Abstract
Circadian rhythms are 24-hour oscillations in physiology and behaviour, which allow organisms to anticipate and adapt to the daily demands associated with the day/night cycle. The currently accepted model of the molecular clockwork is described as a transcriptional process, comprised of negative regulatory feedback loops. However, ample evidence underlines the important contribution of non-transcriptional and metabolic oscillations to cellular timekeeping. Here we will summarize recent evidence pointing to the relationship between the transcriptional oscillator and metabolic redox state, with particular emphasis on the potential nodes of interaction. We highlight the intrinsic difficulty to segregate these two tightly coupled and interdependent processes, in living systems, and how disruption in their synchronicity impacts on physiological and pathological processes as diverse as cardiovascular and metabolic disorders, ageing, and cancer.
Keywords: Circadian rhythms, Redox oscillations, Metabolism, Peroxiredoxins
Co-evolution of the circadian and redox systems
The circadian clock is an intrinsic timekeeping mechanism that provides organisms from bacteria to humans with the means to temporally organise behavioural, physiological and molecular events around the 24-hour day and night cycle. Such clocks likely evolved to enable organisms on Earth to resonate with their environment, such that their internal cycles anticipate and match rhythms externally. In mammals, many physiological processes are under circadian regulation, including the sleep-wake cycle, core body temperature, feeding behavior, glucose homeostasis and various endocrine secretions (e.g. cortisol and melatonin), as well as certain pathologies such as hypertensive crises, myocardial infarctions, asthma and allergy attacks, that occur at specific times of the day [1–3].
Circadian clocks only confer advantage when they beat in harmony with the external environment. Disturbance of circadian timing, as exhibited in rotational shift workers, is linked to significant health issues such as breast and prostate cancer, diabetes and other metabolic complications [4–6]. In mammals, a central oscillator within the hypothalamic suprachiasmatic nuclei (SCN) integrates light signals from the retina and orchestrates tissue/organ functions using neuronal efferents and humoral factors. As almost all cells within tissues exhibit self-sustained oscillations [7–9], a key function of the SCN is to maintain proper phase alignment of peripheral clocks to ensure synchrony in the system [10]. The current model of the molecular clockwork (see glossary) is based on a transcription/translation feedback loop (TTFL) mechanism, whereby a set of transcriptional activators induce the transcription of repressor genes whose protein products then feed back to inhibit their own transcription (Figure 1). However, mounting evidence suggests that transcription-based oscillators are not the only means by which the cells track time. The discovery of the self-autonomous redox rhythms exhibited by the peroxiredoxin (PRX) proteins provided the first convincing example of such transcription-independent oscillations in eukaryotes [11,12]. These highly conserved oscillations, found in organisms from Archaea to man, suggest a strong link between circadian rhythmicity and redox metabolism [13].
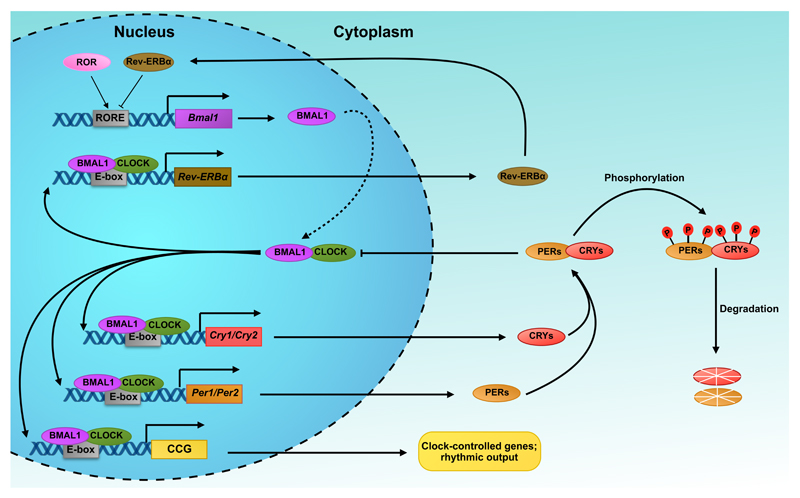
Figure 1. Simplified schematic diagram of the transcription-translation feedback loop (TTFL) in mammals.
The basic helix-loop-helix (bHLH) transcription factors CLOCK and NPAS2 associate with the bHLH transcription factor, BMAL1, to form heterodimeric transcriptional activator complexes. During the day, CLOCK/BMAL1 and BMAL1/NPAS2 bind to E-box enhancer elements and activate transcription of the Period (Per1/2) and Cryptochrome (Cry1/2) genes. Upon accumulation in the cytoplasm, PER and CRY proteins assemble into heterotypic complexes that translocate back to the nucleus, where they repress their own transcriptional by inhibiting BMAL1/CLOCK. During the night, the PER and CRY complexes are gradually phosphorylated (depicted as P) and targeted for degradation in the proteasome, thereby lifting the repression on CLOCK/BMAL1, which can then activate a new cycle of transcription. In addition to the primary feedback loop, the system also incorporates an accessory loop composed of the nuclear orphan receptors, retinoid-related orphan receptor (ROR), and nuclear receptor subfamily 1 group D member 1 and 2 (NR1D1/2 or REV-ERBα/β). The latter are under the direct transcriptional control of the BMAL1/CLOCK activator complex and negatively feed back to inhibit Bmal1 transcription via bindings to retinoic acid-related orphan receptor response elements (ROREs) in the Bmal1 promoter. In contrast, ROR acts as a positive regulator and competes with REV-ERBs for binding to ROREs. The entire cycle takes approximately 24 hours.
Why might redox regulation be tightly linked with circadian biology? The foundations of this coupling may have been laid down around the time of the Great Oxidation Event (GEO) approx. 2.5 billion years ago. The increase in atmospheric oxygen levels as a result of the newly acquired ability of photosynthetic bacteria to use water as the main electron donor are thought to have created a strong selective pressure on anaerobes to evolve defence systems to deal with this harsh and unprecedented oxidising environment [11,14]. Rhythmic photosynthesis, and thus oxygen production as a function of the changing day and night, as well as the generation of reactive oxygen species (ROS) by metabolic reactions, or directly by UV radiation, could have forced the co-evolution of the circadian and redox systems. Thus, the generation of ROS and those processes sensitive to oxidation were temporally segregated, preventing harmful oxidative stress that would otherwise have led to cell dysfunction and death.
Metabolic redox oscillations in mammals
Temporal separation of cellular metabolism might be an adaptation to prevent the simultaneous occurrence of mutually antagonistic reactions that would otherwise result in energetically wasteful futile cycles. Consistent with this, numerous metabolic pathways and metabolite levels are under circadian regulation, and both genetic perturbation (e.g. clock gene mutants) and physiological disturbance (e.g. shift work) of the clock have been shown to increase susceptibility to metabolic stress [4,15]. Reciprocally, growing evidence highlights that core metabolism feeds back to the central oscillator, thereby affecting clock function. For instance, restricted feeding (RF) in mice is capable of entraining the liver’s clock in SCN-lesioned animals [16]. This is further supported by studies showing that RF phase-shifts both locomotor activity rhythms and circadian gene transcription in the periphery but does not affect rhythms in the SCN [17,18]. Furthermore, in mice, high fat diet results in alteration of the locomotor activity and the expression of canonical and clock-controlled genes [19], while in humans three weeks of circadian dyssynchrony is sufficient to induce a decrease in the resting metabolic rate and raise plasma glucose concentration after a meal, symptoms usually associated with the onset of diabetes mellitus [20].
The fact that metabolic changes can directly impact upon the transcriptional oscillator raises the question, what is the molecular mechanism that mediates this coupling. Of all the parameters that change as a function of cellular metabolism, redox poise represents one of the strongest candidates. This is not only due to its crucial role in cellular metabolism, but also because of the tight interplay between redox state and the circadian system. Accumulating evidence in recent years has pointed to the many potential nodes of interaction between the circadian and redox systems (Figure 2). The following sections are going to detail some of the most important discoveries thus far.
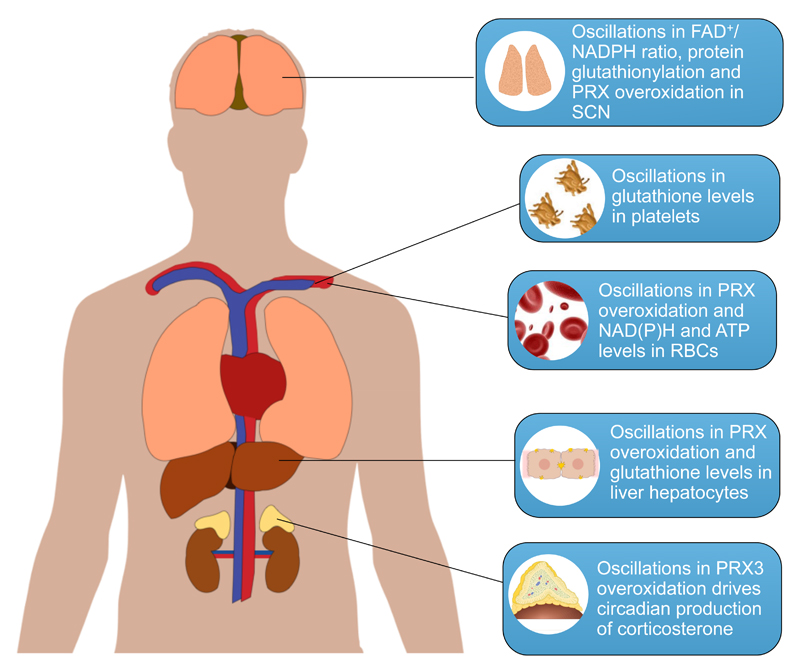
Figure 2. Circadian redox oscillations in mammalian physiology.
The redox status of the SCN as measured by the FAD/NADPH ratio and protein glutathionylation oscillates in rodents, thereby driving rhythmic neuronal firing [57]. Human platelets kept in vitro exhibit rhythmic changes in the glutathione levels [54]. PRX overoxidation, and NAD(P)H and ATP levels oscillate in human and mouse RBCs [11,61]. Oscillations in liver glutathione content [44–47] and PRX oxidation cycles [13] have been demonstrated in the rodent liver. Circadian cycles in the overoxidation of PRX3 are essential for the diurnal pattern of production of corticosterone in the adrenal cortex of mice [79].
Linking metabolic redox rhythms to the transcriptional oscillator
One of the first, and until now, most convincing examples of coupling between the transcriptional circadian oscillator and the redox systems is the finding that the molecular clock directly regulates the abundance of the redox metabolite nicotinamide adenine dinucleotide (NAD+) [21,22]. Binding of the CLOCK:BMAL1 activator complex to the promoter of the rate-limiting enzyme in the NAD+ salvage pathway, nicotinamide phosphoribosyl-transferase (NAMPT), results in rhythmic NAD+ accumulation in mouse liver and in cultured cells [21,22]. Of note, abrogation of the positive limb of the TTFL results in not only lack of oscillations but also markedly decreased NAD+ levels, and conversely absence of the negative regulators, CRY and PER, results in an increase [21,26]. This suggests that BMAL1 and CLOCK might fulfill important functions in basal cytosolic processes beyond their role in the clockwork. Furthermore, attenuation of the NAD+ cycles by genetic ablation of the major NAD+ hydrolase, CD38, disrupts circadian behavior and gene expression, suggesting a reciprocal relationship in which changes in the abundance of NAD+ as a function of altered metabolism could feed back to affect the functions of the transcriptional clock [23]. The transduction of this signal is likely to occur via the class III histone protein deacetylase SIRT1, which in addition to regulating a plethora of metabolic processes has also been demonstrated to control clock function by deacetylating BMAL1 and PER2, and counterbalancing CLOCK-mediated acetylation [24,25]. Another NAD+-dependent deactylase that was recently implicated in the circadian control of metabolism is the mitochondrial SIRT3 deacetylase. As a function of its rhythmic activation by NAD+, SIRT3 mediates acetylation of mitochondrial proteins, thereby generating rhythms in oxidative metabolism [26].
In addition to generating rhythms in redox-active metabolites, a number of clock proteins act as redox sensors, suggesting a reciprocal mode of regulation. The first evidence of redox control over core clock components came from a series of biochemical experiments performed by Rutter and colleagues, when they demonstrated that the DNA binding affinity of CLOCK:BMAL1 and NPAS2:BMAL1 heterodimers could be directly regulated by the redox poise of NAD(P)+/NAD(P)H in vitro [27]. Importantly, the redox response curve demonstrated that the activator complexes display near switch-like behavior, which suggests that even small changes in cellular (or nuclear) redox poise is capable of producing a response. One of the major caveats of these studies was that the concentration of coenzymes utilized in these experiments was much higher than what are likely to be found in vivo. Thus, the results need to be placed in the context of normal physiology. However, the conditions used in the experiments, such as the ionic composition of the reaction buffer, could induce certain conformational changes in the protein complex that would increase the apparent binding constant. Thus, it is highly plausible that in vivo CLOCK/NPAS2:BMAL1 heterodimers respond to much lower NAD(P)H concentrations. In addition, to our knowledge there is no experimental data available that provides us with the local concentration of NAD(P)H in the nucleus. This might be higher than that found in other cellular compartments. The redox dependence of CLOCK/NPAS2:BMAL1 also raises the possibility that NAD+ oscillations generated by the transitional oscillator could directly feed back to affect clock function.
Another redox sensitive mechanism implicated in the control of the clockwork involves haem sensing by a number of clock-associated transcription factors. For instance, haem binding can control NPAS2:BMAL1 activity by inhibiting DNA binding in response to carbon monoxide [28,29,81]. Similarly, the activity of the nuclear hormone receptors REV-ERBα and REV-ERBβ is modulated by haem. Finally, haem also regulates the negative limb of the TTFL because it inhibits the formation of complexes between PER2 and CRY1 [30]. In addition to regulation by redox-active ligands, these metabolic systems also impinge on the molecular clockwork directly by the means of reactive cysteine residues present in a number of clock proteins. For example, haem binding, and thus activity of REV-ERBβ, is governed by the formation of disulphide in the haem binding pocket [31]. Of note, redox regulation by modification of reactive cysteine residues seems to be a common theme in nuclear receptor signaling, as the archetypal glucocorticoid and estrogen receptors harbour reactive cysteine residues that modulate their activity [32].
Recently, two independent studies reported the structure of the PER2:CRY1 protein complex and demonstrated that zinc binding is essential for the stabilisation and activity of this heterodimer. Interestingly, binding of the metal ion was governed by a regulatory disulphide switch embedded in the structure of CRY1 [33,34]. Furthermore, mice carrying mutations in one of the conserved cysteine residues from the regulatory couple display a markedly prolonged period of locomotor activity [35]. Post-translation control of the stability and degradation of clock proteins is an integral part of the clockwork. Interestingly, a number of proteins involved in this mode of regulation have been shown to be redox-regulated. For example, CRY1 degradation is induced by phosphorylation by AMP-activated protein kinase (AMPK) [36], which was recently shown to be under redox regulation – a regulatory disulphide couple was found to be essential for the activation of the enzyme by AMPK [37]. Furthermore, a recent study identified deubiquitinase (DUB) herpesvirus-associated ubiquitin-specific protease (HAUSP) as a major CRY1 interacting partner [38]. Similarly to other DUBs, HAUSP harbours an active site cysteine residue which is controlled by reversible oxidation [39]. One of the recently proposed links between the circadian and redox systems involves the circadian-regulated pineal hormone melatonin. Although melatonin has been shown to possess antioxidant features, its concentration in plasma is much lower than of other known antioxidants, such as serum ascorbate or glutathione[40]. However, melatonin can exert its antioxidant effects by stimulating the NF-kB, AP-1 and Nrf2 pathways through G-protein coupled receptor signaling, thereby up-regulating the expression of antioxidant and detoxification proteins [41].
Circadian Redox Rhythms
In addition to the aforementioned cycles in NAD+ abundance, profiling of energy/redox metabolites in rodent tissues across circadian time has provided ample evidence for oscillations in glutathione, NAD(P)H, ATP and numerous metabolic intermediates [42,43] (Figure 2). Glutathione levels exhibit diurnal fluctuations in the rodent liver [44–47], gut [48], pancreas [47] and lungs [49]. The main driver of these oscillations in liver is feeding, since fasting significantly decreases glutathione levels in this organ [47]. Although partially driven by food intake, the significance of such redox fluctuations in modulating an organ's cytoprotective capability has been clearly demonstrated in the liver, which displays increased sensitivity to toxicity by the analgesic paracetamol [50] and the chemotherapy drug cisplatin [48], during periods of low glutathione levels. Interestingly, the pancreas exhibits robust glutathione rhythms which are unaffected by fasting, suggesting differential regulation of the glutathione system in diverse organs [47]. Furthermore, recent studies suggest that the transcriptional oscillator might in part drive oscillations in the glutathione system.
One of the major cellular antioxidant defense responses is mediated by the transcription factor nuclear factor erythroid-derived 2-like 2 (NRF2). Genes under the transcriptional control of NRF2 include glutathione cysteine ligase (GCL), catalyzing the rate-limiting step in the biosynthesis of glutathione, and glutathione-dependent enzymes such as glutathione S-transferase (GST). Nrf2 transcription is rhythmic in both liver and lung, and the rhythmic accumulation of NRF2 protein drives rhythmic transcription of antioxidant genes such as GCL and GST [49,51]. These rhythms are abolished in Nrf2 null animals, which was paralleled by increased susceptibility to fibrotic injury in the lung [49]. Furthermore, considering the role of melatonin in the activation of the Nrf2 pathway it is possible that hormonal signalling may modulate redox through this mode [52,53]. Glutathione oscillations have also been demonstrated in platelets kept in vitro [54]. This is interesting, as platelets are anucleate and thus these rhythms are not directly driven by the transcriptional clock. In addition, such redox oscillations might be involved in platelet activation, which has recently been shown to occur in circadian manner [55]. Furthermore, diurnal variation in the reduced to oxidised glutathione (GSH/GSSG) and the disulphide to sulfhydryl (SS/SH) ratios have been reported in blood plasma, suggesting that the redox capacity of the blood resonates with day-night cycles [56].
Redox oscillations are not limited to the periphery, since the redox status of the central nervous system pacemaker in the SCN, as measured by abundance of NAD(P)H and FAD+ and protein glutathionylation, oscillates in a circadian manner [57] (figure 2). These rhythms were found to determine the rate of neuronal firing by regulating membrane excitability via redox-regulated potassium (K+) channels. Although the oscillations were abolished in Bmal1 knockout mice, the important role of this transcription factor in maintaining cellular NAD+ availability [21,26] and general redox milieu [58] indicates that the observed effects might be due to non-circadian functions of BMAL1, by virtue of its role as a generic transcription factor. In support of this, constitutive (rather than rhythmic) expression of Bmal1 is capable of restoring molecular rhythms in mutant animals [59]. Furthermore, whereas constitutive expression of Bmal1 in SCN alleviates the behavioural arrhythmicity, it fails to rescue the decreased activity levels and body weight of knockout mice. In contrast, muscle-specific rescue of Bmal1 restores normal activity levels and body weight, yet fails to restore normal locomotor rhythms [60]. Taken together, these observations suggest that interpretations based on clock-relevant transcription factor mutants should be treated carefully due to the potential non-circadian roles of the core “clock” components.
Peroxiredoxins: redox oscillators or redox reporters?
Undoubtedly, the most striking examples of circadian redox oscillations described until now are the redox cycles in the peroxiredoxin proteins, which persist even in the absence of transcription [11,12,61]. Moreover, the high phylogenetic conservation of these redox oscillations suggests that their evolution might have preceded that of the transcriptional clockwork, thereby forcing the reassessment of the current model of cellular timekeeping.
Peroxiredoxins (PRXs) are ubiquitous cellular peroxidases responsible for the scavenging of hydrogen peroxide (H2O2) [62]. Unlike other antioxidant enzymes that rely on cofactors such as metals, heme or selenocysteine for catalysis, PRXs utilize a reactive cysteine residue, which detoxifies H2O2 at the expense of its own oxidation. An intriguing feature of PRXs is that their active site cysteine residue can undergo further oxidation, termed overoxidation. The overoxidised form of the enzyme is catalytically inactive and can be regenerated by the only known sulphinic acid reductase, sulfiredoxin (SRX) (see Box 1).
Box 1. Peroxiredoxins: The Basics.
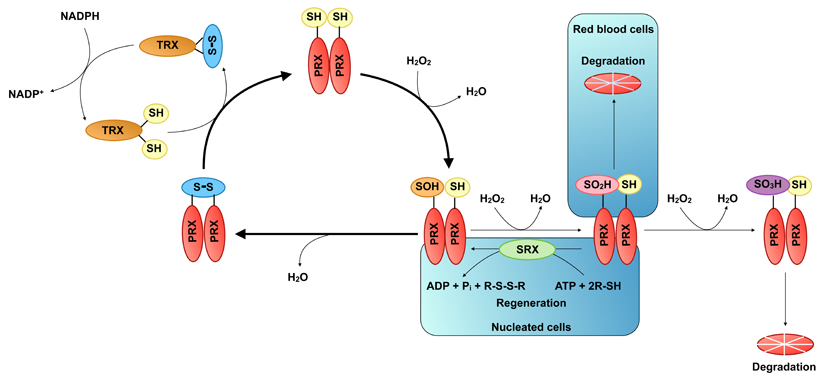
Peroxiredoxins (PRXs) are a highly conserved family of cellular peroxidases found in virtually all aerobic organisms [62]. They detoxify various organic peroxides, including hydrogen peroxide (H2O2) and peroxinitrite (OONO-). Mammals express six PRX isoforms that exhibit discrete subcellular localisation, with PRX 1,2, and 6 being exclusively nucleocytoplasmic, PRX3 located in the mitochondrion, PRX4 in the endoplasmic reticulum (ER), and PRX5 being present in the mitochondrion, peroxisomes, and cytosol [63]. The catalytic center of PRXs is comprised of a highly reactive cysteine residue, termed the peroxidatic cysteine (CP). The first step of the catalytic cycle involves the oxidation of this residue to sulfenic acid (-SOH). In the subclass of typical 2-Cys PRXs, which exist as obligate homodimers, the oxidised CP condenses with another conserved residue, termed the resolving cysteine (CR), which resides on the paired subunit. This results in the formation of intermolecular disulphide (PRX-CP-S-S-CR-PRX), which is reduced by the thioredoxin system in a series of reactions that ultimately consumes reducing equivalents from the ubiquitous redox coenzyme NADPH. During the catalytic cycle, the recycling reaction is occasionally outcompeted by further oxidation of the sulphenic acid intermediate to the enzymatically inactive sulphinic acid (-SO2H) and sulphonic acid (-SO3H) (termed overoxidation and hyperoxidation, respectively). The overoxidised enzyme is regenerated back to its active form by the only known sulphinic acid reductase, sulfiredoxin (SRX), in a reaction that requires energy in the form of ATP and cellular thiols (-SH) [64] (Figure I). Although mass spectrometry experiments have provided evidence for the occurrence of the hyperoxidised form of the enzyme in vitro [80] its occurrence in vivo is still controversial.
Figure I. The catalytic cycle of typical 2-Cys peroxiredoxins (PRXs).
The typical 2-Cys PRXs form obligate homodimers in a head-to-tail configuration so that the peroxidatic cysteine (CP) of one monomer faces the resolving cysteine (CR) of the partner monomer. During the catalytic cycle the highly nucleophilic CP is oxidised to sulphenic acid (-SOH) by incoming peroxide. The oxidised CP then condenses with CR to form an intermolecular disulphide bond, which is subsequently reduced by the thioredoxin system in a series of reactions that use the redox cofactor NADPH as an electron donor. Occasionally, the resolving reaction is outcompeted by further oxidation of the sulphenic acid CP to the chemically inert sulphinic acid (-SO2H) and sulphonic acid (-SO3H), termed over- and hyper-oxidation, respectively. In nucleated cell PRXs are regenerated back to the active state by the only known sulphinic acid reductase, sulfiredoxin (SRX) in a reaction, which consumes cellular thiols and energy in the form of ATP. In red blood cells (RBCs) the overoxidised enzyme is instead targeted for degradation in the proteasome. No mechanism has been identified for the regeneration of the sulphonic acid form of the enzyme, which is probably degraded in the proteasome.
In anucleate human (RBCs) blood cells the overoxidised form of PRX exhibits cyclical accumulation, which fulfills all conditions of a bona fide circadian rhythm, namely, persistence in constant conditions, the ability to respond to synchronisation by external cues, and temperature compensation [11]. Furthermore, the rhythmic accumulation of overoxidised PRX is paralleled by oscillations in a number of redox parameters such as haemoglobin oxidation and NAD(P)H abundance. Similar rhythms were also found in the autophototrophic unicellular alga Ostreococcus tauri, even under conditions of transcriptional arrest as occurs in darkness, as they are, dependent on light for transcription [12]. Interestingly, when the organism is transferred back to light, the phase of the transcriptional rhythm is unchanged, suggesting that the PRX oxidation cycles might act as the primary timekeeping mechanism during periods of attenuated transcription. PRX redox cycles were subsequently observed in variety of model systems including nucleated cells, mouse tissues, plants, nematodes, fruit flies, fungi, cyanobacteria and an archaeon making it the first marker of circadian rhythmicity shared across all domains of life [13].
Recently, work from Rhee and colleagues demonstrated the biochemical basis of PRX oxidation in mouse RBCs [61]. As previously hypothesised [65], PRX oxidation was found to be a result of ROS generated by haemoglobin auto-oxidation, which occurs naturally in RBCs, and is normally buffered by peroxiredoxins [66]. Unexpectedly, these oscillations persisted in cells from Srx knockout mice and were shown to depend on the degradation of the overoxidised enzyme by the proteasome [61]. This mechanism probably reflects the unique environment and demands of RBCs, and it is unlikely to be applicable to nucleated cells, where SRX is expected to play an important role. In agreement with this, SRX is essential for the circadian production of corticosterone (CS) in mouse adrenal cortex [79]. Adrenocorticotropic hormone (ACTH)-induced CS synthesis occurs with a concomitant increase in H2O2 by cytochrome P450 in the mitochondria, which results in the overoxidation of the mitochondrial-specific PRX isoform, PRX3. PRX3 inactivation leads to the accumulation of H2O2 in mitochondria, and its overflow into the cytosol, where it triggers the activation of p38 mitogen-activated protein kinase and consequent down-regulation of both CS synthesis and H2O2 production. In addition, ACTH stimulates the expression of SRX, which translocates to the mitochondrial matrix and mediates the regeneration of PRX3. This SRX-dependent regeneration of inactivated PRX3 thus represents a negative feedback regulatory mechanism for steroidogenesis independently of the hypothalamic-pituitary-adrenal axis. This is also a good example of a delayed negative feedback mechanisms, similar to the TTFL, in which accumulation of H2O2 and subsequent PRX3 inactivation is coupled to the slow reaction kinetics of the regeneration by SRX [67] to generate circadian rhythms in CS production.
Concluding remarks and future perspectives
The studies summarized here underline the numerous nodes of interaction between circadian and redox systems. Despite being highly conserved through evolution, the functional significance of the PRX redox oscillations for cellular timekeeping and general physiology is yet to be completely deciphered. Indeed, the presence of such autonomous oscillations suggests the existence of an underlying redox oscillator. However, whether peroxiredoxins play a functional role in the generation of these rhythms, or are reporters of a more global cellular phenomenon, remains to be determined. Regardless of the exact details, it is highly likely that in a nucleated cell, transcriptional and redox oscillators are tightly coupled in order to generate one unambiguous cellular time base, and that disturbance of one will have an affect on the other. Consistent with this, clock mutants display aberrant PRX rhythms, and conversely, ablation of different PRX isoforms affect transcriptional rhythms [11,68].
The significance of the interplay between the circadian and redox systems is further illustrated by the numerous pathologies that manifest both circadian and redox disturbances such as ageing, neurodegenerative disorders, cancer and various metabolic conditions [69]. The link between circadian timekeeping and ageing has long been recognised and appears to be a reciprocal interaction [70]. It has been convincingly demonstrated that numerous circadian parameters are affected by ageing and that these changes are paralleled by alterations in the neurochemical and electrophysiological output of the SCN, without a decrease in cell number or size. In addition, it appears that ageing affects the ability of the SCN to reset peripheral clocks, whereas transplantation of fetal SCN into aged animals could rescue this effect and increase life span [71]. Conversely, disruption of the circadian system by genetic deletion of clock genes has been shown in some cases to significantly affect the ageing process [70]. The most striking example is provided by Bmal1 mutant animals, which display shortened life span and develop many age-related pathologies such as reduction in muscle and bone mass, cataracts, and organ shrinkage [58]. It should be noted that these effects arise potentially due to non-circadian functions of BMAL1, as constitutive expression of the transcription factor is capable of reversing the phenotype [59,60]. Aging is accompanied by alterations in cellular redox balance, causing increased ROS generation and oxidative damage. Consistently, the imbalance of ROS levels in these animals was proposed to be one of the major drivers of the premature ageing phenotype. In agreement with this, supplementation with the antioxidant N-acetyl-L-cysteine increases the longevity of Bmal1 knockout animals [72]. Interestingly, certain feeding regimens have been shown to affect life span, circadian rhythms and redox balance. For example, caloric restriction entrains the SCN and increases longevity, partially through a decrease in the oxidative burden [73]. A well known example that links the circadian and redox system to aging is SIRT1, the activity of which is increased by caloric restriction and is proportional to lifespan [74].
Another interesting example of the confluence of circadian and redox systems in disease is cancer, in which both aberrant ROS signalling and altered rhythmicity contributing to carcinogenesis [75,76]. SIRT1 function has also been linked to cancer, as its primary targets include p53 and FOXO1, both of which show aberrant regulation in cancer [77,78]. Interestingly, melatonin has been recognised as tumor-suppressor and its anti-cancer effect might be partially due to inhibition of SIRT1 expression and activity [69]. These are only a few examples of the links between the circadian and redox systems in pathological conditions and further research in this area is likely to extend the list. Importantly, the interaction between the two systems opens new avenues of research for the development of therapeutic agents directed against multicomponent diseases, such as the ones outlined above.
Although considerable progress to understand the involvement of redox biology in cellular timekeeping has been made in recent years, there are still a number of important questions that require addressing (Box 2). For instance, the exact mechanism that generates PRX redox rhythms in RBCs is now known; however, it is still unclear what dictates a 24-hour period for these oscillations. In addition, the process remains essentially uncharacterized in nucleated cells. On that note, the use of PRX and “clock” mutants has indicated that the two oscillations might be linked, however, characterization of the molecular basis of this interaction is still pending.
Box 2. Outstanding questions.
What drives the rhythmic oxidation of peroxiredoxins and what is the functional significance of these redox oscillations for cellular time keeping?
Is redox poise a circadian-relevant metabolic cue and, if so, how does it relay timing information to the transcriptional clock?
How is the redox oscillator linked to the transcriptional clock, and does the relative contribution of the two oscillators vary between different model systems, in different tissues and organs, and during the life span of a cell?
Can the interaction between circadian and redox systems be exploited in order to treat disorders that manifest disturbance either or both systems?
Despite the numerous examples in support of the strong link between redox metabolism and circadian rhythmicity there is still lack of direct experimental evidence that the two processes are coupled in vivo. This is mainly due to the intrinsic difficulty involved in teasing apart two processes so fundamental for cellular physiology. In this respect, it would be interesting to see whether disruption of metabolic pathways important for maintenance of cellular redox poise would lead to perturbations in the transcriptional rhythms. Conversely, it would be beneficial to apply more gentle methods than genetic ablation for studying the effect of the transcriptional oscillator on redox rhythms. As pointed out earlier, genetic ablation of clock genes might resulting in pleiotropic effects and these results would thus be hard to interpret.
Acknowledgements
ABR is a Wellcome Trust Senior Fellow in Clinical Sciences. He receives funding from the Wellcome Trust (Grant No. 100333/Z/12/Z), the European Research Council (ERC Starting Grant No. 281348, MetaCLOCK), the European Molecular Biology Organization (EMBO) Young Investigators Programme, and the Lister Institute of Preventative Medicine. NBM is supported by a Wellcome Trust PhD Studentship.
Glossary
- Antioxidant defense
Defense mechanisms, which neutralize cytotoxic oxygen intermediates and maintain redox homeostasis.
- Clockwork
The molecular and biochemical mechanisms, which drive biological clocks.
- Glutathione
A low molecular weight thiol-containing antioxidant, which serves as the main redox buffer in the cell.
- Oscillator
A multicomponent system that never reaches equilibrium and is capable of generating self-sustained oscillations in various parameters such as gene expression and metabolites levels.
- Peroxiredoxins
Small, ubiquitously expressed antioxidant proteins, which serve as the main sink for hydrogen peroxide (H2O2) in the cell.
- Reactive oxygen/nitrogen species (ROS/RNS)
A generic term used to describe reactive molecules and free radicals derived from oxygen. Examples include super oxide (O2-), peroxynitrite (OONO-) and hydrogen peroxide (H2O2).
- Redox metabolism
The ensemble of redox reactions involved in core metabolism such as the reactions involved in glucose oxidation to pyruvate.
- Redox poise
A term synonymous to redox homeostasis that is used to describe the balance in intracellular redox potential.
- Restricted feeding (RF)
A specilised feeding regimen, wherein mice are allowed access to food for a limited period during the light phase.
References
- 1.Shea SA, et al. Existence of an endogenous circadian blood pressure rhythm in humans that peaks in the evening. Circulation Research. 2011;108:980–984. doi: 10.1161/CIRCRESAHA.110.233668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burioka N, et al. Circadian rhythms in the CNS and peripheral clock disorders: function of clock genes: influence of medication for bronchial asthma on circadian gene. J Pharmacol Sci. 2007;103:144–149. doi: 10.1254/jphs.fmj06003x4. [DOI] [PubMed] [Google Scholar]
- 3.Smolensky MH, et al. Chronobiology and chronotherapy of allergic rhinitis and bronchial asthma. Adv Drug Deliv Rev. 2007;59:852–882. doi: 10.1016/j.addr.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Scheer FAJL, et al. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigurdardottir LG, et al. Circadian Disruption, Sleep Loss, and Prostate Cancer Risk: A Systematic Review of Epidemiologic Studies. Cancer Epidemiology Biomarkers & Prevention. 2012;21:1002–1011. doi: 10.1158/1055-9965.EPI-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libra M. Correlation of the risk of breast cancer and disruption of the circadian rhythm (Review) Oncol Rep. 2012 doi: 10.3892/or.2012.1839. [DOI] [PubMed] [Google Scholar]
- 7.Balsalobre A, et al. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 8.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 9.Nagoshi EE, et al. Circadian Gene Expression in Individual Fibroblasts - Cell-Autonomous and Self-Sustained Oscillators Pass Time to Daughter Cells. Cell. 2004;119:13–13. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 11.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowe SA, et al. Atmospheric oxygenation three billion years ago. Nature. 2013;501:535–538. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- 15.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara R, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 17.Satoh Y. Time-restricted feeding entrains daily rhythms of energy metabolism in mice. AJP: Regulatory Integrative and Comparative Physiology. 2005;290:R1276–R1283. doi: 10.1152/ajpregu.00775.2005. [DOI] [PubMed] [Google Scholar]
- 18.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabolism. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Buxton OM, et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4:129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahata Y, et al. Circadian Control of the NAD+ Salvage Pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahar S, et al. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 2011;3:794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asher G, et al. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 25.Nakahata Y, et al. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peek CB, et al. Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science. 2013 doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutter J. Regulation of Clock and NPAS2 DNA Binding by the Redox State of NAD Cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 28.Dioum EM. NPAS2: A Gas-Responsive Transcription Factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 29.Ishida M, et al. Effects of mutations in the heme domain on the transcriptional activity and DNA-binding activity of NPAS2. Biochemical and Biophysical Research Communications. 2008;368:292–297. doi: 10.1016/j.bbrc.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, et al. A Novel Heme-Regulatory Motif Mediates Heme-Dependent Degradation of the Circadian Factor Period 2. Molecular and Cellular Biology. 2008;28:4697–4711. doi: 10.1128/MCB.00236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta N, Ragsdale SW. Thiol-disulfide Redox Dependence of Heme Binding and Heme Ligand Switching in Nuclear Hormone Receptor Rev-erb. Journal of Biological Chemistry. 2011;286:4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter EL, Ragsdale SW. Modulation of nuclear receptor function by cellular redox poise. Journal of Inorganic Biochemistry. 2014;133:92–103. doi: 10.1016/j.jinorgbio.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nangle SN, et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife. 2014;3:e03674. doi: 10.7554/eLife.03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmalen I, et al. Interaction of Circadian Clock Proteins CRY1 and PER2 Is Modulated by Zinc Binding and Disulfide Bond Formation. Cell. 2014;157:1203–1215. doi: 10.1016/j.cell.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 35.Okano S, et al. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neuroscience Letters. 2009;451:246–251. doi: 10.1016/j.neulet.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Lamia KA, et al. AMPK Regulates the Circadian Clock by Cryptochrome Phosphorylation and Degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao D, et al. A redox-dependent mechanism for regulation of AMPK activation by Thioredoxin1 during energy starvation. Cell Metabolism. 2014;19:232–245. doi: 10.1016/j.cmet.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papp SJ, et al. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. Elife. 2015;4 doi: 10.7554/eLife.04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-G, et al. Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nature Communications. 2013;4:1568. doi: 10.1038/ncomms2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardeland R, et al. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int. 2003;20:921–962. doi: 10.1081/cbi-120025245. [DOI] [PubMed] [Google Scholar]
- 41.Luchetti F, et al. Melatonin signaling and cell protection function. The FASEB Journal. 2010;24:3603–3624. doi: 10.1096/fj.10-154450. [DOI] [PubMed] [Google Scholar]
- 42.Robinson JL, et al. Circadian variation of liver metabolites and amino acids in rats adapted to a high protein, carbohydrate-free diet. J Nutr. 1981;111:1711–1720. doi: 10.1093/jn/111.10.1711. [DOI] [PubMed] [Google Scholar]
- 43.Kaminsky YG, et al. Analysis of the circadian rhythm in energy metabolism of rat liver. Int J Biochem. 1984;16:629–639. doi: 10.1016/0020-711x(84)90032-6. [DOI] [PubMed] [Google Scholar]
- 44.Isaacs JT, Binkley F. Cyclic AMP-dependent control of the rat hepatic glutathione disulfide-sulfhydryl ratio. Biochim Biophys Acta. 1977;498:29–38. doi: 10.1016/0304-4165(77)90084-8. [DOI] [PubMed] [Google Scholar]
- 45.Isaacs J, Binkley F. Glutathione dependent control of protein disulfide-sulfhydryl content by subcellular fractions of hepatic tissue. Biochim Biophys Acta. 1977;497:192–204. doi: 10.1016/0304-4165(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 46.Bélanger PM, et al. Temporal variations in microsomal lipid peroxidation and in glutathione concentration of rat liver. Drug Metab Dispos. 1991;19:241–244. [PubMed] [Google Scholar]
- 47.Neuschwander-Tetri BA, Rozin T. Diurnal variability of cysteine and glutathione content in the pancreas and liver of the mouse. Comp Biochem Physiol B, Biochem Mol Biol. 1996;114:91–95. doi: 10.1016/0305-0491(96)83706-0. [DOI] [PubMed] [Google Scholar]
- 48.Li XM, et al. Pharmacologic modulation of reduced glutathione circadian rhythms with buthionine sulfoximine: relationship with cisplatin toxicity in mice. Toxicol Appl Pharmacol. 1997;143:281–290. doi: 10.1006/taap.1996.8088. [DOI] [PubMed] [Google Scholar]
- 49.Pekovic-Vaughan V, et al. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes & Development. 2014;28:548–560. doi: 10.1101/gad.237081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mori K, et al. Evaluation of hepatic damage by reactive metabolites--with consideration of circadian variation of murine hepatic glutathione levels. J Toxicol Sci. 2014;39:537–544. doi: 10.2131/jts.39.537. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y-Q, et al. Diurnal Variation of Hepatic Antioxidant Gene Expression in Mice. PLoS ONE. 2012;7:e44237. doi: 10.1371/journal.pone.0044237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tripathi DN, Jena GB. Effect of melatonin on the expression of Nrf2 and NF-kappaB during cyclophosphamide-induced urinary bladder injury in rat. J Pineal Res. 2010;48:324–331. doi: 10.1111/j.1600-079X.2010.00756.x. [DOI] [PubMed] [Google Scholar]
- 53.Negi G, et al. Melatonin modulates neuroinflammation and oxidative stress in experimental diabetic neuropathy: effects on NF-κB and Nrf2 cascades. 2010 doi: 10.1111/j.1600-079X.2010.00821.x. [DOI] [PubMed] [Google Scholar]
- 54.Radha E, et al. Glutathione levels in human platelets display a circadian rhythm in vitro. Thromb Res. 1985;40:823–831. doi: 10.1016/0049-3848(85)90319-6. [DOI] [PubMed] [Google Scholar]
- 55.Scheer FAJL, et al. The Human Endogenous Circadian System Causes Greatest Platelet Activation during the Biological Morning Independent of Behaviors. PLoS ONE. 2011;6:e24549. doi: 10.1371/journal.pone.0024549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanco RA, et al. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr. 2007;86:1016–1023. doi: 10.1093/ajcn/86.4.1016. [DOI] [PubMed] [Google Scholar]
- 57.Wang TA, et al. Circadian Rhythm of Redox State Regulates Excitability in Suprachiasmatic Nucleus Neurons. Science. 2012;337:839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondratov RV. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes & Development. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu AC, et al. Redundant Function of REV-ERBα and β and Non-Essential Role for Bmal1 Cycling in Transcriptional Regulation of Intracellular Circadian Rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roumagnac P, et al. Evolutionary History of Salmonella Typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho CS, et al. Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci USA. 2014;111:12043–12048. doi: 10.1073/pnas.1401100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood ZA, et al. Structure, mechanism and regulation of peroxiredoxins. Trends in Biochemical Sciences. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 63.Park J, et al. 2-Cys Peroxiredoxins: Emerging Hubs Determining Redox Dependency of Mammalian Signaling Networks. 2014;2014:1–10. doi: 10.1155/2014/715867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhee SG, et al. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007 doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 65.O’Neill JS, Feeney KA. Circadian Redox and Metabolic Oscillations in Mammalian Systems. Antioxidants & Redox Signaling. 2013;20 doi: 10.1089/ars.2013.5582. 131122075437004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Low FM, et al. Peroxiredoxin 2 and peroxide metabolism in the erythrocyte. Antioxidants & Redox Signaling. 2008;10:1621–1630. doi: 10.1089/ars.2008.2081. [DOI] [PubMed] [Google Scholar]
- 67.Chang T-S, et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilking M, et al. Circadian Rhythm Connections to Oxidative Stress: Implications for Human Health. Antioxidants & Redox Signaling. 2013;19:192–208. doi: 10.1089/ars.2012.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Froy O. Circadian rhythms, aging, and life span in mammals. Physiology (Bethesda) 2011;26:225–235. doi: 10.1152/physiol.00012.2011. [DOI] [PubMed] [Google Scholar]
- 71.Hurd MW, Ralph MR. The significance of circadian organization for longevity in the golden hamster. Journal of Biological Rhythms. 1998;13:430–436. doi: 10.1177/074873098129000255. [DOI] [PubMed] [Google Scholar]
- 72.Kondratov RV, et al. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY) 2009;1:979–987. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Froy O, Miskin R. Effect of feeding regimens on circadian rhythms: implications for aging and longevity. Aging (Albany NY) 2010;2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cantó C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends in Endocrinology & Metabolism. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelleher FC, et al. Circadian molecular clocks and cancer. Cancer Letters. Cancer Letters. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 76.Acharya A, et al. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxidative medicine and cellular longevity. 2010;3:23–34. doi: 10.4161/oxim.3.1.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Z, Tindall DJ. Oncogene - Abstract of article: FOXOs, cancer and regulation of apoptosis. Oncogene. 2008 doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 79.Kil IS, et al. Feedback Control of Adrenal Steroidogenesis via H. Molecular Cell. 2012;46:584–594. doi: 10.1016/j.molcel.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 80.Yang K, et al. Inactivation of Human Peroxiredoxin I during Catalysis as the Result of the Oxidation of the Catalytic Site Cysteine to Cysteine-sulfinic Acid. Journal of Biological Chemistry. 2002;41:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 81.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430(6998):467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]