Abstract
Background
Mycobacterial culture is the gold standard test for diagnosing tuberculosis (TB), but it is time-consuming. Polymerase chain reaction (PCR) is a highly sensitive and specific method that can reduce the time required for diagnosis. The diagnostic efficacy of PCR differs, so this study determined the actual sensitivity of TB-PCR in tissue specimens.
Methods
We retrospectively reviewed 574 cases. The results of the nested PCR of the IS6110 gene, mycobacterial culture, TB-specific antigen-induced interferon-γ release assay (IGRA), acid-fast bacilli (AFB) staining, and histological findings were evaluated.
Results
The positivity rates were 17.6% for PCR, 3.3% for the AFB stain, 22.2% for mycobacterial culture, and 55.4% for IGRA. PCR had a low sensitivity (51.1%) and a high specificity (86.3%) based on the culture results of other studies. The sensitivity was higher (65.5%) in cases with necrotizing granuloma but showed the highest sensitivity (66.7%) in those with necrosis only. The concordance rate between the methods indicated that PCR was the best method compared to mycobacterial culture, and the concordance rate increased for the methods using positive result for PCR or histologic features.
Conclusions
PCR of tissue specimens is a good alternative to detect tuberculosis, but it may not be as sensitive as previously suggested. Its reliability may also be influenced by some histological features. Our data showed a higher sensitivity when specimens contained necrosis, which indicated that only specimens with necrosis should be used for PCR to detect tuberculosis.
Keywords: Tuberculosis, Polymerase chain reaction, Mycobacterial culture, Interferon-γ release tests
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (MTB) that remains one of the world’s most important health problems due to its high risk of complications, mortality, and infectivity. TB was initially considered untreatable, but effective drug regimens that have been developed since the 1940s have increased the treatment success rates to 85%. A rapid and reliable diagnosis of TB is essential to provide proper treatment, avoid severe complications, and prevent disease dissemination.
Among the many traditional diagnostic methods, mycobacterial culture is the gold standard due to its high sensitivity and specificity, but results take 2–6 weeks, and the sensitivity varies with the number of organisms cultured [1,2]. Therefore, a less time-consuming method with a high sensitivity and specificity is desirable, such as the nucleic acid amplification technique that has been applied to diagnose TB [3]. Identifying a repetitive sequence in MTB using polymerase chain reaction (PCR) analysis would reduce the time needed to obtain results [4,5].
Clinicians consider invasive procedures to obtain tissue specimens when a proper diagnosis cannot be made using noninvasive techniques [6]. Tissue specimens can be obtained by surgical resection or biopsy, and histological features help to rule out diseases that mimic TB. The typical histological findings of TB are granulomatous inflammation with central caseous necrosis. However, many diseases can cause similar changes, making it difficult to diagnose TB using histological findings alone. In addition, identifying the rod-shaped MTB bacilli using acid-fast bacilli (AFB) staining of a tissue specimen can be helpful, but it has a low sensitivity; the required fixative fluids and organic solvents can lower the sensitivity even further [7]. If bacilli are not identified in the tissue specimen, PCR can be performed. Most specimens are formalin-fixed, which is inadequate for a culture study. In contrast, PCR is carried out to target nucleic acid sequences unique to the bacteria and therefore requires tissue specimens.
The genome of the MTB complex contains various genes, including IS6110, an insertion element. According to a prior study, the amplification of IS6110 reached a higher sensitivity than other genes in paraffin-embedded tissue specimens [8]. IS6110 is exclusively found in MTB but not in other species of mycobacteria, so it has become an important diagnostic tool for the identification of MTB [9-12]. Therefore, nested PCR for IS6110 is now the most effective method to detect TB in formalin-fixed, paraffin-embedded tissue [13] with a high sensitivity (58%–100%) and specificity (75%–100%), but some studies have reported variable PCR sensitivities [14-20]. As a result, our study was planned to determine the clinical usefulness of nested PCR in practically selected cases and also to compare these results to other methods, including culture results.
MATERIALS AND METHODS
Cases
We retrospectively searched cases in which tissue was obtained. We included patients with a clinical suspicion of TB as well as those without. We selected cases that had been investigated for TB using mycobacterial culture, the TB specific antigen-induced interferon-γ release assay (IGRA), and AFB staining and collected their results to compare with the PCR results of the tissue specimens. In total, 574 formalin-fixed paraffin-embedded samples were obtained from Korea University Anam Hospital (Seoul, Korea) between January 2009 and December 2013. This study was approved by the Institutional Review Board of Korea University Anam Hospital (AN15200-002).
All tissues were fixed in 10% formalin or 4% formaldehyde. We reviewed the slides for their histological findings after hematoxylin and eosin staining. Additional AFB staining was performed on some slides to detect Mycobacterium bacilli. We considered the AFB staining results positive when at least one bacillus was found.
Nested PCR
Tissue preprocessing
Ten pieces of 5–7-μm-thick sections were cut from the formalin-fixed, paraffin-embedded block and collected in 1.5-mL microcentrifuge tubes for the nested PCR. Tissue microdissection was not performed. A 1,000 μL aliquot of 1×phosphate buffered saline was added to remove the paraffin; the mixture was vortex mixed for 10 seconds and then allowed to stand at 75°C for 2 minutes before being centrifuged for 2 minutes at 13,000 rpm. The paraffin and supernatant were removed. These steps were repeated twice more after adding 1 mL of sterile distilled water.
DNA extraction
The DNA was extracted and nested PCR was performed using an NTM&MTB PCR Kit (BioSewoom Inc., Seoul, Korea) according to the manufacturer’s instructions. A 50-μL aliquot of a DNA extraction buffer was added to the preprocessed specimen, and then the mixture was vortex mixed and allowed to stand at 56°C for 15 minutes, followed by thorough mixing. Next, the mixture was placed on a heating block at 100°C for 8 minutes and cooled by mixing for 20 seconds. The mixture was centrifuged for 2 minutes at 13,000 rpm, and the 4-μL upper portion of the supernatant was used.
Nested PCR for IS6110
A 0.5-μL aliquot of enzymes and a mixture of Taq polymerase and buffer were added to 15.5 μL of a mixture of the primers of both MTB and NTM, dNTPs, buffer, and pink dye. Next, 4.0 μL of the extracted DNA were added to the reaction mixture for the first PCR round. Positive and negative controls were used. After the first PCR round, 18.0 μL of the first PCR round product and 0.5 μL of the second PCR round enzyme were mixed with 1.5 μL of the first PCR round product. Both PCR cycles were conducted with an initial 4-minute denaturation step at 95°C, three repeating cycles of 30 seconds each at 94°C, 65°C, and 72°C, followed by 35 cycles of 30 seconds at 94°C, 68°C, and 72°C, and 5 minutes of the final extension at 72°C for 30 seconds. The third cycle was repeated 25 times instead of the 35 cycles that took place during the second PCR round.
PCR products
A 5–10-μL aliquot of the PCR product was resolved on 1.5%–2% agarose gels. The products were visualized by ultraviolet transillumination and compared to the positive control.
Other methods used to detect TB
Different fluid specimens were obtained, decontaminated with 4% NaOH, and used for mycobacterial culture. Whole blood that had been obtained from a patient was used for the IGRA, and the interferon-gamma level was assessed via an enzyme immunoassay.
Statistical analysis
We considered patients with culture-positive results to be patients with TB. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for PCR as well as the other methods. We compared the results from the surgical specimens with those of the biopsy specimens to detect the influence of sampling. Concordance between the results from different methods was assessed using the к coefficient (к>0.75, excellent agreement; 0.75≥к≥0.40, fair to good agreement; к<0.40, poor agreement). Analyses were performed using the SPSS ver. 20.0 (IBM Co., Armonk, NY, USA).
RESULTS
Clinicopathological characteristics of the cases
The median patient age was 50.3 years (range, 2 to 89 years); 293 cases (51.0%) were male, and 281 participants (49.0%) were female. A total of 239 specimens (41.6%), were obtained by surgical resection, while 335 (58.4%) were procured through biopsy, including 198 (34.5%) from the gastrointestinal tract (colon, stomach, terminal ileum, and duodenum), 108 (18.8%) from the bone, joints, or soft tissue, 86 (15.0%) from the lymph nodes, 73 (12.7%) from the lung or bronchus, 29 (5.1%) from the pleura, and 31 (5.4%) from the skin (Table 1).
Table 1.
Clinicopathological characteristics of the patients
| Clinicopathological characteristic | No. (%) |
|---|---|
| Sex | |
| Male | 293 (51.0) |
| Female | 281 (49.0) |
| The way obtaining tissue | |
| Surgical resection | 239 (41.6) |
| Biopsy | 335 (58.4) |
| Histology | |
| Necrotizing granulomatous inflammation | 158 (27.5) |
| Granulomatous inflammation without necrosis | 95 (16.6) |
| Necrosis without granuloma | 45 (7.8) |
| No granuloma or necrosis | 276 (48.1) |
| Site | |
| Gastrointestinaltract | 198 (34.5) |
| Bone and soft tissue | 108 (18.8) |
| Lymph node | 86 (15.0) |
| Lung and pleura | 102 (17.8) |
| Skin | 31 (5.4) |
| Peritoneum | 18 (3.1) |
| Head and neck | 14 (2.4) |
| Brain | 3 (0.5) |
| Adnexa, breast, pericardium, spleen, testis | Each 2 (0.3) |
| Liver, kidney, bladder, thymus | Each 1 (0.17) |
Analysis of PCR and other methods compared to culture
PCR was performed on tissue specimens from all cases (n=574), and 17.6% (n=101) were positive for TB, whereas 82.4% (n=473) were negative (Fig. 1C). Among the 574 tissue specimens, 233 underwent AFB staining; only 3.3% (n=8) had a positive result. In all, 405 cases were cultured, with a positivity rate of 22.2%. The IGRA-positive rate was 55.4% (Table 2).
Fig. 1.
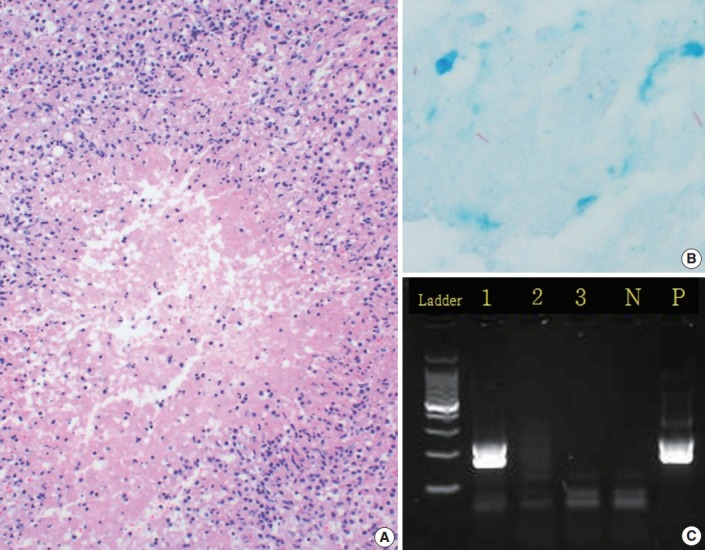
A representative case of tuberculosis. (A) A typical tuberculosis histological finding of granulomatous inflammation with central caseous necrosis. (B) Acid-fast bacilli stain highlights rod-shaped bacilli (acid-fast bacilli). (C) Agarose gel electrophoresis of Mycobacterium tuberculosis by nested polymerase chain reaction. There is a positive result (lane 1) for IS6110 and two negative results (lanes 2 and 3). Lane P contains the positive control, while lane N is the negative control.
Table 2.
Positive and negative rates for each method
| Positive | Negative | Total | |
|---|---|---|---|
| PCR | 101 (17.6) | 473 (82.4) | 574 (100) |
| AFB stain | 8 (3.3) | 225 (96.7) | 233 (100) |
| IGRA | 77 (55.4) | 62 (44.6) | 139 (100) |
| Mycobacterial culture | 90 (22.2) | 315 (77.8) | 405 (100) |
Values are presented as number (%).
PCR, polymerase chain reaction; AFB, acid-fast bacilli; IGRA, tuberculosis-specific antigen-induced interferon gamma release assay.
As mycobacterial culture is the gold standard for TB diagnosis, we calculated the accuracy of each method compared to the mycobacterial culture results to determine the diagnostic yield (Table 3). The PCR sensitivity and specificity were 51.1% and 86.3%, respectively. The PPV was 51.7%, and the NPV was 86.1%, indicating low sensitivity and high specificity compared to prior data [14-20]. The sensitivity of AFB-stained specimens was 12.5%, and the specificity was 98.4%. The PPV was 75.0%, while the NPV was 72.4%.
Table 3.
Statistics of each method based on the mycobacterial culture results
| Method | Culture |
No. of cases | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||
|---|---|---|---|---|---|---|---|---|
| (+) | (–) | |||||||
| PCR | (+) | 46 | 43 | 405 | 51.1 | 86.3 | 51.7 | 86.1 |
| (–) | 44 | 272 | ||||||
| AFB stain | (+) | 3 | 1 | 91 | 12.5 | 98.5 | 75.0 | 75.9 |
| (–) | 21 | 66 | ||||||
| IGRA | (+) | 21 | 30 | 91 | 80.8 | 53.8 | 41.2 | 87.5 |
| (–) | 5 | 35 | ||||||
PPV, positive predictive value; NPV, negative predictive value; PCR, polymerase chain reaction; AFB, acid-fast bacilli; IGRA, tuberculosis-specific antigen-induced interferon gamma release assay.
A comparison of PCR and AFB staining of tissue specimens according to their histological features
PCR and AFB staining were performed on the same tissue specimens, so we compared the results of the two methods. Among the eight AFB stain–positive cases (Fig. 1B), six (75.0%) were PCR-positive and two (25.0%) were PCR-negative. Out of the 225 AFB stain–negative cases, 31 (13.8%) were PCR-positive while 194 (86.2%) were PCR-negative.
A review of the hematoxylin and eosin slides (Fig. 1A) revealed the histological findings of necrotizing granulomatous inflammation, which suggests the possibility of TB, in 158 cases (27.5%). Among these, 95 cases (16.6%) had non-necrotizing granulomas, while 45 (7.8%) were diagnosed with necrosis without granuloma. In total, 276 cases (48.1%) had no granuloma or necrosis (Table 1). Only 405 of the 574 cases were used to compare the histological findings (Table 4). Based on the culture results, our data showed different diagnostic values depending on the histologic findings.
Table 4.
Statistical analysis of PCR-based on the histological findings
| (+) | PCR | Culture |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|
| (+) | (–) | ||||||
| Necrotizing granuloma (n = 131) | (+) | 36 | 32 | 65.5 | 57.9 | 52.9 | 69.8 |
| (–) | 19 | 44 | |||||
| Non-necrotizing granuloma (n = 61) | (+) | 5 | 8 | 26.3 | 81.0 | 38.5 | 70.8 |
| (–) | 14 | 34 | |||||
| Necrosis only (n = 38) | (+) | 2 | 3 | 66.7 | 91.4 | 40.0 | 97.0 |
| (–) | 1 | 32 | |||||
| No granuloma and necrosis (n = 175) | (+) | 3 | 2 | 23.1 | 98.8 | 60.0 | 94.2 |
| (–) | 10 | 162 | |||||
PCR, polymerase chain reaction; PPV, positive predictive value; NPV, negative predictive value.
Specimens with necrotizing granulomatous inflammation had a sensitivity of 65.5%, a specificity of 57.9%, a PPV of 52.9%, and an NPV of 69.8%. For specimens with non-necrotizing granuloma only, the sensitivity was 26.3%, the specificity was 81.0, the PPV was 38.5%, and the NPV was 70.8%, while those specimens with necrosis only had a sensitivity of 66.7%, a specificity of 91.4%, a PPV of 40.0%, and an NPV of 97.0%. The sensitivity was 23.1%, the specificity was 98.8%, the PPV was 60.0%, and the NPV was 94.2% for specimens without granuloma or necrosis. Specimens with necrotizing granulomatous inflammation and with necrosis only showed a higher sensitivity than all others. The latter revealed a higher specificity, while the former had the lowest specificity. The highest specificity was found in specimens without granuloma or necrosis; these cases demonstrated not only the highest PPV but also a high NPV. Specimens with necrotizing granulomatous inflammation were found to have a high PPV, and NPV was also high in specimens with necrosis only.
Concordance rate between methods
The concordance rate between the methods was assessed using the к coefficient. None of the methods had excellent agreement with the cultures. When correlating was carried out based on the culture results, PCR was the best method (k=0.376), and the concordance rate increased for the method that used a positive result for the PCR or histologic features (k=0388). However, PCR alone had a better concordance rate (k=0.442) with the histological findings (Table 5).
Table 5.
Concordance rates between methods
| Method | к coefficient | p-value |
|---|---|---|
| PCR and culture | 0.376 | <.001 |
| PCR and IGRA | 0.126 | .034 |
| PCR and AFB stain | 0.235 | <.001 |
| PCR and histopathologic features | 0.442 | <.001 |
| Culture and positive result for both PCR and histopathologic features | 0.288 | <.001 |
| Culture and positive result for PCR or histopathologic features | 0.388 | <.001 |
PCR, polymerase chain reaction; IGRA, tuberculosis-specific antigen-induced interferon gamma release assay; AFB, acid-fast bacilli.
DISCUSSION
TB remains an important health problem, even though the cure rate has increased and the incidence has dropped [6]. As a rapid and sensitive method to detect TB, PCR was able to eliminate the disadvantages of mycobacterial culture [3-5]. However, efficacy assessments for diagnosing TB by PCR differ. Our study evaluated the use of PCR in paraffin-embedded tissue specimens to diagnose TB.
The sensitivity of PCR for the IS6110 insertion sequence was 51.1% in tissue specimens, which is lower than that reported by previous studies (58%–100%) (Table 6) [14-20] and suggests that PCR alone may have had a relatively lower sensitivity for the patients in our study than any previous results in TB-suspected patients.
Table 6.
PCR statistics values of previous reports
| Reference | Positive criteria | PCR methods | No. of cases | Type of specimens | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|
| Park et al. [14] | Culture | Nested | 152 | FFPE, lung | 85 | 99 | 98 | 88 |
| Frevel et al. [15] | Histology, culture, clinical finding | NA | 229 | FFPE, various organs | 90 | 92 | 81 | 96 |
| Mishra et al. [16] | Culture | Real time | 28 | FFPE, gastrointestinal specimens | 96.4 | 100 | 100 | 96 |
| Seo et al. [17] | Culture | Nested | 88 | FFPE, various organs | 87.5 | 98.2 | 96.6 | 93.2 |
| Diaz et al. [18] | Clinical and microbiological | Nested | 43 | FFPE, liver | 58 | 96 | NA | NA |
| Salian et al. [19] | Culture | NA | 60 | FFPE, various organs | 100 | 93 | 76.9 | 100 |
| Lee et al. [20] | Culture | Nested | 129 | FFPE, various organ | 68.3 | 98.5 | NA | NA |
| Present study | Culture | Nested | 405 | FFPE, various organ | 51.1 | 86.3 | 51.7 | 86.1 |
PCR, polymerase chain reaction; PPV, positive predictive value; NPV, negative predictive value; FFPE, formalin fixed, paraffin embedded tissue; NA, not available.
The low sensitivity of PCR in our study can be explained in several ways. First, the quantity of DNA in the tissue specimens may have been insufficient for a proper evaluation. If a specimen contained a smaller number of bacteria, the bacilli may not have been included in the section used for PCR. The possibility of bacilli being contained in a biopsy specimen was lower because of the small sample size. In our study, isolating TB using the PCR resulted in a significantly different sensitivity between the surgical and biopsy specimens (59.5% vs 43.8%), which supports this notion and is consistent with previous reports [14,21], indicating that false-negative PCR results may occur due to an inadequate amount of biopsy tissue. Additionally, surgical specimen samples may contain various components, including normal tissue and various inhibitors of PCR amplification [22]. These inhibitors can be removed by dilution, but this process also reduces the amount of target DNA, resulting in a low possibility of detecting DNA [22]. Therefore, obtaining a pure DNA extent is important; so tissue microdissection to isolate the necrotizing granulomatous inflammatory area can overcome it. According to Ryan et al. [23], 40% of cases that use microdissected tissue detected 155 bp fragments in the IS900 region, while 20% of cases employed entire sections of paraffin-embedded tissue blocks. However, we did not perform the tissue microdissection, so the possibility of detecting TB-DNA becomes lower [22].
Second, low DNA quality can lead to low sensitivity. The extent of formalin penetration and fixation time are important factors when recovering DNA from fixed tissues [21]. A long formalin exposure time will cause DNA fragmentation and decrease the DNA’s quality. Studies on the effect of a longer fixation time on DNA amplification products in tissue specimens have revealed that DNA has a greater chance of damage and fragmentation, so shorter DNA fragments are typically extracted from formalin-fixed tissue as time goes on [24,25]. Successful amplification occurred after 2–24-hour formalin exposure times [25].
Third, some cases may not have contained the IS6110 element. The target was the IS6110 sequence [12,26], which is specific for most MTB complex bacilli (Mycobacterium bacilli, Mycobacterium bovis, Mycobacterium bovis bacillus Calmette-Guerin, Mycobacterium africanum, and Mycobacterium microti). However, the copy number of the sequence inserted into each bacillus genome varied from 0 to 30 depending on the organism [27,28]. PCR can detect 10 fg or two bacilli in a tissue specimen [13]; however, specimens without the IS6110 element will always be negative [2,27,28].
Furthermore, a false-positive nested PCR result can occur in a tissue specimen. Nested PCR requires additional steps compared to PCR conducted in a single tube [29], so it is very sensitive even for paraffin-embedded tissue specimens. However, the extra steps may also result in contamination, causing a false-positive result [5,30].
The presence of a false-negative culture result can also affect the diagnostic efficiency of PCR. Nested PCR is so sensitive that it can detect residual DNA in a tissue specimen even after medical therapy. Salian et al. [19] suggested that healed TB granulomas can be PCR-positive, even though the culture and AFB staining were negative. Hernandez-Pando et al. [31] also suggested that MTB DNA is located in the macrophages and in other phagocytic cells, which can be important for eliminating latent and persistent bacilli. Among our 315 cases with negative culture results, 45 were PCR-positive and 32 cases showed necrotizing granulomatous inflammation. This finding suggests that the bacteria were not successfully isolated for culture and that additional PCR may help detect the TB bacteria. Miller et al. [32] explained that the higher sensitivity of PCR than culture is due to the increased number of viable organisms, whereas the presence of viable bacteria is crucial for culture. Therefore, they recommended combining bacterial culture and PCR to effectively diagnose TB.
As the diagnostic criteria include the histological features of TB [6], it is essential to identify necrotizing granulomatous inflammation. Yum and Choi [33] showed that histological features are significantly associated with the TB-positive rate in pleural tissue biopsies (71.4% vs 15.8%). Jambhekar et al. [34] reported a good correlation between typical granulomas and PCR results. According to their findings, specimens with no granuloma have a 20% positive rate, whereas the presence of a granuloma results in 72.5% positivity (84.6% in well-formed granuloma cases). The histological findings were correlated with a higher PCR sensitivity, which increases in cases of necrotizing granulomatous inflammation and decreases in cases of nonspecific inflammation (65.5% vs 23.1%, respectively). Thus, only one of these features increased the sensitivity. One other study [35] reported that specimens with a granuloma have a 2.8-times higher possibility of detecting bacilli than those without, and our data support this finding (11.6 times, 35.0% vs 3.0%).
Our data showed the highest sensitivity for specimens with necrosis but no granuloma. According to Nopvichai et al. [2], necrotizing non-granulomatous lymphadenitis had an overall PCR-positive rate of 28% (10 of 35) with a sensitivity of 33% compared to the culture results. Therefore, necrotizing specimens without granuloma are typically attributed to TB, so PCR should be performed in these cases. However, our results were limited to three culture-positive cases.
Among the 90 specimens from patients with positive culture results, 13 cases (14.4%) had no histological features of TB, which may have been due to differences between the specimens; however, three cases (1.7%) were PCR-positive. To explain these cases, a patient with TB may have had neither granuloma nor necrosis, or the patients could have had recurrent TB or were taking TB medications, which would suggest that Mycobacterium remain even when no histological changes are present.
Although the PCR sensitivity in selected patients was not high, we found that PCR was the best alternative to culture for detecting TB. The concordance rate between PCR and the histological findings was high, suggesting that typical histological findings can be used to identify Mycobacterium by PCR. In addition, the culture and the combination of PCR and the histological findings showed the highest concordance rate, so we can recommend that the histopathologic findings be added to help detect TB when the PCR result is negative.
Generally, the results of our study indicated that the IGRA was the most sensitive method to diagnose TB. Blood-based in vitro IGRAs have been developed as alternatives to the tuberculin skin test for identifying a TB infection. Previous data [36] revealed that the sensitivity and the specificity of IGRA in serum for diagnosing active TB are 80%–81% and 59%–79%, respectively. These authors reported that IGRA has a higher sensitivity than the tuberculin skin test, but it was not high enough to rule out TB because the NPV was not sufficiently high. In addition, its specificity was much lower, which is likely due to the inclusion of samples from patients with latent TB infections, which was a limitation in differentiating active from latent TB.
Some limitations of this study should be mentioned. First, this was a retrospective study, so the data were limited. We did not assess the symptoms, history of medication use or radiological findings or correlate them with our results. No additional studies were performed, so the number of positive cases for the AFB stain was small. Second, the cultures were not performed on the same tissues that were used for the nested PCR because a culture study is not usually carried out on a tissue specimen; therefore, we could not determine the exact sensitivity and specificity of the PCR. Third, we did not evaluate the differences between adults and children or between immunosuppressed and nonimmunosuppressed patients.
In conclusion, PCR was a good alternative to detect TB, but it may not be sufficiently sensitive in selected patients. These results correlate with the histological features, suggesting that specimens with necrosis should be used to identify TB with PCR. IGRA was actually the most sensitive method, but it had a low specificity. To overcome the lower diagnostic sensitivity of PCR, large-sized tissue samples with typical histological changes detected in TB, a proper fixation time, care to reduce contamination, and tissue microdissection are required, as well as performing additional studies to detect MTB in samples without the IS6110 sequence and collecting histopathologic features. According to our data, the IGRA was also sensitive but had a low specificity, making it difficult to use to rule out TB.
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC) Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009;58:7–10. [PubMed] [Google Scholar]
- 2.Nopvichai C, Sanpavat A, Sawatdee R, et al. PCR detection of Mycobacterium tuberculosis in necrotising non-granulomatous lymphadenitis using formalin-fixed paraffin-embedded tissue: a study in Thai patients. J Clin Pathol. 2009;62:812–5. doi: 10.1136/jcp.2008.062828. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Nucleic acid amplification tests for tuberculosis. MMWR Morb Mortal Wkly Rep. 1996;45:950–2. [PubMed] [Google Scholar]
- 4.Eisenach KD, Sifford MD, Cave MD, Bates JH, Crawford JT. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dis. 1991;144:1160–3. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- 5.Chawla K, Gupta S, Mukhopadhyay C, Rao PS, Bhat SS. PCR for M. tuberculosis in tissue samples. J Infect Dev Ctries. 2009;3:83–7. doi: 10.3855/jidc.53. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization . Global tuberculosis report 2014. Geneva: World Health Organization; 2014. [Google Scholar]
- 7.Fukunaga H, Murakami T, Gondo T, Sugi K, Ishihara T. Sensitivity of acid-fast staining for Mycobacterium tuberculosis in formalin-fixed tissue. Am J Respir Crit Care Med. 2002;166:994–7. doi: 10.1164/rccm.2111028. [DOI] [PubMed] [Google Scholar]
- 8.Marchetti G, Gori A, Catozzi L, et al. Evaluation of PCR in detection of Mycobacterium tuberculosis from formalin-fixed, paraffin-embedded tissues: comparison of four amplification assays. J Clin Microbiol. 1998;36:1512–7. doi: 10.1128/jcm.36.6.1512-1517.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEvoy CR, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2007;87:393–404. doi: 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Thierry D, Cave MD, Eisenach KD, et al. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coros A, DeConno E, Derbyshire KM. IS6110, a Mycobacterium tuberculosis complex-specific insertion sequence, is also present in the genome of Mycobacterium smegmatis, suggestive of lateral gene transfer among mycobacterial species. J Bacteriol. 2008;190:3408–10. doi: 10.1128/JB.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thierry D, Brisson-Noël A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon JL, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–73. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho MS, Lee SN, Sung SH, Han WS. Comparison of Ziehl-Neelsen stain and TB-PCR on detection of Mycobacterium tuberculosis in formalin-fixed, paraffin-embedded tissues of chronic granulomatous inflammation. Korean J Pathol. 2003;37:379–83. [Google Scholar]
- 14.Park JS, Kang YA, Kwon SY, et al. Nested PCR in lung tissue for diagnosis of pulmonary tuberculosis. Eur Respir J. 2010;35:851–7. doi: 10.1183/09031936.00067209. [DOI] [PubMed] [Google Scholar]
- 15.Frevel T, Schäfer KL, Tötsch M, Böcker W, Dockhorn-Dworniczak B. PCR based detection of mycobacteria in paraffin wax embedded material routinely processed for morphological examination. Mol Pathol. 1999;52:283–8. doi: 10.1136/mp.52.5.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra PK, Gorantla VR, Bhargava A, Varshney S, Vashistha P, Maudar KK. Molecular detection of Mycobacterium tuberculosis in formalin-fixed, paraffin-embedded tissues and biopsies of gastrointestinal specimens using real-time polymerase chain reaction system. Turk J Gastroenterol. 2010;21:129–34. doi: 10.4318/tjg.2010.0070. [DOI] [PubMed] [Google Scholar]
- 17.Seo AN, Park HJ, Lee HS, et al. Performance characteristics of nested polymerase chain reaction vs real-time polymerase chain reaction methods for detecting Mycobacterium tuberculosis complex in paraffin-embedded human tissues. Am J Clin Pathol. 2014;142:384–90. doi: 10.1309/AJCP2QZRH4ZNPRDD. [DOI] [PubMed] [Google Scholar]
- 18.Diaz ML, Herrera T, Lopez-Vidal Y, et al. Polymerase chain reaction for the detection of Mycobacterium tuberculosis DNA in tissue and assessment of its utility in the diagnosis of hepatic granulomas. J Lab Clin Med. 1996;127:359–63. doi: 10.1016/s0022-2143(96)90184-5. [DOI] [PubMed] [Google Scholar]
- 19.Salian NV, Rish JA, Eisenach KD, Cave MD, Bates JH. Polymerase chain reaction to detect Mycobacterium tuberculosis in histologic specimens. Am J Respir Crit Care Med. 1998;158:1150–5. doi: 10.1164/ajrccm.158.4.9802034. [DOI] [PubMed] [Google Scholar]
- 20.Lee HS, Park KU, Park JO, Chang HE, Song J, Choe G. Rapid, sensitive, and specific detection of Mycobacterium tuberculosis complex by real-time PCR on paraffin-embedded human tissues. J Mol Diagn. 2011;13:390–4. doi: 10.1016/j.jmoldx.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulimood AB, Peter S, Rook GW, Donoghue HD. In situ PCR for Mycobacterium tuberculosis in endoscopic mucosal biopsy specimens of intestinal tuberculosis and Crohn disease. Am J Clin Pathol. 2008;129:846–51. doi: 10.1309/DKKECWQWMG4J23E3. [DOI] [PubMed] [Google Scholar]
- 22.Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. Methods of mycobacterial DNA isolation from different biological material: a review. Vet Med. 2006;51:180–92. [Google Scholar]
- 23.Ryan P, Bennett MW, Aarons S, et al. PCR detection of Mycobacterium paratuberculosis in Crohn's disease granulomas isolated by laser capture microdissection. Gut. 2002;51:665–70. doi: 10.1136/gut.51.5.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer CE, Peterson SL, Kiviat NB, Manos MM. PCR amplification from paraffin-embedded tissues: effects of fixative and fixation time. Am J Clin Pathol. 1991;95:117–24. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]
- 25.Greer CE, Lund JK, Manos MM. PCR amplification from paraffin-embedded tissues: recommendations on fixatives for long-term storage and prospective studies. PCR Methods Appl. 1991;1:46–50. doi: 10.1101/gr.1.1.46. [DOI] [PubMed] [Google Scholar]
- 26.Bascuñana CR, Belák K. Detection and identification of mycobacteria in formalin-fixed, paraffin-embedded tissues by nested PCR and restriction enzyme analysis. J Clin Microbiol. 1996;34:2351–5. doi: 10.1128/jcm.34.10.2351-2355.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sajduda A, Dziadek J, Dela A, Zalewska-Schonthaler N, Zwolska Z, McFadden J. DNA fingerprinting as an indicator of active transmission of multidrug-resistant tuberculosis in Poland. Int J Infect Dis. 1998;3:12–7. doi: 10.1016/s1201-9712(98)90089-3. [DOI] [PubMed] [Google Scholar]
- 28.Moatter T, Mirza S, Siddiqui MS, Soomro IN. Detection of Mycobacterium tuberculosis in paraffin embedded intestinal tissue specimens by polymerase chain reaction: characterization of IS6110 element negative strains. J Pak Med Assoc. 1998;48:174–8. [PubMed] [Google Scholar]
- 29.Baba K, Pathak S, Sviland L, et al. Real-time quantitative PCR in the diagnosis of tuberculosis in formalin-fixed paraffin-embedded pleural tissue in patients from a high HIV endemic area. Diagn Mol Pathol. 2008;17:112–7. doi: 10.1097/PDM.0b013e31814ceac3. [DOI] [PubMed] [Google Scholar]
- 30.Rish JA, Eisenach KD, Cave MD, Reddy MV, Gangadharam PR, Bates JH. Polymerase chain reaction detection of Mycobacterium tuberculosis in formalin-fixed tissue. Am J Respir Crit Care Med. 1996;153(4 pt 1):1419–23. doi: 10.1164/ajrccm.153.4.8616575. [DOI] [PubMed] [Google Scholar]
- 31.Hernández-Pando R, Jeyanathan M, Mengistu G, et al. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet. 2000;356:2133–8. doi: 10.1016/s0140-6736(00)03493-0. [DOI] [PubMed] [Google Scholar]
- 32.Miller JM, Jenny AL, Payeur JB. Polymerase chain reaction detection of Mycobacterium tuberculosis complex and Mycobacterium avium organisms in formalin-fixed tissues from culture-negative ruminants. Vet Microbiol. 2002;87:15–23. doi: 10.1016/s0378-1135(02)00027-5. [DOI] [PubMed] [Google Scholar]
- 33.Yum HK, Choi SJ. Detection of mycobacterial DNA using nested polymerase chain reaction of pleural biopsy specimens: compared to pathologic findings. Korean J Intern Med. 2003;18:89–93. doi: 10.3904/kjim.2003.18.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jambhekar NA, Kulkarni SP, Madur BP, Agarwal S, Rajan MG. Application of the polymerase chain reaction on formalin-fixed, paraffin-embedded tissue in the recognition of tuberculous osteomyelitis. J Bone Joint Surg Br. 2006;88:1097–101. doi: 10.1302/0301-620X.88B8.17625. [DOI] [PubMed] [Google Scholar]
- 35.Park DY, Kim JY, Choi KU, et al. Comparison of polymerase chain reaction with histopathologic features for diagnosis of tuberculosis in formalin-fixed, paraffin-embedded histologic specimens. Arch Pathol Lab Med. 2003;127:326–30. doi: 10.5858/2003-127-0326-COPCRW. [DOI] [PubMed] [Google Scholar]
- 36.Sester M, Sotgiu G, Lange C, et al. Interferon-gamma release assays for the diagnosis of active tuberculosis: a systematic review and meta-analysis. Eur Respir J. 2011;37:100–11. doi: 10.1183/09031936.00114810. [DOI] [PubMed] [Google Scholar]


