/mL ampicillin, and varying final concentrations of BV (0, 5, 10, 20, 40, and 80 µM). Aliquots of these cultures (1 mL) were added into 96-well deep well plates and incubated for 5 hours at 37°C while shaking at 250 rpm in the dark. Fractions (200 µL) of each culture were transferred to three wells within clear polystyrene 96-well v bottom plates (Rainin), cells were pelleted by centrifuging plates at 3,000 × g for 5 minutes, supernatant containing excess BV was removed, and whole cell absorbance (600 nm) and fluorescence (λex = 690 nm; λem = 700–800 nm) was measured using a Tecan M1000 plate reader. Control experiments analyzing the signal from cells expressing iRFP with varying levels of BV yielded a lower coefficient of variance when fluorescence was measured in cells pelleted in V-bottom plates compared with cells resuspended in liquid medium.
Translation initiation calculations
The effect of circular permutation on protein expression from the P4 permuteposon was analyzed using a thermodynamic model for translation initiation.27 The rate of translation initiation at the start codon within P4 was calculated by using sequence that encompasses 47 base pairs before the intended start codon and 96 base pairs following that codon.
Western immunoblots
E. coli XL1 Blue transformed with pBAD-derived vectors were grown as described for whole cell fluorescence measurements, with the exception that growth medium contained 100 µg/mL ampicillin and 1 mM arabinose. After resuspending cultures to identical optical densities, cells from each culture (10 µL) were analyzed using sodium dodecyl sulfate (SDS)-PAGE under reducing conditions using NuPAGE 12% Bis-Tris Gels (Life Technologies) and MOPS running buffer. Total protein was transferred to a Protran nitrocellulose membrane (Whatman) using a TE 22 Mini Tank Transfer Unit (GE Healthcare) at 200 mA for 45 minutes. Membranes were washed with 20 mL TBST buffer (100 mM Tris pH 7.5, 150 mM NaCl, and 0.1% Tween 20) for 5 minutes, and blocked for 1 hour with 30 mL 10% dry milk in TBST. Membranes were incubated for 1 hour with a His-tag polyclonal antibody (Qiagen) diluted 1:1000 in 20 mL TBST, incubated for 1 hour in 20 mL TBST containing a secondary goat anti-rabbit IgG conjugated to peroxidase conjugate (Calbiochem) at a dilution of 1:1000. A visual signal was generated using the ECL western blotting substrate (GE Healthcare) and imaged using autoradiographic film (BioExcell).
HeLa cells transiently transfected with different plasmids were collected by trypsinization and lysed in Complete lysis-M buffer containing a protease inhibitor cocktail (Roche Applied Science) for 8 minutes with intermittent vortexing. Lysed cells were then centrifuged for 5 min at 15,000 rpm at 4°C, and the supernatant was collected. Protein concentrations were determined by Bradford assay. Total protein (5 µg) from each sample was separated by 12% SDS-PAGE and transferred to PVDF membrane (Bio-Rad). Membranes were blocked using 5% dry milk in TBST overnight at 4°C. Primary antibody incubations were performed for 3 hours at 23°C using rabbit anti-HA (Santa Cruz Biotechnology, 1:8,000), chicken anti-GFP (AnaSpec; 1:8,000), and mouse anti-α-Tubulin (Sigma Aldrich, 1:10,000) diluted in 1% milk in TBST. Secondary antibody incubations were conducted for 1 hour at room-temperature using horseradish peroxidase-conjugated goat anti-rabbit, goat anti-chicken and goat anti-mouse (Santa Cruz Biotechnology; 1:12,000). Chemiluminescent visualization was done using Luminata Forte Western HRP substrate (Millipore) on an LAS4000 imager (GE Healthcare). Quantification of all protein bands was performed using ImageJ.
Protein purification
E. coli JW2509-2 transformed with a plasmid that constitutively expresses Synechocystis PCC 6803 hemeoxygenase (pSR34-Bvd) and arabinose-inducible vectors expressing different iRFP variants were grown on LB-agar plates containing 100 µg/mL streptomycin and 100 µg/mL ampicillin at 37°C. Single colonies were used to inoculate fresh LB cultures (50 mL) containing 100 µg/mL streptomycin and 100 µg/mL ampicillin. After overnight growth, 1 mL culture was used to inoculate fresh 50 ml LB containing 100 µg/mL streptomycin and 100 µg/mL ampicillin. After 3.5 hours of growth at 37°C while shaking at 250 rpm, cells (10 mL) were harvested by centrifugation and used to inoculate 1 L LB containing the same antibiotics. When these cultures reached an OD ~0.5, arabinose was added to a final concentration of 1 mM to induce protein production. Cells were grown for 19 hours at 37°C while shaking at 250 rpm in dark. Cells harvested by centrifugation (4,000×g for 10 minutes at 4°C) were resuspended in lysis buffer (50 mM phosphate buffer pH 7.0, 300 mM NaCl, 10 mM Imidazole, 1 mM MgCl2, 0.5 mg/mL lysozyme, and 0.04 mg/mL DNAse I) and frozen at −80°C. Cells were thawed, centrifuged at 45,000×g for 1 hour at 4°C, the soluble fraction was loaded onto a Ni-NTA column (Qiagen), and washed with NTA wash buffer (50 mM phosphate buffer pH 7.0, 300 mM NaCl, and 10 mM Imidazole). Permuted iRFP were eluted using NTA elution buffer (50 mM phosphate buffer pH 7.0, 300 mM NaCl, and 250 mM Imidazole). Fractions were analyzed using SDS-PAGE, and those containing permuted iRFP were pooled. Ammonium sulfate was added to a final concentration of 40%, protein was loaded onto a HiTrap Phenyl HP (GE Healthcare) that had been equilibrated in a similar buffer using an AKTA protein purification system (GE Healthcare), and permuted iRFP were eluted using a linear ammonium sulfate gradient from 40 to 0% in 20 mM Tris buffer. Protein appearing homogeneous by SDS-PAGE was pooled, dialyzed against phosphate buffered saline (PBS) pH 7.5 (137 mM NaCl, 2.7 mM KCl, 4.3mM Na2HPO4, and 1.47 mM KH2PO4), and stored at −80°C. All purification steps were performed under green or no light conditions.
Gel filtration chromatography
A Superdex 200 10/300 GL (GE Healthcare) run using PBS and a flow rate of 0.5 ml/min at 4°C was used to assess the size of each permuted iRFP. A standard curve generated using the elution volumes of five standards (β-amylase, alcohol dehydrogenase, bovine serum albumin, carbonic anhydrase, and cytochrome C) was used to estimate the weight of each protein.
Native gel electrophoresis
Purified proteins (5 µg) were loaded on native gel and run using a non-denaturing running buffer (25 mM Tris, 192 mM glycine) with 30 mA for 45 minutes. The native gel was composed of a separating gel (10% acrylamide pH 8.8) and a stacking gel (3.9% acrylamide pH 6.8).
Quantum yield determination
Spectral measurements used purified iRFP variants diluted in PBS to a maximum absorbance <0.1 and Nile blue dye in ethanol with an absorbance of 0.1. For each sample, absorbance spectra (205 to 750 nm) were collected using a Cary 50 UV-visible spectrophotometer. Fluorescence emission (669 to 800 nm) arising from excitation at 659 nm was acquired using a Tecan M1000 plate reader. The quantum yield was calculated using the equation [ϕF(x)=(As/Ax)(Fx/Fs)(nx/ns)2·ϕF(S)], where ϕF is fluorescence quantum yield of the standard (s) and unknown (x), A is absorbance at the excitation wavelength for each sample, F is area of the emission spectrum for each sample, and n is refractive index of the solvents.28 Extinction coefficients were determined by calculating the ratio of absorbance at 694 nm to 391 nm and multiplying this value with 39,900, which represents the extinction coefficient of the free BV.22
Equilibrium unfolding
A 6 M stock of GdnHCl was mixed with PBS to make solutions containing a range of GdnHCl concentrations (0, 0.5, 1, 1.5, 2, 2.25, 2.5, 2.75, 3, 3.25, 3.5, 4 and 4.5 M). Aliquots of each protein were mixed with 325 µL of each GndHCl solution, incubated at room temperature for 2 hours in dark, and transferred into transparent flat-bottom 96-well plate (Corning) where fluorescence (λex = 690 nm and λem = 700 to 800 nm) was measured. The fluorescence signal at 715 nm was used for comparison.
Mammalian tissue culture and flow cytometry
HeLa cells (American Type Culture Collection) were cultured in DMEM (Dulbecco modified eagle medium), Lonza, supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin–streptomycin–glutamine (Hyclone) and maintained at 37 °C and 5% CO2. Cells were plated in 12-well plates at 8×104 cells per well. 24 hours after seeding, cells were transiently transfected using JetPrime (Polyplus transfection) according to the manufacturer’s protocol with 0.5 ug of plasmid expressing the iRFP variant along with 0.05 ug of pcDNA4-GFP plasmid as transfection control per well. The culture media of transfected cells was replaced with fresh media 16 hours post transfection. Cells were harvested for analysis at 48 hours post transfection by trypsinization (TrypLE, GIBCO Invitrogen). Cells were analyzed with a FACSCanto II flow cytometer (BD, San Jose, CA) to measure fluorescence intensity of iRFP variants in the APC-Cy7 (Allophycocyanin conjugated with cyanine dye) channel (633 nm laser, 780/60 nm emission filter). At least 10,000 cells were recorded in each sample for analysis. For transient transfection experiments, the APC-Cy7 signal changes were monitored within GFP-positive cells (488 nm laser, 530/30 emission filter) to monitor changes within transfected cells. The reported output signal was calculated by normalizing APC-Cy7 intensity by GFP intensity in order to eliminate differences arising from transfection efficiencies.
RESULTS
Mapping knotted protein tolerance to permutation
As a model system for studying BphP tolerance to random circular permutation, we chose iRFP,24 a mutant of the photosensory core from Rhodopseudomonas palustris BphP2 whose native structure displays enhanced near-infrared fluorescence upon biliverdin (BV) binding. iRFP was targeted because it displays enhanced stability and fluorescence compared with the other bacteriophytochromes.24 We hypothesized that these properties would maximize: (i) the reliable detection of folded variants and (ii) the fraction of variants that retain an iRFP-like structure upon permutation. Previous studies have shown that stability increases the fraction of folded variants in libraries encoding proteins subjected to random mutation and fission.29,30 A modified PERMUTE protocol was used to construct a library of vectors that express circularly permuted iRFP.26 With PERMUTE, an artificial transposon containing all of the attributes of a vector (called a permuteposon) is randomly inserted into a second vector harboring the gene of interest, and a multistep procedure is used to convert the product of this reaction into a library. To allow for regulated protein expression in PERMUTE libraries, we synthesized a new transposon, permuteposon P4 (Figure S1a), and we randomly inserted P4 into a circular iRFP gene (Figure 1b). Restriction digestion of the library yielded a DNA pattern consistent with random insertion (Figure S1b).
Our vector library was transformed into Escherichia coli, and individual colonies were screened at either 23 or 37°C for near-infrared fluorescence. Two temperatures were used for screening to increase the likelihood of identifying marginally-stable variants. Screening a total of 1760 colonies identified 38 circularly permuted iRFP (cp-iRFP) with emission >5σ over that observed with the parental strain transformed with a vector lacking an iRFP gene. Assuming that each cp-iRFP had an equal probability of occurring in our library, this level of screening was predicted to sample 58% of the possible cp-iRFP.31 Sequencing vectors from these colonies revealed 27 unique in frame cp-iRFP that retained near-infrared fluorescence with signals ≥5σ of cells lacking an iRFP (Table S1). To visualize how the structures of these permuted proteins relate to one another, the first iRFP residue that is translated in each cp-iRFP was mapped onto the structure of the Deinococcus radiodurans BphP photosensory core (Figure 1c),20 a BphP that displays 39% identity with iRFP.24 This analysis revealed the highest density of variants arising from backbone fission proximal to the domain linker and near the termini of the protein. To determine how this tolerance relates to iRFP folding, we also evaluated the contact order of fluorescent cp-iRFP. Previous studies have shown that CO is inversely correlated with protein folding rates,32 suggesting that increases in CO may add an additional burden to achieving a knotted protein topology. Fluorescent variants had CO values that are both higher and lower than native iRFP. However, the highest density of permuted variants had new termini that clustered in a region with CO values that are lower than iRFP.
Comparison of circularly permuted iRFP in bacteria
Distinct sets of cp-iRFP were discovered at the different temperatures where screening was performed (23 and 37°C). This finding suggested that some cp-iRFP displayed temperature-dependent fluorescence. To test this hypothesis, we compared the emission and excitation spectra of each cp-iRFP with those of native iRFP. Initial spectroscopic measurements at 23°C revealed that all of the cp-iRFP displayed similar peak excitation and emission as native iRFP, albeit with a range of peak intensities (Table S1). At 23°C, five cp-iRFP yielded whole cell emission that were greater than that observed with iRFP, three cp-iRFP yielded intensities that were comparable to that of iRFP, and the remaining cp-iRFP showed emission significantly lower than that of iRFP (Figure 2a). When measurements were performed at 37°C, the majority of the cp-iRFP displayed a signal that was lower than iRFP; only three variants yielded a signal that matched or exceeded iRFP.
Figure 2.
Effect of temperature on fluorescence. (A) The fluorescence of cells expressing each cp-iRFP was measured at 23°C and 37°C, normalized to cell density, and compared to the fluorescence of cells expressing parental iRFP and lacking iRFP (−ctrl). All values are reported as the fraction of the signal obtained with the variant that yielded the highest signal at each temperature. All cp-iRFP except variants 59, 182, 198 and 291 had significantly higher signal than background at 37°C, and all variants except 59, 122, 129, 182 and 198 had signals that were significantly higher than background 23°C (two-tailed t test; p<0.05). (B) The ratio of the fluorescence signals acquired at 37°C to the signals observed at 23°C. Blue and red symbols represent the variants identified in screens at 23°C and 37°C, respectively. White symbols represent cp-iRFP discovered at both temperatures, and the dashed line represents the value obtained for iRFP. Error bars represent ±1σ calculated using three or more independent measurements.
The iRFP fluorescence signal increased ~2.5-fold as temperature was increased from 23 to 37°C (Figure 2b). To determine whether any of our cp-iRFP display enhanced signals as temperature increases, we calculated the ratio of fluorescence emission for each variant expressed in cells grown at these different temperatures. This analysis revealed that 52% of the cp-iRFP displayed higher emission intensity at 37°C compared with 23°C like iRFP (ratio >1), 37% had similar fluorescence ratios as iRFP (ratio ≥ 2.5), and 11% displayed decreased emission as temperature was increased (ratio <1). Among the cp-iRFP with ratios ≤1, 80% were identified when screening was performed at 23°C.
The variability in fluorescence intensity across our different cp-iRFP could have arisen because permutation differentially affected protein expression, folding, cofactor binding, stability, degradation, or quantum yield. In a previous study, we found that backbone fission increases the concentration of BV required for fluorescence in IFP, a homolog of iRFP.18 Because permutation involves backbone fission, this finding suggested that permutation could have altered permuted protein fluorescence by decreasing BV affinity to levels that decreased the fraction of cofactor bound under the conditions of the measurements. To test this hypothesis, we evaluated whether near-infrared fluorescence varied with the amount of BV added to the cells that express each cp-iRFP (Figure S2). Under the conditions of the fluorescence measurements conducted in this study, a majority of the unmodified iRFP was expected to have BV bound, since the BV concentration required for half-maximal fluorescence in cells expressing the unmodified iRFP (11 µM) was ~7-fold lower than the concentration used in our screening protocol. A majority of our cp-iRFP (85%) required a similar (or lower) amount of BV for half-maximal fluorescence, 4 to 15 µM. The remaining variants, which arose from backbone fission in the GAF domain, required between 18 and 25 µM BV for half-maximal fluorescence. These values are lower than the concentration (80 µM) used in our whole cell analysis of fluorescence that yielded signal differences. These findings suggest that the large variability in whole cell fluorescence among our variants, which varied by as much as 98-fold, was not due to differences in the BV-binding affinity and attachment.
To determine if the cp-iRFP arising from backbone fission within the N-terminal tail (e.g., cp-iRFP-11 and cp-iRFP-12) require the residues from this tail for maturation into a fluorescent protein, we created a truncated iRFP that encodes residues 12–316 but lacks residues 1–11. This truncated iRFP displayed similar fluorescence as cp-iRFP-12 (data not shown), suggesting that the initial residues in iRFP are not required for chromophore binding or stability.
Translation initiation variability
Previous studies have shown that the genetic context of a ribosomal binding site (RBS) can alter the strength of translation initiation in bacteria by more than an order of magnitude,27 and vectors in our library express cp-iRFP with gene sequences that change the context of the RBS in our permuteposon. To examine whether permuted gene sequences influence protein expression from this RBS, we used a thermodynamic model for translation initiation to estimate how the strength of our RBS site varies in vectors expressing different cp-iRFP.27 This model uses the sequence and the genetic context of an RBS to quantify the strength of the mRNA-ribosome interaction that is required for translation initiation and competing off-pathway RNA folding reactions that inhibit formation of a productive mRNA-ribosome complex. Experimental validation of this model through forward engineering of RBS sites revealed that the predictions are accurate within a factor of 2.3-fold over a range of translation initiative values spanning five orders of magnitude.27
We found that the calculated translation initiation rates for all possible cp-iRFP varied by three orders of magnitude when expressed using the RBS in permuteposon P4 (Figure 3a). To test whether the fluorescent signals obtained with our original clones correlate with translation initiation rates, we compared the calculated rates for each variant to the whole cell fluorescence signals (Figure 3b). This analysis revealed significant correlations between calculated translation rates and fluorescence intensity. These findings suggest that translation initiation variability contributed to the dispersion in signals in whole cell measurements.
Figure 3.
Thermodynamic analysis of protein expression. (A) Calculated translation rates of all cp-iRFP (line) are compared with the calculated rates for fluorescent variants (●). (B) The relative whole cell fluorescent signals of cp-iRFP at 23°C and 37°C are compared with calculated rates. Calculation of the Spearman's rank correlation using our data acquired at 23°C (RSR = 0.403) and 37°C (RSR = 0.384) yielded values that were significant (two-tailed t test, p < 0.05).
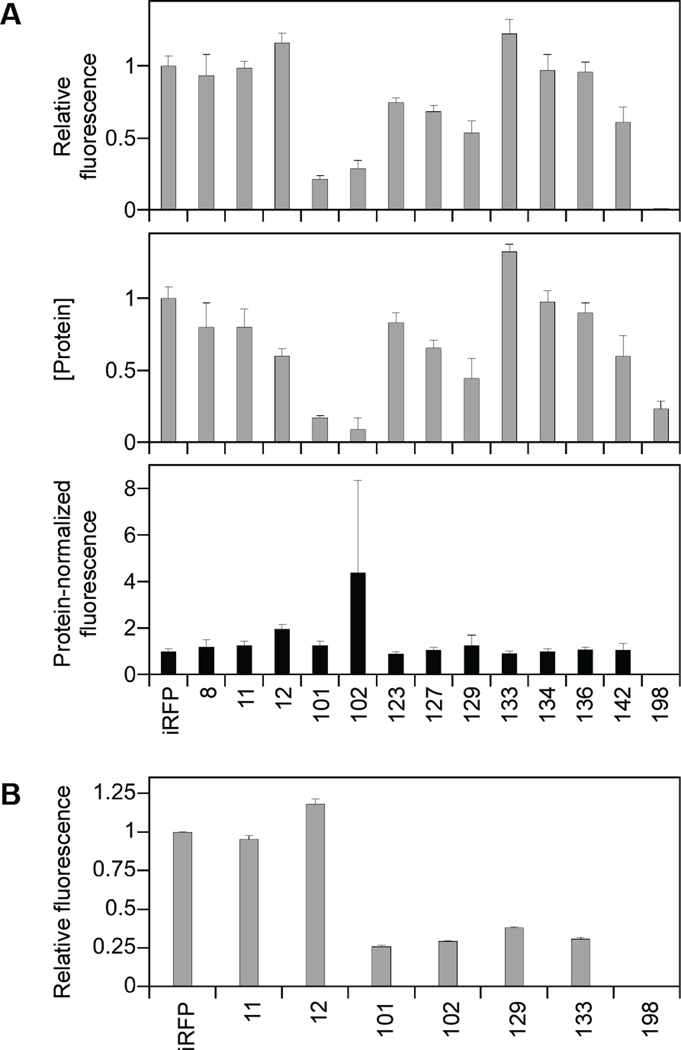
To minimize differences in protein synthesis that arose from changes in RBS genetic context, we created vectors that expressed each of our cp-iRFP with a NH2-terminal His tag connected through a 27 residue polypeptide. When expressed from these vectors, cp-iRFP with NH2 and COOH-termini proximal to the domain linker in unmodified iRFP displayed the highest whole cell fluorescence (Figure S3). The fluorescence of each variant was also compared in the presence and absence of the affinity tag. A majority of the cp-iRFP (85%) displayed an increased signal in the presence of the His tag, indicating that this expression strategy yielded higher levels of each fluorescent BV-bound variant, consistent with predictions from thermodynamic calculations. To better understand the signal variability observed when proteins are expressed under conditions that yield more consistent calculated translation initiation rates, we analyzed the levels of a series of selected cp-iRFP (Figure S4). Specifically, we focused on the subset of cp-iRFP that displayed enhanced fluorescence at 37°C compared with 23°C. Immunoblot analysis revealed that the levels of these cp-iRFP varied up to 10-fold from iRFP (Figure 4a). When whole cell fluorescence was normalized to protein expression in matched samples, a majority of the cp-iRFP displayed a signal that was similar to that measured using cells expressing native iRFP, with the exception of cp-iRFP-198, which displayed a signal 25-fold lower than iRFP, and cp-iRFP-12 and cp-iRFP-102, which exhibited average signals that were 1.9- and 4.4-fold higher than iRFP, respectively. While the signal from cp-iRFP-12 was significantly higher than iRFP, the signal from cp-iRFP-102 displayed a large error and was not significantly different from iRFP.
Figure 4.
Protein-normalized whole cell fluorescence. (A) The whole cell fluorescence of His-tagged cp-iRFP at 37°C in E. coli was normalized to cell density, and immunoblot analysis was used to quantify the levels of each cp-iRFP. The ratio of fluorescence to the level of each protein was calculated using data acquired from matched samples. Error bars represent ±1σ calculated using three or more independent measurements. cp-iRFP-12 displayed protein-normalized fluorescence that was significantly higher than iRFP (two tailed t test; p<0.05), while the cp-iRFP-102 signal was not significantly higher. (B) Fluorescence of circularly permuted iRFP in tissue culture. HeLa cells were transiently transfected with vectors that express different cp-iRFP and a vector that expresses GFP, green and red fluorescence was measured using flow cytometry, and the near-infrared fluorescence signal for each cp-iRFP was normalized to the GFP transfection control. Cells were grown in the medium that had not been supplemented with BV. Error bars represent ±1σ calculated from two independent measurements. Variant 12 displayed a GFP-normalized signal that was significantly higher than iRFP (two tailed t test; p<0.05).
In vitro analysis of circularly permuted iRFP
The variation in protein-normalized fluorescence signals suggested that permutation affected the fraction of cp-iRFP that accumulated in a fluorescence-competent conformation and/or quantum yield. To directly evaluate the effects of permutation on spectroscopic properties, we expressed, purified, and characterized four cp-iRFP. Two circularly permuted iRFP were purified that displayed improved signals (cp-iRFP-12 and cp-iRFP-102), and two were purified that exhibited iRFP-like signals (cp-iRFP-129 and cp-iRFP-133). Proteins were purified using a combination of affinity and reverse phase chromatography. During purification, we found that the reverse phase column yielded two protein populations, one that eluted as a monodisperse peak at ~5% (NH4)2SO4 and one that eluted when the column was washed with buffer lacking (NH4)2SO4. In the case of iRFP, <50% of the total recombinant iRFP eluted at 5% ammonium sulfate, while the remaining iRFP eluted in the absence of ammonium sulfate. All of our biochemical characterization was performed using the protein from the former peak, since it contained holophytochrome as previous observed.25
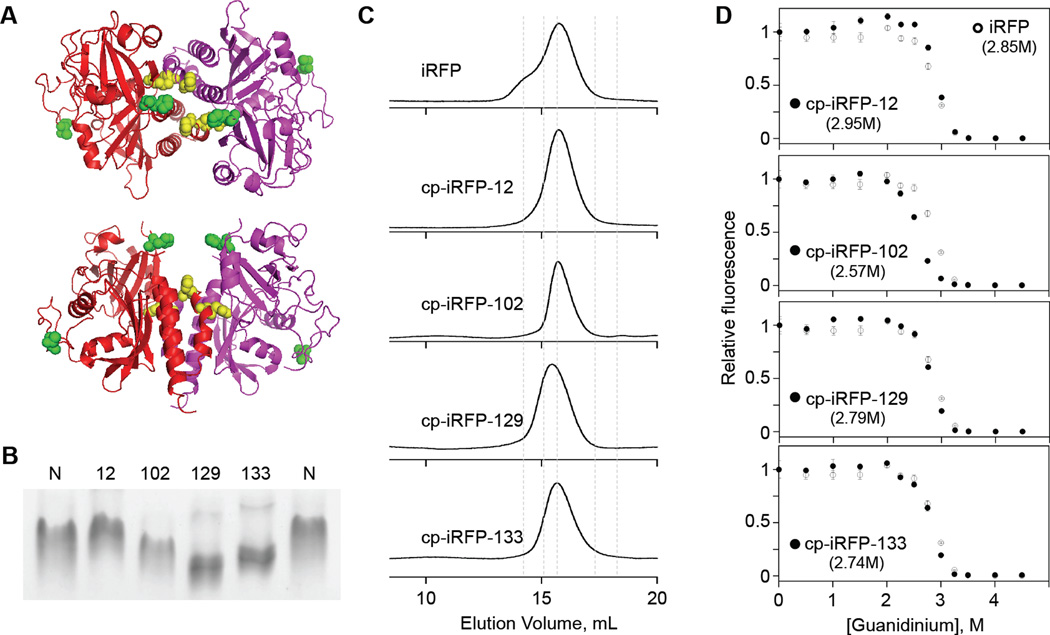
All of the cp-iRFP had three absorbance peaks with maxima at similar wavelengths (280, 391, and 693 nm) to those observed with iRFP (Figure S5).24 In addition, all four cp-iRFP displayed maximal emission wavelengths and quantum yields that were not significantly different from those obtained with iRFP. These findings suggest that permutation does not disrupt the iRFP structure sufficiently to alter the chromophore environment. We next sought to determine how permutation affects protein structure, since the introduction of NH2- and COOH-termini within the iRFP dimerization interface could disrupt oligomerization. cp-iRFP-129 and cp-iRFP-133 have termini that map onto the dimer interface in the BphP crystal structure (Figure 5a), while cp-iRFP-12 and cp-iRFP-102 have termini that map to sites distal from the interface. To test whether these cp-iRFP have structures that differ from iRFP, we compared their electrophoretic mobility using native PAGE (Figure 5b). This analysis revealed that cp-iRFP-102, cp-iRFP-129 and cp-iRFP-133 migrated farther than iRFP. Two of these variants have termini that map onto the dimer interface. To test whether the mobilities of these cp-iRFP were altered because oligomerization was disrupted, analytical gel filtration was used to evaluate their oligomeric state (Figure 5c). As previously observed, iRFP eluted as a monodisperse peak with an apparent MW (72,443 Da) that was 1.92-fold greater than the calculated MW of the iRFP polypeptide. All of the cp-iRFP eluted at volumes consistent with oligomerization. However, the cp-iRFP-129 displayed an apparent molecular weight that was greater than iRFP (91,201 Da). This cp-iRFP also displayed the largest change in electrophoretic mobility with native polyacrylamide gel electrophoresis (PAGE). Taken together, these findings suggest that circular permutation affects the radius of gyration but does not disrupt oligomerization.
Figure 5.
Effect of circular permutation on iRFP structure. (A) Protein termini created by permutation mapped onto the structure of the D. radiodurans BphP chromophore-binding domain. A comparison of the subunits (red and purple) with the residues at the termini of variants 12 and 102 (green) and 129 and 133 (yellow) reveal that only the latter pair of cp-iRFP create termini proximal to the dimer interface. (B) Native PAGE shows how the migration of unmodified iRFP (N) relates to each cp-iRFP. (C) Gel filtration analysis reveals how the elution profiles of iRFP and the four cp-iRFP compare with the elution of standards (dashed lines) having molecular weights of 12.4, 29, 66, 150, and 200 kDa. (D) The effect of GdnHCl concentrations on cp-iRFP fluorescence (●) is compared with unmodified iRFP (○). The concentration required for the unfolding midpoint is indicated for each variant. For cp-iRFP-12, GdnHCl concentrations >1 M and <3.25 displayed signals that were significantly higher than iRFP (two-tailed t-test; P<0.05). Error bars represent ±1σ calculated using three or more measurements.
Since permutation has been shown to alter the stability of other proteins,13 we compared the equilibrium unfolding of cp-iRFP with iRFP, which was previous shown to be irreversible.33 Unfolding was monitored by measuring fluorescence as a function of the chemical denaturant guanidine (GdnHCl). With iRFP, GdnHCl-induced equilibrium unfolding yielded a single cooperative transition with a midpoint at 2.85 M guanidine. All of the variants were destabilized compared with iRFP with the exception of cp-iRFP-12 (Figure 5d), which displayed enhanced stabilization.
Fluorescence in mammalian cells
The rate of protein folding varies between prokaryotes and eukaryotes, with bacterial proteins folding post-translationally due to faster translation rates and eukaryotic proteins folding cotranslationally.34,35 To evaluate whether the nature of the expression system (i.e., prokaryotic or eukaryotic) affects the folding and cofactor binding of cp-iRFP, which are both required for maturation into a fluorescent protein, we assessed the fluorescence signal of seven variants in mammalian cells. Five of these cp-iRFP (11, 12, 101, 102, and 129) were selected because they display the highest fluorescence signals among the iRFP variants discovered in E. coli. The remaining two variants initiate translation within the domain linker (cp-iRFP-133) and within the GAF domain (cp-iRFP-198). The signals from these permuted iRFP were evaluated using HeLa cells that had been transiently transfected with a plasmid encoding each HA-tagged cp-iRFP (or unmodifiied iRFP) and a plasmid constitutively expressing green fluorescent protein (GFP) as a transfection control. Cellular fluorescence was measured using flow cytometry 48-hours post-transfection (Figure S6a). Relative cp-iRFP fluorescence intensities were calculated by normalizing the absolute iRFP fluorescence by the absolute GFP fluorescence to account for differences due to transfection efficiency (Figure 4b). Comparison of the relative near-infrared fluorescence values revealed that cp-iRFP-12 displayed near-infrared fluorescence that was 18% higher than native iRFP. Additionally, one variant displayed a similar intensity as iRFP (cp-iRFP-11), four cp-iRFP showed fluorescence approximately 3-fold lower than iRFP (101, 102, 129 and 133) and one variant presented near undetectable fluorescence (cp-iRFP-198).
To compare the expression levels of cp-iRFP, we evaluated the levels of HA-tagged native and permuted iRFP using Western immunoblots. Experiments were performed using total protein from HeLa cells transfected as described for cytometry analyses (Figure S6b). The intensity of all of the protein bands detected using an HA-specific antibody were normalized to the intensity of an α-tubulin loading control, since our goal was to normalize the cellular fluorescence signal by the total protein expressed. Immunoblot detection of GFP was used to confirm transfection. The relative iRFP fluorescence values obtained by cytometry were normalized to the relative intensity of HA bands, which had been normalized to the loading control (Figure S6c). cp-iRFP-101, cp-iRFP-102 and cp-iRFP-133 presented modest increases (22 to 50%) in protein-normalized fluorescence compared to iRFP, while cp-iRFP-11 and cp-iRFP-12 exhibited modest (8 to 31%) decreases in signal. cp-iRFP-129 presented only 28% of the signal obtained with iRFP, and variant 198 displayed fluorescence that was ~2% of the iRFP signal. While a subset of the variants displayed a similar protein-normalized signal in tissue culture and bacterial cells, several variants displayed weaker signals in tissue culture, whose underlying causes are not known.
Biophysical implications
The results from this study show that knotted BphP are tolerant to a wide range of sequence rearrangements that alter contact order. High-throughput screening revealed that 9% of all possible cp-iRFP display detectable near-infrared fluorescence. Since our screening was predicted to sample only 58% of all possible cp-iRFP, this number likely represents a lower bound on the fraction of permuted iRFP that fold and fluoresce. Our findings also suggest that many of these cp-iRFP display BphP-like structures, because retention of fluorescence requires a bound cofactor within a similar structural environment as iRFP. However, it is not clear which of these cp-iRFP acquire a knot. Our in vitro biochemical studies revealed parent-like stability and spectral properties, suggesting that the BV chromophore environment is not disrupted by permutation, while native gel analysis suggested that some variants may display subtle structural differences from iRFP. Some members of the BphP family lack protein knots, suggesting that our cp-iRFP might not require a knot for fluorescence. Studies of phytochromes having sensory modules that are made up of tandem GAF domains have revealed structures that lack the knot found in PAS-GAF modules.36 Sequence comparisons have also shown that the cyanobacteriochromes AnPixJ and TePixJ lack the loop that serves as the knot lasso,37,38 although the structures of these proteins have not yet been reported. In addition, a recent study revealed that the BphP knot can be removed through truncation without disrupting cofactor binding, provided that a new cysteine residue is incorporated into the GAF domain to covalently attach the cofactor.39 Structural studies are needed to determine how the topology of our cp-iRFP relate to iRFP, and if some of our these variants must thread a peptide through the loop that is larger than the peptide in iRFP to mature into folded and fluorescent proteins.
All of our cp-iRFP displayed excitation and emission maxima that are similar to iRFP. However, at each temperature, we observed a large variation in fluorescence intensities even though each variant was transcribed and translated using the same promoter and RBS. Circular permutation has been shown to alter protein folding,40 ligand binding,12 quantum yield,41 and expression.42 Our results suggest that variability in fluorescence signals do not arise from changes in BV binding affinity. For each variant, the BV concentration required for half-maximal fluorescence was uniformly lower than the concentration used in our assays. Variability in protein expression, on the other hand, is thought to make larger contributions to variability in cellular fluorescence. Thermodynamic calculations analyzing the effects of different cp-iRFP genes on the RBS in our permuteposon revealed a correlation between fluorescence and calculated translation initiation rates. In addition, expression of each cp-iRFP with the same peptide tag at the NH2 terminus, which is thought to improve the consistency of translation initiation across the different variants,27 differentially altered the relative signals obtained with each cp-iRFP from variants expressed without tags. These observations emphasize the importance of considering protein levels when using whole cell assays to screen for circularly permuted proteins with improved fluorescence.
Among the fluorescent variants discovered in our screen, cp-iRFP-12 displayed significantly higher protein-normalized fluorescence compared with iRFP in bacteria. Biochemical characterization of this variant did not reveal any differences in spectral properties compared to the parental iRFP, implicating improved maturation as the underlying cause of the enhanced signal. Our biochemical results also suggest that only a fraction of the iRFP translated in E. coli matures into a fluorescent protein. When we subjected recombinant iRFP to reverse phase chromatography, it eluted at two different salt concentrations, which differed in their fluorescent signals as previously observed.25 cp-iRFP-12 also eluted at two different salt concentrations, although the results from whole cell fluorescence measurements suggest that a greater fraction of this variant fully matures into a fluorescent protein. Mapping cp-iRFP-12 onto the D. radiodurans BphP structure suggests a possible mechanism for enhanced maturation in E. coli.20 Because this permuted protein arises from fission within the NH2-terminal tail, near the BV attachment site at residue fifteen, it has fewer residues to thread through the loop that serves as the iRFP knot lasso. The effects of permutation on protein maturation could be evaluated in future studies by analyzing protein folding and fluorescence within prokaryotic and eukaryotic cell free transcription and translation systems. A recent study demonstrated that this approach can be used to study cofactor binding to a protein using the heme-binding myoglobin.43 A comparison of cofactor incorporation into myoglobin orthologs with varying stability revealed correlation between the levels of holo-myoglobin and the protein stability in the absence of cofactor.43 This trend is consistent with our finding that cp-iRFP-12 displays the highest stability among the proteins characterized using in vitro studies, although our analysis was performed with a holophytochrome rather than an apoprotein.
Applications of circularly permuted iRFP
The permuted proteins discovered have implications for protein design. Circularly permuted proteins are increasingly being used to construct protein switches through domain insertion, where an ensemble of permuted protein variants are randomly inserted at different locations within the backbone of a second protein to create new proteins.44 Domain insertion studies have shown that a subset of the variants created in such libraries display switching behavior, where ligand binding within one domain modulates the structure and function of the second domain.14,15 In fact, high signal-to-noise switches that couple the ligand-binding to the fluorescence of different GFP family members have been created by inserting circularly permuted fluorescent proteins into periplasmic binding proteins.45 The cp-iRFP discovered should be useful for creating similar biosensors, albeit with emission in the near-infrared region which is ideally suited for tissue imaging.24 All of the cp-iRFP displayed parent-like BV affinity, which was sufficient to fluoresce in tissue culture without the addition of biliverdin. This finding suggests that biosensors created using these variants will exhibit sufficient BV affinity to fluoresce in mammalian cells without requiring the addition of exogenous BV. The extent to which each of our cp-iRFP will fold, bind BV, and fluoresce upon insertion into other proteins will require further studies. If knot formation is required for cp-iRFP fluorescence, then it is possible that some cp-iRFP could experience folding challenges upon insertion into other domains.
By using a circular iRFP gene for library construction, rather than a vector-encoded iRFP, we were able to create a library that encodes different cp-iRFP in a single step. This modified approach is quicker than the original protocol described for PERMUTE because it avoids manipulations required after the initial MuA reaction.26 The modified protocol described herein will simplify future studies that seek to create vector libraries that express circularly permuted variants of other proteins. Our results also showed that PERMUTE can be used with a permuteposon that contains a hybrid transposase and ribosomal binding site that only amends two extra amino acids to protein termini.46 The simplified PERMUTE protocol should also be compatible with other permuteposons that have been developed for PERMUTE, including those that amend peptides of varying size to the NH2-termini of permuted proteins.26 Beyond simplifying the construction of permuted protein libraries, this approach has the advantage that it creates permuted proteins with well-defined sequence diversity and avoids random deletions that arise with other methods.47 Applications of deep sequencing to PERMUTE libraries will be useful in future studies to improve our understanding of library diversity and sampling of individual variants, similar to that described for libraries created using random mutation.48
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation [MCB-1150138] (to J.J.S.), the Robert A Welch Foundation [C-1614 and C-1824] (to J.J.S. and L.S.), the John S. Dunn Collaborative Research Award (to J.J.S.), and National Science Foundation (CBET-1336053 and CBET-1254318) (to L.S.).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Detailed experimental procedures, supplementary figures, and a supplementary table. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Liang C, Mislow K. Topological Features of Protein Structures: Knots and Links. J. Am. Chem. Soc. 1995;117:4201–4213. [Google Scholar]
- 2.Mansfield ML. Are there knots in proteins? Nat. Struct. Biol. 1994;1:213–214. doi: 10.1038/nsb0494-213. [DOI] [PubMed] [Google Scholar]
- 3.Jamroz M, Niemyska W, Rawdon EJ, Stasiak A, Millett KC, Sułkowski P, Sulkowska JI. KnotProt: a database of proteins with knots and slipknots. Nucleic Acids Res. 2015;43:D306–D314. doi: 10.1093/nar/gku1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallam AL, Jackson SE. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nat. Chem. Biol. 2012;8:147–153. doi: 10.1038/nchembio.742. [DOI] [PubMed] [Google Scholar]
- 5.Sułkowska JI, Sulkowski P, Szymczak P, Cieplak M. Stabilizing effect of knots on proteins. Proc Natl Acad Sci USA. 2008;105:19714–19719. doi: 10.1073/pnas.0805468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO. Structure and folding of a designed knotted protein. Proc Natl Acad Sci USA. 2010;107:20732–20737. doi: 10.1073/pnas.1007602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner J, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- 8.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 9.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat. Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y, Lutz S. Improved triglyceride transesterification by circular permuted Candida antarctica lipase B. Biotechnol. Bioeng. 2010;105:44–50. doi: 10.1002/bit.22471. [DOI] [PubMed] [Google Scholar]
- 11.Daugherty AB, Govindarajan S, Lutz S. Improved biocatalysts from a synthetic circular permutation library of the flavin-dependent oxidoreductase old yellow enzyme. J. Am. Chem. Soc. 2013;135:14425–14432. doi: 10.1021/ja4074886. [DOI] [PubMed] [Google Scholar]
- 12.Guntas G, Kanwar M, Ostermeier M. Circular permutation in the Ω-loop of TEM-1 β-lactamase results in improved activity and altered substrate specificity. PLoS ONE. 2012;7:e35998. doi: 10.1371/journal.pone.0035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead TA, Bergeron LM, Clark DS. Tying up the loose ends: circular permutation decreases the proteolytic susceptibility of recombinant proteins. Protein Eng. Des. Sel. 2009;22:607–613. doi: 10.1093/protein/gzp034. [DOI] [PubMed] [Google Scholar]
- 14.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Creation of a zymogen. Nat. Struct. Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.To T-L, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci USA. 2015;112:3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey N, Nobles CL, Zechiedrich L, Maresso AW, Silberg JJ. Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein-protein interactions. ACS Synth Biol. 2015;4:615–624. doi: 10.1021/sb5002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockwell NC, Su Y-S, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 21.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 22.Quail PH. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 23.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science (New York, N.Y.) 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filonov GS, Piatkevich KD, Ting L-M, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auldridge ME, Satyshur KA, Anstrom DM, Forest KT. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. Journal of Biological Chemistry. 2012;287:7000–7009. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta MM, Liu S, Silberg JJ. A transposase strategy for creating libraries of circularly permuted proteins. Nucleic Acids Res. 2012;40:e71–e71. doi: 10.1093/nar/gks060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Würth C, Grabolle M, Pauli J, Spieles M, Resch-Genger U. Relative and absolute determination of fluorescence quantum yields of transparent samples. Nat Protoc. 2013;8:1535–1550. doi: 10.1038/nprot.2013.087. [DOI] [PubMed] [Google Scholar]
- 29.Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. U.S.A. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segall-Shapiro TH, Nguyen PQ, Santos, Dos ED, Subedi S, Judd J, Suh J, Silberg JJ. Mesophilic and hyperthermophilic adenylate kinases differ in their tolerance to random fragmentation. J. Mol. Biol. 2011;406:135–148. doi: 10.1016/j.jmb.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 31.Bosley AD, Ostermeier M. Mathematical expressions useful in the construction, description and evaluation of protein libraries. Biomol. Eng. 2005;22:57–61. doi: 10.1016/j.bioeng.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 33.Stepanenko OV, Bublikov GS, Stepanenko OV, Shcherbakova DM, Verkhusha VV, Turoverov KK, Kuznetsova IM. A knot in the protein structure - probing the near-infrared fluorescent protein iRFP designed from a bacterial phytochrome. FEBS J. 2014;281:2284–2298. doi: 10.1111/febs.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 35.Agashe VR, Guha S, Chang H-C, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 36.Anders K, Daminelli-Widany G, Mroginski MA, Stetten, von D, Essen L-O. Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. Journal of Biological Chemistry. 2013;288:35714–35725. doi: 10.1074/jbc.M113.510461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narikawa R, Fukushima Y, Ishizuka T, Itoh S, Ikeuchi M. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 2008;380:844–855. doi: 10.1016/j.jmb.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 38.Ishizuka T, Narikawa R, Kohchi T, Katayama M, Ikeuchi M. Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007;48:1385–1390. doi: 10.1093/pcp/pcm106. [DOI] [PubMed] [Google Scholar]
- 39.Rumyantsev KA, Shcherbakova DM, Zakharova NI, Emelyanov AV, Turoverov KK, Verkhusha VV. Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence. Sci Rep. 2015;5:18348. doi: 10.1038/srep18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindberg M, Tångrot J, Oliveberg M. Complete change of the protein folding transition state upon circular permutation. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- 41.Topell S, Hennecke J, Glockshuber R. Circularly permuted variants of the green fluorescent protein. FEBS Lett. 1999;457:283–289. doi: 10.1016/s0014-5793(99)01044-3. [DOI] [PubMed] [Google Scholar]
- 42.Jones AM, Mehta MM, Thomas EE, Atkinson JT, Segall-Shapiro TH, Liu S, Silberg JJ. The structure of a thermophilic kinase shapes fitness upon random circular permutation. ACS Synth Biol. 2016 doi: 10.1021/acssynbio.5b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuel PP, Smith LP, Phillips GN, Olson JS. Apoglobin Stability Is the Major Factor Governing both Cell-free and in Vivo Expression of Holomyoglobin. Journal of Biological Chemistry. 2015;290:23479–23495. doi: 10.1074/jbc.M115.672204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostermeier M. Designing switchable enzymes. Curr. Opin. Struct. Biol. 2009;19:442–448. doi: 10.1016/j.sbi.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marvin JS, Schreiter ER, Echevarría IM, Looger LL. A genetically encoded, high-signal-to-noise maltose sensor. Proteins. 2011;79:3025–3036. doi: 10.1002/prot.23118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segall-Shapiro TH, Meyer AJ, Ellington AD, Sontag ED, Voigt CA. A “resource allocator” for transcription based on a highly fragmented T7 RNA polymerase. Molecular Systems Biology. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graf R, Schachman HK. Random circular permutation of genes and expressed polypeptide chains: application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firnberg E, Ostermeier M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS ONE. 2012;7:e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.