SUMMARY
Artificial transcription factors (ATFs) are designed to mimic natural transcription factors in the control of gene expression and are comprised of domains for DNA binding and gene regulation. ATF domains are modular, interchangeable, and can be composed of protein-based or nonpeptidic moieties, yielding DNA-interacting regulatory molecules that can either activate or inhibit transcription. Sequence-specific targeting is a key determinant in ATF activity, and DNA-binding domains such as natural zinc fingers and synthetic polyamides have emerged as useful DNA targeting molecules. Defining the comprehensive DNA binding specificity of these targeting molecules for accurate manipulations of the genome can be achieved using cognate site identifier DNA microarrays to explore the entire sequence space of binding sites. Design of ATFs that regulate gene expression with temporal control will generate important molecular tools to probe cell-and tissue-specific gene regulation and to function as potential therapeutic agents.
Keywords: Artificial transcription factor, Polyamide, DNA-binding domain, Transcriptional activation domain, Cognate site identifier arrays
1. Introduction
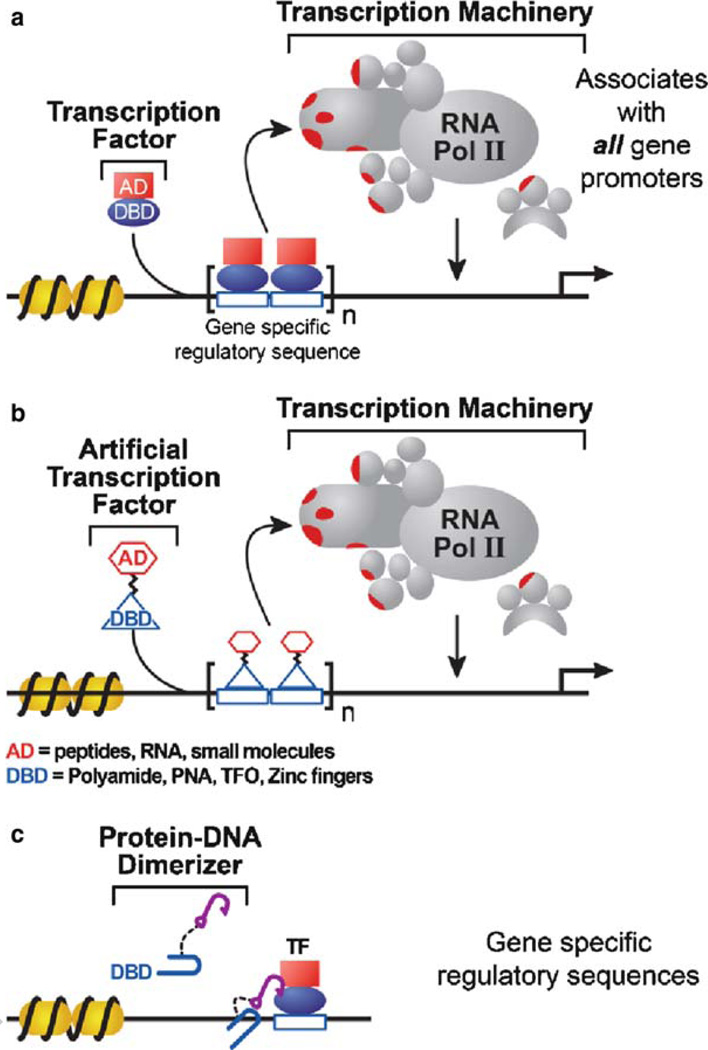
Natural transcription factors are nuclear proteins that bind to specific DNA sequences localized at a gene or set of genes and recruit enzymes to modify chromatin structure and/or synthesize messenger RNA (mRNA) (Fig. 1a). Transcription factors are responsible for choreographing the complex cascade of signaling events that give rise to specific cell types and tissues. Sequencing of the human genome has indicated that nearly 6% of the genome (~2,000 genes) encodes transcription factors, yet the genome-wide role of these factors in controlling gene expression has been well characterized for only a small subset (1–6). Transcription factors are capable of activating or repressing gene expression, and their aberrant activity has been linked to an array of disorders including cancer, obesity, diabetes, and inflammation. Artificial transcription factors (ATFs) that can be engineered to target specific genomic sites and modulate gene activity will be useful as novel therapeutics for disease treatment and for mechanistic studies of gene expression.
Fig. 1.
Role of ATFs and protein–DNA dimerizers in transcription. (a) Transcription factors are modular proteins composed of a DNA-binding domain (DBD) that recognizes gene-specific regulatory sequences and an activation domain (AD) that recruits RNA polymerase II and/or associated proteins. (b) Artificial transcription factors (ATFs) can be created using transcriptional activation domains such as VP16 or small molecules and DBDs such as protein-based DNA-binding modules or synthetic molecules including polyamides, triplex-forming oligonucleotides (TFOs), or peptide nucleic acids (PNAs). (c) Protein–DNA dimerizers contain a DBD linked to a molecular “hook” such as a short peptide or small molecule, which facilitates binding of a natural transcription factor at an adjacent DNA site.
ATFs mimic the modular design of natural transcription factors and are comprised of a DNA-binding domain (DBD), a regulatory domain, and, in some cases, a linker region (Fig. 1b) (7, 8). ATFs can also target a natural transcription factor to an adjacent DNA-binding site by use of a molecular “hook” such as a peptide or small molecule that specifically recruits the transcription factor (Fig. 1c). Early design of ATFs combined the DBD of one protein with the regulatory domain of another, resulting in chimeric proteins with altered DNA binding specificity. Common activating domains include the herpes simplex viral transactivator VP16 or a 20-residue peptide known as amphipathic helix (AH), with the sequence PEFPGIELQELQELQALLQQ (9). Despite decades of research to uncover a specific structure or sequence associated with transcriptional regulatory domains, the only preferences that emerged were acid-rich, glutamine-rich, or proline-rich activation domains and alanine-rich or positively charged repression domains (10–14). These studies indicated that regulatory domain determinants such as structural folds or specific motifs were not clearly defined, whereas the DBDs could be more accurately described by structure, and their binding specificity could be mimicked by natural or synthetic molecules.
DBDs direct specific interaction with DNA sites to deliver the regulatory domain to a target gene or set of genes. Protein-based DBDs are often categorized by their structural fold such as a helix-turn-helix or coordination molecules such as a zinc finger. Zinc fingers of the type Cys2His2 are ~30-amino acid domains that fold into two β-strands and one α-helix, with the N-terminal residues of the α-helix making specific base contacts in the major groove of DNA. Cys2His2 zinc fingers are useful as ATF DBDs because they can be tailored to recognize any 3-bp DNA site with precise specificity and high affinity (15–17). Although empirical rules for zinc finger recognition of all DNA 9-mers theoretically exist, some zinc finger domains make base contacts outside of their three target nucleotides, preferring a 4-bp rather than a 3-bp site. Computational approaches, including sequential and bipartite selection strategies, and use of predictive design tools have improved zinc finger design (15,17) but also highlight the need to define and refine DBD binding specificity. Some limitations of using protein-based ATFs for disease therapy include the low efficiency of cellular delivery, poor nuclear localization, significant potential for antigenicity, and protein degradation (8).
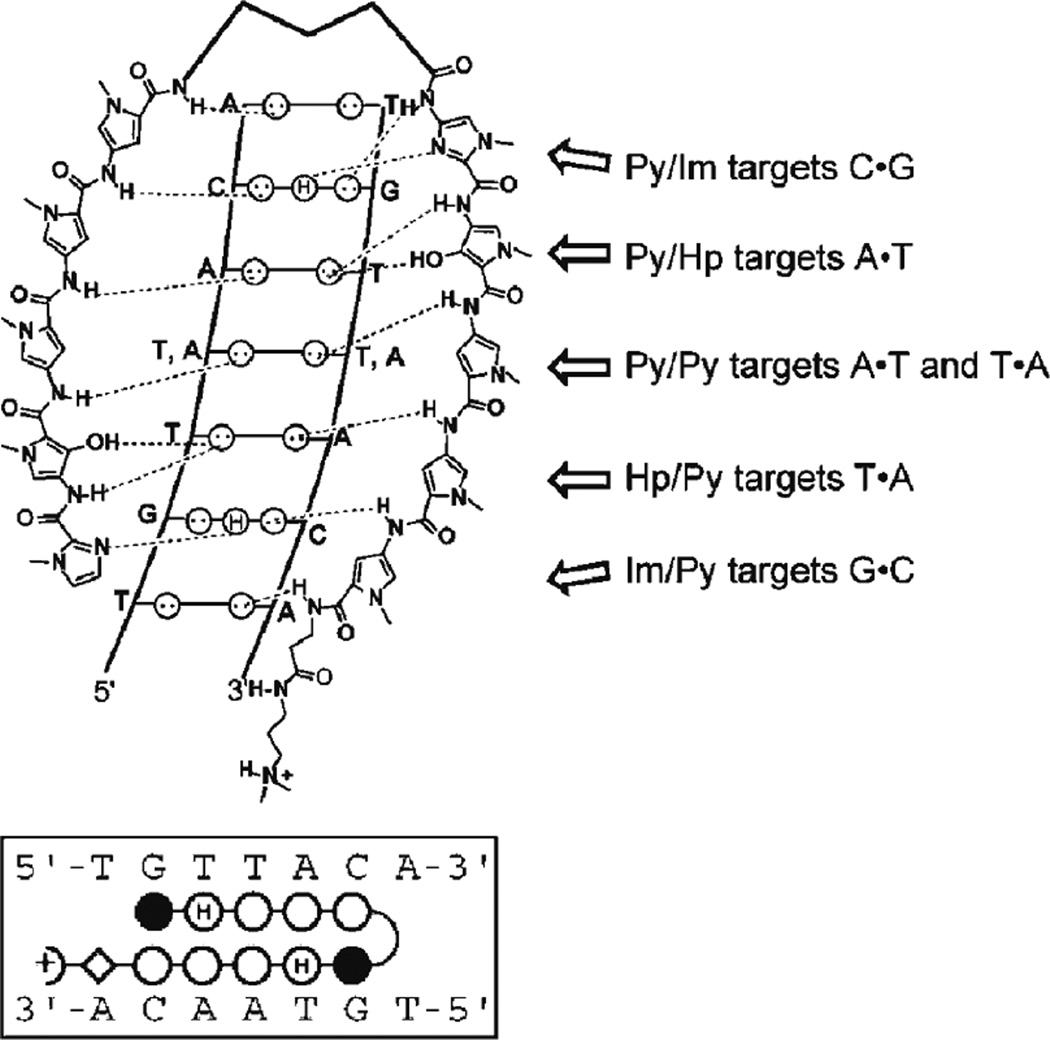
Synthetic DBDs were designed to address some of these limitations of cellular uptake and to provide a chemical approach to selectively alter gene expression; examples include polyamides, triplex-forming oligonucleotides (TFOs), and peptide nucleic acids (PNAs). Polyamides bind in the minor groove of DNA and are composed of N-methylpyrrole (Py) and N-methylimidazole (Im) amino acids or derivatives such as hydroxypyrrole (Hp). The advantage of polyamides is that the combination of the Py and Im aromatic rings in a side-by-side arrangement can be exploited, according to a set of pairing rules, to generate a DBD that targets a specific DNA sequence (Fig. 2). Important aspects of DNA binding include the specificity of DNA recognition and the binding affinity or efficacy for a particular site. For some DNA targets, polyamides possess improved specificity and affinity if polyamide sequence recognition rules are extended to include polyamide pairings such as ImPy and PyPy (i.e., two pairs of rings) to target Watson-Crick base pairs (19). Especially promising in ATF design, polyamides are permeable to mammalian cells and can affect gene expression. Characterization of polyamide binding in colon cancer cells indicated that expression of particular genes was affected but some of the expected signaling pathways were not down-regulated, although it was unclear whether this occurred because of altered DNA recognition, chromatin structure, or interplay with other transcription factor signaling pathways (20). Thus, sequence-specific targeting and refining this DNA recognition remain critical areas of ATF development.
Fig. 2.
Polyamide pairing rules. (a) Polyamides target DNA based on a set of pairing rules. An Im/Py ring pair recognizes G·C; Py/Py pair targets A·T or T·A; and Py/Hp binds to A·T (18). (b) Polyamides can be represented by the following abbreviations and symbols:Im, N-methylimidazole, filled circle; Py, N-methylpyrrole, open circle; Hp, hydroxypyrrole, open circle with H; β, β-alanine, diamond; Dp, dimethylaminopripylamide, half circle with positive charge; γor turn, γ-aminobutyric acid.
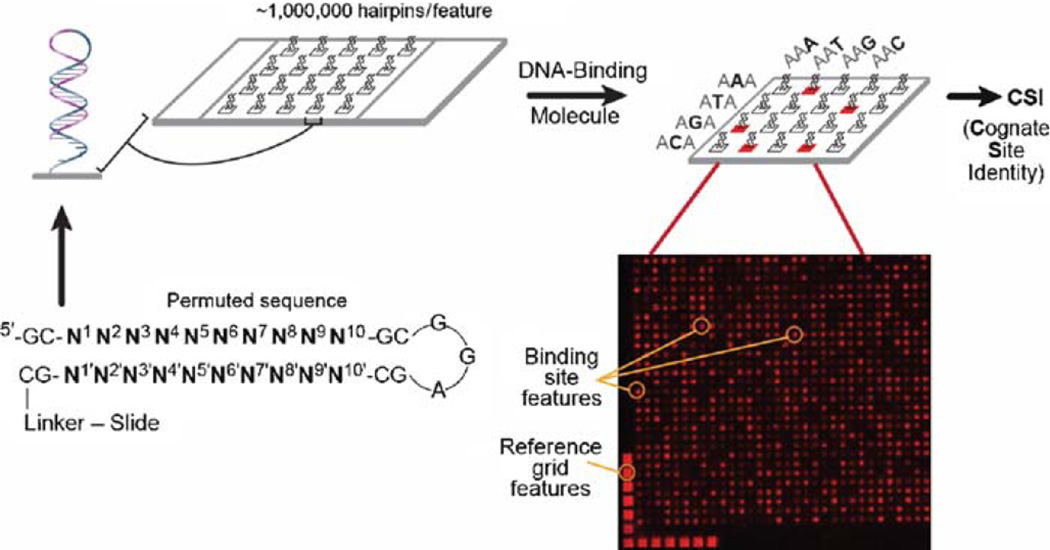
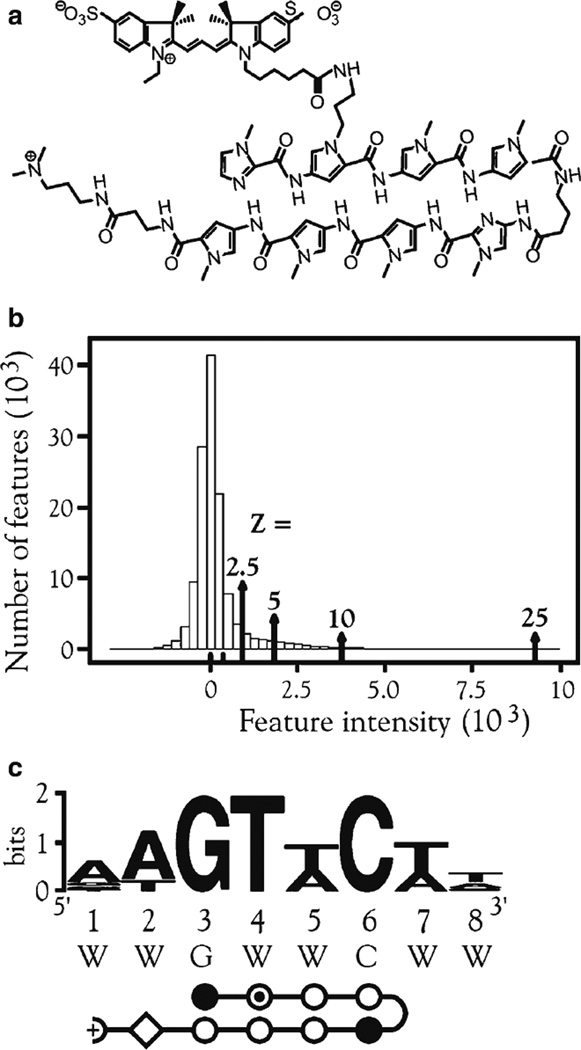
The specific binding of ATFs can be evaluated using cognate site identifier (CSI) arrays bearing every permutation of an 8-to 10-bp DNA that is double stranded and B-form (Fig. 3) (21). DNA is synthesized on the array using maskless array synthesizer (MAS) technology with every feature containing multiple copies of a particular sequence (22). DNA-binding molecules are applied to the array and are detected using fluorescence by direct labeling of the molecule of interest or with a fluorescent antibody. CSI analysis was applied to determine the binding specificity of a Cy3-labeled polyamide, PA1 (Fig. 4a), which was engineered to target the sequence 5'-WWGWWCWW-3 '(W = A or T). Most features were not bound by PA1, as shown by the peak centered at zero, whereas a subset of features displayed high intensity (Fig. 4b). The binding motif was very similar to the expected binding sequence for PA1 (Fig. 4c). Further CSI analysis revealed subtle and unobvious contributions of the core 6 bp of the PA1-binding site to DNA binding specificity, highlighting the usefulness of CSI arrays in determining DNA specificity (21).
Fig. 3.
Illustration of a CSI microarray. Every permutation of a 10 bp (N1–N10) sequence is displayed in a hairpin probe containing a GC stem and GGA turn. The array is incubated with a fluorescently labeled DNA-binding molecule. The fluorescent features are identified and used to determine DNA-binding preferences.
Fig. 4.
Binding of PA1. (a) Structure of Cy3-conjugated polyamide PA1 (ImPy*PyPy-γ-ImPyPyPy-β-Dp). (b) Histogram of averaged intensities of all replicate features. Z scores (see Methods) are noted. (c) Logo obtained from top Z-score bin of 25. Abbreviations: Py*, N-methylpyrrole ring with Cy3 dye attached, open circle with inner dot; W = A or T.
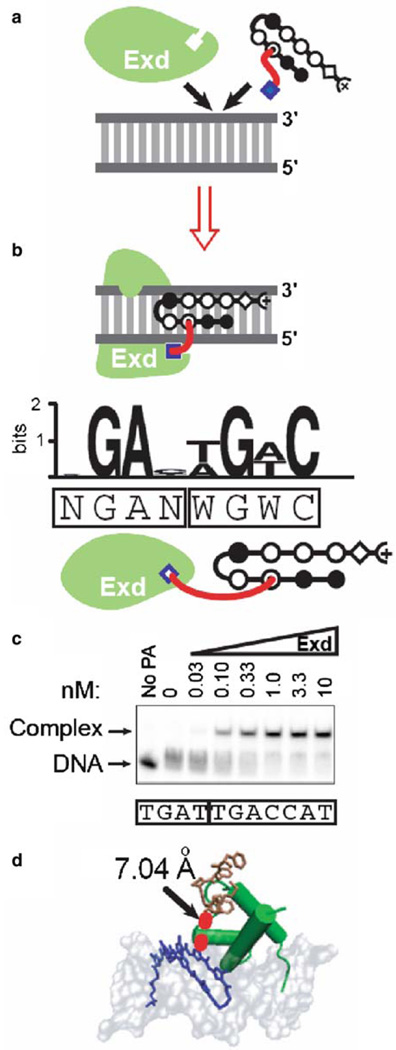
DNA binding specificity and affinity can also be engineered using cooperative complexes. The Drosophila transcription factors, Hox and Extradenticle (Exd), bind DNA with lower specificity and affinity as individual proteins, but their binding specificity and affinity dramatically increase as a cooperatively bound complex (23). Polyamides have been used in the creation of bifunctional molecules, known as protein-DNA dimerizers, comprised of a protein-interacting molecule linked to the polyamide DBD (Fig. 1c) (24, 25). The protein-interacting molecule, also referred to as a “hook,” can stabilize or enhance the binding of a natural transcription factor to an adjacent DNA site. A polyamide bearing the Hox peptide YPWM has been shown to recruit Exd in a cooperative complex on DNA, mimicking the developmental regulator Hox-Exd heterodimeric complex (Fig. 5a) (25). The length of the linker between the protein-interacting moiety and the polyamide DBD determines the effectiveness of the protein-DNA dimerizer in recruiting the natural transcription factor (24, 31). The binding specificity of the polyamide-Exd complex was comprehensively defined using CSI arrays (Fig. 5b) and verified by electrophoretic mobility shift assays (EMSAs) (Fig. 5c) and molecular modeling (Fig. 5d).
Fig. 5.
Polyamide–Exd cooperative complex. (a) Schematic of polyamide–Exd cooperative complex on DNA. (b) Logo (26–28) obtained from CSI analysis of polyamide–Exd binding. Boxed sequence displays binding sites for Exd and polyamide. (c) The polyamide-peptide conjugate at 50 nM was incubated with increasing concentrations of Exd (nM). Arrows indicate free DNA (lower arrow) and DNA–polyamide complex (upper arrow). Boxed sequence denotes the Exd and polyamide-binding sites. (d) Molecular modeling (29, 30) of Exd (green) and polyamide (blue) bound to consensus DNA site, with peptide hook (brown) and linker (red). Models were obtained by aligning crystal structures of the DNA complexed with Exd or hairpin polyamide (Protein Data Bank files 1B8I and 1M18).
This review chapter focuses on methods to precisely and comprehensively define the DNA targeting specificity of ATFs, or any DNA-binding molecule or protein, using CSI microarrays. Biochemical/biophysical tools to validate the accuracy of the CSI platform are also described.
2. Materials
2.1. Polyamide Synthesis
Succinimidyl ester of Cy3 dye (Amersham, Piscataway, NJ) for dye labeling.
Phenylacetamidomethyl (PAM) resin (Peptides International, Louisville, KY).
Chemical reagents: dimethylaminopropylamine, trifluoro-acetic acid (TFA), acetonitrile, methanol, toluene, dimethylformamide (DMF), diisopropylethylamine (DIEA).
N-methylpyrrole and N-methylimidazole
2.2. DNA Array Synthesis
ArrayIt SuperClean microscope slides (TeleChem, Inc., Sunnyvale, CA).
UV-protected desiccator (Secador™) and Drierite desiccant (Fisher Scientific, Pittsburgh, PA).
Microarraying facility with MAS technology.
N-(3-Triethoxysilylpropyl)-4-hydroxy-butyramide (Gelest, Morrisville, PA).
Slide buffer: 95% ethanol, 0.1% glacial acetic acid (see Note 1).
Silane buffer: 95% ethanol, 0.1% glacial acetic acid, 1.6% N-(3-triethoxysilylpropyl)-4-hydroxy-butyramide.
Acetone.
Metal rack slide holder (Wheaton, Millville, NJ) and glass container (Pyrex or comparable) large enough to hold metal slide holder; prewashed with acetone.
Standard lab oven with vacuum drying capability (Lab-Line/Barnstead International, Dubuque, IA).
200-Proof ethanol.
Forceps.
Adjustable speed platform shaker (Fisher Scientific).
Glass slide holder, ~100 mL size (Fisher Scientific).
Ethylenediamine (Fisher Scientific).
50-mL Conical tubes.
Methanol (Fisher Scientific).
7 M Urea in 1× phosphate-buffered saline (PBS).
5× PBS: 685 mM NaCl, 13.5 mM KCl, 50 mM Na2HPO4, 9 mM KH2PO4. Adjust to pH 7.4 with HCl if necessary. Filter-sterilize or autoclave and store at room temperature.
Nonstringent wash buffer (0.9 M NaCl, 60 mM NaH2PO4, 7.6 mM EDTA, 0.01% v/v Tween-20).
Final wash buffer (NimbleGen Systems, Inc., Madison, WI).
ArrayIt array dryer (TeleChem, Inc., Sunnyvale, CA) (see Note 2).
Axon 4000B 5-µm m microarray scanner (Molecular Devices, Sunnyvale, CA), or comparable, connected to an IBM-compatible computer with 1.6-GHz dual-core processor or faster; Windows XP or Vista (32-bit) operating system; 1-GB RAM; 40-GB hard drive.
2.3. Measurement of Microarray Signal Intensities
NimbleScan 2.4 or GenePix® Pro 6.0 microarray analysis software.
Excel software (Microsoft).
Access software (Microsoft).
2.4. Data Analysis
MEME/MAST System Motif Discovery and Search (http://meme.sdsc.edu/meme/intro.html).
2.5. Polyamide Binding by CSI Arrays
Hybridization buffer (100 mM MES, 1 M NaCl, 20 mM EDTA pH 7.5, 0.01% v/v Tween-20).
Secure-seal™ hybridization chamber (Grace Bio-Labs, Bend, OR).
Blocking buffer (2.5% nonfat dried milk in dH2O).
2.6. Biochemical/Biophysical Validationof CSI Arrays
2.6.1. Electrophoretic Mobility Shift Assays
10 µm M DNA stock (Integrated DNA Technologies, Coralville, IA, or comparable), in dH2O or TE buffer.
TE buffer (10 mM Tris-Cl pH 7.5, 1 mM EDTA).
5× forward buffer (Invitrogen, Carlsbad, CA).
Kinase T4 (Invitrogen).
γ32P-ATP (6,000 Ci/mmoL, PerkinElmer, Waltham, MA).
0.6-mL, 1.5-mL, and 2-mL Eppendorf tubes.
G25 spin column (GE Healthcare, Piscataway, NJ).
Binding buffer (150 mM potassium glutamate, 50 mM HEPES pH 7.5, 2 mM DTT, 100 ng/µL bovine serum albumin (BSA), 10% DMSO, 10% glycerol).
10% Acrylamide/3% glycerol gel.
1× TBE: 90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA pH 8.3; can be prepared as 10× stock, kept at room temperature, and diluted one part in nine parts dH2O.
Geiger counter.
Gel electrophoresis voltage box (Pharmacia/GE Healthcare).
Typhoon PhosphorImager System (GE Healthcare) or comparable.
ImageQuant software (GE Healthcare).
2.6.2. Fluorescence Polarization
Cy3-labeled polyamide (synthesized as in Subheading 3.3.1).
Double-stranded DNA (Integrated DNA Technologies or comparable) in dH2O or TE buffer.
Instrument capable of fluorescence polarization (Tecan, Inc., Durham, NC or comparable).
2.6.3. Nuclease Protection Assay/Footprinting
Footprinting binding buffer: 10 mM KCl, 10 mM Tris-HCl (pH 7.5), 2 mM CaCl2, 2 mM MgCl2, 5% glycerol, 100 ng/µL BSA.
DNaseI (Invitrogen; diluted 1:50,000 before use).
Stop buffer: 10 mM EDTA, 10 mM NaOH, 80% forma-mide, 0.01% xylene cyanol, and 0.01% bromphenol blue.
8% Acrylamide/7 M urea gel.
Software for nonlinear regression such as SigmaPlot (Systat Software, San Jose, CA) or Prism (GraphPad, San Diego, CA).
3. Methods
3.1. Polyamide Synthesis of Cy3-Conjugated PA1 (Fig. 4)
Use orthogonally protected N-methylpyrrole or N-methylimidazole building blocks in standard Boc-based solid-phase synthesis (see Note 3).
Cleave the polyamide from PAM resin (100 mg) by treatment with 1 mL dimethylaminopropylamine to remove the phthalimide protecting group and make the free base accessible.
Dilute the crude cleavage mixture with 0.1% TFA (aq) and acetonitrile to a final volume of 5 mL and load onto a preconditioned solid-phase extraction column (C18-bonded phase).
Wash the column with a 4:1 (v:v) solution of 0.1% TFA (aq) and acetonitrile.
Elute the product with methanol. Remove solvents by azeo-tropic distillation from toluene.
The resulting product is the aminopropyl precursor of PA1 and should be a slightly yellow solid. Verify the identity and purity of the product using analytical high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS).
Dissolve 0.5 m mol of the intermediate free base in 0.45 mL anhydrous DMF and 0.050 mL DIEA. Add 1 mg of prepackaged amine-reactive Cy3 fluorophore and agitate in the absence of light, at ambient temperature, for 4 h.
Purify the crude product by preparative HPLC using C18-bonded phase silica with 0.1% TFA and acetonitrile as mobile phases and confirm the purity and identity of product by analytical HPLC and MALDI-TOF MS.
Store polyamides in small aliquots at −80°C. Do not subject to multiple freeze-thaw cycles.
3.2. DNA Array Synthesis
3.2.1. Slide Derivatization
Immerse the slides arranged in the metal slide holder in silane buffer for 4 h with gentle agitation.
Wash slides twice in stock slide buffer for 20 min each wash with gentle agitation (see Note 4).
Bake slides in metal rack in oven for 1 h at 120°C with no vacuum. Then bake overnight at 120°C with vacuum applied (see Note 5).
3.2.2. Microarray Synthesis
Synthesize arrays using MAS technology (22). Covalently attach homopolymer (T5) linkers to monohydroxysilane glass slides. Synthesize oligonucleotides on the homopolymers to create a high-density DNA microarray (see Notes 5 and 6).
Deprotect arrays by incubating in 50% ethanol, 50% ethylenediamine for 2 h at room temperature, protected from light. Rinse the slide in a 50-mL conical tube of dH2O for 30 s and then in a 50-mL conical tube of methanol for 30 s.
As an alternative, DNA arrays may be obtained from Nimble-Gen Systems, Inc. (Madison, WI) and are ready for hairpin induction (step 4).
To induce hairpins, incubate the slide in a 50-mL conical tube of 7 M urea prepared in 1× PBS for 30 min in a 65°C water bath, with a back-and-forth shake of the tube every 10 min. Next, immerse the slide in preheated 1× PBS and incubate in a 65°C water bath for 15 min with one back-and-forth shake during the incubation (see Note 7).
Incubate slide in nonstringent wash buffer for 5–10 min at room temperature.
Wash the array in final wash buffer for 10–20 s.
Dry the array for 20 s with an ArrayIt slide centrifuge (see Note 2).
Scan the microarray to check for low background with an Axon 4000B, ScanArray 5000 (GSI Lumionics, Billerica, MA) or comparable 5-µM scanner. Settings on the Axon 4000B at 532 nm (Cy3) are initially set at a photomultiplier tube (PMT) gain setting of 470 and 100% lamp power. Pixel size is 5, lines to average is 1, and focus position is 0.
Examine data with GenePix Pro version 6.0 or comparable for background.
3.3. Polyamide Binding by CSI Arrays
3.3.1. Polyamide Binding
Attach a hybridization chamber to the array. Rinse the chamber on the array by filling it with dH2O and then withdrawing all dH2O using a pipettor. Add 2.5% nonfat dried milk to fill the chamber and then withdraw enough such that the chamber is slightly more than half full (~200 µL). Incubate for 1.5 h at room temperature with rotation (approximately three to four rotations per minute).
Wash the hybridization chamber twice with hybridization buffer.
Dilute the PA1 to 5 nM in hybridization buffer and add to the hybridization chamber for incubation (1–16 h) with rotation (see Note 8).
Remove the PA1 solution and wash the array with hybridization buffer.
Wash the array with final wash buffer for 10 s and dry.
Scan the microarray with a GenePix 4000B scanner (Molecular Devices). Settings on GenePix 4000B at 532 nm for Cy3 are initially set at a PMT gain setting of 470 and 100% lamp power. Pixel size is 5, lines to average is 1, and focus position is 0. Settings at 635 nm for Cy5 are initially set at a PMT of 710 and 100% lamp power. If features are saturated (=65,535 intensity), the PMT gain setting should be decreased until no saturated features are detected. Save data as a single-channel image TIF for data extraction (see Note 9).
Extract data using NimbleScan 2.1 (or comparable extraction software such as GenePix) and open the output file in an appropriate spreadsheet, such as Microsoft Excel. Data can be further sorted using Microsoft Access.
3.4. Data Analysis/Normalization
Perform global mean normalization for each of three to four replicates to verify that the mean intensity of the replicate arrays is similar and to correct differences in array brightness from experimental variation (32–34).
Perform local mean normalization to correct artifacts caused by uneven distribution and spatial abnormalities using a Loess function spanning 0.5–0.02 as necessary (35).
Determine and remove outliers between replicate features using a Q test at 90% confidence (36, 37).
Average the intensities of duplicate features from the same array.
Perform quantile-normalization of the replicates to eliminate any possible nonlinearity between arrays (38).
Average the intensities of replicate features (different arrays) to give a single intensity for every sequence.
Subtract the center of the histogram peak of the averaged features to correct for background.
Calculate Z scores as |signal|/standard deviation to determine the signal-to-noise ratio and to indicate the probability (P-value) that a given sequence is preferentially bound by the DNA-interacting molecule (39).
Graph the feature intensity (x-axis) versus the number of features (y-axis). If specific binding is observed, a peak around zero intensity (no binding) will be present with a right-handed tail indicating features with higher intensity.
Use MEME and WebLogo (26) or other algorithms to determine the specific DNA sequence recognized by PA1.
3.5. Biochemical/Biophysical Validationof CSI Arrays
3.5.1. Electrophoretic Mobility Shift Assays
Incubate 1 µL of 10 µM double-stranded or hairpin DNA with 4 µL of 5× forward buffer, 1 µL kinase T4, 9 µL dH2O, and 5 µL γ32P-ATP in a 0.6-mL microfuge tube for 1 h at 37°C.
To purify radioactive probe using G25 spin column, centrifuge the spin column from the supplier in a 2-mL centrifuge tube at 735 × g for 75 s in a standard microcentrifuge and dispose of liquid. Add the radioactive reaction to the G25 spin column, place the spin column in a 1.5-mL microfuge tube, and centrifuge for 1 min at. 735 × g Quantitate 1 µL of eluted radioactive DNA probe using a Geiger counter.
Mix 50 nM polyamide-peptide conjugates with a 32P-DNA dilution (150–500 cpm/lane) in binding buffer for 30 min at 4°C. Titrate in partner transcription factor (example is Exd at final concentrations of 0.033, 0.1, 0.33, 10, 33, and 100 nM) for a final reaction volume of 20 m L, and incubate for 1 h at 4°C.
Pour 10% acrylamide/3% glycerol gel in 1× TBE and prerun the electrophoresis for 20 min.
Load 15 µL of reaction onto gel and run for 2–3 h at 220 V. Place the gel on Whatman filter paper, cover with plastic wrap, and dry. Expose the dried gel overnight on a phosphorimager screen and visualize the bound DNA versus free DNA using a Typhoon phosphorimager or comparable instrument (see Note 10).
3.5.2. Fluorescence Polarization
Depending on the fluorescence polarization instrument, final volumes for each sample will be 20–100 µL.
Label polyamide with FITC, as described in Subheading 3.1.
Titrate DNA from 0.1 nM to 1 µM.
Add 1 or 5 nM FITC-polyamide in binding buffer to each sample. The concentration of FITC-polyamide may need to be optimized.
Measure fluorescence polarization at the appropriate wavelength (fluorescein excitation: 485 nm; emission: 530 nm).
3.5.3. Nuclease Protection Assay/Footprinting
Use standard PCR with one 32P-labeled primer and one unlabeled primer to prepare labeled double-stranded DNA.
Prepare several dilutions of polyamide in footprinting binding buffer.
Incubate polyamide dilutions with 10,000 cpm of 32P-DNA for 1 h at room temperature, in a final volume of 10 µL.
Add 1 µL DNaseI and incubate for 1 min.
Add 10 µL stop buffer and heat to 95°C for 5 min.
Chill samples on ice and then load immediately on an 8% acrylamide/7 M urea gel.
Run gel at 2,000 V in 0.5× TBE buffer until the bromophenol blue is run off the gel.
Dry gel, expose to phosphorimager screen overnight, and visualize using a Typhoon imager.
Run experiment in triplicate and determine binding constants using ImageQuant and appropriate software for nonlinear regression.
Acknowledgments
The authors thank Karl Hauschild and Clayton Carlson for reviewing the manuscript and Laura Vanderploeg for help with figures. This work was supported by the National Institutes of Health grant R01 GM069420 to (AZA), University of Wisconsin Industrial and Economic Development Research Program (AZA), National Foundation—March of Dimes (AZA), US Department of Agriculture—Hatch/McIntire/Stennis grant (AZA), the Greater Milwaukee Foundation—Shaw Scientist Award (AZA), and Computation and Informatics in Biology and Medicine Training Grant T15LM007359 (CLW).
Footnotes
All solutions are prepared in Nanopure distilled water (dH2O) with a resistivity of 18 MΩ cm.
As an alternative, arrays may be dried using an argon gas stream applied in a back-and-forth motion evenly across the top and bottom of the array.
All polyamide synthesis work was done in collaboration with Dervan and colleagues, and the reader is referred to their publications for additional details of synthesis (40, 41).
Clean slides are critical for efficient array synthesis. All buffers should be made fresh, and all containers should be cleaned with acetone and rinsed with dHSO before use.
Store derivatized slides and synthesized DNA arrays in a UV-protected desiccator, when not being used.
Each array can be synthesized with a distinct “reference” sequence synthesized at the edges for quality control and to align the grid for data extraction.
Hairpin percentage formation can be assessed by including two distinct features on the arrays: one that forms a hairpin (5′-CGC-TTAGTTCA-CGC-TCCT-GCG-TGAACTAA-GCG-3′) and a single-stranded version (5′-CGC-TTAGTTCACGC-3′). Cy3-labeled 5′-GCG-TGAACTAA-GCG-3′was added to the array at 50 nM and annealed to both the hairpin and single-stranded features. The fluorescence intensity of the hairpin sequence was divided by the fluorescence intensity of the single-stranded sequence, averaged, and background-subtracted, yielding a hairpin formation of 95.6%.
Protein binding to the array can be done using similar techniques as for PA1. We recommend storing protein aliquots at −80°C and minimizing freeze-thaw cycles. Binding buffer and protein concentration will need to be optimized for each protein. Other blocking agents such as 5–10% fetal bovine serum (FBS) or 1–5% BSA may be tested. Protein incubation with the array may also be stabilized at 4°C, versus room temperature.
If the PMT is reduced too much to eliminate signal saturation, this may indicate that DNA saturation is occurring. The concentration of fluorescent DNA-binding molecule may need to be reduced to address this issue.
To validate the DNA microarray results, a subset of DNA sequences that yielded various fluorescent intensities on the DNA microarray can be tested with polyamide/ATF titrations by EMSA to determine affinity values. The association constant (Ka) values form a linear relationship with the fluorescence intensities obtained by CSI arrays. This linear relationship can be used to estimate the affinity of the polyamide for any DNA sequence on the array. Other techniques such as fluorescence polarization may also be used to determine the affinity of the ATF-DNA interaction.
References
- 1.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Wells J, Graveel CR, Bartley SM, Madore SJ, Farnham PJ. The identification of E2F1-specific target genes. Proc. Natl Acad. Sci. U. S. A. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian ChIP-chip analysis. Proc. Natl Acad. Sci. U. S. A. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, Weissman S, Snyder M. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc. Natl Acad. Sci. U. S. A. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Ansari AZ, Mapp AK. Modular design of artificial transcription factors. Curr. Opin. Chem. Biol. 2002;6:765–772. doi: 10.1016/s1367-5931(02)00377-0. [DOI] [PubMed] [Google Scholar]
- 8.Mapp AK, Ansari AZ. A TAD further: exogenous control of gene activation. ACS Chem. Biol. 2007;2:62–75. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 9.Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Activation of gene expression by small molecule transcription factors. Proc. Natl Acad. Sci. U. S. A. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptashne M, Gann A. Genes & Signals. New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 11.Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 12.Courey AJ, Holtzman DA, Jackson SP, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 13.Mermod N, O’Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 14.Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 15.Mandell JG, Barbas CF., III Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat. Biotech. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 17.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotech. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrroleimidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 19.Buchmueller KL, Staples AM, Howard CM, Horick SM, Uthe PB, Le NM, Cox KK, Nguyen B, Pacheco KA, Wilson WD, Lee M. Extending the language of DNA molecular recognition by polyamides: unexpected influence of imidazole and pyrrole arrangement on binding affinity and specificity. J. Am. Chem. Soc. 2005;127:742–750. doi: 10.1021/ja044359p. [DOI] [PubMed] [Google Scholar]
- 20.Supekova L, Pezacki JP, Su AI, Loweth CJ, Riedl R, Geierstanger B, Schultz PG, Wemmer DE. Genomic effects of polyamide/DNA interactions on mRNA expression. Chem. Biol. 2002;9:821–827. doi: 10.1016/s1074-5521(02)00174-6. [DOI] [PubMed] [Google Scholar]
- 21.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. U. S. A. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotech. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 23.Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 24.Hauschild KE, Metzler RE, Arndt HD, Moretti R, Raffaelle M, Dervan PB, Ansari AZ. Temperature–sensitive protein–DNA dimerizers. Proc. Natl Acad. Sci. U. S. A. 2005;102:5008–5013. doi: 10.1073/pnas.0501289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arndt HD, Hauschild KE, Sullivan DP, Lake K, Dervan PB, Ansari AZ. Toward artificial developmental regulators. J. Am. Chem. Soc. 2003;125:13322–13323. doi: 10.1021/ja0371395. [DOI] [PubMed] [Google Scholar]
- 26.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 28.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotech. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 29.Guex N, Peitsch MC. SWISSMODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Stafford RL, Dervan PB. The reach of linear protein-DNA dimerizers. J. Am. Chem. Soc. 2007;129:14026–14033. doi: 10.1021/ja075247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 33.Quackenbush J. Microarray data normalization and transformation. Nat. Gen. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 34.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colantuoni C, Henry G, Zeger S, Pevsner J. Local mean normalization of microarray element signal intensities across an array surface: quality control and correction of spatially systematic artifacts. Bio Techniques. 2002;32:1316–1320. doi: 10.2144/02326mt02. [DOI] [PubMed] [Google Scholar]
- 36.Dixon W. Analysis of extreme values. Ann. Math. Stat. 1950;21:488–506. [Google Scholar]
- 37.Rorabacher D. Statistical treatment for rejection of deviant values: critical values of Dixon Q parameter and related subrange ratios at the 95 percent confidence level. Anal. Chem. 1991;83:139–146. [Google Scholar]
- 38.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 39.Adbi H. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage; 2007. [Google Scholar]
- 40.Wurtz NR, Turner JM, Baird EE, Dervan PB. Fmoc solid phase synthesis of polyamides containing pyrrole and imidazole amino acids. Org. Lett. 2001;3:1201–1203. doi: 10.1021/ol0156796. [DOI] [PubMed] [Google Scholar]
- 41.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]