Abstract
The enzymes of the thiosulfate dehydrogenase (TsdA) family are wide-spread diheme c-type cytochromes. Here, redox carriers were studied mediating the flow of electrons arising from thiosulfate oxidation into respiratory or photosynthetic electron chains. In a number of organisms, including Thiomonas intermedia and Sideroxydans lithotrophicus, the tsdA gene is immediately preceded by tsdB encoding for another diheme cytochrome. Spectrophotometric experiments in combination with enzymatic assays in solution showed that TsdB acts as an effective electron acceptor of TsdA in vitro when TsdA and TsdB originate from the same source organism. Although TsdA covers a range from −300 to +150 mV, TsdB is redox active between −100 and +300 mV, thus enabling electron transfer between these hemoproteins. The three-dimensional structure of the TsdB-TsdA fusion protein from the purple sulfur bacterium Marichromatium purpuratum was solved by X-ray crystallography to 2.75 Å resolution providing insights into internal electron transfer. In the oxidized state, this tetraheme cytochrome c contains three hemes with axial His/Met ligation, whereas heme 3 exhibits the His/Cys coordination typical for TsdA active sites. Interestingly, thiosulfate is covalently bound to Cys330 on heme 3. In several bacteria, including Allochromatium vinosum, TsdB is not present, precluding a general and essential role for electron flow. Both AvTsdA and the MpTsdBA fusion react efficiently in vitro with high potential iron-sulfur protein from A. vinosum (Em +350 mV). High potential iron-sulfur protein not only acts as direct electron donor to the reaction center in anoxygenic phototrophs but can also be involved in aerobic respiratory chains.
Keywords: crystal structure, cytochrome c, enzyme kinetics, heme, protein chemistry, respiratory chain, TsdA, electron acceptor, thiosulfate dehydrogenase
Introduction
The bifunctional thiosulfate dehydrogenase/tetrathionate reductase TsdA is present in various organisms of different proteobacterial genera (1). In the diheme cytochrome c from the purple sulfur bacterium Allochromatium vinosum, an axial histidine/cysteine ligation of the central iron atom has been firmly established for the active site heme (2). This type of ligation is rare among prokaryotes, usually leads to a low redox potential of the corresponding heme (3–6), and appears to be of special importance in sulfur-based energy metabolism. TsdA proteins catalyze the reversible formation of a sulfur-sulfur bond between the sulfane atoms of two thiosulfate molecules, yielding tetrathionate and releasing two electrons. TsdA enzymes of various source organisms exhibit different catalytic bias (7). Although the enzyme from the sulfur oxidizer A. vinosum is strongly adapted to catalyzing thiosulfate oxidation (2), TsdA from Campylobacter jejuni acts primarily as a tetrathionate reductase and enables the organism to use tetrathionate as an alternative electron acceptor for anaerobic respiration (8).
At present, it is largely unclear which redox carriers mediate the flow of electrons arising from thiosulfate oxidation into respiratory or photosynthetic electron chains. In several organisms, including Thiomonas intermedia, Sideroxydans lithotrophicus, and Pseudomonas stutzeri, tsdA is immediately preceded by a gene encoding another diheme cytochrome, TsdB (1). TsdB itself is not reactive with thiosulfate but accepts electrons from TsdA even when TsdA and TsdB do not originate from the same organism (1). Kinetic data that quantitatively describe the interaction between TsdA and TsdB have not been published so far. In the anoxygenic phototrophic purple sulfur bacterium Marichromatium purpuratum, TsdA and TsdB form a fusion protein with TsdB constituting the N-terminal domain (7). TsdBA fusion proteins are also encoded in other members of the family Chromatiaceae, i.e. Thiorhodococcus sp. AK35 (D779_1816), Thiocystis violascens (Thivi_3993), Thiorhodococcus drewsii (ThidrDRAFT_3922), and Thioflaviococcus mobilis (Thimo_0460). However, TsdBA fusions are not a common trait in purple sulfur bacteria. In A. vinosum, a tsdB gene is not present (1).
In A. vinosum, the protein with the closest relationship to T. intermedia or P. stutzeri TsdB is Alvin_2879. This cytochrome c4 (previously cytochrome c553(550)) is membrane-bound (possibly via the hydrophobic protein Alvin_2880) and has a positive redox potential of +330 mV (9). Another candidate for accepting electrons from TsdA in purple anoxygenic phototrophic bacteria is the high potential iron-sulfur protein (HiPIP).11 A. vinosum and M. purpuratum produce HiPIP, and as this protein has a quite positive reduction potential (+350 mV (10)) it would be well suited as an electron acceptor for TsdA. This proposal is corroborated by a previous report where a protein preparation with thiosulfate dehydrogenase activity from A. vinosum reduced HiPIP in vitro (11).
Here, we study Tsd(B)A enzymes from dedicated sulfur oxidizers and characterize in detail the interaction of TsdA with TsdB. Additionally, we pose the following question. Which proteins serve as immediate electron acceptors for either TsdA alone or the TsdBA fusion protein (when present)? It is furthermore intended to derive models for the electron flow involved.
Results
Characterization of TsdA and TsdB
UV-visible electronic absorbance spectroscopy, X-ray diffraction, and activity assays have revealed a number of characteristic features of AvTsdA (1, 2, 12). However, the electrochemical window in which the hemes are redox active remained unknown. To gain insight into this property, we mapped out the redox activity of AvTsdA adsorbed as an electroactive film on optically transparent mesoporous nanocrystalline SnO2 electrodes. The spectrum of the enzyme-coated electrode equilibrated at +302 mV contained features typical of ferric c-type hemes superimposed on a small contribution from light scattering by the electrode material (Fig. 1). The Soret maximum at 406 nm and broad lower intensity features in the αβ-region are typical of those displayed by solutions of oxidized AvTsdA (1). When the electrode potential was lowered to −648 mV in 50-mV steps with a spectrum recorded after a 60-s pause at each desired potential (Fig. 1), those features were replaced with peaks having maxima at 418, 523, and 553 nm, which are typical of dithionite-reduced enzyme (1). Variation of the Soret intensity at 418 nm with electrode potential revealed that the hemes were reduced between approximately +150 and −350 mV (Fig. 1, inset, closed squares).
FIGURE 1.
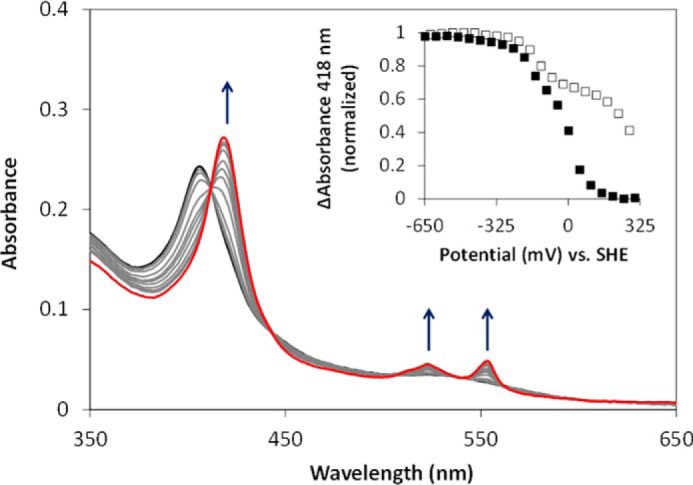
Redox activity of AvTsdA adsorbed on a mesoporous nanocrystalline SnO2 electrode. Electronic absorbance was recorded with the electrode poised at +302 mV (black), +152 to −298 mV at 50-mV intervals (gray), and −648 mV (red). All potentials are quoted versus the standard hydrogen electrode (SHE). The arrows indicate increases in absorbance as the electrode potential was lowered. Inset shows the normalized change in absorbance at 418 nm against the applied potential as the enzyme was reduced (closed squares) and re-oxidized (open squares).
The response of reduced AvTsdA to a stepwise increase of the electrode potential was assessed in a similar manner. The potential was raised in 50-mV steps, and the spectra were recorded after a 60-s pause at each potential (Fig. 1, inset, open squares). Between −648 and −98 mV, the variation in Soret intensity with the applied potential was very similar to that recorded on reduction. However, further increase of potential revealed significantly less oxidation than anticipated from the behavior seen on reduction of the enzyme. Importantly, spectra typical of the fully oxidized enzyme were measured after the electrode was poised at +302 mV for ∼30 min. It was concluded that electrodic redox cycling of adsorbed AvTsdA was fully reversible but that full reduction occurred more quickly than complete re-oxidation. Further experiments confirmed that this behavior persisted over multiple rounds of reduction and re-oxidation. The spectral changes induced by variation of potential between −648 and −98 mV were rapidly reversed and accounted for ∼35% of the change in absorbance at 418 nm when spectra of the fully oxidized and fully reduced forms of the enzyme were compared. By contrast, variations of electrode potential between −98 and +302 mV showed rapid reduction and much slower reoxidation, and the associated changes in absorbance accounted for ∼65% of the total seen on full redox cycling of the enzyme. It was concluded that the slow reoxidation associated with higher potential redox event(s) was not a consequence of reversible redox events that occurred at lower potentials. Detailed inspection of the spectra provided no indication for the presence of high-spin ferric- or ferrous-heme.
The hysteretic nature of the plot of absorbance versus potential prevented Nernstian analysis to define the heme reduction potentials. Nevertheless, some further conclusions regarding the redox activity of AvTsdA can be proposed in light of the crystal structures reported (2, 12). The fully oxidized diheme cytochrome AvTsdA contains His/Cys coordination in heme 1 and His/Lys in heme 2. His/Cys-ligated hemes are typically distinguished from other low-spin c-type hemes by having much lower reduction potentials and smaller changes in extinction coefficient associated with the Fe(III)/(II) couple (4, 13). As a consequence, we propose that reduction of His/Cys-ligated heme 1 occurs reversibly between approximately −100 and −350 mV. Reduction of His/Lys-ligated heme 2 is proposed to occur between approximately +150 and −100 mV. AvTsdA X-ray structures reveal that this reduction is accompanied by a switch of Lys by Met as axial distal ligand to ferrous-heme 2 (2). Such a change of ligation would be expected to raise the reduction potential of heme 2. If the Met ligand is replaced slowly by Lys upon enzyme oxidation, this would account for the hysteretic nature of the redox behavior displayed by AvTsdA.
In contrast to TsdAs, TsdB proteins are not well characterized. The most closely related characterized cytochromes on a sequence level belong to the diheme cytochromes of the c4 family (1). The recombinant TsdB protein from T. intermedia indeed binds two heme groups of 616.5 Da as the mass of 21,792.5 Da determined by MALDI-TOF mass spectrometry almost exactly matched the mass of 21,783.3 Da predicted for the mature recombinant protein, including Strep tag and two hemes. The same held true for recombinant S. lithotrophicus TsdB (measured mass, 22,864 Da, and predicted mass, 22,835 Da). UV-visible spectroscopy of pure recombinant TsdB from S. lithotrophicus is shown in Fig. 2. The spectrum conforms to that of TiTsdB (1).
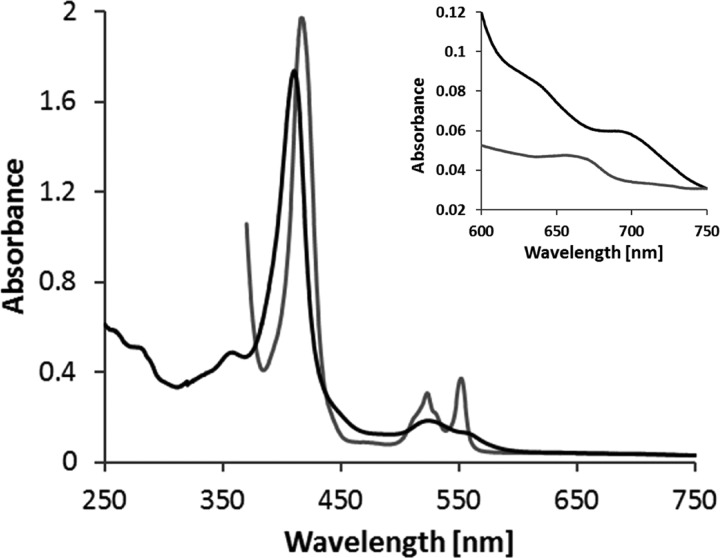
FIGURE 2.
UV-visible spectra of TsdB from S. lithotrophicus. As the protein is partly reduced in the “as isolated” state, up to 170 μm ferricyanide were added to record the oxidized spectrum (black line). For full reduction of the protein, sodium dithionite was added (gray line). 100 mm Tris buffer, pH 8.0, with 150 mm NaCl and 2.5 mm desthiobiotin was used, and spectra are normalized to 750 nm. The oxidized spectrum exhibits a 700-nm peak indicating methionine as heme iron ligand. Protein concentration was 6 μm in the overview and 29 μm in the blowup.
A sequence alignment of various TsdB proteins (Fig. 3) reveals two conserved methionine residues but no conserved histidines or cysteines indicating that both hemes of TsdB have axial coordination by His/Met. This is underpinned by the 700-nm peak in the spectrum, which is characteristic for methionine as the sixth axial heme iron ligand (14) and corroborated by the MpTsdBA crystal structure determined herein (see below). A shift of the Soret band from 411 to 417 nm upon reduction was observed. Moreover, there is a distinct δ band at 359 nm in the oxidized protein spectrum. In the reduced state, the α band was detected at 552 nm and the β band at 523 nm. A split α band characteristic for a number of c4-type cytochromes, including those from purple sulfur bacteria, was not observed (9, 15). The UV-visible spectrum for oxidized TsdB exhibited a low intensity high spin feature at 620 nm similar to that noted for cytochrome c4 from P. stutzeri (16). Obviously, the ferric form of TsdB holds a small fraction of high-spin heme probably caused by weakening of the Fe-S bond at one of the two hemes with concomitant partial dissociation of the methionine and formation of an Fe-aquo bond just as outlined for the P. stutzeri cytochrome (16).
FIGURE 3.
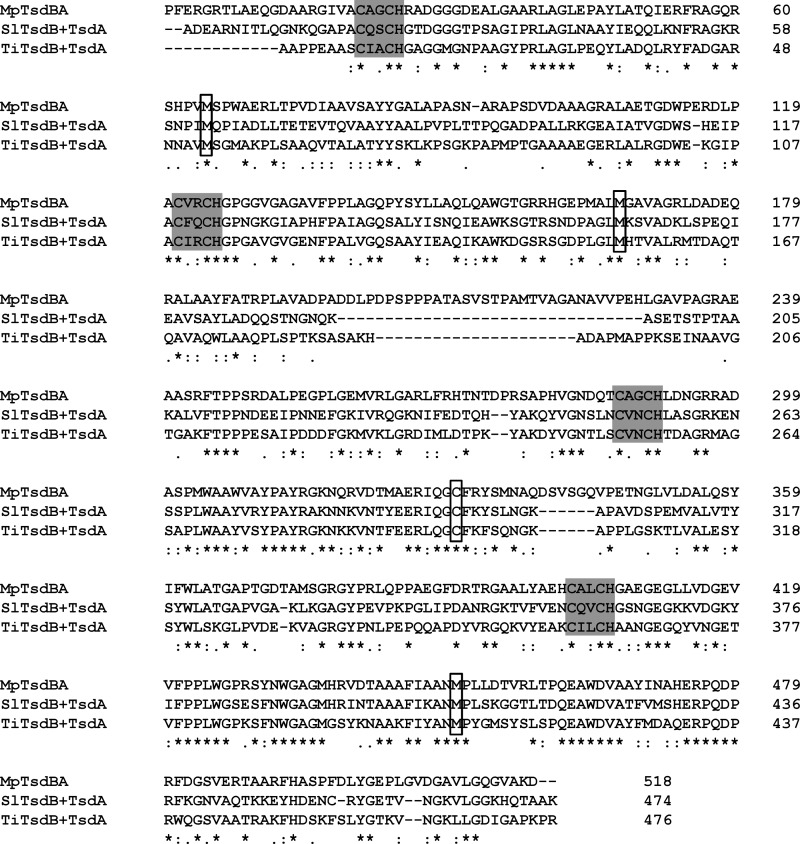
Sequence alignment of MpTsdBA and SlTsdB + TsdA as well as TiTsdB + TsdA. Sequence comparison of TsdBA fusion protein of M. purpuratum (MARPU_02550) with the combined sequence of TsdB and TsdA from S. lithotrophicus (Slit_1877 and Slit_1878) and T. intermedia (Tint_1893 and Tint_2892). All signal peptide sequences were removed. Heme-binding motifs are indicated by gray boxes, and putative distal heme ligands are marked by black edging. Strictly conserved residues are marked with asterisks. TsdA sequences of S. lithotrophicus and T. intermedia start after the gap at amino acids 195 and 189, respectively.
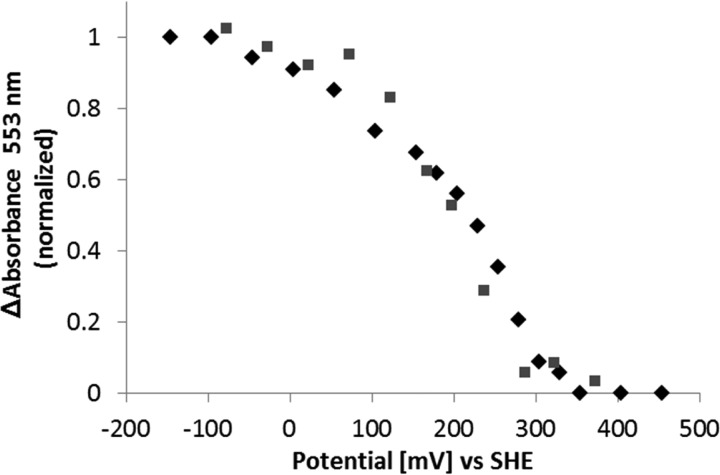
The reduction potential of TiTsdB was determined by potentiometry with a gold electrode (Fig. 4). The potential changes determined upon reduction and reoxidation of TsdB match well. The two hemes are redox active between −100 and +300 mV. Hysteresis was not observed, consistent with both hemes being His/Met-ligated. Ligand changes are not apparent.
FIGURE 4.
Determination of TiTsdB reduction potential by potentiometry with a gold electrode. Potentiometric determination of redox potentials of both TsdB hemes. Applied potential according to normalized values of the α-peak (553 nm) is shown. Reduction of TsdB (black diamonds) and re-oxidation of the protein (gray squares) was measured. 10 μm TsdB in phosphate buffer, pH 5.0, was used. SHE, standard hydrogen electrode.
TsdB Is an Electron-transferring Unit Tightly Interacting with TsdA
The interaction of TsdA and TsdB proteins was analyzed for the recombinant proteins from S. lithotrophicus. Upon analytical gel permeation chromatography SlTsdA alone eluted at a volume corresponding to a molecular mass of 65 kDa, indicating dimerization of the protein under the conditions applied (monomer, 33,042 Da). In contrast, SlTsdB behaved as a 22-kDa monomer (predicted molecular mass, 22,835 Da). When both proteins were produced simultaneously in Escherichia coli and purified employing the Strep tag attached to TsdA, a preparation was obtained that exhibited two heme stainable polypeptides. In size, these exactly matched SlTsdB and SlTsdA (Fig. 5). The co-purification of the two proteins is evidence for significant interaction between them. Upon size exclusion chromatography, SlTsdA and SlTsdB co-eluted again in fractions corresponding to a mass of 108 kDa, indicating formation of an α2β2 heterodimer.
FIGURE 5.
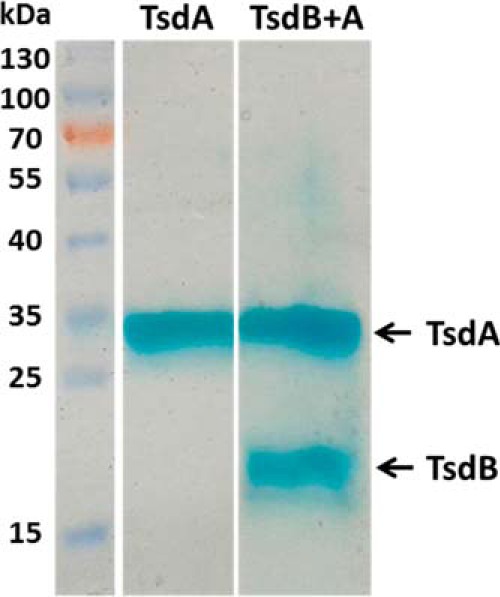
Analysis of purified SlTsdA and SlTsdB + A by SDS-PAGE. 10–15 μg of SlTsdA and SlTsdB + A obtained after Strep tag affinity chromatography were loaded per lane on a 12.5% gel and stained for presence of heme. In case of SlTsdB + A, both proteins were produced simultaneously in E. coli and purified on the basis of a Strep tag attached to TsdA.
Characterization of the TsdBA Fusion Protein from M. purpuratum
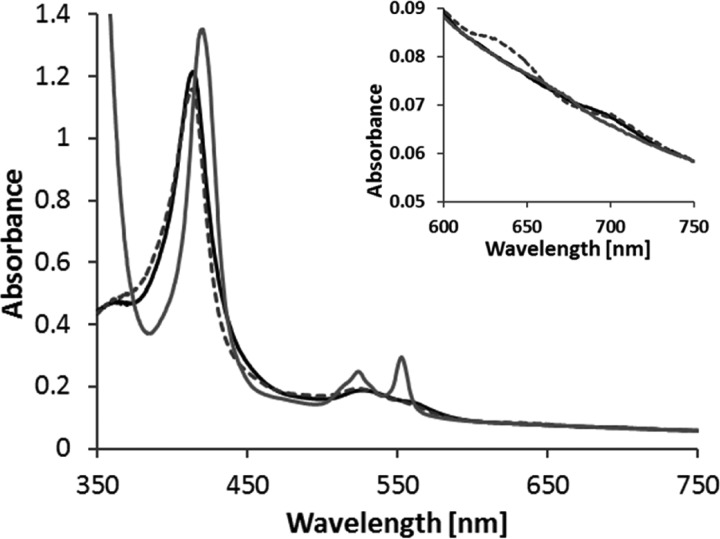
The protein encoded by tsdB-tsdA gene fusion in M. purpuratum provides an exceptional possibility to study the complete tetraheme cytochrome for catalytic properties and internal electron transfer. A sequence alignment of MpTsdBA with combined TsdB and TsdA sequences from S. lithotrophicus and T. intermedia shows significant similarity between the N-terminal region of MpTsdBA and TsdB (amino acids 1–199 of MpTsdBA and TiTsdB share 45% sequence identity) and between the C-terminal region of MpTsdBA and TsdA (amino acids 224–518 of MpTsdBA compared with AvTsdA or TiTsdA, 39 and 50% sequence identity, respectively). The heme distal ligands cysteine and methionine of TsdA as well as the two putative heme-ligating methionines are strictly conserved (Fig. 3). Therefore, we predicted MpTsdBA to contain three His/Met ligated and one His/Cys ligated heme. UV-visible spectroscopy of MpTsdBA protein is shown in Fig. 6. The presence of His/Met-ligated hemes in MpTsdBA is substantiated by the 700-nm peak in the spectrum of the oxidized protein. This absorption band is characteristic for methionine as heme iron ligand (14). A shift of the Soret band from 413 to 420 nm was observed upon reduction. Moreover, there is a distinct δ band at 363 nm in the oxidized protein spectrum. The α band is located at 553 nm, and the β band resides at 524 nm. The partly reduced spectrum exhibits a low intensity high spin feature at 620 nm similar to AvTsdA (2). The A413 nm/A280 nm for pure oxidized MpTsdBA is 3.4.
FIGURE 6.
UV-visible spectra of MpTsdBA. As the protein is slightly reduced in the “as isolated” state, 60 μm ferricyanide were added to record the oxidized spectrum (black line). For partial (gray broken line) and full reduction (gray line) of the protein, 0.33 and 5 mm sodium dithionite were added, respectively. 100 mm ammonium acetate buffer, pH 5, with 200 mm NaCl was used, and spectra are normalized to 750 nm. The oxidized spectrum exhibits a 700-nm peak indicating methionine as heme iron ligand and the partially reduced protein exhibits a feature at 630 nm. Protein concentration was 3.3 μm.
Crystal Structure Determination and Model Quality of MpTsdBA
To compare structural features of TsdA and TsdBA, to get a closer look into ligation of the four heme groups and to see how TsdA and TsdB domains are linked to each other, we have determined the X-ray structure of MpTsdBA by Fe-SAD. Crystals belong to the hexagonal space group H32 with cell dimensions a = b = 159.75 Å and c = 393.09 Å. There are two molecules in the asymmetric unit corresponding to a Matthews coefficient (17) of 2.8 Å3 Da−1 and a solvent content around 56%. The “as isolated” structure was refined to 2.75 Å resolution with Rcryst of 15.7% and Rfree of 19.8% using a 141°-sweep of data with overall better statistics. Data collection and refinement statistics are depicted in Table 1. The high Rmeas values observed for both sweeps are due, first, to the fine binning that autoPROC implements during data reduction, and second, to the fact that the crystal has some regions in the rotational space that are of bad quality. This is perfectly clear in the plots Rmeas versus image number output by autoPROC (data not shown), highlighting that the high Rmeas values are due to the crystal quality and not radiation damage. No evidence for radiation damage is also observed in a m|Fo| − m|Fo| map calculated using both data sweeps (data not shown). MpTsdBA is numbered without taking into account its 23-amino acid signal peptide that is removed upon transport into the periplasm. The model comprises the following residues of the mature recombinant protein: Pro1–Leu191 and Arg237–Val515 (chain A) and Pro1–Ala192 and Ala240–Ala516 (chain B), eight heme molecules, two thiosulfate ions, two chloride ions, seven ethylene glycols, two 1,2-propanediols and one triethylene glycol, and 113 water molecules. Electron density maps are of good quality except for the C-terminal His tag and the disordered loop connecting TsdB and TsdA domains. This region (45 amino acid residues long in chain A and 47 in chain B) has a predicted loop-like secondary structure and is not included in the final model, because no electron density was observed. Moreover, some parts of the map are somewhat “noisy” with positive and negative difference map peaks in the solvent region that could not be modeled.
TABLE 1.
Data reduction and refinement statistics for MpTsdBA structure
| 142.65°-sweep data set (refinement) | Full 360° data set (Fe-SAD phasing) | |
|---|---|---|
| PDB code | 5LO9 | |
| Data collection | ||
| Synchrotron | ESRF (Grenoble, France) | |
| Beamline | ID-29 | |
| Wavelength (Å) | 1.7236 | |
| Space group | H32 | |
| Unit cell | ||
| a, b, c (Å) | 159.75, 159.75, 393.09 | 159.89, 159.89, 392.99 |
| α, β, γ (o) | 90.0, 90.0, 120.0 | |
| Resolution rangea (Å) | 113.13–2.75 (2.76–2.75) | 130.99–2.82 (2.83–2.82) |
| Total no. of reflections | 377,526 (2187) | 891,650 (6064) |
| No. of unique reflections | 50,199 (461) | 49,983 (442) |
| Completeness (%) | 99.0 (87.8) | 99.6 (94.2) |
| Anomalous completeness (%) | 98.7 (84.8) | 99.6 (94.1) |
| Multiplicity | 7.5 (4.7) | 19.0 (13.7) |
| Anomalous multiplicity | 3.9 (2.5) | 9.9 (7.0) |
| 〈I/σ(I)〉 | 14.9 (2.0) | 15.4 (2.4) |
| Rmeasb (%) | 11.7 (70.6) | 25.5 (184.7) |
| Rpim (%) | 5.7 (41.8) | 8.1 (69.2) |
| CC1/2c (%) | 99.7 (63.8) | 99.4 (75.7) |
| Refinement | ||
| Rcrystd (%) | 15.7 (25.9) | |
| Rfreee (%) | 19.8 (30.0) | |
| No. of non-H atoms | ||
| Protein | 7028 | |
| Ligands | 330 | |
| Waters | 113 | |
| r.m.s.d. bonds (Å) | 0.013 | |
| r.m.s.d. angles (°) | 1.50 | |
| Protein residues | Pro1–Leu191 and Arg237–Val515 (chain A), Pro1–Ala192 and Ala240–Ala516 (chain B) | |
| Ramachandran plot | ||
| Most favored (%) | 97.6 | |
| Allowed (%) | 2.4 | |
| Outliers (%) | 0 | |
| Rotamer outliers (%) | 0.9 | |
| Clashscore | 2.69 | |
| MolProbity scoref | 1.28 | |
| B-Factors (Å2) | ||
| Protein | 52.04 | |
| Ligands/ions | 50.43 | |
| Waters | 45.06 | |
a Information in parentheses refers to the last resolution shell.
b Rmeas = ΣhΣl|Ihl − 〈Ih〉|/ΣhΣl 〈Ih〉, where Ihl is the Ith observation of reflection h and 〈Ih〉.
c CC1/2 is as described previously (57).
d Rcryst = Σh‖Fobs(h)| − |Fcalc(h)‖/Σh|Fobs(h), where Fobs(h) − Fcalc(h) are the observed and calculated structure factors for reflection h, respectively.
e Rfree was calculated as Rfactor but using only 5% of reflections randomly selected and omitted from refinement.
f MolProbity score provides a single number that represents the central MolProbity protein quality statistics; it is a log-weighted combination of clashscore, Ramachandran not favored and bad side-chain rotamers, giving one number that reflects the crystallographic resolution at which those values would be expected.
Overall Fold and Similar Structures of MpTsdBA
MpTsdBA is organized into two domains, an N-terminal TsdB domain and a C-terminal TsdA domain (Fig. 7A). Each domain includes two subdomains that are related by a pseudo-2-fold symmetry axis. Each subdomain consists of four α-helices surrounding a heme group, the typical class I c-type cytochrome topology. This has been previously reported for the AvTsdA crystal structure (2, 12) and is also observed for the M. purpuratum N-terminal TsdB domain. The four subdomains superimpose with r.m.s.d. of 1.3–3.0 Å for ∼70 aligned Cα atoms corresponding to sequence similarities between 10 and 38%.
FIGURE 7.
X-ray structure of MpTsdBA and heme arrangement. A, overall fold with TsdB N-terminal domain (residues 1–193) depicted in blue and TsdA C-terminal domain (residues 240–516) in red; the two shades in each domain represent the respective sub-domains (1–90 and 91–193 for TsdB and 240–377 and 378–516 for TsdA). The heme prosthetic groups are colored by atom type (orange or yellow for carbon, blue for nitrogen, red for oxygen, and dark red for iron). B, Fe-to-Fe distances; C, closest edge-to-edge distances.
The final model coordinates were submitted to the DALI server (18), showing no similar structures to the complete MpTsdBA arrangement. However, several hits structurally match each domain of the enzyme separately. The highest matches to MpTsdB domain were cytochrome c4 from P. stutzeri (PDB code 1M6Z), cytochrome c552 from Acidithiobacillus ferroxidans (PDB code 1H1O), the cytochrome subunit of flavocytochrome c sulfide dehydrogenase from A. vinosum (PDB code 1FCD), and flavocytochrome c from Thermochromatium tepidum (PDB code 3VRD), with Z-scores of 22.7 to 17.5, r.m.s.d. of 2.0–2.4 Å, and sequence identities between 30 and 23%.
The highest hit for the MpTsdA domain was AvTsdA (4V2K and 4WQ9) with a Z-score of 30.6 (with 20% sequence identity and r.m.s.d. of 1.4 and 1.2 Å, respectively), followed by SoxAX from Rhodovulum sulfidophilum (2OZ1), the SoxD subunit of SoxCD from Paracoccus pantotrophus (2XTS), and the SoxA subunits of SoxAX from Starkeya novella (3OA8) and P. pantotrophus (2C1D). Z-scores ranged from 12–7.4 and r.m.s.d.s from 2.1 to 4.7 Å.
Heme Coordination in the “As Isolated” MpTsdBA
The MpTsdBA crystal structure shows four heme groups per chain packed as a wire with closest iron-to-iron distances between 15 and 19 Å and shortest edge-to-edge distances of 3.5 to 6.6 Å (Fig. 7, B and C). This agrees well with other multiheme cytochrome structures that show edge-to-edge distances of 4–8 Å (19, 20).
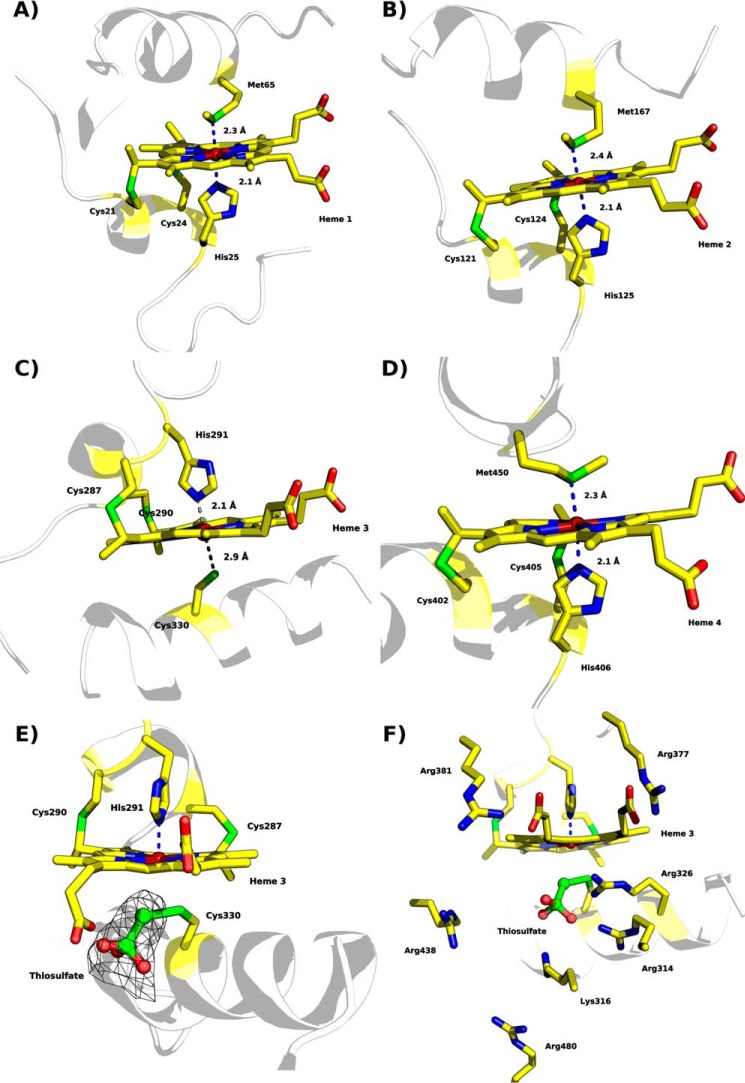
The four hemes are covalently bound to the polypeptide chain through thioether bonds formed by cysteine residues Cys21 and Cys24 for heme 1, Cys121 and Cys124 for heme 2, Cys287and Cys290 for heme 3, and Cys402 and Cys405 for heme 4 (Fig. 8, A–D). Moreover, the structure confirmed the spectroscopic evidence gathered showing that this tetraheme cytochrome c has three hemes (hemes 1, 2, and 4) with His/Met coordination (Fig. 8, A, B, and D). Axial ligation by histidine and cysteine is typical for the active site of TsdA proteins (2, 12). Indeed, heme 3 exhibits axial ligation by His291, and the Sγ atom of Cys330 is located in close vicinity to the heme iron such that it could serve as the sixth ligand. However, the 2.9 Å distance between the sulfur and the iron atom precludes direct ligation (Fig. 8, C, E, and F). It has been shown earlier that the Sγ atom of the corresponding active site cysteine (Cys96) in TsdA from A. vinosum can adopt two different conformations by rotation of the cysteine Cα–Cβ bond. Therefore, the sulfur atom switches between iron-ligating and iron-non-ligating states (2). The non-ligating conformation has been proposed as an essential intermediate step in the catalytic cycle, possibly involving covalent attachment of a substrate molecule (2, 12).
FIGURE 8.
Heme coordination of “as isolated” MpTsdBA (PDB code 5LO9). A, heme 1 is coordinated by His25 and Met65. B, heme 2 is coordinated by His125 and Met167. C, heme 3 is ligated to His291 but not to Cys330. The distance of Sγ to the heme iron is 2.9 Å and thus not close enough for direct ligation. Thiosulfate covalently bound to Sγ of Cys330 is not shown here for clarity. Presence of thiosulfate is illustrated in detail in E and F. D, Heme 4 is ligated by His406 and Met450. E, heme 3 with Sγ of Cys330 covalently bound to thiosulfate, displayed in ball and stick, and polder map electron density contoured at 6σ level depicted as a black mesh. F, heme 3 in a similar view as in E but with positively charged residues surrounding the substrate cleft depicted as sticks. Scheme representation is shown in pale gray with heme moieties and coordinating amino acid residues shown as sticks; color code as in Fig. 7 with sulfur atoms in green.
Remarkably, in the “as isolated” structure of MpTsdBA a thiosulfate ion is indeed covalently bound to Cys330. A polder map supporting the modeling of the thiosulfate ion is depicted in Fig. 8E. The thiosulfate is oriented such that the S1-S2 plane points toward the heme plane, and the S2 atom lies 2.06 Å away from the Sγ atom of Cys330, thus being within covalent bond distance. Thiosulfate was refined to 66% occupancy in chain A and 72% in chain B. The Sγ of Cys330 (full occupancy) superposes well with the Sγ atom of Cys96 in AvTsdA with bisulfite (PDB code 4WQB). Here, the Sδ of persulfurated Cys96 superimposes with S2 of thiosulfate in the MpTsdBA structure. In both structures, these ligand-bound cysteines are not coordinating the heme. Noteworthy, some continuity in the electron density maps is still seen on heme 3, even though Cys330 is not ligated to the heme iron. We expect MpTsdBA Cys330 to coordinate the heme when no ligand/substrate is present, similar to what is observed in AvTsdA crystal structures (2, 12).
The thiosulfate substrate lies in a cleft accessible from the solvent to Cys330 and heme 3. This cavity is delineated by the side chains of positively charged residues Arg314, Lys316, Arg326, Arg438, and Arg480 (Fig. 8F), which have been previously proposed to be involved in the orientation and stabilization of the substrate for catalysis (2, 12). Some positive electron density (m|Fo| − D|Fc|) is present near the substrate, although no density is observed in 2m|Fo| − D|Fc| maps (even at low contours). This electron density is observed between the Nζ atom of Lys316 and the plane formed by the three oxygen atoms of the thiosulfate ion (although independent from the density observed for the thiosulfate ion itself). Because this electron density was not amenable to refinement, nothing was included in the final 3D structure. Furthermore, heme 3 seems to display another residual conformation with one of the propionates alternating between this cavity and a cleft above the heme plane delineated by Arg377, Arg381, and the N main chain atoms of Gly378 and Tyr379. This motion is illustrated by the different conformation modeled for heme 3 in both chains, either pointing toward the active site cavity, in chain A (Fig. 8E), or toward the cleft above the heme plane, in chain B (Fig. 8, C and F). However, this possible alternate conformation could not be properly refined, and therefore it was not added to the crystallographic model.
Reactivity with External Electron Acceptors
To test different electron acceptors for TsdA and TsdBA, we performed enzyme activity assays with TsdB from T. intermedia, A. vinosum cytochrome c4 (Alvin_2879), and A. vinosum HiPIP as electron acceptors. All three potential electron acceptors were produced as recombinant proteins in E. coli.
Previously, it had been shown that TsdB from T. intermedia is not reactive with thiosulfate but that it is instantaneously reduced when TsdA is added in the presence of thiosulfate (1). Here, we succeeded in obtaining quantitative kinetic data for a homologous system by analyzing electron transfer between TsdA and TsdB from S. lithotrophicus. Just as TiTsdB, SlTsdB alone is not reduced by addition of thiosulfate. An enzyme activity assay with SlTsdA as the catalyst and SlTsdB as the electron acceptor resulted in a specific activity of 6.3 units mg−1 for SlTsdA. S0.5 for thiosulfate was determined to be 0.04 mm. This unambiguously identified SlTsdB as an effective electron acceptor for TsdA from the same organism. However, a heterologous approach yielded a different result; with 0.6 units mg−1 the specific activity of AvTsdA with TsdB from T. intermedia amounted to only one-tenth that determined for the homologous system, whereas S0.5 for thiosulfate (0.03 mm) resided in a similar range.
In a further series of experiments, AvTsdA activity was tested with Cyt c4 originating from the same host. It should be noted that the recombinant cytochrome was electrophoretically pure and that it exhibited exactly the same spectral features as Cyt c4 purified from A. vinosum cells, including the characteristic split α-band (9). The specific activity of AvTsdA with AvCyt c4 as the electron acceptor amounted to 0.6 units mg−1 and was thus not found to be higher than with TsdB from a different source organism. Therefore, we exclude those diheme cytochromes as efficient electron acceptors for AvTsdA in vitro as well as in vivo.
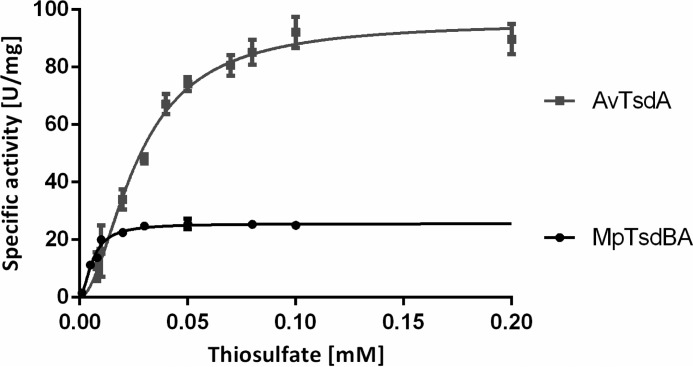
HiPIP from A. vinosum, a protein with a positive reduction potential (+350 mV (10)), was tested as another potential candidate for accepting electrons from AvTsdA as well as from MpTsdBA. Indeed, both thiosulfate dehydrogenases reacted efficiently with A. vinosum HiPIP (Fig. 9 and Table 2). AvTsdA exhibited a higher Vmax with HiPIP as electron acceptor, whereas MpTsdA featured an especially low S0.5 value for thiosulfate when the reaction was measured with HiPIP as electron acceptor. In both cases, S0.5 for thiosulfate was much lower with HiPIP than with ferricyanide as the electron acceptor indicating cooperativity between the electron-transferring heme 2 and the active site heme 1. A. vinosum and M. purpuratum both encode HiPIP in their genome, and both thiosulfate dehydrogenases exhibit substantial specific activity with HiPIP as electron acceptor in vitro, leading us to conclude that HiPIP also serves as an efficient in vivo electron acceptor for Tsd(B)A in both organisms.
FIGURE 9.
Thiosulfate oxidation catalyzed by AvTsdA and MpTsdBA with HiPIP as electron acceptor. Enzyme assays with AvTsdA were performed in 100 mm ammonium acetate buffer, pH 5, at 30 °C with 8 nm enzyme. Activity measurements with MpTsdBA were performed in 100 mm ammonium acetate buffer, pH 5.2, with 200 mm NaCl at 25 °C and with 3.9 nm enzyme. In both assays 10 μm HiPIP and 40 μm ferricyanide were used. A change in absorbance was measured at 480 nm. v versus [S] plots were fitted to the Hill equation.
TABLE 2.
Thiosulfate oxidation of AvTsdA and MpTsdBA with ferricyanide and HiPIP
Enzyme assays with AvTsdA were performed in 100 mm ammonium acetate buffer, pH 4, at 30 °C with 8 nm enzyme. Activity measurements with MpTsdBA were performed in 100 mm ammonium acetate buffer, pH 5.2, with 200 mm NaCl at 25 °C and with 3.9 nm enzyme. In assays with HiPIP as electron acceptor, 10 μm HiPIP and 40 μm ferricyanide were used, and absorbance at 480 nm was followed. In activity assays with ferricyanide as electron acceptor, 1 mm ferricyanide was used, and the absorbance at 420 nm was measured. The units for Vmax are μmol min−1 mg protein−1. v versus [S] plots were fitted to the Hill equation.
| Electron acceptor | Enzyme | Vmax | S0.5 | kcat | cat/S0.5 |
|---|---|---|---|---|---|
| units mg−1 | μm | s−1 | mm−1 s−1 | ||
| Ferricyanide | AvTsdA | 31,419 ± 2408 | 835 ± 119 | 14,091 | 16,875 |
| MpTsdBA | 3011 ± 108 | 179 ± 21 | 2794 | 15,611 | |
| HiPIP | AvTsdA | 96 ± 3 | 27 ± 2 | 43 | 1595 |
| MpTsdBA | 26 ± 1 | 6 ± 0 | 24 | 4000 |
Discussion
In our approach to find suitable electron acceptors for TsdA-type thiosulfate dehydrogenases, we first focused on TsdB, a diheme cytochrome encoded upstream of TsdA in a number of different organisms. As demonstrated here for the proteins from S. lithotrophicus and earlier for those from T. intermedia (1), TsdA and TsdB enzymes interact strongly with each other and form an α2β2 heterodimer. The same arrangement has been described for thiosulfate dehydrogenase from Halothiobacillus neapolitanus (21), which consists of heme c binding subunits of 27 and 33 kDa conforming in size with TsdA and TsdB, respectively.
In this work, a redox range of −300 to +150 mV was determined for AvTsdA, whereas TiTsdB is redox active between −100 and +300 mV. Generalizing this finding, we state that the overall reduction potential of TsdB is more positive than that of TsdA, which should enable electron flow from TsdA to TsdB. Indeed, reduction of TiTsdB by AvTsdA had been shown previously (1) and was verified here for the proteins from S. lithotrophicus. Enzyme activity assays further revealed SlTsdB as an effective electron acceptor for SlTsdA but not for AvTsdA. This was not surprising as A. vinosum does not contain a gene encoding TsdB (Table 3).
TABLE 3.
Occurrence of genes encoding TsdA and putative electron acceptors in the genome sequenced organisms relevant to this study
| Organism | TsdA | TsdB | Cyt c4 | HiPIP |
|---|---|---|---|---|
| A.vinosum DSM 180T | Alvin_0091 | Alvin_2879 | Alvin_2274 | |
| M. purpuratum 984 (DSM 1591T) | Marpu_02550 (TsdBA fusion) | Marpu_15750 | Marpu_11560 | |
| S. lithotrophicus ES-1 (ATCC 700298T) | Slit_1878 | Slit_1877 | ||
| T. intermedia K12 (DSM 18155T) | Tint_2892 | Tint_2893 |
In A. vinosum, the gene with strongest similarity to tsdB is Alvin_2879. The encoded high potential diheme cytochrome c4 has been suggested to play a role in transferring electrons to the photosynthetic reaction center (9). The M. purpuratum genome also encodes a protein (Marpu_15750) with high similarity to A. vinosum Cyt c4 (78% identity on the sequence basis; see Table 3). In the anoxygenic phototroph Rubrivivax gelatinosus, a related cytochrome c4 indeed has an established function as an alternative electron donor to the photosynthetic reaction center (22). It was therefore feasible to assume that electrons generated by thiosulfate oxidation could be shuttled to the reaction center via cytochrome c4 in purple sulfur bacteria. However, the very low specific activity of AvTsdA with A. vinosum cytochrome c4 essentially precludes such a role (Fig. 10).
FIGURE 10.
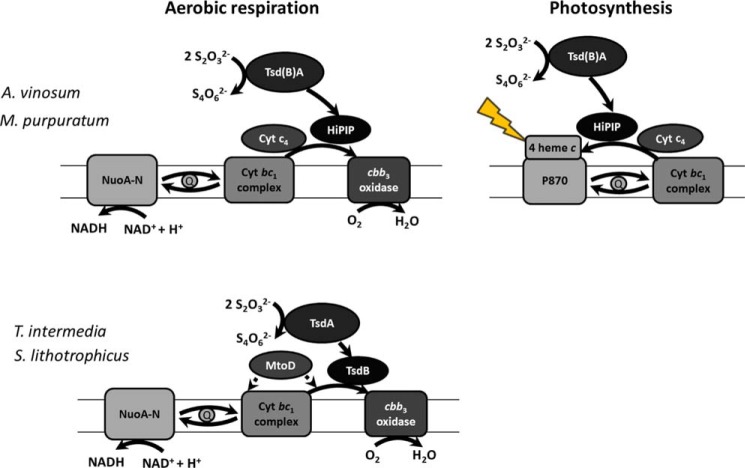
Role of periplasmic electron transfer proteins in aerobic respiration or photosynthesis of A. vinosum, M. purpuratum, S. lithotrophicus, and T. intermedia. All organisms contain genes for NuoA-N (Alvin_2418–2430 + Alvin_2412, Marpu_04365–04430, Slit_1070–1083, and Tint_2255–2268), the cytochrome bc1 complex (Alvin_0068–0070, Marpu_01465–01475, Slit_0130–0132, and Tint_2192–2194), and cbb3 oxidase (Alvin_0781–0784, Marpu_02795–02810, Slit_0411–0414, and Tint_1070–1073). Moreover, A. vinosum and M. purpuratum can gain energy by photosynthetic growth. HiPIP can transfer electrons to the photosynthetic reaction center (23, 24) as well as to cbb3 oxidase (29, 30). Cytochrome c4 also is known to transfer electrons to the photosynthetic reaction center (22) as well as to cbb3 oxidase (32–34) in some bacteria. For S. lithotrophicus, it is assumed that MtoD (Slit_2498) can transfer electrons to cbb3 oxidase and the cytochrome bc1 complex (31).
In many anoxygenic photosynthetic bacteria, the periplasmic high potential iron-sulfur protein is well known to shuttle electrons between the cytochrome bc1 complex and the photosynthetic reaction center during cyclic electron flow (23–26). This function has also been firmly established for the protein from A. vinosum (24, 27). Here, we demonstrate that HiPIP is a suitable electron acceptor for Tsd(B)A from A. vinosum and M. purpuratum in vitro, identifying this protein as the most likely electron carrier between the thiosulfate-oxidizing enzyme and the reaction center during growth in the light. It should be noted that a direct interaction between Tsd(B)A and the photosynthetic reaction center cannot be completely excluded so far.
Many purple sulfur bacteria, including A. vinosum, are capable of chemolithotrophic growth on reduced sulfur compounds and oxygen under microaerobic conditions (28). Accordingly, cbb3 as well as ubiquinol oxidases are encoded in their genomes. Although the standard reduction potential of the thiosulfate/tetrathionate couple (+198 mV (7)) appears too positive to feed electrons directly into the quinone pool, and from there to oxygen, delivery of electrons originating from the thiosulfate to tetrathionate conversion to cbb3 oxidase is certainly feasible. In fact, HiPIP has been reported to be involved in bacterial respiratory chains (29, 30) and is a prime candidate for electron transport between Tsd(B)A and the terminal oxidase in those organisms where it is present. However, in chemotrophs like T. intermedia or S. lithotrophicus, the situation must be different because these bacteria do not contain HiPIP. In fact, for these organisms, it is not exactly established so far which periplasmic proteins deliver electrons to cbb3 oxidase, regardless of the electron donor oxidized. For S. lithotrophicus it is assumed that the c-type cytochrome MtoD (Slit_2498) can transfer electrons stemming from iron oxidation to cbb3 oxidase and the cytochrome bc1 complex (31). In the T. intermedia genome, there are two c-type cytochromes (Tint_2575 and Tint_3060) with 36 and 42% sequence identity to S. lithotrophicus MtoD, respectively, which may serve a similar function. It may be possible that thiosulfate dehydrogenase delivers electrons to the MtoD(-like) cytochrome, which then shuffles the electrons to the terminal oxidase.
Nevertheless, an alternative scenario is also possible when we consider the similarity between TsdB and cytochromes of the c4-type (about 49% sequence identity between TiTsdB and Cyt c4 from Achromobacter xylosoxidans or Pseudomonas protegens). Cytochromes of the c4-type have been reported to donate electrons to cbb3-type cytochrome c oxidases in various oxygen-respiring bacteria (32–34), and we therefore consider the possibility that TsdB serves as a direct electron donor to cbb3 oxidase at least in tetrathionate-forming thiosulfate oxidizers that neither contain HiPIP nor any cytochrome c4 homolog except TsdB.
We have determined the first three-dimensional structure of M. purpuratum TsdBA, where TsdA is fused with its electron acceptor TsdB. It showed heme arrangement with characteristic class I c-type cytochrome topology, unveiling their relative heme spatial disposition and providing insights into the electron flow during enzymatic reaction. In the MpTsdBA structure, a thiosulfate ion is covalently bound to Sγ of Cys330 in heme 3, although the protein was produced in and purified from E. coli without the addition of thiosulfate to media or buffers. This implies high affinity of the enzyme to thiosulfate, which is possibly present in the complex growth medium in very low concentrations. It should be noted that recombinant AvTsdA and also several SoxA proteins have been isolated with the active site cysteine in a partially or fully persulfurated state (2, 35–37). This has been interpreted as indication for temporary binding of thiosulfate and subsequent incomplete catalysis. Just as proposed here, thiosulfate was assumed to originate from the E. coli growth medium (2).
Regardless of its source, the covalent attachment of a complete thiosulfate molecule to the MpTsdBA active site cysteine strongly supports the hypothesis that tetrathionate formation from two thiosulfate molecules proceeds via a rhodanese-like reaction mechanism involving a thiosulfate transfer reaction with a thiosulfate molecule covalently bound to the active site cysteine as an essential intermediate in the catalytic cycle (2, 12). This type of mechanism has been illustrated in detail by Grabarczyk et al. (12) for TsdA from A. vinosum. A rhodanese-like reaction cycle has also repeatedly been depicted and discussed for the closely related SoxXA protein, but it could not be unambiguously proven before (4, 35). The MpTsdBA structure provides conclusive evidence that the reactions catalyzed by TsdA as well as SoxXA enzymes indeed involve a cysteine S-thiosulfonate intermediate that is formed once the first thiosulfate molecule is positioned in the substrate binding pocket by positively charged amino acid side chains (Arg314, Lys316, Arg326, and Arg438 in MpTsdBA, Fig. 8F). The latter also stabilized the cysteine S-thiosulfonate group once it had formed. Formation of the cysteine S-thiosulfonate releases two electrons that reduce the iron atoms of the two hemes in TsdA to the Fe(II) state. Heme reoxidation by an external electron acceptor is then likely to be followed by a thiol-disulfide exchange reaction that proceeds via an attack of the sulfane atom of a second thiosulfate molecule on the thiosulfonate group (12).
We conclude that catalysis of thiosulfate oxidation by Tsd(B)A enzymes and very probably also that by SoxXA proteins involves formation of a covalent adduct between the sulfane sulfur atom of thiosulfate and the Sγ of the active site cysteine. When present, TsdB is the immediate electron acceptor of TsdA. TsdB is very likely able to transfer electrons directly to the cbb3 terminal oxidase. In organisms containing HiPIP, this electron carrier is likely to act as an additional shuttle not only between Tsd(B)A and the terminal oxidase during oxygen respiration but also between Tsd(B)A and the photosynthetic reaction center during photolithotrophic growth in the light.
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions
Table 4 lists the bacterial strains and plasmids used for this study. E. coli BL21 (DE3) was used for recombinant protein production and was grown in LB medium. E. coli DH5α was used for molecular cloning.
TABLE 4.
E. coli strains and plasmids used in this study
| Strains and plasmids | Description | Ref. or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− ϕ80d lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1hsdR17 (rk− mk+) supE44 λ−thi-1 gyrA relA1 | 56 |
| E. coli BL21 (DE3) | F− ompT hsdSB (rB−mB−) gal dcm met(DE3) | Novagen |
| Plasmids | ||
| pEC86 | Cmr, product from pEC66 and pACYC184 with E. coli ccmABCDEFGH genes | 40 |
| pET-22b (+) | Apr, T7 promoter, lac operator, C-terminal His tag, pelB leader | Novagen |
| pASK-IBA3 plus | Apr, tetA promoter/operator, C-terminal Strep tag | IBA (Göttingen) |
| pET_ MarpuDRAFT_1194 | Apr, tsdA from M. purpuratum (Marpu_02550) was cloned into pET-22b (+) with NdeI and XhoI, C-terminal His tag | 7 |
| pASK-IBATint_tsdB | Apr; tsdB from T. intermedia cloned into pASK-IBA3 plus with BsaI, C-terminal Strep tag | 1 |
| pASK-IBA3plus-slit1877 | Apr; tsdB from S. lithotrophicus (Slit_1877) cloned into pASK-IBA3 plus with BsaI, C-terminal Strep tag | This study |
| pASK-IBA3plus-slit1878 | Apr; tsdA from S. lithotrophicus (Slit_1878) cloned into pASK-IBA3 plus with BsaI, C-terminal Strep tag | This study |
| pASK-IBA3plus-slit1877-slit1878 | Apr; tsdBA from S. lithotrophica (Slit_1877-Slit_1878) cloned into pASK-IBA3 plus with BsaI, C-terminal Strep tag | This study |
| pET-soxXAK-Strep | Apr, soxXAK, including fragment cloned into pET-22b(+) together with C-terminal Strep tag by use of XbaI and HindIII | This study |
| pET-Alvin2274-C-Strep | Apr, hip gene coding for HiPIP from A. vinosum (Alvin_2274) cloned into pET-soxXAK-Strep with NdeI and NcoI, C-terminal Strep tag | This study |
| pPR-IBAAvtsdA | Apr; tsdA from A. vinosum (Alvin_0091) cloned into pASK-IBA3, C-terminal Strep tag | 1 |
| pASK-IBA3plus_Alvin_2879 | Apr; Alvin_2879 from A. vinosum cloned into pASK-IBA3 plus with BsaI, C-terminal Strep tag | This study |
Recombinant DNA Techniques
All general molecular genetics techniques were described earlier (38). Restriction enzymes, T4 ligase, and Pfu DNA polymerase were obtained from Thermo Scientific (Schwerte, Germany) and used according to the manufacturer's instructions. Oligonucleotides for cloning were obtained from Eurofins MWG (Ebersberg, Germany).
Construction of Expression Plasmids
A. vinosum tsdA and T. intermedia tsdB genes coding for the mature proteins without the signal peptides were amplified and cloned as described earlier (1). The tsdA gene (Slit_1878), the tsdB gene (Slit_1877), and the tsdBA gene combination (Slit_1877-Slit1878) from S. lithotrophicus ES-1 (ATCC 700298T) were amplified from genomic DNA with primers Slit1877_fw/Slit1877_rev, Slit1878_fw/Slit1878_rev, and Slit1877_fw/Slit1878_rev (Table 5), respectively. The native signal peptides encoding sequences were included in all three cases. The tsdBA gene fusion (Marpu_02550) from M. purpuratum 984 (DSM 1591T) was amplified without its original signal peptide encoding sequence using primers MarpuDR1194_for/MarpuDR1194_rev. The gene Alvin_2879, encoding a cytochrome c4 with similarity to TsdB, was amplified with primers 2879+Sp-for and 2879-rev such that the signal peptide-encoding sequence was included. The HiPIP-encoding hip (Alvin_2274) gene (39) was amplified without the signal peptide-encoding sequence applying primers Alvin2274-C-strep_for/Alvin2274-C-strep_rev. Chromosomal DNA from A. vinosum DSM 180T served as the template. For cloning of SltsdB, SltsdA, SltsdB-tsdA, and Alvin_2879, primers included BsaI restriction sites, and the digested PCR products were cloned into BsaI-digested pASK-IBA3plus (IBA, Göttingen) resulting in vectors pASK-IBA3plus-slit1877, pASK-IBA3plus-slit1878, pASK-IBA3plus-slit1877-slit1878, and pASK-IBA3plus_Alvin_2879. For the cloning of MptsdBA into pET-22b(+) (Novagen), the restriction enzymes NdeI and XhoI were used yielding plasmid pET_MarpuDRAFT_1194. Plasmid pET-Alvin2274 was constructed by cloning Avhip into a modified pET-22b(+) vector (pET-soxXAK-strep) encoding a C-terminal Strep tag. This vector had previously been constructed in the course of cloning the A. vinosum soxXAK genes. PCR primers with NdeI (XAK-NdeI-for) and NcoI (XAK-NcoI-rev) sites were used, and the resulting fragment was cloned into pET-22b(+). The XbaI/NcoI fragment of the resulting plasmid was cloned into pASK-IBA3. XbaI and HindIII served for excising the soxXAK genes together with the Strep tag-encoding sequence. The XbaI/HindIII fragment was cloned between the XbaI and HindIII sites of pET22b(+) giving pET-soxXAK-strep. Replacing the soxXAK genes in this construct by Avhip yielded A. vinosum HiPIP fused to a C-terminal Strep tag.
TABLE 5.
Primers used in this study
| Primer | Sequence 5′–3′ | Ref. |
|---|---|---|
| MarpuDR1194_for | CGGAGGGATCCTCATATGACGCATCTC | 7 |
| MarpuDR1194_rev | GACCTGCTCGAGATCCTTGGC | 7 |
| Slit1877_fw | ATGGTAGGTCTCAAATGAAGCAAATATTACTAGCAGCATTAAC | This study |
| Slit1877_rev | ATGGTAGGTCTCAGCGCTTTTCTGGTTTCCATTGGTTGATTGT | This study |
| Slit1878_fw | ATGGTAGGTCTCAAATGAAGAATCCCATCGCTATCGCCAT | This study |
| Slit1878_rev | ATGGTAGGTCTCAGCGCTCTTTGCTGCAGTCTGGTGCTTTC | This study |
| XAK-NdeI-for | GGAGATTTCATATGCCGTTGAACGTCTCACACCG | This study |
| XAK-NcoI-rev | ATGGCTCCATGGTATCGAGACCGATCGAGC | This study |
| Alvin2274-C-strep_for | GCCCATATGTCCGCTCCCGCCAAT | This study |
| Alvin2274-C-strep_rev | CAACGGCCCATGGCCGGCCTTCAG | This study |
| 2879+Sp-for | ATGGTAGGTCTCAAATGAAGAAGACTTGGCTGACAACGGT | This study |
| 2879-rev | ATGGTAGGTCTCAGCGCTCTTCGACAGGCCCTGGATGTAC | This study |
Overproduction, Purification, and Preparation of Recombinant Proteins
AvTsdA and TiTsdB were produced as described before (1). For production of S. lithotrophicus TsdA, SlTsdB and the simultaneous production of SlTsdB and SlTdsA E. coli BL21(DE3) cells containing pASK-IBA3plus-slit1877, pASK-IBA3plus-slit1878, or pASK-IBA3plus-slit1877-slit1878 and pEC86 (40) were cultured in 700 ml of LB media supplemented with 100 μg ml−1 ampicillin and 25 μg ml−1 chloramphenicol at 37 °C and 180 rpm after inoculation with an overnight pre-inoculum in a (1:50) dilution. At an OD600 nm of 0.4 to 0.6, 200 ng ml−1 anhydrotetracycline were added, and the appropriate culture was switched to 25 °C and 90 rpm in case of TsdB production. Cells were harvested after 18 h. MpTsdBA and HiPIP were produced in E. coli BL21(DE3) cells containing pET_MarpuDRAFT_1194 or pET-Alvin2274 and pEC86 (40). After 2 or 0.5% inoculation with a pre-culture, the cells were grown in 700 ml of LB medium containing 100 μg ml−1 ampicillin and 25 μg ml−1 chloramphenicol at 37 °C and 180 rpm. At an OD600 nm of 0.5 to 0.6, the cultures were switched to 25 °C and 120 rpm for about 18 h. For production of AvCyt c4 (Alvin_2879), E. coli BL21(DE3) cells containing pASK-IBA3plus_Alvin_2879 and pEC86 (40) were cultured in 400 ml of LB medium, 100 μg ml−1 ampicillin, and 25 μg ml−1 chloramphenicol at 37 °C and 180 rpm after 2% inoculation with a preculture. At OD600 nm ∼0.5, 200 ng ml−1 anhydrotetracycline were added, and the culture was switched to 25 °C and 90 rpm for 18 h. Harvested cells were resuspended in 100 mm Tris-HCl buffer, pH 8.0, containing 150 mm NaCl and lysed by sonication. After removal of insoluble cell material by centrifugation (10,000 × g for 25 min at 4 °C), SlTsdA, SlTsdB, SlTsdB + TsdA, A. vinosum cytochrome c4 (Alvin_2879), and AvTsdA were purified by Strep-Tactin (IBA, Göttingen, Germany) affinity chromatography according to the manufacturer's instructions. MpTsdBA was purified by nickel-chelate (Qiagen, Hilden, Germany) affinity chromatography according to the manufacturer's instructions and then subjected to a size-exclusion chromatography step performed on a HiLoad 16/60 Superdex 75-pg column (GE Healthcare) using an ÄKTApurifier system (GE Healthcare). The column was equilibrated with 20 mm Tris-HCl buffer, pH 7.5, and 150 mm NaCl. TsdB and TsdA from S. lithtrophicus were analyzed by the same procedure either separately or as a mixture of both proteins. In this case, the Superdex 75 column was equilibrated with 100 mm Tris-HCl buffer, pH 7.5, and 150 mm NaCl. The column was calibrated with the molecular weight marker kit MW-GF-70 (GE Healthcare). All purified proteins were desalted with 5 ml of HiTrap Desalting columns (GE Healthcare) and concentrated with Amicon Ultra-15 centrifugal filter units (Merck Millipore). Recombinant S. lithotrophicus proteins were stored in 100 mm sodium acetate buffer pH 5 at −70 °C, A. vinosum Cyt c4 in 20 mm Tris-HCl buffer, pH 7.5, at 4 °C, MpTsdBA in 20 mm Tris-HCl buffer, pH 7.5, with 150 mm NaCl at −70 °C, and HiPIP in 20 mm Tris-HCl, pH 7, at 4 °C. The concentration of purified proteins was determined with the BCA kit from Pierce. For assessment of purity, SDS-PAGE was performed, and the proteins were visualized either by Coomassie or heme staining techniques.
UV-visible Spectroscopy with TsdA in Solution
UV-visible spectra were recorded between 250 and 750 nm with an Analytik Jena Specord 210 (Analytik Jena, Jena, Germany).
Assay of Thiosulfate Oxidase Activity with Ferricyanide
Thiosulfate-dependent ferricyanide reduction was measured by following the decrease in absorbance at 420 nm (ϵ = 1.09 mm−1 cm−1). Enzyme activity measurements with AvTsdA at pH 4 are described in Ref. 2. Activity measurements with MpTsdBA were performed with 1 mm ferricyanide at 25 °C in 100 mm ammonium acetate buffer, pH 5.2, with 200 mm NaCl. Assays were started by addition of TsdA, and data were recorded in a Specord 210 spectrophotometer (Analytik Jena, Jena, Germany). Activity is expressed as micromoles of tetrathionate produced per min and milligram of protein on the basis of one tetrathionate formed per two ferricyanides reduced. In the case of enzymes that use two molecules of the same substrate (here thiosulfate) primary v versus [S] plots provide the best way to examine the data (41). Data were fitted to the empirical Hill equation (Equation 1) using GraphPad Prism (version 6; GraphPad).
| (Eq. 1) |
The Hill equation resembles the classical Henri-Michaelis-Menten equation; however, the n term allows accounting for non-hyperbolic shapes. A substrate concentration [S]0.5 can be reported that yields half-maximal velocity and is characteristic of the process. The constant K, which is not equivalent to Km, characterizes enzyme-substrate interaction. The relationship between K and [S]0.5 is K = [S]0.5n.
Assay of Thiosulfate Oxidase Activity with HiPIP
For assays of electron transfer from thiosulfate to the electron acceptor HiPIP, 10 μm HiPIP preoxidized with 40 μm ferricyanide were used. The reaction was started by addition of enzyme and followed by the absorbance decrease at 480 nm. A molar extinction coefficient at 480 nm of 10.7 mm−1 cm−1 (10) was used. Measurements with AvTsdA were performed in 100 mm ammonium acetate buffer, pH 5, at 30 °C and with MpTsdBA in 100 mm ammonium acetate buffer, pH 5.2, with 200 mm NaCl at 25 °C.
Assay of Thiosulfate Oxidase Activity with TsdB or AvCyt c4
Thiosulfate-dependent reduction of T. intermedia TsdB or A. vinosum Cyt c4 was measured by following the increase in absorbance at 417 nm (ϵΔ417 nm = 99 mm−1 cm−1) for TiTsdB or SlTsdB and at 420 nm (ϵΔ420 nm = 55 mm−1 cm−1 (9)) for AvCyt c4. The extinction coefficient for TsdB was calculated with the help of the Beer-Lambert law using distinct concentrations of TiTsdB and the differences in absorbance at 417 nm in the reduced and oxidized spectra. A value averaged from measurements with three different protein concentrations was derived. Assays of SlTsdA activity with SlTsdB as electron acceptor were carried out in 100 mm ammonium acetate buffer, pH 4, at 25 °C. AvTsdA activity with TiTsdB was assayed in 100 mm ammonium acetate buffer, pH 5, at 30 °C, whereas AvTsdA activity with AvCyt c4 was determined in 100 mm ammonium acetate buffer, pH 5.5, at 25 °C.
Determination of Redox Properties of AvTsdA Adsorbed on a Mesoporous Nanocrystalline SnO2 Electrode
An optically transparent mesoporous nanocrystalline SnO2 electrode coated with AvTsdA was prepared using the previously described method (42) with adsorption from a solution of 10 μm AvTsdA, 2 mm neomycin, 50 mm NaCl, 50 mm HEPES, pH 7. The enzyme-coated electrode was rinsed with 2 mm neomycin, 50 mm NaCl, 50 mm HEPES, pH 7, to remove unbound protein, taken into a N2-filled chamber (atmospheric O2 < 2 ppm) and immersed in an anaerobic solution of the same composition within a previously described spectroelectrochemical cell (42). The cell was sealed, removed from the anaerobic chamber, and inserted into a Jasco V650 UV-visible spectrophotometer thermostated at 4 °C and flushed with argon to maintain anaerobic status. Spectral contributions from light scattering by the electrode were minimized by placing a bare SnO2 electrode (i.e. having no adsorbed enzyme) in the reference beam of the spectrophotometer. The electronic absorbance of the as prepared enzyme-coated electrode revealed features indicative of a mixture of ferric and ferrous hemes. After the electrode had been poised at +302 mV for 45 min, the spectrum revealed that the enzyme had been converted to the fully oxidized all ferric state. To determine the redox activity of AvTsdA, the electrode potential was swept from +302 to −648 mV at a scan rate of 5 mV s−1 with a pause of 150 s every 50 mV. At 60 s into each pause, a spectrum was measured before the scan continued. Reoxidation of the sample was performed in a similar manner. Spectra are presented after equating absolute absorbance at 600 nm to account for potential dependent changes in the spectral contributions that arise from scattering by the electrode material.
Redox Potentiometry with TsdB in Solution Measured with a Gold Electrode
The reduction potential of TiTsdB was measured with help of a gold-platinum electrode system under anoxic conditions. The electrode extended into a cuvette containing the protein solution (20 μm) and redox mediators (N,N-dimethyl-1,4-phenylenediamine, p-benzoquinone, trimethylhydroquinone, phenazine, 1,4-naphthoquinone, and 1,2-naphthoquinone at 2 μm each) in 20 mm MOPS buffer, pH 6, and was connected to a potentiometer. TiTsdB was reduced by changing the applied potential from −150 to 450 mV. Subsequential decrease of the potential again to −150 mV led to reoxidation of the protein. A spectrum was recorded every 2 min and potentials converted to values versus standard hydrogen electrode.
Crystallization, Data Collection, Structure Determination, and Refinement
MpTsdBA at a concentration of 3.2 mg ml−1 in 20 mm BisTris-HCl, pH 6.5, and 150 mm NaCl was crystallized in 10% (w/v) PEG 8000, 0.1 m Tris-HCl, pH 7.0, 0.2 m MgCl2, and 10 mm trimethylamine hydrochloride (as additive) by vapor-diffusion hanging-drop method at 20 °C. Crystallization droplets contained 1.0 μl of protein, 0.8 μl of precipitant, and 0.2 μl of additive and were equilibrated against a 200-μl reservoir solution (26% (w/v) PEG 3350). Crystals were cryoprotected with No. 2 solution of CryoProtX screen (Molecular Dimensions), consisting of 25% (v/v) diethylene glycol, 25% (v/v) 1,2-propanediol, and 25% (v/v) glycerol. X-ray diffraction data were collected at a wavelength of 1.7236 Å on beamline ID-29 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Data were indexed and integrated with XDS (43), and the space group was determined with POINTLESS (44) and scaled with AIMLESS (45, 46), all within the autoPROC (47) data processing pipeline. An Rfree flag was created at this stage corresponding to 5% of the measured reflections of the data set. The structure was determined by single wavelength anomalous dispersion method around the iron edge (Fe-SAD), employing a high multiplicity data collection strategy using the autoSHARP module (48), within the SHARP package (49). Iterative model building and refinement cycles were performed with COOT (50) and BUSTER-TNT (51) (at early stages of refinement), followed by phenix.refine (52), until a complete model was built and refinement convergence achieved. Friedel mates were kept separately and refinement was carried out against I(+)/SIGI(+),I(−)/SIGI(−). A polder map (an omit map that excludes the bulk solvent around the omitted region) and m|Fo| − m|Fo| map were calculated within the PHENIX package of programs. The Ramachandran diagram was assessed with RAMPAGE (53), and the model was validated with MolProbity (54) as implemented in PHENIX. All figures were rendered with PyMOL, Schrödinger LLC (55).
Author Contributions
J. M. K., J. A. B., J. N. B., M. A., and C. D. wrote the manuscript. C. D. conceived and coordinated all experiments except the MpTsdBA crystallization and determination of reduction potentials. J. M. K. analyzed and compiled the data for those experiments. J. R. constructed the vector for production of MpTsdBA and T. F. the vector for production of HiPIP. A. F. and J. R. performed activity assays with MpTsdBA, AvTsdA, and HiPIP (Fig. 9; Table 2) and recorded UV-visible spectra of MpTsdBA (Fig. 6). T. K. produced AvCyt c4 and measured activity of AvTsdA with this protein. E. K. and J. M. K. produced and analyzed proteins from S. lithotrophicus (Figs. 2 and 5). K. D. determined the redox activity of TiTsdB (Fig. 4) under the supervision of I. A. C. P. and S. F. R. examined the redox activity of AvTsdA (Fig. 1) under the supervision of J. N. B. J. A. B. crystallized MpTsdBA, processed the X-ray data, determined the crystal structure, and performed model building and refinement (Figs. 7 and 8; Table 1). J. A. B. and M. A. analyzed the crystal structure.
Acknowledgments
We acknowledge Isabel Bento and Ana Maria Gonçalves for collecting the X-ray diffraction data. We also acknowledge Susana Gonçalves and the ID-29 beamline staff at the European Synchrotron Radiation Facility (ESRF; Grenoble, France) for providing assistance in using the beamline. S. lithotrophicus DNA was kindly provided by David Emerson, Bigelow Laboratory for Ocean Sciences, West Boothbay, ME. MALDI-TOF mass spectrometry of TiTsdB and SlTsdB was kindly performed by Michaele Josten and Hans Georg Sahl, Institute for Medical Microbiology, Immunology and Parasitology, University of Bonn, Germany. We thank James Durrant (Imperial College London) for the SnO2 electrodes.
This work was supported by Deutsche Forschungsgemeinschaft Grant Da 351/7-2; Fundação para a Ciência e Tecnologia through iNOVA4Health Research Unit, LISBOA-01-0145-FEDER-007344, co-funded by Fundação para a Ciência e a Tecnologia do Ministério para a Ciência e Ensino Superior (FCT/MCES) and Fonds Européen de Developpement Économique et Régional (FEDER) under the PT2020 Partnership Agreement through the Research and Development Unit; UID/Multi/04551/2013 (GreenIT); Bio-Struct-X Proposal 1493; MostMicro Unit by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), and by National Funds through FCT.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 5LO9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HiPIP
- high potential iron-sulfur protein
- PDB
- Protein Data Bank
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.d.
- root mean square deviation.
References
- 1. Denkmann K., Grein F., Zigann R., Siemen A., Bergmann J., van Helmont S., Nicolai A., Pereira I. A., and Dahl C. (2012) Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688 [DOI] [PubMed] [Google Scholar]
- 2. Brito J. A., Denkmann K., Pereira I. A., Archer M., and Dahl C. (2015) Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pires R. H., Venceslau S. S., Morais F., Teixeira M., Xavier A. V., and Pereira I. A. (2006) Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex–a membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry 45, 249–262 [DOI] [PubMed] [Google Scholar]
- 4. Bradley J. M., Marritt S. J., Kihlken M. A., Haynes K., Hemmings A. M., Berks B. C., Cheesman M. R., and Butt J. N. (2012) Redox and chemical activities of the hemes in the sulfur oxidation pathway enzyme SoxAX. J. Biol. Chem. 287, 40350–40359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reijerse E. J., Sommerhalter M., Hellwig P., Quentmeier A., Rother D., Laurich C., Bothe E., Lubitz W., and Friedrich C. G. (2007) The unusual redox centers of SoxXA, a novel c-type heme-enzyme essential for chemotrophic sulfur-oxidation of Paracoccus pantotrophus. Biochemistry 46, 7804–7810 [DOI] [PubMed] [Google Scholar]
- 6. Kappler U., Bernhardt P. V., Kilmartin J., Riley M. J., Teschner J., McKenzie K. J., and Hanson G. R. (2008) SoxAX cytochromes, a new type of heme copper protein involved in bacterial energy generation from sulfur compounds. J. Biol. Chem. 283, 22206–22214 [DOI] [PubMed] [Google Scholar]
- 7. Kurth J. M., Dahl C., and Butt J. N. (2015) Catalytic protein film electrochemistry provides a direct measure of the tetrathionate/thiosulfate reduction potential. J. Am. Chem. Soc. 137, 13232–13235 [DOI] [PubMed] [Google Scholar]
- 8. Liu Y.-W., Denkmann K., Kosciow K., Dahl C., and Kelly D. J. (2013) Tetrathionate stimulated growth of Campylobacter jejuni identifies TsdA as a new type of bi-functional tetrathionate reductase that is widely distributed in bacteria. Mol. Microbiol. 88, 173–188 [DOI] [PubMed] [Google Scholar]
- 9. Cusanovich M. A., and Bartsch R. G. (1969) A high potential cytochrome c from Chromatium vinosum chromatophores. Biochim. Biophys. Acta 189, 245–255 [DOI] [PubMed] [Google Scholar]
- 10. Bartsch R. G. (1978) Purification of (4Fe-4S)1–2− ferredoxins (high-potential iron-sulfur proteins) from bacteria. Methods Enzymol. 53, 329–340 [DOI] [PubMed] [Google Scholar]
- 11. Fukumori Y., and Yamanaka T. (1979) A high-potential nonheme iron protein (HiPIP)-linked, thiosulfate-oxidizing enzyme derived from Chromatium vinosum. Curr. Microbiol. 3, 117–120 [Google Scholar]
- 12. Grabarczyk D. B., Chappell P. E., Eisel B., Johnson S., Lea S. M., and Berks B. C. (2015) Mechanism of thiosulfate oxidation in the SoxA family of cysteine-ligated cytochromes. J. Biol. Chem. 290, 9209–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du J., Sono M., and Dawson J. H. (2011) The H93G myoglobin cavity mutant as a versatile scaffold for modeling heme iron coordination structures in protein active sites and their characterization with magnetic circular dichroism spectroscopy. Coord. Chem. Rev. 255, 700–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miles C. S., Manson F. D., Reid G. A., and Chapman S. K. (1993) Substitution of a haem-iron axial ligand in flavocytochrome b2. Biochim. Biophys. Acta 1202, 82–86 [DOI] [PubMed] [Google Scholar]
- 15. Branca R. M., Bodó G., Várkonyi Z., Debreczeny M., Osz J., and Bagyinka C. (2007) Oxygen and temperature-dependent structural and redox changes in a novel cytochrome c4 from the purple sulfur bacterium Thiocapsa roseopersicina. Arch. Biochem. Biophys. 467, 174–184 [DOI] [PubMed] [Google Scholar]
- 16. Nissum M., Karlsson J.-J., Ulstrup J., Jensen P. W., and Smulevich G. (1997) Resonance Raman characterization of the di-heme protein cytochrome c4 from Pseudomonas stutzeri. J. Biol. Inorg. Chem. 2, 302–307 [Google Scholar]
- 17. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 18. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Igarashi N., Moriyama H., Fujiwara T., Fukumori Y., and Tanaka N. (1997) The 2.8 Å structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea. Nat. Struct. Biol. 4, 276–284 [DOI] [PubMed] [Google Scholar]
- 20. Taylor P., Pealing S. L., Reid G. A., Chapman S. K., and Walkinshaw M. D. (1999) Structural and mechanistic mapping of a unique fumarate reductase. Nat. Struct. Biol. 6, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 21. Visser J. M., de Jong G. A., Robertson L. A., and Kuenen J. G. (1996) Purification and characterization of a periplasmic thiosulfate dehydrogenase from the obligately autotrophic Thiobacillus sp. W5. Arch. Microbiol. 166, 372–378 [DOI] [PubMed] [Google Scholar]
- 22. Ohmine M., Matsuura K., Shimada K., Alric J., Verméglio A., and Nagashima K. V. (2009) Cytochrome c4 can be involved in the photosynthetic electron transfer system in the purple bacterium Rubrivivax gelatinosus. Biochemistry 48, 9132–9139 [DOI] [PubMed] [Google Scholar]
- 23. Bartsch R. G. (1991) The distribution of soluble metallo-redox proteins in purple phototrophic bacteria. Biochim. Biophys. Acta 1058, 28–30 [DOI] [PubMed] [Google Scholar]
- 24. Kennel S. J., Bartsch R. G., and Kamen M. D. (1972) Observations on light-induced oxidation reactions in the electron transport system of Chromatium. Biophys. J. 12, 882–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagashima K. V., Matsuura K., Shimada K., and Verméglio A. (2002) High-potential iron-sulfur protein (HiPIP) is the major electron donor to the reaction center complex in photosynthetically growing cells of the purple bacterium Rubrivivax gelatinosus. Biochemistry 41, 14028–14032 [DOI] [PubMed] [Google Scholar]
- 26. Lieutaud C., Nitschke W., Verméglio A., Parot P., and Schoepp-Cothenet B. (2003) HiPIP in Rubrivivax gelatinosus is firmly associated to the membrane in a conformation efficient for electron transfer towards the photosynthetic reaction centre. Biochim. Biophys. Acta 1557, 83–90 [DOI] [PubMed] [Google Scholar]
- 27. Verméglio A., Li J., Schoepp-Cothenet B., Pratt N., and Knaff D. B. (2002) The role of high-potential iron protein and cytochrome c8 as alternative electron donors to the reaction center of Chromatium vinosum. Biochemistry 41, 8868–8875 [DOI] [PubMed] [Google Scholar]
- 28. Kämpf C., and Pfennig N. (1980) Capacity of Chromatiaceae for chemotrophic growth. Specific respiration rates of Thiocystis violacea and Chromatium vinosum. Arch. Microbiol. 127, 125–135 [Google Scholar]
- 29. Hochkoeppler A., Jenney F. E. Jr., Lang S. E., Zannoni D., and Daldal F. (1995) Membrane-associated cytochrome cγ of Rhodobacter capsulatus is an electron carrier from the cytochrome bc1 complex to the cytochrome c oxidase during respiration. J. Bacteriol. 177, 608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonora P., Principi I. I., Monti B., Ciurli S., Zannoni D., and Hochkoeppler A. (1999) On the role of high-potential iron-sulfur proteins and cytochromes in the respiratory chain of two facultative phototrophs. Biochim. Biophys. Acta 1410, 51–60 [DOI] [PubMed] [Google Scholar]
- 31. Beckwith C. R., Edwards M. J., Lawes M., Shi L., Butt J. N., Richardson D. J., and Clarke T. A. (2015) Characterization of MtoD from Sideroxydans lithotrophicus: a cytochrome c electron shuttle used in lithoautotrophic growth. Front. Microbiol. 6, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arai H., Kawakami T., Osamura T., Hirai T., Sakai Y., and Ishii M. (2014) Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J. Bacteriol. 196, 4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barco R. A., Emerson D., Sylvan J. B., Orcutt B. N., Jacobson Meyers M. E., Ramírez G. A., Zhong J. D., and Edwards K. J. (2015) New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl. Environ. Microbiol. 81, 5927–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang H. Y., Ahn Y., Pace L. A., Lin M. T., Lin Y. H., and Gennis R. B. (2010) The diheme cytochrome c4 from Vibrio cholerae is a natural electron donor to the respiratory cbb3 oxygen reductase. Biochemistry 49, 7494–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bamford V. A., Bruno S., Rasmussen T., Appia-Ayme C., Cheesman M. R., Berks B. C., and Hemmings A. M. (2002) Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 21, 5599–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dambe T., Quentmeier A., Rother D., Friedrich C., and Scheidig A. J. (2005) Structure of the cytochrome complex SoxXA of Paracoccus pantotrophus, a heme enzyme initiating chemotrophic sulfur oxidation. J. Struct. Biol. 152, 229–234 [DOI] [PubMed] [Google Scholar]
- 37. Kilmartin J. R., Maher M. J., Krusong K., Noble C. J., Hanson G. R., Bernhardt P. V., Riley M. J., and Kappler U. (2011) Insights into structure and function of the active site of SoxAX cytochromes. J. Biol. Chem. 286, 24872–24881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dahl C., Schulte A., Stockdreher Y., Hong C., Grimm F., Sander J., Kim R., Kim S.-H., and Shin D. H. (2008) Structural and molecular genetic insight into a wide-spread bacterial sulfur oxidation pathway. J. Mol. Biol. 384, 1287–1300 [DOI] [PubMed] [Google Scholar]
- 39. Brüser T., Trüper H. G., and Dahl C. (1997) Cloning and sequencing of the gene encoding the high potential iron-sulfur protein (HiPIP) from the purple sulfur bacterium Chromatium vinosum. Biochim. Biophys. Acta 1352, 18–22 [DOI] [PubMed] [Google Scholar]
- 40. Arslan E., Schulz H., Zufferey R., Künzler P., and Thöny-Meyer L. (1998) Overproduction of Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251, 744–747 [DOI] [PubMed] [Google Scholar]
- 41. Segel I. H. (1993) Enzyme Kinetics: Behaviour and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, Wiley-Interscience, New York [Google Scholar]
- 42. Marritt S. J., Kemp G. L., Xiaoe L., Durrant J. R., Cheesman M. R., and Butt J. N. (2008) Spectroelectrochemical characterization of a pentaheme cytochrome in solution and as electrocatalytically active films on nanocrystalline metal-oxide electrodes. J. Am. Chem. Soc. 130, 8588–8589 [DOI] [PubMed] [Google Scholar]
- 43. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 46. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vonrhein C., Blanc E., Roversi P., and Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 49. de la Fortelle E., and Bricogne G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 50. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., and Bricogne G. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 52. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lovell S. C., Davis I. W., Arendall W. B. 3rd., de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 54. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Delano W. L. (2002) The PyMOL Molecular Graphics System, version 1.7.2.1, DeLano Scientific, San Carlos, CA [Google Scholar]
- 56. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 57. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]