Abstract
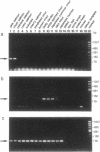
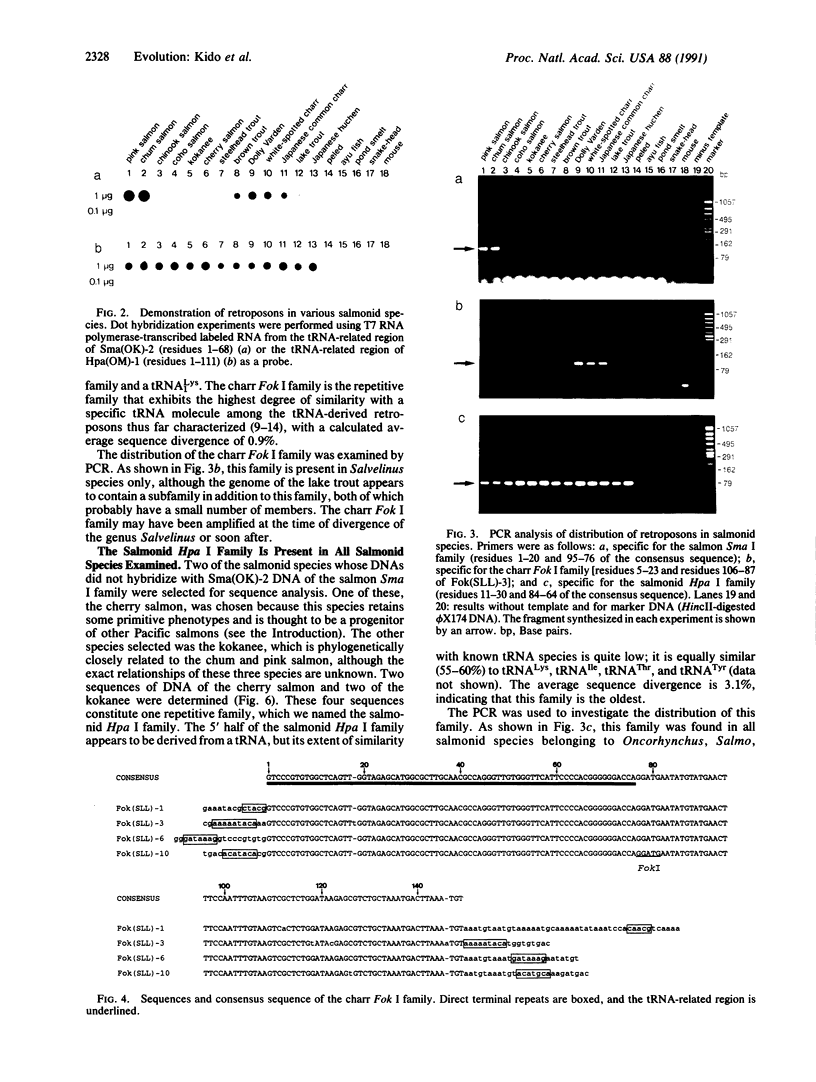
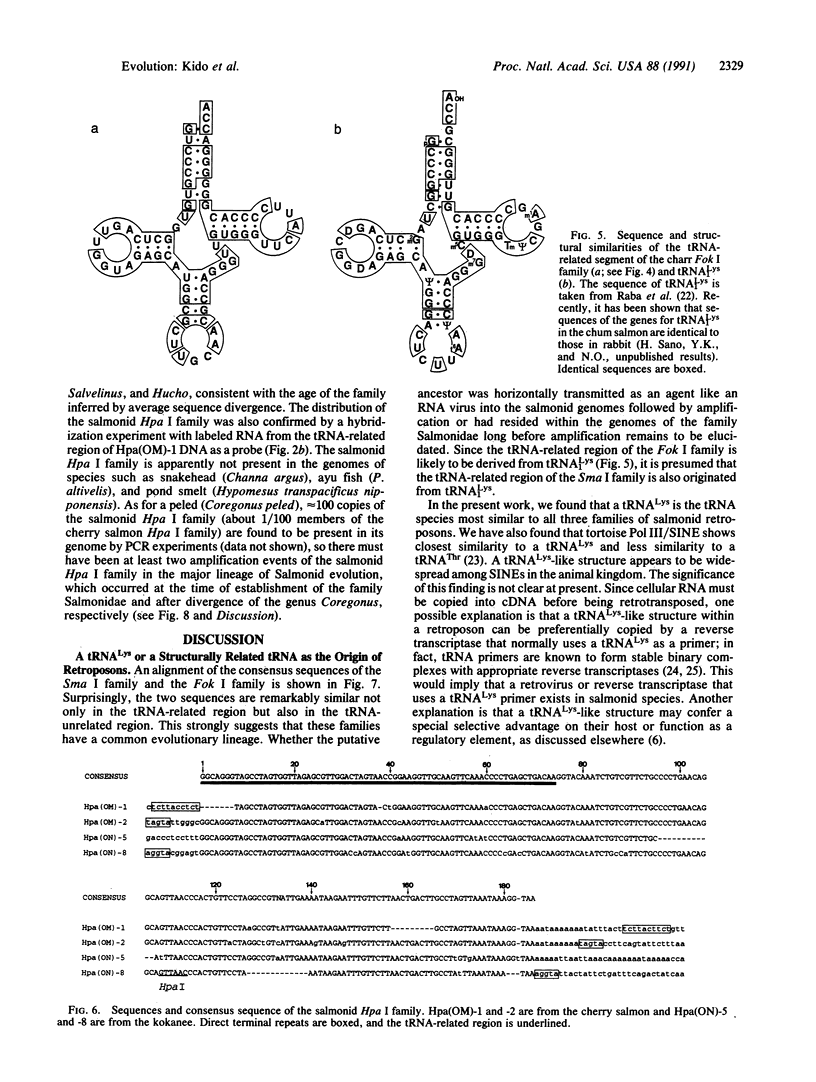
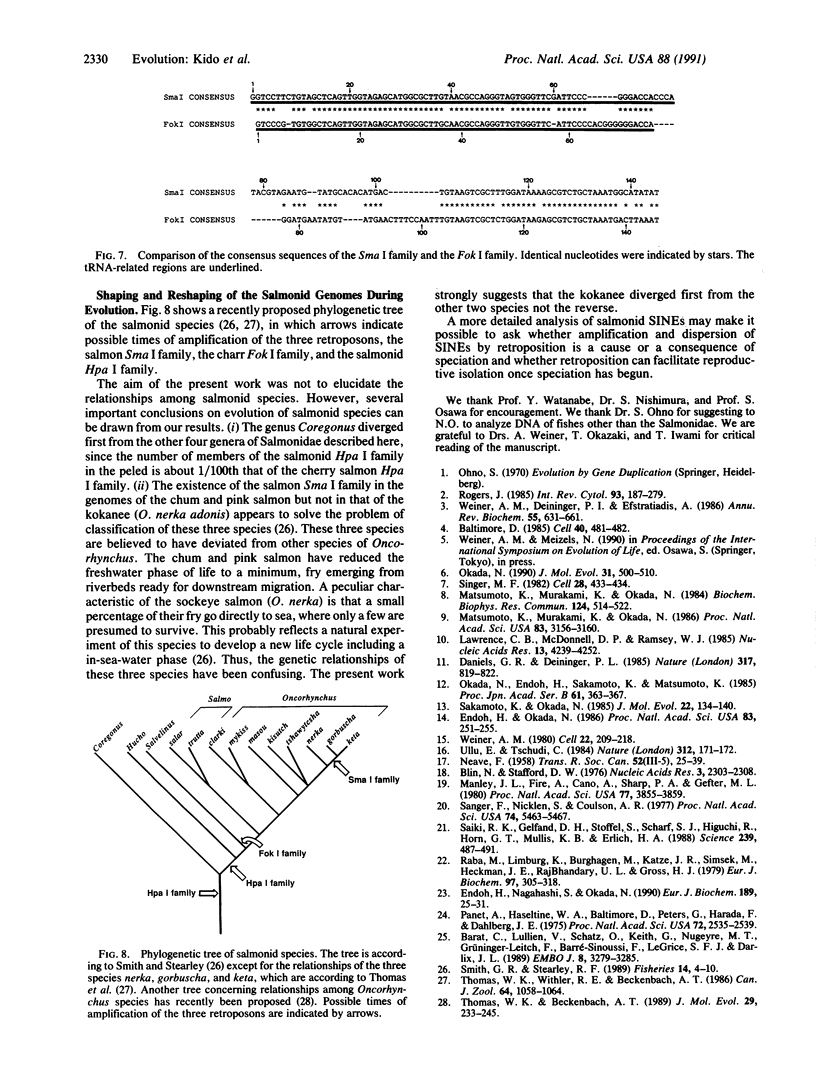
Three families of tRNA-derived repeated retroposons in the genomes of salmonid species have been isolated and characterized. These three families differ in sequence, but all are derived from a tRNA(Lys) or from a tRNA species structurally related to tRNA(Lys). The salmon Sma I family is present in the genomes of two species of the genus Oncorhynchus but not in other species, including five other species of the same genus. The charr Fok I family is present only in four species and subspecies of the genus Salvelinus. The third family, the salmonid Hpa I family, appears to be present in all salmonid species but is not present in species that are not members of the Salmonidae. Thus, the genome of proto-Salmonidae was originally shaped by amplification and dispersion of the salmonid Hpa I family and then reshaped by amplification of the Sma I and Fok I families in the more recently evolved species of salmon and charr, respectively. We speculate that amplification and dispersion of retroposons may have played a role in salmonid speciation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Endoh H., Nagahashi S., Okada N. A highly repetitive and transcribable sequence in the tortoise genome is probably a retroposon. Eur J Biochem. 1990 Apr 20;189(1):25–31. doi: 10.1111/j.1432-1033.1990.tb15455.x. [DOI] [PubMed] [Google Scholar]
- Endoh H., Okada N. Total DNA transcription in vitro: a procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci U S A. 1986 Jan;83(2):251–255. doi: 10.1073/pnas.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B., McDonnell D. P., Ramsey W. J. Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res. 1985 Jun 25;13(12):4239–4252. doi: 10.1093/nar/13.12.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Murakami K., Okada N. Gene for lysine tRNA1 may be a progenitor of the highly repetitive and transcribable sequences present in the salmon genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3156–3160. doi: 10.1073/pnas.83.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Murakami K., Okada N. Pseudouridylic modification of a 6S RNA transcribed in vitro from highly repetitive and transcribable (Hirt) sequences of salmon total DNA. Biochem Biophys Res Commun. 1984 Oct 30;124(2):514–522. doi: 10.1016/0006-291x(84)91584-5. [DOI] [PubMed] [Google Scholar]
- Okada N. Transfer RNA-like structure of the human Alu family: implications of its generation mechanism and possible functions. J Mol Evol. 1990 Dec;31(6):500–510. doi: 10.1007/BF02102077. [DOI] [PubMed] [Google Scholar]
- Panet A., Haseltine W. A., Baltimore D., Peters G., Harada F., Dahlberg J. E. Specific binding of tryptophan transfer RNA to avian myeloblastosis virus RNA-dependent DNA polymerase (reverse transcriptase). Proc Natl Acad Sci U S A. 1975 Jul;72(7):2535–2539. doi: 10.1073/pnas.72.7.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raba M., Limburg K., Burghagen M., Katze J. R., Simsek M., Heckman J. E., Rajbhandary U. L., Gross H. J. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem. 1979 Jun;97(1):305–318. doi: 10.1111/j.1432-1033.1979.tb13115.x. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Okada N. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol. 1985;22(2):134–140. doi: 10.1007/BF02101691. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Thomas W. K., Beckenbach A. T. Variation in salmonid mitochondrial DNA: evolutionary constraints and mechanisms of substitution. J Mol Evol. 1989 Sep;29(3):233–245. doi: 10.1007/BF02100207. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980 Nov;22(1 Pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]