Abstract
Colorectal cancer (CRC) arises from mutations in a subset of genes. We investigated the germline and somatic mutation spectrum of patients with CRC in Taiwan by using the AmpliSeq Cancer Hotspot Panel V2. Fifty paired freshly frozen stage 0–IV CRC tumors and adjacent normal tissue were collected. Blood DNA from 20 healthy donors were used for comparison of germline mutations. Variants were identified using an ion-torrent personal genomic machine and subsequently confirmed by Sanger sequencing or pyrosequencing. Five nonsynonymous germline variants on 4 cancer susceptible genes, CDH1, APC, MLH1, and NRAS, were observed in 6 patients with CRC (12%). Among them, oncogene NRAS G138R variant was identified as having a predicted damaging effect on protein function, which has never been reported by other laboratories. CDH1 T340A variants were presented in 3 patients. The germline variants in the cancer patients differed completely from those found in asymptomatic controls. Furthermore, a total of 56 COSMIC and 21 novel somatic variants distributed in 20 genes were detected in 44 (88%) of the CRC samples. High inter- and intra-tumor heterogeneity levels were observed. Nine rare variants located in the β-catenin binding region of the APC gene were discovered, 7 of which could cause amino acid frameshift and might have a pathogenic effect. In conclusion, panel-based mutation detection by using a high-throughput sequencing platform can elucidate race-dependent cancer genomes. This approach facilitates identifying individuals at high risk and aiding the recognition of novel mutations as targets for drug development.
Keywords: next-generation sequencing, colorectal cancer, ampliSeq cancer hotspot panel, NRAS, APC
INTRODUCTION
Internationally, more than 1 million people are diagnosed for colorectal cancer (CRC) annually, and it is the third leading cause of cancer mortality in Taiwan [1]. CRC arises from a series of sequentially mutated genes that can transform normal epithelial cells into adenoma, eventually becoming adenocarcinoma [2, 3]. The polyp-to-cancer transition takes several years and involves alteration of adenomatous polyposis coli (APC), Kirsten-RAS(KRAS), TP53, and other genes that have a role in controlling the cell proliferation process [4, 5]. Except in cases of somatic genomic alteration, CRC can be inherited. The estimated fraction of CRC attributed to inherited predisposition ranges between 10% and 30%. Most cancer-susceptible genes are involved in restraining cell proliferation, DNA repair, and genetic stability [6]. In inherited CRC, defects in the tumor suppressor gene APC is a well-known cause of familial adenomatous polyposis (FAP), and hereditary nonpolyposis CRC (HNPCC) is due to germline mutations on DNA mismatch repair (MMR) genes such as MLH1 and MSH2 [7, 8].
In the past decade, the success of the Human Genome Project has engendered unprecedented advancements in “precision medicine” [9]. Mutation-derived individual cancer therapy improves patient cure rates. In metastatic CRC, cetuximab or panitumumab, which are monoclonal antibodies and can specifically block epidermal growth factor receptor (EGFR) signal pathways, are effective only in patients with wild-type KRAS and BRAF [10]. Currently, KRAS (Codons 12 and 13) and BRAFV600E mutation detection are routine molecular companion tests in clinical laboratories before the administration of monoclonal antibody therapy. However, a considerable body of evidence shows that other mutations occurring in genes on EGFR pathways, such as NRAS [11] and KRAS Codons 61 and 146 [12], are associated with a poor response rate for monoclonal antibody therapy. In addition, the Food and Drug Administration has approved almost 30 types of targeted cancer drugs to specific indications [13], and hundreds of clinical trials are ongoing to develop new drugs targeting certain genes exhibiting specific mutations [14]. Prognosis prediction also relies on specific gene mutation patterns for stratifying patients. For instance, CRC patients with TP53 mutation, particularly in Codon 175, have a shorter survival period compared with those with wild-type TP53 [15, 16]. Therefore, developing a high-throughput screening platform that can cover most cancer–related genes is warranted in order to improve the management of patient care.
Recently, targeted next-generation sequencing (NGS) has provided unprecedented potential for detecting underlying changes in the genetic architecture of cancer in a comprehensive and economically feasible manner [17]. The Cancer Genome Atlas study [18] as well as several hospital- and commercial company-based study groups [17, 19, 20] have used NGS platforms to seek specific germline and somatic mutation signatures in CRC, confirming the existence of population-specific mutation patterns [21]. Moreover, race-dependent differences could influence the survival rate of CRC patients under certain conditions [22]. In the present study, we aimed to discover germline and somatic variants in paired tumors and adjacent normal tissues from patients with CRC in Taiwan. Through extensive sequencing on 50 cancer-related genes, a novel germline mutation on oncogene NRAS, instead of on well-known cancer-predisposing genes, was observed in our population. Rare somatic variants with frameshift mutations on APC and cetuximab resistance mutation on KRAS were identified in the tumor tissues. The findings obtained from this type of research can alter the design of gene contents either for screening high-risk individuals with a family history of CRC, or for candidate selection for target therapy.
RESULTS
One novel germline variant on NRAS and 4 variants on 3 cancer susceptible genes detected in the CRC-adjacent normal tissue samples but not in the asymptomatic controls
To elucidate which genomic alterations were inherited or acquired, paired tumor and adjacent normal tissue samples were collected from 50 patients with CRC. Table 1 lists the clinical features of all enrolled participants. Sixty percent of CRC patients received a diagnosis at early stage (stages 0–II), and the location of tumors was evenly distributed among the right site, left of the large intestine, and the rectum. All patients received surgery to remove tumor lesions. Only one patient with metastatic cancer received target therapy of anti-EGFR monoclonal antibody cetuximab. For an advanced comparison of germline variants between cancer patients and the general population, PBMC DNA samples were collected from 20 young asymptomatic controls with normal CEA and iFOBT lab data and no self-reported cancer history who were recruited for this study.
Table 1. Clinical characteristics of the 50 CRC patients and 20 asymptomatic controls.
| Clinical features | CRC patient | Asymptomatic control | |
|---|---|---|---|
| (n=50) | (n=20) | ||
| Sex | Male | 27 (54%) | 9 (45%) |
| Female | 23 (46%) | 11 (55%) | |
| Age | Median (range) | 64 (37-86) | 30 (25-48) |
| CEA | Positive rate | 16% | 0% |
| iFOBT | Positive rate | 50% | 0% |
| Tumor Stage | 0+I | 13 (26%) | NA |
| II | 17 (34%) | NA | |
| III | 19 (38%) | NA | |
| IV | 1 (2%) | NA | |
| Tumor site | Right* | 14 (28%) | NA |
| Left** | 21 (42%) | NA | |
| Rectum | 15 (30%) | NA | |
| Treatment | Surgery | 50 (100%) | NA |
| Chemotherapy | 23 (56%) | NA | |
| Radiotherapy | 3 (6%) | NA | |
| Target therapy | 1 (2%) | NA |
Right site = cancer located in the cecum, ascending colon, transverse colon, and hepatic flexture
Left site = cancer located in the sigmoid, descending colon, and splenic flexture
NA = not available
Six (12%, 6/50) patients with CRC were identified as carrying germline mutations after being subjected to an analysis pipeline for nonsynonymous variants. Table 2 summarizes the location, annotation, and frequency of the germline and somatic variants in the 6 CRC patients. A total of 5 germline variants were detected on 4 genes, among which a missense variant in the E-cadherin gene (CDH1) T340A (COSMIC19821) was present in 3 patients with CRC (Sample ID 1307, 1705, 1738), which represented a germline mutation hotspot in our population (6%, 3/50). The other 2 variants (V1125A and V1352A) on APC and one variant (R291Q) on MLH1 were observed in 3 patients with a family history of CRC. Notably, one novel germline mutation, NRAS G138R was observed in Sample ID 1736. In this case, SIFT software selects 122 sequences which are closely related to Homo sapiens NRAS and calculate the effect of G138R substitution on NRAS function. It generates a score of 0.01 which is predicted to be deleterious to affect NRAS function. On the other hand, PolyPhen2 software aligns 75 amino acids sequences surrounding the 138th Glycine position from 206 species and found 91.6% of identity in Glycine which means high conservation in this position among species. According to the phylogenetic and structural information of this substitution, this NRAS G138R mutation is predicted to be Possibly Damaging with a score of 0.764 (sensitivity: 0.85; specificity: 0.92). This germline variant harbored an oncogene instead of the tumor suppressor gene or DNA repair gene, which are typical in cases of inherited CRC.
Table 2. Clinical information of 6 CRC patients and their genomic alterations in paired tumor and adjacent normal tissue samples. Dark blocks highlight the same germline heterozygous variants detected in both compartments.
| Patient no | Sample ID | Gender | Age at diagnosis | Family history | Adjacent normal | Tumor | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Locus | a. a. change | Frequency | Gene | Locus | a. a. change | Frequency | |||||
| 1 | 1307 | Male | 64 | Yes | CDH1 | chr16:68846047 | p.T340A | 51% | CDH1 | chr16:68846047 | p.T340A | 48% |
| MLH1 | chr3:37067255 | p.R291Q | 50% | MLH1 | chr3:37067255 | p.R291Q | 51% | |||||
| NRAS | chr1:115256529 | p.Q61R | 67% | |||||||||
| 2 | 1423 | Male | 78 | Yes | APC | chr5:112175346 | p.V1352A | 50% | APC | chr5:112175346 | p.V1352A | 50% |
| KRAS | chr12:25398284 | p.G12D | 39% | |||||||||
| 3 | 1461 | Male | 47 | Yes | APC | chr5:112174665 | p.V1125A | 49% | APC | chr5:112174665 | p.V1125A | 49% |
| HNPCC himself | ERBB2 | chr17:37881426 | p.D873G | 22% | ||||||||
| FGFR1 | chr8:38285950 | p.A32D | 23% | |||||||||
| EGFR | chr7:55211101 | p.N115T | 19% | |||||||||
| KDR | chr4:55980297 | p.S265L | 18% | |||||||||
| ATM | chr11:108236087 | p.R3008H | 20% | |||||||||
| PIK3CA | chr3:178916876 | p.R88Q | 17% | |||||||||
| PIK3CA | chr3:178916946 | p.K111N | 17% | |||||||||
| TP53 | chr7:7578212 | p.R213* | 43% | |||||||||
| APC | chr5:112174631 | p.R1114* | 38% | |||||||||
| 4 | 1705 | Female | 73 | Yes | CDH1 | chr6:68846047 | p.T340A | 48% | CDH1 | chr16:68846047 | p.T340A | 53% |
| 5 | 1736 | Female | 53 | NO | NRAS | chr1:115252228 | p.G138R | 48% | NRAS | chr1:115252228 | p.G138R | 47% |
| KRAS | chr12:25398284 | p.G12V | 35% | |||||||||
| APC | chr5:112175775 | p.S1495I | 73% | |||||||||
| APC | chr5:112175777 | p.1496_1498fs | 73% | |||||||||
| APC | chr5:112175792 | p.S1501A | 73% | |||||||||
| 6 | 1738 | Male | 84 | NO | CDH1 | chr6:68846047 | p.T340A | 52% | CDH1 | chr6:68846047 | p.T340A | 53% |
| TP53 | chr5:7578212 | p.R213* | 49% | |||||||||
| APC | chr7:112175213 | p.1309fs* | 20% | |||||||||
Four patients with germline mutations had a family history of CRC (Table 2), among whom Sample ID 1461 with APC V1125A variant had received a diagnosis of HNPCC. Notably, APC is highly associated with FAP and rarely reported in cases of HNPCC. However, this patient developed HNPCC at the age of 47 years, which was younger than most patients with germline mutations in our study.
In general, germline mutations exhibit a mutation frequency of approximately 50% in both normal and tumor tissues. Except for germline variants, 5 of the 6 patients acquired at least one additional somatic mutation in their paired tumor tissues. Notably, Sample ID 1736 gained 4 somatic mutations and Sample ID 1461 gained 9 additional somatic mutations in the tumor samples (except for the inherited NRAS G138R and APC V1125A variant, respectively). Apart from these 5 patients, only one patient (Sample ID 1705) developed cancer with only one heterozygous germline CDH1 mutation with the absence of other somatic mutations.
To investigate the germline variants in the general population, 20 PBMC DNA samples were collected from the asymptomatic controls and examined. Figure 1 depicts the germline variants obtained from the normal tissues of the CRC patients and controls. Notably, 5 independent variants were observed in each group. Compared with the variants observed in the CRC group, the set of germline alterations in the control group were completely distinct. SMO R199W was the only COSMIC variant; the other 4 nonsynonymous variants were predicted as having “tolerated” or “benign” impacts on protein function, and sporadically presented in one individual.
Figure 1. Five unique independent nonsynonymous variants were identified in the 50 normal tissue samples from the CRC patients (left half) and 20 PBMC DNA from the asymptomatic controls (right half).
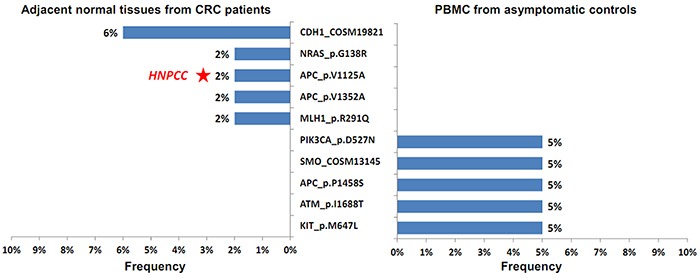
The frequency of each mutant is shown. The red asterisk indicates that the APC V1125A germline variant is present in one patient (Sample ID 1461) with diagnosed HNPCC.
Twenty-one novel somatic mutations detected in the CRC tumor tissue samples
Initially, 830 variants were identified in the 50 tumor tissue samples. After annotation, only nonsynonymous, frameshift, or stopgain variants affecting the amino acid constitution or function(s) of encoded proteins were filtered. Those variants were further filtered with variants found in adjacent normal tissue samples. Finally, a total of 77 distinct variants on 20 genes were identified and confirmed by Sanger sequencing or pyrosequencing. Among these, 21 (27%) were novel variants not in the COSMIC database. Table 3 ranks the 77 somatic variants by frequency in the 44 CRC patients and provides the serial number of each variant for each gene. According to the variant effect, 56 missense mutations (73%), 13 indel mutations (17%), and 8 nonsense mutations (10%) were identified. Missense mutations of Codons 12 and 13 on KRAS (including G12V, G12D, G12S, and G13D) were the most frequently observed variants in the CRC samples (36%, 18/50). Indel and nonsense mutations, which can lead to truncated proteins, were distributed mostly on TP53 and APC genes.
Table 3. Seventy-seven somatic variants distributed on 20 genes in CRC tumors. The 21 novel variants not recorded in the COSMIC database are shaded block.
| Gene | Variant serial no | Coding sequence | Transcript | Variant type | Variant effect | Patient no |
|---|---|---|---|---|---|---|
| TP53 | 1 | c.524G>A | NM_000546 | COSM10648_p.R175H | nonsynonymous missense | 4 |
| 2 | c.637C>T | NM_000546 | COSM10654_p.R213* | stop gain | 3 | |
| 3 | c.844C>T | NM_000546 | COSM10704_p.R282W | nonsynonymous missense | 1 | |
| 4 | c.818G>A | NM_000546 | COSM10660_p.R273H | nonsynonymous missense | 1 | |
| 5 | c.814G>A | NM_000546 | COSM10891_p.V272M | nonsynonymous missense | 1 | |
| 6 | c.797delG | NM_000546 | COSM44187_p.G266fs | frameshift | 1 | |
| 7 | c.796G>A | NM_000546 | COSM10794_p.G266R | nonsynonymous missense | 1 | |
| 8 | c.743G>A | NM_000546 | COSM10662_p.R248Q | nonsynonymous missense | 1 | |
| 9 | c.742C>T | NM_000546 | COSM10656_p.R248W | nonsynonymous missense | 1 | |
| 10 | c.592G>T | NM_000546 | COSM44241_p.E198* | stop gain | 1 | |
| 11 | c.586C>T | NM_000546 | COSM10705_p.R196* | stop gain | 1 | |
| 12 | c.536A>G | NM_000546 | COSM10889_p.H179R | nonsynonymous missense | 1 | |
| 13 | c.476C>T | NM_000546 | COSM11148_p.A159V | nonsynonymous missense | 1 | |
| 14 | c.469G>T | NM_000546 | COSM10670_p.V157F | nonsynonymous missense | 1 | |
| 15 | c.455C>T | NM_000546 | COSM10790_p.P152L | nonsynonymous missense | 1 | |
| 16 | c.406C>T | NM_000546 | COSM11166_p.Q136* | stop gain | 1 | |
| 17 | c.379T>C | NM_000546 | COSM44687_p.S127P | nonsynonymous missense | 1 | |
| 18 | c.294_297delTTCC | NM_000546 | COSM278467_p.S99fs*23 | frameshift +stop gain | 1 | |
| 19 | c.277_278insCCTGGCCCCT | NM_000546 | p.L93fs | frameshift | 1 | |
| KRAS | 1 | c.35G>T | NM_033360 | COSM520_p.G12V | nonsynonymous missense | 8 |
| 2 | c.38G>A | NM_033360 | COSM532_p.G13D | nonsynonymous missense | 5 | |
| 3 | c.35G>A | NM_033360 | COSM521_p.G12D | nonsynonymous missense | 4 | |
| KRAS | 4 | c.436G>A | NM_033360 | COSM19404_p.A146T | nonsynonymous missense | 2 |
| 5 | c.34G>A | NM_033360 | COSM517_p.G12S | nonsynonymous missense | 1 | |
| 6 | c.179G>A | NM_033360 | COSM87290_p.G60D | nonsynonymous missense | 1 | |
| 7 | c.183A>C | NM_033360 | COSM554_p.Q61H | nonsynonymous missense | 1 | |
| APC | 1 | c.2626C>T | NM_000038 | COSM18852_p.R876* | stop gain | 3 |
| 2 | c.3340C>T | NM_000038 | COSM13125_p.R1114* | stop gain | 2 | |
| 3 | c.3921_3925delAAAAG | NM_000038 | COSM18764_p.E1309fs*4 | frameshift+stop gain | 1 | |
| 4 | c.3964G>T | NM_000038 | COSM18702_p.E1322* | stop gain | 1 | |
| 5 | c.4330C>T | NM_000038 | COSM19021_p.Q1444* | stop gain | 1 | |
| 6 | c.4484G>T | NM_000038 | COSM99778_p.S1495I | nonsynonymous missense | 1 | |
| 7 | c.4661dupA | NM_000038 | p.E1554fs | frameshift | 2 | |
| 8 | c.3360delA | NM_000038 | p.G1120fs | frameshift | 1 | |
| 9 | c.4282delG | NM_000038 | p.G1428fs | frameshift | 1 | |
| 10 | c.4285delC | NM_000038 | p.Q1429fs | frameshift | 1 | |
| 11 | c.4313_4314insCACCT | NM_000038 | p.T1438fs | frameshift | 1 | |
| 12 | c.4348_4357delCGAGAAGTAC | NM_000038 | p.1450_1453fs | frameshift | 1 | |
| 13 | c.4475C>T | NM_000038 | p.A1492V | nonsynonymous missense | 1 | |
| 14 | c.4486_4493delACTCCAGA | NM_000038 | p.1496_1498fs | frameshift | 1 | |
| 15 | c.4501T>G | NM_000038 | p.S1501A | nonsynonymous missense | 1 | |
| PIK3CA | 1 | c.1633G>A | NM_006218 | COSM763_p.E545K | nonsynonymous missense | 4 |
| 2 | c.263G>A | NM_006218 | COSM746_p.R88Q | nonsynonymous missense | 2 | |
| 3 | c.1035T>A | NM_006218 | COSM754_p.N345K | nonsynonymous missense | 2 | |
| 4 | c.331A>G | NM_006218 | COSM13570_p.K111E | nonsynonymous missense | 1 | |
| 5 | c.333G>T | NM_006218 | COSM27505_p.K111N | nonsynonymous missense | 1 | |
| 6 | c.1258T>C | NM_006218 | COSM757_p.C420R | nonsynonymous missense | 1 | |
| PIK3CA | 7 | c.3073A>G | NM_006218 | COSM771_p.T1025A | nonsynonymous missense | 1 |
| 8 | c.248T>C | NM_006218 | p.F83S | nonsynonymous missense | 1 | |
| 9 | c.3073A>C | NM_006218 | p.T1025P | nonsynonymous missense | 1 | |
| SMAD4 | 1 | c.1082G>A | NM_005359 | COSM14122_p.R361H | nonsynonymous missense | 2 |
| 2 | c.353C>T | NM_005359 | COSM14215_p.A118V | nonsynonymous missense | 1 | |
| 3 | c.1081C>T | NM_005359 | COSM14140_p.R361C | nonsynonymous missense | 1 | |
| 4 | c.1496G>A | NM_005359 | COSM14221_p.C499Y | nonsynonymous missense | 1 | |
| 5 | c.344G>A | NM_005359 | p.C115Y | nonsynonymous missense | 1 | |
| 6 | c.1586T>C | NM_005359 | p.L529S | nonsynonymous missense | 1 | |
| FBXW7 | 1 | c.1154G>A | NM_018315 | COSM117308_p.R385H | nonsynonymous missense | 2 |
| 2 | c.1273C>T | NM_018315 | COSM74637_p.R425C | nonsynonymous missense | 1 | |
| 3 | c.1504T>A | NM_018315 | COSM1427667_p.S502T | nonsynonymous missense | 1 | |
| 4 | c.562_563delAT | NM_018315 | COSM1052123_p.M188fs*18 | frameshift+stop gain | 1 | |
| NRAS | 1 | c.182A>G | NM_002524 | COSM584_p.Q61R | nonsynonymous missense | 2 |
| 2 | c.181C>A | NM_002524 | COSM580_p.Q61K | nonsynonymous missense | 1 | |
| BRAF | 1 | c.1799T>A | NM_004333 | COSM476_p.V600E | nonsynonymous missense | 1 |
| 2 | c.1780G>A | NM_004333 | COSM27639_p.D594N | nonsynonymous missense | 1 | |
| CTNNB1 | 1 | c.121A>G | NM_001904 | COSM5664_p.T41A | nonsynonymous missense | 1 |
| 2 | c.131_133delCTT | NM_001904 | COSM33668_p.S45del | frameshift | 1 | |
| GNAS | 1 | c.602G>A | NM_001077489 | COSM27895_p.R201H | nonsynonymous missense | 1 |
| AKT1 | 1 | c.49G>A | NM_001014432 | COSM33765_p.E17K | nonsynonymous missense | 1 |
| ATM | 1 | c.9023G>A | NM_000051 | COSM21626_p.R3008H | nonsynonymous missense | 1 |
| ERBB4 | 1 | c.1825G>A | NM_001042599 | COSM131772_p.D609N | nonsynonymous missense | 1 |
| FGFR3 | 1 | c.1153T>G | NM_001163213 | p.F383V | nonsynonymous missense | 1 |
| ERBB2 | 1 | c.2618A>G | NM_004448 | p.D873G | nonsynonymous missense | 1 |
| FGFR1 | 1 | c.95C>A | NM_023106 | p.A32D | nonsynonymous missense | 1 |
| PDGFRA | 1 | c.2470G>A | NM_006206 | p.V824I | nonsynonymous missense | 1 |
| EGFR | 1 | c.344A>C | NM_005228 | p.N115T | nonsynonymous missense | 1 |
| KDR | 1 | c.794C>T | NM_002253 | p.S265L | nonsynonymous missense | 1 |
| PTEN | 1 | c.71A>T | NM_000314 | p.D24V | nonsynonymous missense | 1 |
Figure 2 illustrates the distribution of these variants on 20 genes stratified according to each CRC patient. The number of somatic variants in each CRC tumor sample ranged from 0 to 9 with an average of 2.2. The most frequently mutated gene was TP53 (46%), followed by KRAS (44%), APC (32%), PIK3CA (24%), SMAD4 (14%), FBXW7 (10%), and NRAS (6%), all of which accounted for 88% of CRC patients. The variant frequency in one tumor can be quantified and marked according to the size and color of the circle in Figure 2. The mutation frequency of each variant in one tumor (in one column) can be compared to clarify the possible clonal expansion history. Most tumors can exert stepwise mutation on various genes. Moreover, we observed multiple mutants in one gene. For example, in the tumor of Sample ID 1736, 3 mutations were observed in the APC gene including one 8 nucleotide deletion (Variant Serial Number 14 in Table 3) and 2 missense alterations (Variant Serial Numbers 6 and 15). Other evidence of multiple mutations in one gene can be discovered in the TP53 and PIK3CA genes in Sample IDs 1459, 1461, and 1801.
Figure 2. Distribution of 77 somatic variants on 20 genes in the 50 CRC patients.
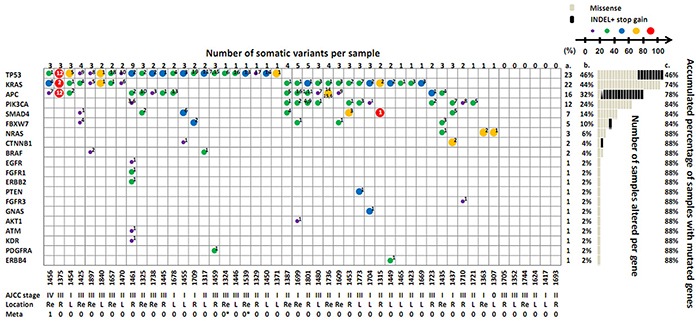
The number on the uppermost layer represents the number of somatic variants per sample. The size and color of the circle in the cell represents the individual variant frequency. For interpreting the meaning of each symbol, please refer to the scale bar. The number labeled in the cell indicates the designated variant serial number of each gene, which are listed in Table 3. More than one number in one cell indicates multiple mutations in the same gene. The gray line at the right part of the figure indicates a missense mutation, and the black bar denotes an indel or nonsense mutation. Columns a and b denote the number and percentage of samples altered per gene; Column c denotes the accumulated percentage of samples with mutated genes. The patient information denoted at the bottom of the figure includes the tumor ID, AJCC stage, location, and metastasis condition. “Re” = rectum; “R” = right site; “L” = left site; 1 = metastasis; 0 = no evidence of metastasis; * = follow-up observation of metastasis events 1 year after surgery in 2 stage III patients.
Correlation of mutation rate and variant frequency with clinicopathological factors
To investigate the possible correlation between the mutants and disease status, Table 4 summarizes the mutation rate of the top 4 mutated genes and the average variant frequency in the mutated tumors stratified by cancer stage and tumor location. Except for the correlation between the TP53 mutation rate and the advanced tumor stage (65% vs 33.3%, P = .027), tumors at a higher stage (stage III and IV) do not exhibit higher mutation rates relative to the total gene mutation rate (85% vs 90% in late stage vs early stage). Patients with tumors in the right site appeared to have a higher KRAS mutation frequency (right site vs left site vs rectum, 62% ± 21.6% vs 38% ± 17.6%, 31.9% ± 12.5%, P = .025), although only a small portion of tumors exhibited KRAS mutation in the right site (28.5%). No particular tendency in the mutation pattern at different locations of the large intestine lumen was observed.
Table 4. Correlation of the mutation rate and variants frequency of top-4 mutated genes stratified by tumor AJCC stage and tumor location.
| AJCC stage | Tumor location | |||||||
|---|---|---|---|---|---|---|---|---|
| Stage 0-II | Stage III-IV | p value | Rectum | Left | Right | p value | ||
| n=30 | n=20 | n=15 | n=21 | n=14 | ||||
| Total genes | Tumor with mutation (%) | 90 | 85 | 0.594 | 93.3 | 85.7 | 85.7 | 0.749 |
| Variants frequency (%) | 40.2 ± 18.74 | 32 ± 20.4 | 0.046 | 33 ± 16.8 | 37.6 ± 19.0 | 40 ± 22.7 | 0.226 | |
| TP53 | Tumor with mutation (%) | 33.3 | 65 | 0.027 | 53.3 | 42.8 | 42.8 | 0.793 |
| Variants frequency (%) | 42.7 ± 18.8 | 41 ± 20.8 | 0.815 | 32 ± 12.5 | 48.4 ± 18.8 | 43 ± 24.9 | 0.239 | |
| KRAS | Tumor with mutation (%) | 46.6 | 40 | 0.641 | 53.3 | 47.6 | 28.5 | 0.369 |
| Variants frequency (%) | 42 ± 15.7 | 38 ± 25.4 | 0.692 | 31.9 ± 12.5 | 38 ± 17.6 | 62 ± 21.6 | 0.025 | |
| APC | Tumor with mutation (%) | 30 | 40 | 0.464 | 53.3 | 14.2 | 35.7 | 0.043 |
| Variants frequency (%) | 41.5 ± 20.1 | 35.1 ± 25.7 | 0.638 | 39.6 ± 24.4 | 25.6 ± 11.6 | 45.16 ± 20.8 | 0.421 | |
| PIK3CA | Tumor with mutation (%) | 23.3 | 25 | 0.892 | 13.3 | 19 | 42.8 | 0.139 |
| Variants frequency (%) | 31.8 ± 8.67 | 21.5 ± 5.9 | 0.031 | 29.5 ± 7.8 | 25.7 ± 8.3 | 27.7 ± 10.9 | 0.902 | |
Spatial distribution of the variants in the 4 most frequently mutated genes in the CRC patients and 78% of the novel variants on APC can result in frameshifting and early protein termination
Figure 3 illustrates the spatial distribution of the variants according to protein function domain in the 4 most frequently mutated genes in the CRC patients. Reported mutations and their frequency extracted from the COSMIC database are also shown under the protein domain bar for comparison. R175H on TP53, mutation of Codons 12 and 13 on KRAS, and E545K on PIK3CA were the highest frequency mutations in our population (detected in at least 4 patients, marked with a green triangle in Figure 3). All of these mutations also contributed to the most frequently reported mutations in the COSMIC database (11%, 97%, and 31% in each gene, indicated by the line length in the figure). Otherwise, the remaining mutations were distributed widely among the cancer-related genes with no obvious hotspot. However, all mutations on the TP53 gene were located in the DNA-binding domain, and the variants on the APC gene were distributed in the β-catenin-binding domain. Both domains are central parts of the TP53 and APC proteins and govern the tumor suppression function by binding the downstream ligands. Notably, we unveiled 21 novel variants in 11 genes, 9 of which were observed in the APC gene (marked by a red triangle in Figure 3) and were located in the mutation cluster region (MCR). Furthermore, 78% (7/9) of these novel variants on the APC gene can result in a frameshift and early protein termination and may lead to deleterious consequences.
Figure 3. Spatial distribution of somatic variants on the protein function domains of the top-4 mutated genes.
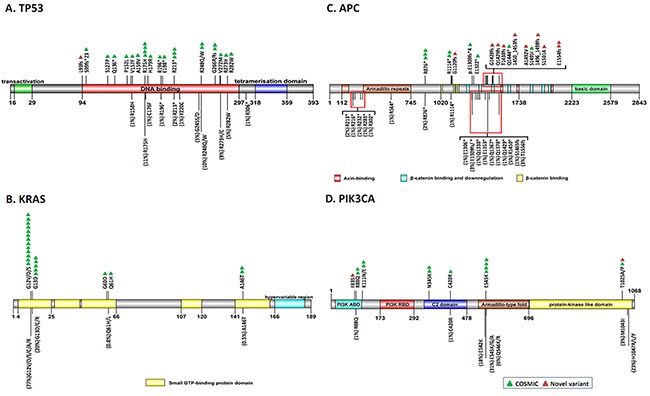
One triangle represents one variant found in one tumor (green = COSMIC mutation; red = novel variant). Color blocks illustrate the different functional domains of each protein. For comparison, the variants of each gene with a frequency of ≥1% recorded in the COSMIC database are marked with a black line below the domain bar, and the mutation rate is indicated in parentheses and indicated by the length of the line. A. TP53; B. KRAS; C. APC; D. PIK3CA.
DISCUSSION
In this study, we established an analysis pipeline for a well-designed cancer hotspot panel on an IT-PGM platform and comprehensively investigated the germline and somatic variants in CRC patients in a Taiwanese population. The results reveal the existence of 5 germline variants specifically in cancer patients but not in the general population. Among the 5 germline mutants, NRAS G138R is a novel mutation that has never been reported, and CDH1 T340A was the most frequently occurring mutation in the CRC group (Figure 1). In addition, 21 novel somatic variants were identified in the tumor samples, among which 7 mutations were frameshift alterations located in the central part of the APC gene, which may alter the function of the APC protein (Table 3).
In studying the germline mutations, we unexpectedly determined a novel NRAS G138R in one CRC patient. In well-investigated inherited cancers, such as breast cancer and colon cancer, germline mutations frequently occur in tumor suppressor genes (eg, APC, STK11, and PTEN) and DNA repair genes (eg, MLH1, MSH2, BRCA1, and BRCA2) [3, 27]. Inherited mutations in oncogenes, which can be predisposed to cancer, have only been reported on missense mutations of the RET proto-oncogene for multiple endocrine neoplasia type 2A cancer syndrome [28]. For our understanding, this is the first study to report the finding of germline NRAS mutations in CRC patients. NRAS belongs to the RAS oncogene family (KRAS, HRAS, and NRAS) and Ras protein functions as a GTPase, which can conduct signal transduction from the outside of the cell to the nucleus. Previous literatures have reported that the 138th residue is located in the allosteric lobe of Ras protein (residues 87-171) and is responsible for the interaction with GTPase-activating proteins (GAPs) [29]. GAPs interact with Ras-GTP by insertion of the arginine finger into the active site to switch “on” the GTP hydrolysis. Based on this model, the Glycine to Arginine substitution found in this study would possibly enhance downstream signaling. Moreover, the mutation was predicted to have a damaging effect on Ras protein function, as determined by SIFT and Polyphen2 software. However, in vitro function test of this mutant is needed to prove the cancer-prone property.
The remaining germline mutations were located in 3 cancer-susceptible genes: CDH1, APC, and MLH1. Among these, CDH1 mutation has been recognized as a risk for early onset diffuse gastric cancer in Western countries [30]. However, CDH1 T340A was reported to have a high association only with inherited and sporadic colon cancer in a Korean population [31]. As many as 3 patients in our CRC group also carried this specific heterozygous germline mutation (Table 2), indicating the existence of an Asian-specific genome structure. Another finding of interest is the role of APC in inherited CRC. Although APC is a known causal gene for FAP with high penetrance, the specific V1125A substitution and another V1352A mutation observed in our study was previously reported in a cohort study comprising 480 non-FAP patients [32]. Compared with CRC, most de novo germline mutations in the general population are not related to cancer development and have been designated to benign alteration by protein function prediction software. Recently, a commercial laboratory in the United States reported descriptive findings from screening inherited CRC from 586 patients by using the ColoNext™ NGS panel, which recruits 14 genes [20]. That study showed that 71% of patients with pathogenic mutations on CRC-susceptible genes met the syndrome-based testing criteria. However, their panel did not include NRAS and no Asian patients had positive findings. In summary, the result of this study highlight the importance of establishing a population-specific screening panel to maximize the detection rate of germline mutation for cancer prevention.
Regarding somatic mutation, an average of 2.2 variants can be identified in each tumor. However, 12 patients harbored only one aberrant mutation in the TP53, KRAS, PIK3CA, or NRAS gene in their tumor tissues (Figure 2), emphasizing the dominant role of these genes in carcinogenesis. Among all the variants, 21 somatic variants (27%) were unique to Taiwanese CRC patients (Table 3). Furthermore, 9 of these 21 variants were located in the APC gene, 7 of which belong to the indel-type, which can cause truncation of the encoded APC protein. The full length of APC acts as a tumor suppressor protein, which can disconnect the Wnt signal pathway by forming a multiprotein complex with Axin and β-catenin and promoting the phosphorylation and subsequent degradation of β-catenin [33]. More than 90% of the reported APC mutations found in sporadic and inherited CRC are nonsense or frameshift mutations located in the β-catenin-binding region (MCR: Codons 1267-1529), and the resulting shorter protein may lose its ability to bind to β-catenin, thereby activating cell proliferation and migration [34]. In the present study, 9 novel mutations on the APC gene were distributed from Codons 1120 to 1554 and might contribute to carcinogenesis. Moreover, a small-molecule compound was identified recently as selectively poisoning cancer cell lines with truncated APC [35], which might benefit the patients with APC novel frameshift mutations in our population. Other non-COSMIC mutations were involved in EGFR-RAS, PI3K, and P53 signaling pathways.
The variant profile in each patient provided an opportunity to inspect the inter- and intra-heterogeneous nature of CRC tumors [36] by examining 77 variant types distributed in 44 patients with CRC. We observed only 2 patients (Sample IDs 1423 and 1669) had the same mutation profile with KRAS G12D alteration. The remaining 42 patients had unique mutation signatures (Figure 2). This high inter-tumor heterogeneity may be induced by the stochastic nature of genome damage during passage through differences in tumor-initiating insults, immune surveillance, and factors influencing cancer progression [37]. The heterogeneity explains the loose correlation between genotype and phenotype (Table 4). However, concordance to our finding, higher levels of TP53 mutant DNA [38] or mutant p53 protein [39] can be found in late AJCC stage of CRC tumors. One large cohort study which recruited 1110 Chinese CRC patients revealed that mutant KRAS and BRAF were associated with right-sited tumors [40] and it correlated with the poor response to epidermal growth factor receptor (EGFR) inhibition with cetuximab [41].
On the other aspect, intra-tumor heterogeneity can be demonstrated by observing the highly extreme variant frequency in one tumor and the discrepant finding of common serial mutation order from APC to TP53 which advised by Bert Vogelstein in 1988 [3]. According to the theory, the earliest event in the colorectal cancer involves the mutation of APC gene. Acquiring and accumulating more somatic mutations on specific genes is essential for malignant transformation (Supplementary Figure S1A). However, the variant frequency on APC gene in each tumor is not always the largest one in our study. This phenomenon provides a clue that the APC may not be the necessary driver gene. Another subclones which carry the essential mutations gain growth advantage as tumor progression (Supplementary Figure S1B). Both conditions can occur in one tumor and cause the intra-tumor heterogeneity. The nature of intra-tumor heterogeneity may hinder the correct mutation profile detection and the subsequent choice for personalized target therapy. Consequently, it could be at the risk of introducing the propagation of minor clones in the original tumor after incomplete therapy. However, the impact and clinical correlation of intra-tumor heterogeneity needs larger sample cohort and long-term follow-up study.
The only one stage IV patient (Sample ID 1456) who received palliative chemotherapy and cetuximab in this study showed progression of bone metastasis 1 year later. After examination of the mutation profile of this patient by using NGS Cancer Panel, one rare KRAS mutation was observed at A146T. This mutation accounts for only 0.5% of all KRAS mutations in the COSMIC database. However, a recent study indicated that this mutation reduces the sensitivity to anti-EGFR antibody therapy [12]. The examination of other drug-actionable targets through NGS cancer panel shows promise for cancer patients to receive the newest therapy, even for drugs undergoing clinical trials. Park applied NGS to 2221 clinical cases and observed clinically actionable alterations in 76% of tumors, which is 3-fold the number of actionable alterations detected by conventional diagnostic tests [42]. These findings strengthen the necessity of implementing an NGS cancer panel in clinical settings instead of conventional PCR strategies for detecting hotspots.
Collectively, the unexpected germline oncogene mutation and high frequency of novel variants observed in our cancer group may have an ethnic impact [43]. The clinical implementation of the NGS cancer panel, either in germline or somatic genome detection, is currently under development in our laboratory with the aim of improving the screening rate of cancer for high-risk individuals with a family history of CRC and providing more actionable information for physicians to improve medical care for CRC patients.
MATERIALS AND METHODS
Study patients
CRC patients
Fifty patients with untreated CRC diagnosed in 2013 at Chang Gung Memorial Hospital in Taiwan were enrolled in this study. Cancer was staged according to the 2009 American Joint Committee on Cancer staging criteria (7th edition) [23]. Clinicopathological factors, including age, sex, plasma carcinoembryonic antigen (CEA) data, immuno-occult blood test (iFOBT) data, and the anatomic subsite of tumors in the intestine lumen, were recorded at enrollment. Tumor location was classified into 3 parts: right site (tumors at the cecum, ascending colon, hepatic flexure, and transverse colon); left site (tumors at the splenic flexure, descending colon, and sigmoid colon); and the rectum.
Asymptomatic controls
For the control group, we recruited 20 volunteers from staffs at the Department of Laboratory Medicine at Linkou Chang Gung Memorial Hospital. All controls had no family history of CRC and had negative serum CEA and iFOBT results. All patients and healthy individuals were provided with a form of written informed consent, and the study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (101-4609A3).
Sample preparation and routine laboratory test
Fresh tumors and adjacent normal tissue samples (at least 5-cm from the tumor site) were collected on the day of operation from 50 patients with CRC. For the control group, the EDTA blood samples were collected from 20 asymptomatic controls. Genomic DNAs were extracted using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) and stored at −80°C until use. CEA and iFOBT were respectively determined using an ADVIA Centaur® Analyzer (WI, USA) with a cutoff of 5 ng/mL and OC-Sensor Diana Latex Reagent (Eiken Chemical, Tokyo, Japan) with a cutoff of 100 ng/mL.
Ion-torrent personal genome machine (IT-PGM) sequencing
AmpliSeq Cancer Hotspot Panel Version 2 (Life Technologies, CA, USA) specifically targets 50 cancer-related genes, most of which are tumor suppressor genes and oncogenes, and harbors 2855 COSMIC (Catalogue of somatic mutations in cancer) [24] hotspots (for detailed information, see Supplementary Table S1). The AmpliSeq Library was prepared according to the manufacturer's Ion AmpliSeq Library Kit protocol. In brief, 10 ng of genomic DNA was extracted from the samples and PCR was conducted to amplify 207 amplicons, with sizes ranging from 49 to 140 bp, in one primer pool. After AMPure bead purification, barcoded adapter-ligated products were nick-translated, and the resulting library concentration was determined using an Agilent 2100 bioanalyzer and adjusted to 10 pmole. Emulsion PCR and enrichment were performed on an Ion One Touch System by using the Ion OneTouch™ 200 Template Kit Version 2.0 (Life Technologies) according to the manufacturer's instructions. The samples were then sequenced using the IT-PGM 200 Sequencing Kit Version 2.0 protocol. To obtain an average depth of 1500 for each amplicon, 6 samples were pooled on one 316 chip, and 12 samples were pooled on one 318 chip.
Bioinformatics analysis
Base sequences were processed initially by using IT-PGM pipeline software (Torrent Suite Version 4.2), and the sequences were aligned to human genome build 19 reference genome (hg19). Identification of variants was facilitated by using the IT Variant Caller software plugin (Life Technologies), and advanced annotation was performed by uploading an exported VCF file from Variant Caller to Vanno (developed by the Department of Bioinformatics at Chang Gung University) [25]. Initially, variants were selected by mutation type if they belonged to nonsynonymous or frameshift or stopgain at the exonic region. Variant frequencies >3% in the dbSNP-Asian database were further filtered. To enhance the reliability of these variants, only those mutations with a frequency of >5% and variant coverage of >30 were considered candidate variants for further analysis. Integrative Genomics Viewer was employed to visualize the variants by confirming the presence of possible strand biases and alignment errors. Nomenclature of novel variants followed the rules from Human Genome Variation Society (http://www.hgvs.org/mutnomen/). Variants with amino acid changes were further examined for whether the changes were potentially damaging alterations by using Sorting Tolerant From Intolerant (SIFT) and Polymorphism Phenotyping v2 (PolyPhen2) software, which can predict the possible impact of an amino acid substitution on the structure and function of a protein [26]. SIFT calculates conservation value and scales probability for each position. The SIFT score ranges from 0.0 (deleterious) to 1.0 (tolerated). The PolyPhen-2 score ranges from 0.0 (tolerated) to 1.0 (deleterious).
Variant confirmation
Mutations that met the filtering criteria were further confirmed by Sanger sequencing when the variant frequency was above 20%, or by pyrosequencing when the variant frequency was 5%–20%.
Statistical analysis
Descriptive statistics were summarized in percentages, ranges, means, and standard deviations. Between-group comparisons were conducted using the Student t test, one-way ANOVA, and chi-square test for each marker. A P value less than 0.05 (2-tailed) was considered statistically significant. All statistical tests were conducted using PASW Statistics 18. Protein domain structure and distribution of variants on specific proteins were plotted using DOG Version 1.0 (http://dog.biocuckoo.org).
SUPPLEMENTARY FIGURE AND TABLE
Footnotes
CONFLICTS OF INTERESTS
The authors declare no conflict of financial or nonfinancial competing interest.
GRANT SUPPORT
This study was funded by Chang Gung Memorial Hospital (CMRPG3C1371 and CMRPG3C1372).
Author's contributions
JJL and ECC developed the concept, initiated the study, and revised the manuscript. PYC designed the experiments and wrote the manuscript. NCC, MCW, andSHT performed the experiments and organized the figures and tables. SCC and YHW helped to analyze the data and review the patient charts. JSC and WST collected the clinical samples, collected patient information, and obtained consent from the enrolled patients.
REFERENCES
- 1.Cancer Statistic Report. Taiwan Department of Health, Available from https://cris.hpa.gov.tw/ Released July 6, 2010.
- 2.Cho KR, Vogelstein B. Genetic alterations in the adenoma—carcinoma sequence. Cancer. 1992;70:1727–1731. doi: 10.1002/1097-0142(19920915)70:4+<1727::aid-cncr2820701613>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Bert Vogelstein, Fearon ER, Stanley R H, Kern SE, Preisinger AC, Leppert M, Smits AMM, Bos JL. Genetic Alterations during Colorectal-Tumor Development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Niko Beerenwinkel TA, Dingli David, Traulsen Arne, Kinzler Kenneth W, Velculescu Victor E, Vogelstein Bert, Nowak Martin A. Genetic Progression and the Waiting Time to Cancer. PLoS Computational Biology. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature Medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 6.H L, A B, SL Z. Molecular Cell Biology. 4. New York: W H Freeman; 2000. Section 24. 5, Mutations Affecting Genome Stability. [Google Scholar]
- 7.Grover S, Syngal S. Genetic testing in gastroenterology: Lynch syndrome. Best Practice & Research Clinical Gastroenterology. 2009;23:185–196. doi: 10.1016/j.bpg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 9.Varmus FSCaH. A New Initiative on Precision Medicine. The New England Journal of Medicine. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers Predicting Clinical Outcome of Epidermal Growth Factor Receptor-Targeted Therapy in Metastatic Colorectal Cancer. Journal of the National Cancer Institute. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peeters M, Oliner KS, Parker A, Siena S, Van Cutsem E, Huang J, Humblet Y, Van Laethem JL, Andre T, Wiezorek J, Reese D, Patterson SD. Massively Parallel Tumor Multigene Sequencing to Evaluate Response to Panitumumab in a Randomized Phase III Study of Metastatic Colorectal Cancer. Clinical Cancer Research. 2013;19:1902–1912. doi: 10.1158/1078-0432.CCR-12-1913. [DOI] [PubMed] [Google Scholar]
- 12.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilsky RL. Implementing personalized cancer care. Nature reviews Clinical oncology. 2014;11:432–438. doi: 10.1038/nrclinonc.2014.54. [DOI] [PubMed] [Google Scholar]
- 14.Abramson RG. Overview of Targeted Therapies for Cancer. My Cancer Genome. 2015 http://www.mycancergenome.org/content/molecular-medicine/overview-of-targeted-therapies-for-cancer/ Updated February 24. [Google Scholar]
- 15.Wanda M, Krajewska MS, Bryś Magdalena, Młynarski Wojciech, Witas Henryk, Okruszek Andrzej, Kiliańska Zofia M. Genotyping of p53 codon 175 in colorectal cancer. Medical Science Monitor. 2003;9:BR228–231. [PubMed] [Google Scholar]
- 16.Hak-Su Goh JY, Smith Duncan R. p53 Point Mutation and Survival in Colorectal Cancer Patients'. Cancer research. 1995;55:5217–5221. [PubMed] [Google Scholar]
- 17.Rechsteiner M, von Teichman A, Ruschoff JH, Fankhauser N, Pestalozzi B, Schraml P, Weber A, Wild P, Zimmermann D, Moch H. KRAS, BRAF, and TP53 deep sequencing for colorectal carcinoma patient diagnostics. JMD. 2013;15:299–311. doi: 10.1016/j.jmoldx.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Lee KH, Won JK, Kim TY, Oh DY, Im SA, Bang YJ, Jeong SY, Park KJ, Park JG, Kang GH, Seo JS, et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS One. 2013;8:e64271. doi: 10.1371/journal.pone.0064271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cragun D, Radford C, Dolinsky JS, Caldwell M, Chao E, Pal T. Panel-based testing for inherited colorectal cancer: a descriptive study of clinical testing performed by a US laboratory. Clinical genetics. 2014;86:510–520. doi: 10.1111/cge.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Wu WKK, Li X, He J, Li XX, Ng SSM, Yu C, Gao Z, Yang J, Li M, Wang Q, Liang Q, Pan Y, Tong JH, To KF, Wong N, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64:636–645. doi: 10.1136/gutjnl-2013-306620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon HH, Shi Q, Alberts SR, Goldberg RM, Thibodeau SN, Sargent DJ, Sinicrope FA, Alliance for Clinical Trials in O Racial Differences in BRAF/KRAS Mutation Rates and Survival in Stage III Colon Cancer Patients. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edge S, Byrd D.R, Compton C.C, Fritz A.G, Greene F.L, Trotti A. AJCC Cancer Staging Manual. 7. 2010. p. 649. pages. [Google Scholar]
- 24.COSMIC v65: Catalogue Of Somatic Mutations In Cancer. http://cancer.sanger.ac.uk/cosmic
- 25.Huang PJ, Yeh YM, Gan RC, Lee CC, Chen TW, Lee CY, Liu H, Chen SJ, Tang P. CPAP: Cancer Panel Analysis Pipeline. Human mutation. 2013;34:1340–1346. doi: 10.1002/humu.22386. [DOI] [PubMed] [Google Scholar]
- 26.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human mutation. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet. 2008;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan LM, Kwok JBJ, Healey CS, Elsdon MJ, Eng C, Gardner E, Love DR, Mole SE, Moore JK, Papi L, Ponder MA, Telenius H, Tunnacliffe A, Ponder BAJ. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature. 1993;363:458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 29.Buhrman G, O'Connor C, Zerbe B, Kearney BM, Napoleon R, Kovrigina EA, Vajda S, Kozakov D, Kovrigin EL, Mattos C. Analysis of binding site hot spots on the surface of Ras GTPase. Journal of molecular biology. 2011;413:773–789. doi: 10.1016/j.jmb.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teresa SHPKHL-CMWJSHPKASDFSKSGZ Hereditary Diffuse Gastric Cancer Syndrome CDH1 Mutations and Beyond. JAMA Oncology. 2015;1:23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 31.Kim HC, Wheeler JMD, Kim JC, Ilyas M, Beck NE, Kim BS, Park KC, Bodmer WF. The E-cadherin gene (CDH1) variants T340A and L599V in gastric and colorectal cancer patients in Korea. Gut. 2000;47:262–267. doi: 10.1136/gut.47.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzopardi D, Dallosso AR, Eliason K, Hendrickson BC, Jones N, Rawstorne E, Colley J, Moskvina V, Frye C, Sampson JR, Wenstrup R, Scholl T, Cheadle JP. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–363. doi: 10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 33.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Human Molecular Genetics. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 34.Taketo KAaMM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. Journal of cell science. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Kim SB, Eskiocak U, Posner B, Das P, Wright WE, De Brabander JK, Shay JW. Therapeutic targeting truncated adenomatous polyposis coli (APC) proteins for the selective killing of colorectal cancer cells. BMC Genomics. 2014;15:O3. [Google Scholar]
- 36.Kogita A, Yoshioka Y, Sakai K, Togashi Y, Sogabe S, Nakai T, Okuno K, Nishio K. Inter- and intra-tumor profiling of multi-regional colon cancer and metastasis. Biochemical and biophysical research communications. 2015;458:52–56. doi: 10.1016/j.bbrc.2015.01.064. [DOI] [PubMed] [Google Scholar]
- 37.Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nature reviews Clinical oncology. 2012;9:621–630. doi: 10.1038/nrclinonc.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lleonart ME, Garcia-Foncillas J, Sanchez-Prieto R, Martin P, Moreno A, Salas C, Ramon y Cajal S. Microsatellite instability and p53 mutations in sporadic right and left colon carcinoma: different clinical and molecular implications. Cancer. 1998;83:889–895. [PubMed] [Google Scholar]
- 39.Zhang Z, Deng X, Ren X, Zhang B, Chen X, Yang J, Ding H, Sui J, Song X. Expression of mutant p53 and of the multidrug resistant proteins P-glycoprotein and glutathione S-transferase-pi correlated in colorectal adenocarcinoma. Scand J Gastroenterol. 2010;45:925–934. doi: 10.3109/00365521003734117. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu T, Sun J, Jiang H, Zhu Y, Zheng Y, Liang Z, Liu T. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep. 2015;5:18678. doi: 10.1038/srep18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brulé SY, Jonker DJ, Karapetis CS, O'Callaghan CJ, Moore MJ, Wong R, Tebbutt NC, Underhill C, Yip D, Zalcberg JR, Tu D, Goodwin RA. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO. 17. European Journal of Cancer. 2015;51:1405–1414. doi: 10.1016/j.ejca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Park JY, Kricka LJ, Fortina P. Next-generation sequencing in the clinic. Nature biotechnology. 2013;31:990–992. doi: 10.1038/nbt.2743. [DOI] [PubMed] [Google Scholar]
- 43.Jing L, Su L, Ring BZ. Ethnic background and genetic variation in the evaluation of cancer risk: a systematic review. PLoS One. 2014;9:e97522. doi: 10.1371/journal.pone.0097522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


