Abstract
Mammalian cells express three Class II nonmuscle myosins (NM): NM2A, NM2B, and NM2C. The three NM2s have well established essential roles in cell motility, adhesion, and cytokinesis and less well defined roles in vesicle transport and other processes that would require association of NM2s with cell membranes. Previous evidence for the mechanism of NM2-membrane association includes direct interaction of NM2s with membrane lipids and indirect interaction by association of NM2s with membrane-bound F-actin or peripheral membrane proteins. Direct binding of NM2s to phosphatidylserine-liposomes, but not to phosphatidylcholine-liposomes, has been reported, but the molecular basis of the interaction between NM2s and acidic phospholipids has not been previously investigated. We now show that filamentous, full-length NM2A, NM2B, and NM2C and monomeric, non-filamentous heavy meromyosin bind to liposomes containing one or more acidic phospholipids (phosphatidylserine, phosphatidylinositol 4,5-diphosphate, and phosphatidylinositol 3,4,5-triphosphate) but do not bind to 100% phosphatidylcholine-liposomes. Binding of NM2s to acidic liposomes occurs predominantly through interaction of the liposomes with the regulatory light chain (RLC) binding site in the myosin heavy chain with concomitant dissociation of the RLC. Phosphorylation of myosin-bound RLC by myosin light chain kinase substantially inhibits binding to liposomes of both filamentous NM2 and non-filamentous heavy meromyosin; the addition of excess unbound RLC, but not excess unbound essential light chain, competes with liposome binding. Consistent with the in vitro data, we show that endogenous and expressed NM2A associates with the plasma membrane of HeLa cells and fibrosarcoma cells independently of F-actin.
Keywords: cell biology, cell motility, lipid-binding protein, liposome, myosin, phosphatidylserine
Introduction
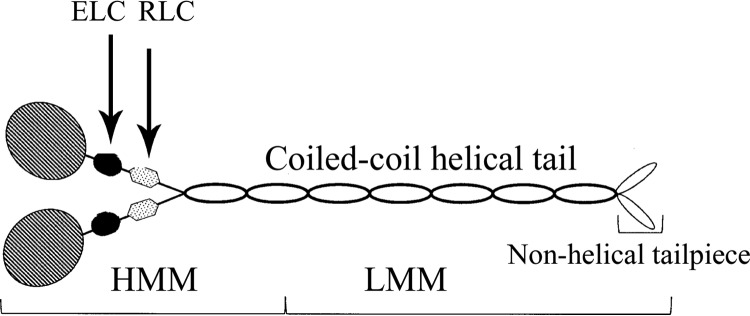
Mammalian nonmuscle cells express three Class II myosins, NM2A,3 NM2B, and NM2C, which, like all Class II myosins (1), consist of six polypeptides: two identical heavy chains, a pair of essential light chains, and a pair of regulatory light chains (see Fig. 1). Each heavy chain has an N-terminal globular motor domain with an ATPase site and an F-actin-binding site followed by a lever arm that binds both an essential light chain and a regulatory light chain. The HC terminates in a long α-helical tail with a short nonhelical tailpiece (1, 2). The helical tails of the two HCs form a stable coiled-coil (2, 3) creating the six-polypeptide molecule, hereafter referred to as the monomer. Monomers assemble via their helical tails forming short (∼300 nm) antiparallel, bipolar filaments consisting of a mean value of 29, 30, and 14 molecules for NM2A, NM2B, and NM2C, respectively (3). The coiled-coil helical tail of NM2 can be cleaved (Fig. 1), producing heavy meromyosin (HMM), a non-polymerizable monomer with actin-activated ATPase activity, and light meromyosin (LMM), which can polymerize but differently than full-length NM2.
FIGURE 1.
Schematic representation of nonmuscle myosin II. NM2 consists of two heavy chains and two pairs of light chains. The two heavy chains have an N-terminal globular head, a helical neck followed by a coiled-coil helical tail, and a C-terminal non-helical tailpiece. The ELCs and the RLCs bind to the neck domains of the heavy chains. NM2 can be proteolytically cleaved into HMM, the two heads and ∼⅓ of the tail domain, and LMM, the C-terminal ∼⅔ of the tail domain.
Interaction of actin filaments with the motor domains of myosin filaments and HMM activates myosin's ATPase activity. The energy produced by ATP hydrolysis is converted into mechanical force by coupled lever movement of the light chain binding regions of the HCs, which allows the motor domains both to translocate actin filaments and move along actin filaments. As described originally for smooth muscle myosin (4–7), actin-activated ATPase activity of NM2s is stimulated by phosphorylation of Thr-18 and/or Ser-19 of the RLC (8–11).
NM2s have well documented essential roles in migration, adhesion, and cytokinesis of nonmuscle cells (12, 13). The functions of NM2s in vesicle transport are less well established, although there is evidence that NM2s are the motors for vesicle transport along actin filaments in clam oocytes (14) and for the involvement of NM2s in the formation and transport of Golgi vesicles (15–20), squid axoplasmic organelles (21), brain neuronal membranes (22, 23), and epithelial membranes (24, 25). However, it is not known whether the associations of NM2s and membranes are mediated by the interaction of NM2s with membrane-associated actin and/or peripheral membrane proteins or if NM2s bind directly to the lipid bilayer. For example, there are reports that binding of NM2 to membranes of mouse forebrains requires interaction of the motor domain of NM2 with F-actin (23), that the tail (and not the motor domain) of single NM2 molecules may bind to the peripheral membrane proteins of Golgi stacks (19) and lytic granules from lymphocytes (26), and, to the contrary, that the association of NM2 with plasma membranes is independent of F-actin and other proteins that can be extracted by 0.1 m Na2CO3 (27) or 0.6 m KI (22).
NM2A and two other proteins from lysates of human breast cancer cells have been shown to bind to PIP3-coated beads in vitro, and NM2A has been shown to co-localize with PIP3 in the plasma membrane of breast cancer cells in situ (28). However, direct interaction between NM2A and PIP3 was not shown in those experiments. The possibility of direct binding of NM2 to membrane anionic lipids is suggested, however, by the observation (22) that purified bovine brain NM2, but neither skeletal nor cardiac muscle myosin II, binds to 100% phosphatidylserine liposomes but not to 100% phosphatidylcholine liposomes. The molecular basis of the binding of NM2 to the PS-liposomes was not explored in those experiments.
In this paper we report that recombinant NM2A, NM2B, and NM2C bind to negatively charged liposomes containing either PS, PIP2, or PIP3 but not to liposomes composed only of PC, that liposome binding depends substantially, but perhaps not exclusively, the RLC-binding site of the HC with concomitant dissociation of the bound RLC, and that NM2A localizes at the plasma membrane of HeLa cells and HT1080 fibrosarcoma cells even when F-actin is depolymerized by latrunculin A.
Results
Full-length NM2 Filaments Bind to Phosphatidylserine Liposomes
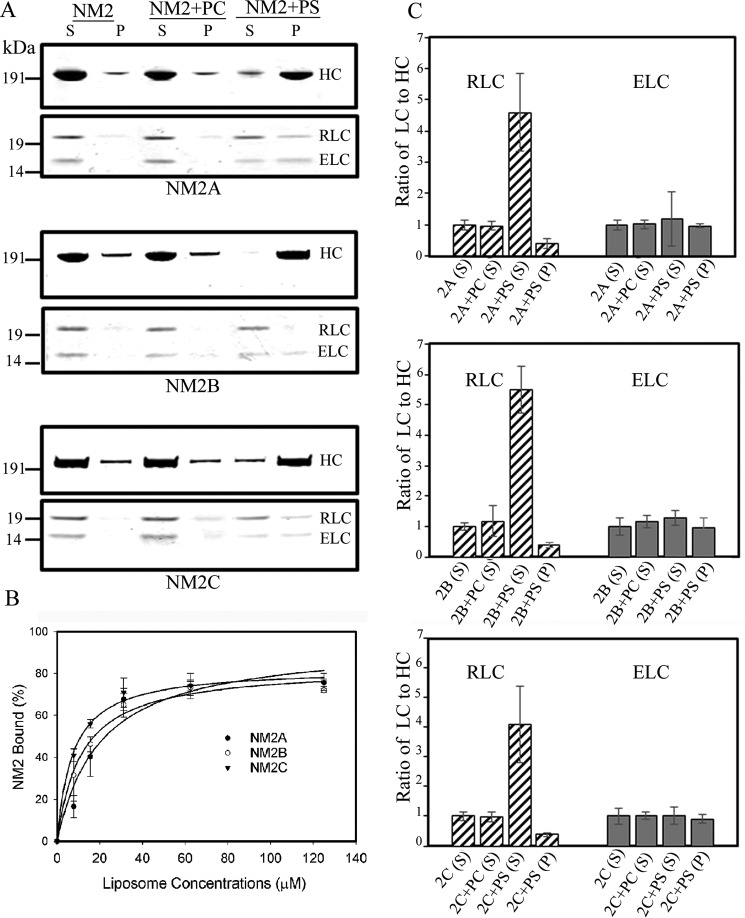
Filaments of full-length NM2A, NM2B, and NM2C were incubated with and without liposomes consisting of 100% PS or 100% PC. The mixtures were centrifuged at 17,000 × g for 20 min (see “Experimental Procedures”), and the amounts of NM2 HC in the supernatants and pellets were determined by SDS-PAGE. None of the three NM2s pelleted when incubated with 100% PC-liposomes; the small amount of HC in the pellet was the same as in the absence of liposomes (Fig. 2A). However, all three myosins pelleted when incubated with 100% PS-liposomes (Fig. 2A) with a maximum of ∼80% of the 100 nm NM2s binding to 60–125 μm PS-liposomes (Fig. 2B).
FIGURE 2.
Filaments of full-length recombinant NM2A, NM2B, and NM2C bind to PS-liposomes displacing the RLC. A, SDS-PAGE of supernatants (S) and pellets (P) after centrifugation for 20 min at 13,000 × g of 100 nm NM2s alone or mixed with 125 μm 100% PC-liposomes (PC) or 100% PS-liposomes (PS). B, binding of NM2s to 100% PS-liposomes as a function of liposome concentration. C, quantitative analysis of the ratios of RLC and ELC to HC in centrifugal supernatants (S) and pellets (P) of NM2s alone or mixed with 100% PC- or PS-liposomes. The ratios of RLC and ELC to HC in the liposome supernatants and pellets are normalized to 1:1 for the ratios of the intensities of the respective bands on SDS-PAGE for myosin in the supernatant of NM2 centrifuged without liposomes.
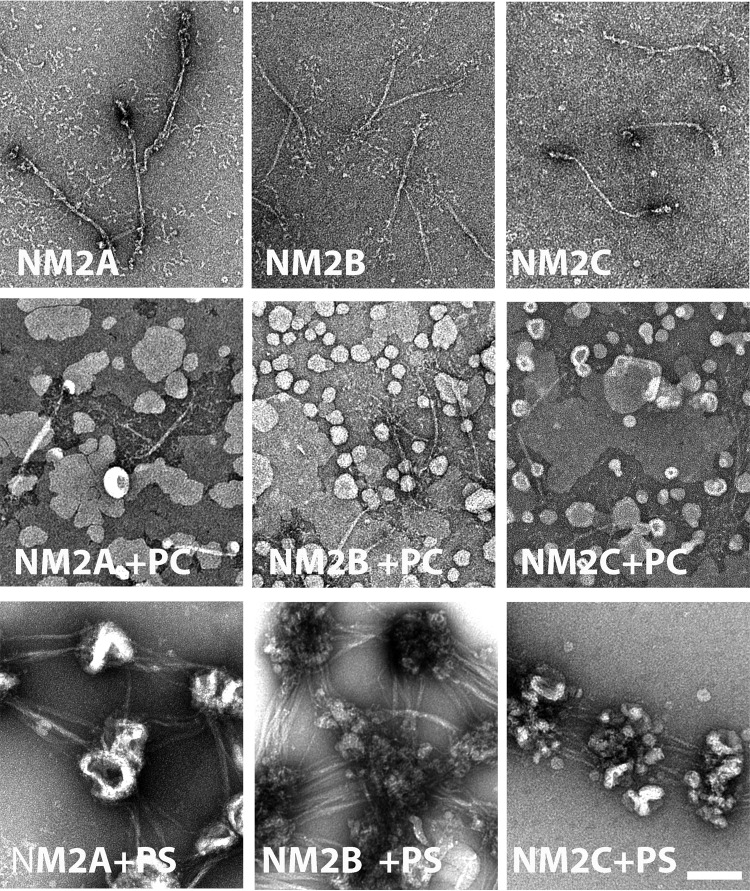
Notably, binding of the NM2s to PS-liposomes was accompanied by substantial displacement of the RLC from the myosin HC (Fig. 2A). The ratios of RLC:HC for NM2s bound to PS-liposomes in the centrifugal pellet were 0.4–0.5:1 (Fig. 2C), and the RLC:HC ratios for the unbound proteins in the centrifugal supernatant were 4–5.5:1 (Fig. 2C), demonstrating the presence of excess free RLC in the supernatant. The ELC:HC ratios remained 1:1 in both the supernatant and pellet. These results suggested that full-length filamentous NM2s bind to PS-liposomes via the RLC-binding site on the HC with the liposomes displacing the RLC but not the ELC. Electron micrographs of mixtures of NM2s and PS-liposomes before centrifugation (Fig. 3) were consistent with this interpretation showing clusters of PS-liposomes bound to the ends of the bipolar NM2 filaments.
FIGURE 3.
Negative staining electron microscopy of liposomes mixed with NM2s. Top row, bipolar filaments of NM2A, NM2B, and NM2C. Middle row, mixtures of NM2s with 100% PC-liposomes (PC) show no liposome-bound filaments. Bottom row, mixture of NM2s with 100% PS-liposomes (PS) show clusters of liposomes bound to the heads of bipolar filaments of NM2A, NM2B, and NM2C. The scale bar represents 100 nm.
Heavy Meromyosin Binds to PS-, PIP2-, and PIP3-liposomes
The average RLC:HC ratio of 0.4–0.5 for liposome-bound full-length NM2s (Fig. 2C) indicated that 50–60% of the heads of the NM2 heavy chains had bound to the 100% PS-liposomes by displacement of the RLC. However, because full-length NM2s are filaments of 14–30 monomers (3), not all of the myosin molecules in the filaments that pelleted with the liposomes were necessarily individually bound to the liposomes. More quantitative data were obtained from the binding to liposomes of monomeric HMM, which does not form filaments (Fig. 4).
FIGURE 4.
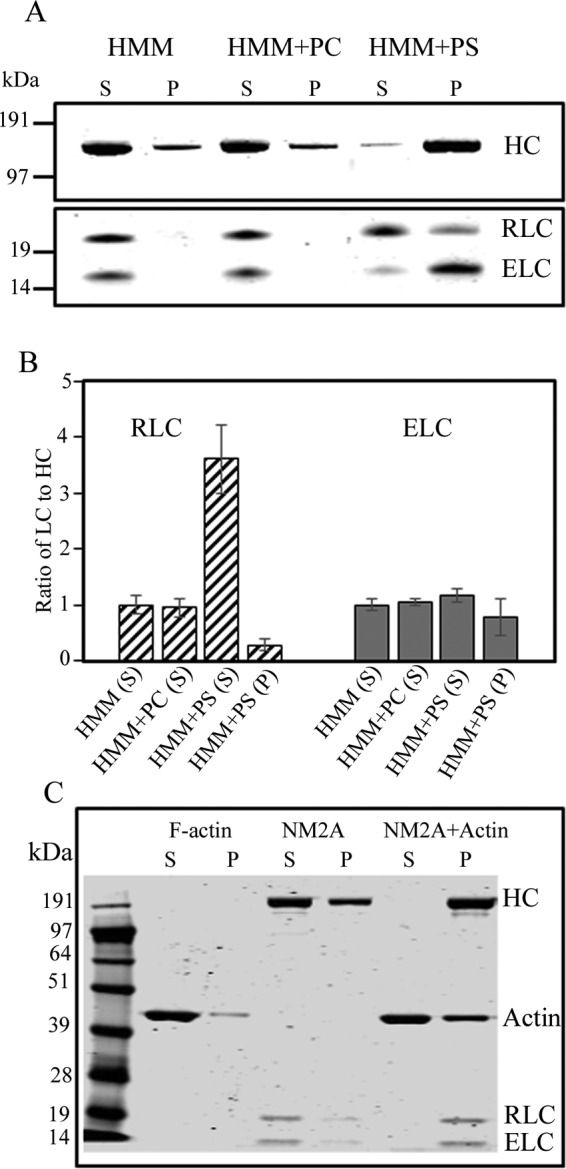
Recombinant NM2A-HMM binds to PS-liposomes with displacement of the RLC. A, purified 100 nm NM2A-HMM (HMM) was incubated with 125 μm 100% PC-liposomes and 125 μm 100% PS-liposomes. The mixtures and NM2-HMM alone were centrifuged at 189,000 × g to separate liposome-bound HMM (P) from unbound HMM (S). B, ratios of RLC and ELC to HC in the supernatants and pellets. The ratios are normalized to 1:1 for HMM alone. The data are the average and S.D. of at least three experiments. C, as a control, full-length NM2A was mixed with muscle F-actin (without liposomes) in a molar ratio of 1:5, and the actomyosin complex pelleted at 13,000 × g for 20 min.
As observed for filaments of full-length myosin (Fig. 2), SDS-PAGE analysis showed no detectable binding of NM2A-HMM to 100% PC-liposomes but substantial NM2A-HMM bound to 100% PS-liposomes (Fig. 4A). The decrease in the RLC:HC ratio to 0.29:1 in the liposome-bound HMM (Fig. 4B, HMM+PS) indicated that both RLCs were displaced from ∼40% of the HMMs. RLC was not displaced when NM2A was bound to, and sedimented with, F-actin (Fig. 4C) showing that displacement of RLC was specific for liposome binding.
Binding of HMM to PS-liposomes increased with both the concentration of liposomes and the percentage of PS in the liposomes (Fig. 5A). As with filaments of full-length NM2A (Fig. 2B), maximum binding of NM2A-HMM occurred at about 120 μm concentrations of PS-liposomes (Fig. 5A). In contrast to HMM, NM2A-LMM (Fig. 1) bound weakly to 100% PS-liposomes, a maximum of ∼20% for LMM compared with ∼80% for HMM, as analyzed by the sedimentation assay (Fig. 5B). No, or very little, binding of LMM was observed by electron microscopy (Fig. 5C).
FIGURE 5.
Quantification of binding of NM2A-HMM and NM2A-LMM to PS-liposomes. A, binding of 100 nm HMM to PS-liposomes as a function of liposome concentration and percentage of PS in liposomes composed of PS and PC. B, 100 nm LMM bound to 100% PS-liposomes much more weakly than 100 nm HMM. C, negatively stained EM images of a mixture of 100 nm LMM filaments and 125 μm PS-liposomes show only a few possibly liposome-bound LMM (arrow). The data in A and B are the average and S.D. of at least three experiments. The bar is 100 nm.
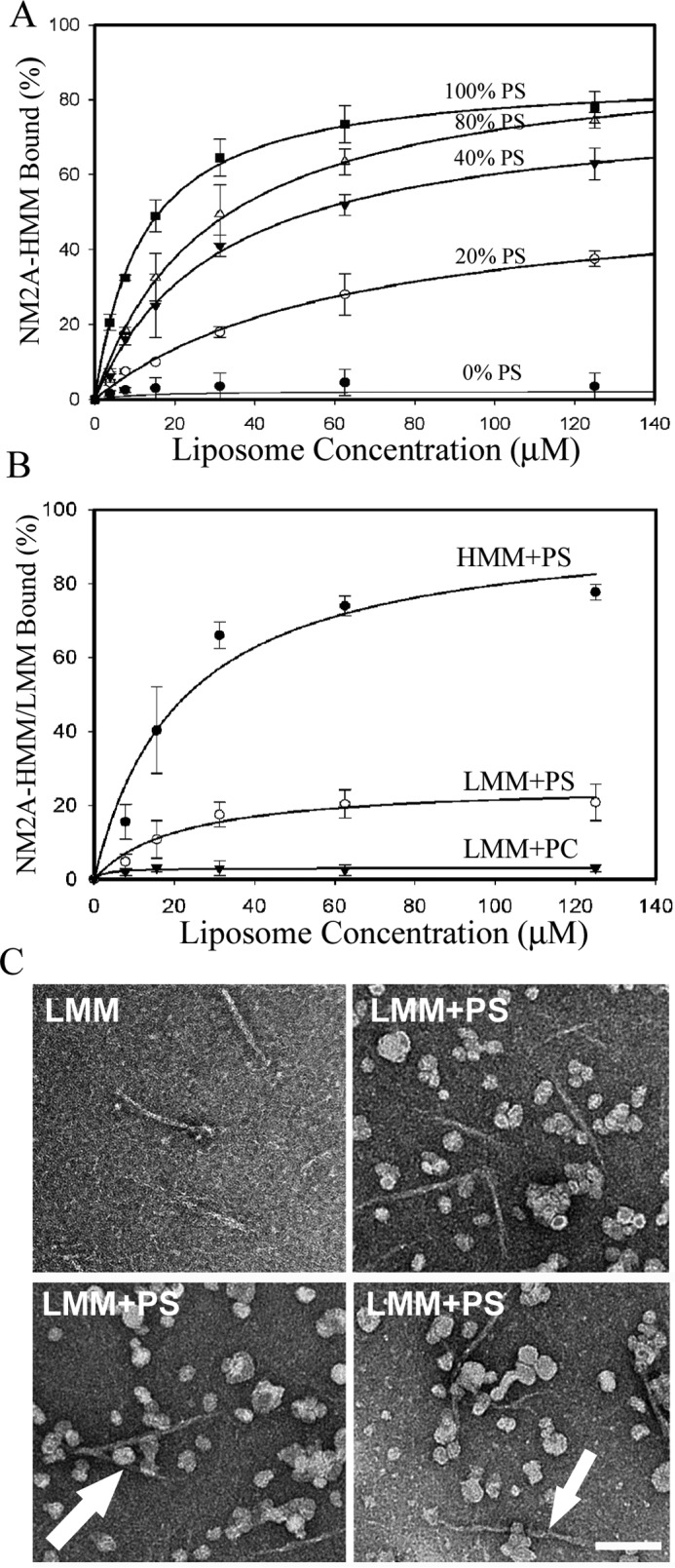
The affinities of NM2A-HMM for PS-, PIP2-, and PIP3-liposomes were proportional to the net negative charges of the phospholipids, assuming that PIP2 has four times and PIP3 six times the negative charge of PS (Fig. 6, A and B). Similarly, NM2A bound to liposomes with the lipid composition of plasma membranes (29, 30) in proportion to their net negative charge (Fig. 7A) and with displacement of the RLC (Fig. 7, B and C).
FIGURE 6.
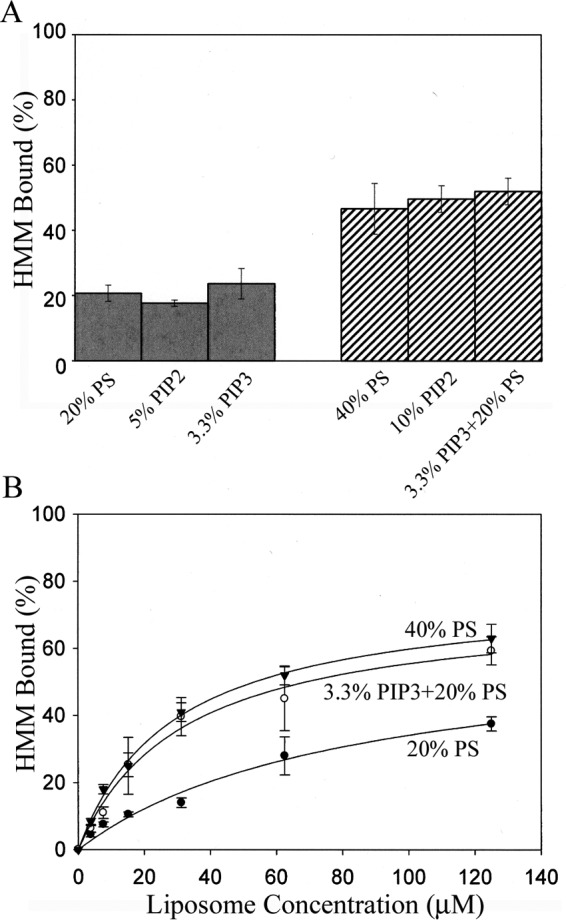
NM2A-HMM binds equally to PS-, PIP2- and PIP3-liposomes with equivalent net negative charges. A, binding of 100 nm NM2A-HMM to 50 μm acidic liposomes with different percentages of PS, PIP2, and PIP3 in PC-liposomes. B, binding of 100 nm HMM to PC-liposomes that contained 20% PS, 40% PS, or 20% PS + 3.3% PIP3 as a function of liposome concentration. The data are the average and S.D. of at least three experiments.
FIGURE 7.
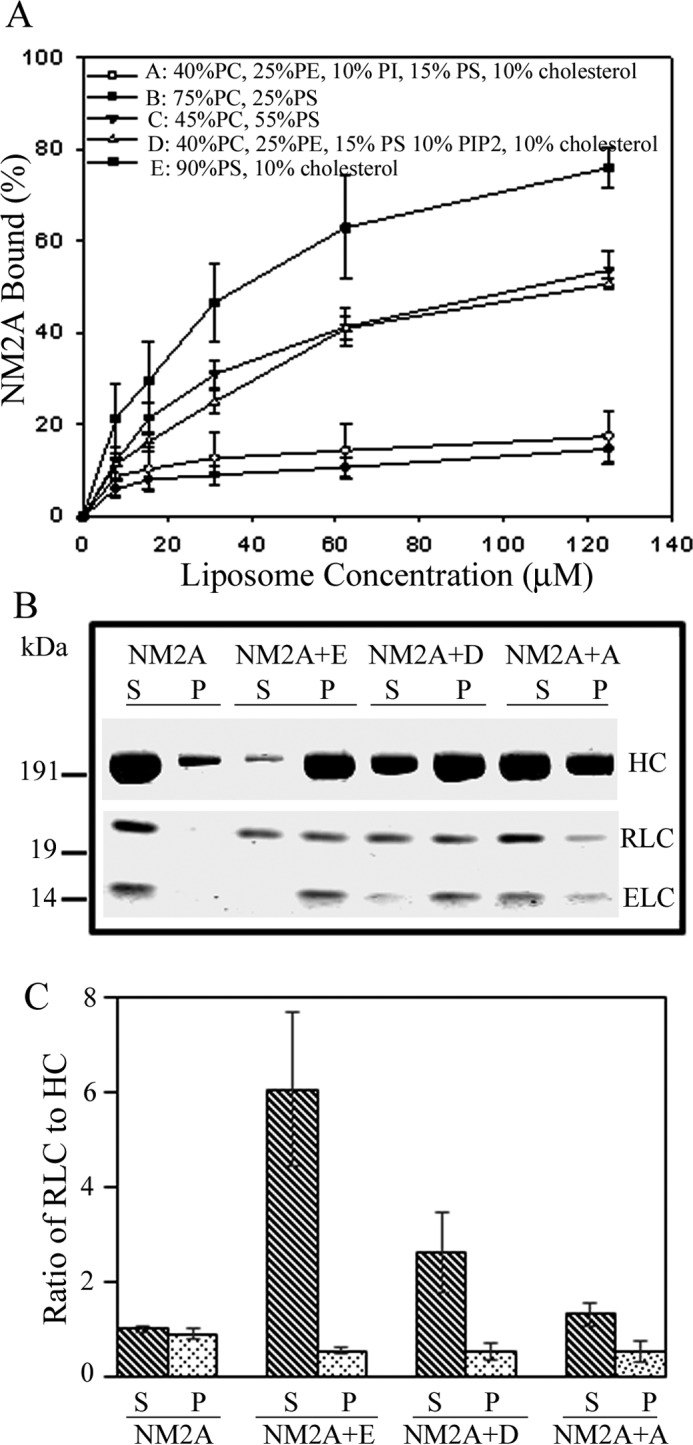
Liposomes with compositions of plasma membrane lipids bind to NM2A with the same affinity as PS-liposomes of similar negative charge and displace RLC. A, binding of 150 nm NM2A to liposomes of the compositions shown in the figure as a function of liposome concentration. B, SDS-PAGE of supernatants (S) and pellets (P) of 150 nm NM2A and samples E, D, and A from panel A centrifuged for 20 min at 13,000 × g. C, ratios of RLC to HC in supernatants (S) and pellets (P) of samples in panel B. The ratios of RLC to HC were normalized to NM2A alone as 1:1. The data in A and C are the average and S.D. of at least three experiments.
Deletion, Addition, and Phosphorylation of RLC Affect Binding of NM2A-HMM to Liposomes
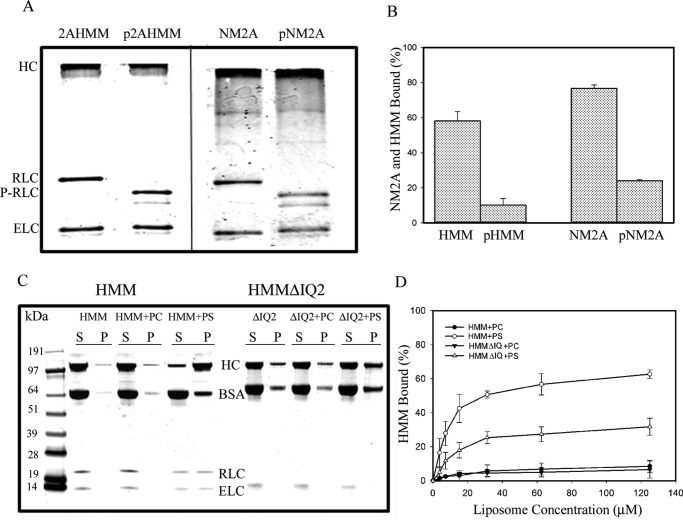
We found that binding of NM2A-HMM to PS-liposomes was enhanced ∼20% when HMM was expressed without the RLC (Fig. 8, A and C). The addition of excess free RLC inhibited binding of HMM to liposomes ∼45%, but the addition of excess free ELC had no effect on HMM-binding to PS-liposomes (Fig. 8, B and C). As neither free RLC nor free ELC bound to PS-liposomes (Fig. 8D), these results are consistent with liposomes competing with RLC for binding to the RLC-binding site on the HC. Phosphorylation of the RLC bound to HMM and to full-length NM2A (Fig. 9A) inhibited binding of both HMM and NM2A to PS-liposomes (Fig. 9B), indicative of pRLC having a higher affinity than RLC for the RLC-binding site, which is consistent with the basic charge of the RLC-binding site (see “Discussion”). Deletion of the RLC-binding site (IQ2 motif) of NM2A-HMM (HMMΔIQ2) reduced by ∼60% (Fig. 9, C and D) but did not eliminate binding of HMM to PS-liposomes (see “Discussion”).
FIGURE 8.
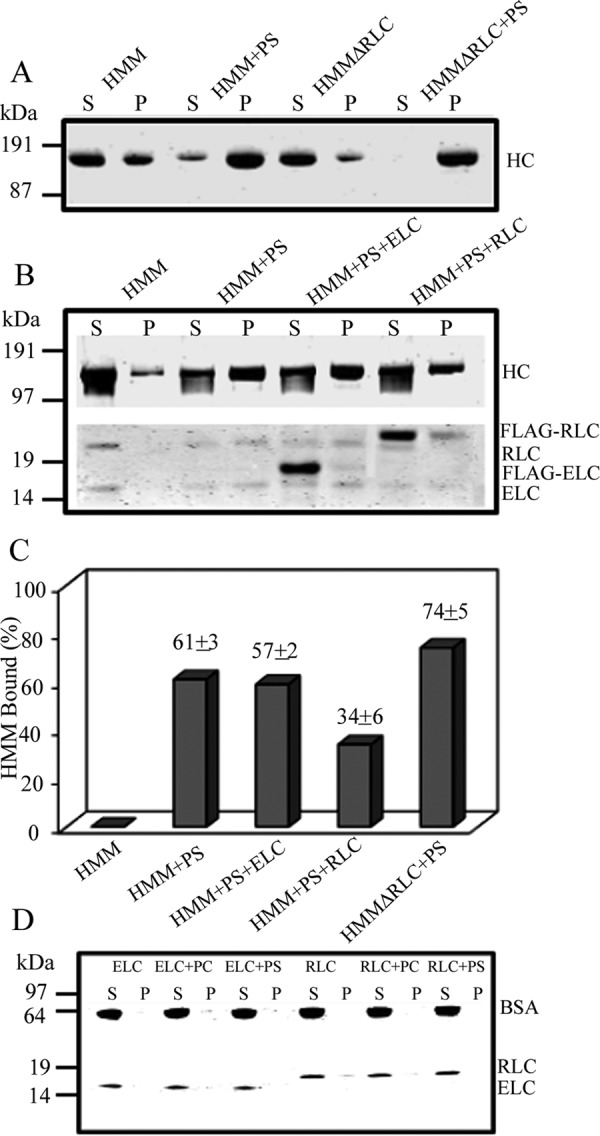
Effect of deletion of RLC or addition of external RLC on binding of HMM (100 nm) to 100% PS-liposomes (30 μm). Deletion of RLC from the recombinant HMM (HMMΔRLC) increased binding by ∼20% (A and C). S, supernatant; P, pellets. The addition of 1 μm unbound external RLC inhibited binding of 100 nm HMM by ∼40%, and the addition of 1 μm ELC did not affect HMM binding (B and C, neither 400 nm recombinant ELC nor 400 nm recombinant RLC alone binds to PS-liposomes (D).
FIGURE 9.
Phosphorylation of myosin-bound RLC and deletion of RLC-binding site affects binding of NM2A-HMM and NM2A to 100% PS-liposomes. A, urea-glycerol gel electrophoretic analysis of phosphorylation of NM2A and NM2A-HMM by myosin light chain kinase. The RLC of both pHMM and pNM2A was fully phosphorylated (P-RLC). The upper pRLC band is monophosphorylated RLC, and the lower band is diphosphorylated RLC. B, RLC phosphorylation almost completely inhibited the binding of 100 nm NM2A and 100 nm NM2A-HMM to 30 μm PS-liposomes. C and D, deletion of the RLC-binding site (HMMΔIQ2) inhibited binding of HMM to PS-liposomes by ∼50%. C, SDS-PAGE of centrifugal supernatant and pellets of 100 nm HMM and HMMΔIQ2 incubated with 125 μm PC- or PS-liposomes. D, 100 nm HMM and HMMΔIQ2 incubated with 125 μm PC- or PS-liposomes. The data in B and D are the average and S.D. of at least three experiments.
NM2A Binds to the Plasma Membrane in Situ
Immunofluorescence microscopy of formaldehyde-fixed cells showed that endogenous NM2A localizes with F-actin in stress fibers and at the plasma membrane of HeLa cells (Fig. 10A). Plasma membrane association of NM2A was independent of actin, however, as the myosin remained associated with the plasma membrane, including on blebs, when actin filaments were depolymerized by latrunculin A and actin was aggregated in the cytoplasm (Fig. 10B). Moreover, expressed NM2A-Tomato also associated with both the plasma membrane and cytoplasmic stress fibers when expressed in live HeLa cells (Fig. 10C, 0 s) and concentrated at the plasma membrane when the actin was depolymerized by latrunculin A (Fig. 10C, 1200 s). Furthermore, as shown in sequential images of live cells (Fig. 11), expressed NM2A localized at the plasma membrane of HT1080 cells, especially after the cells were exposed to latrunculin A for 1000 s. PIP2 was also present in the plasma membrane of these cells (Fig. 11), in addition to its expected occurrence (31) in endosomal vesicles.
FIGURE 10.
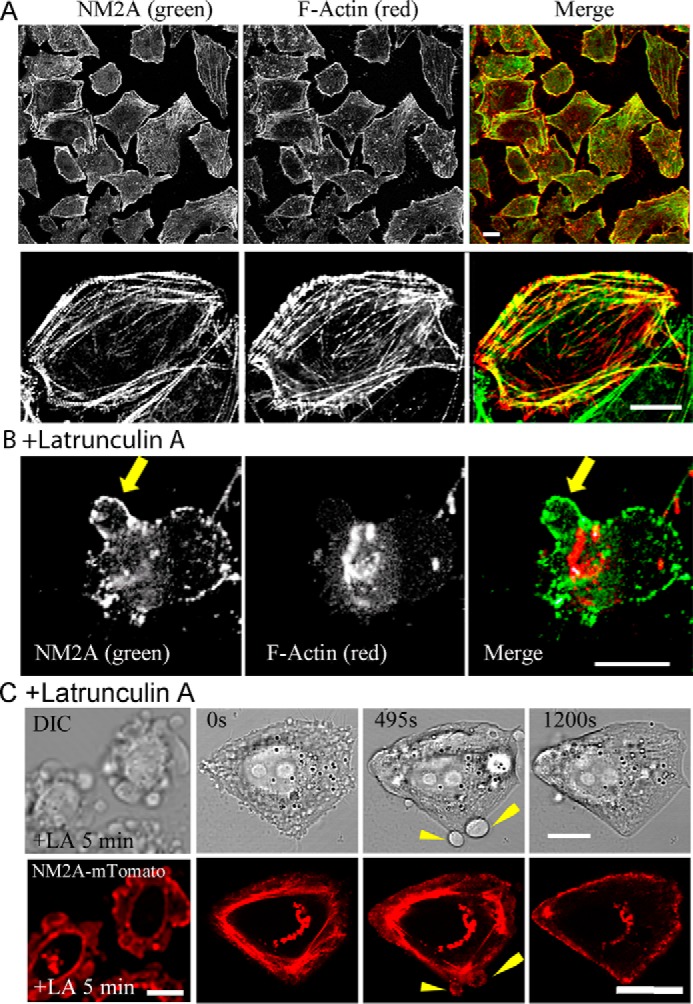
Immunofluorescence localization of NM2A in HeLa cells. A, endogenous NM2A localizes at the plasma membrane and stress fibers. The top row is a field image of multiple cells; the bottom row is a single cell at higher magnification. B, HeLa cells were treated with 4 μm latrunculin A for 15 min before formaldehyde fixation. Although F-actin was depolymerized and aggregated, NM2A still localized at the plasma membrane. The arrow indicates bleb. C, time course of the localization of expressed NM2A-mTomato localization in HeLa cells treated with 4 μm latrunculin A for 5 min before beginning live imaging. At 0 s, NM2A localized at stress fibers (majority) and cell membrane; at 495 s, NM2A localized at plasma membranes of blebs and less at stress fibers; at 1200 s, most of the NM2A had moved to the cell plasma membrane. All bars are 10 μm.
FIGURE 11.
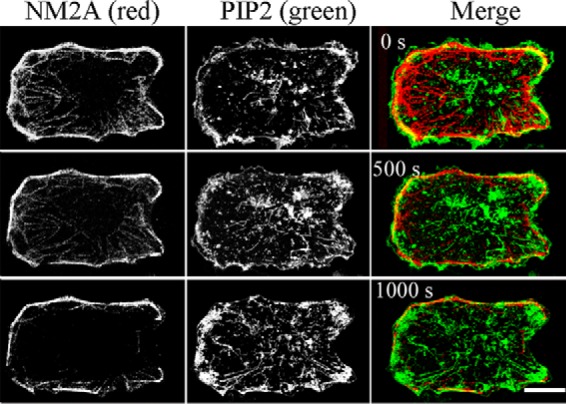
Immunofluorescence localization of NM2A in HT1080 fibrosarcoma cells. HT1080 cells were transfected with NM2A-mTomato and PLCδ1-PH-GFP (identifies PIP2), and treated with 4 μm latrunculin A at time 0 to depolymerize actin filaments. Live images show NM2A localized at the plasma membrane by 1000 s and PIP2 at the plasma membrane and endogenous endosomes. Bar, 10 μm.
Discussion
As outlined in the Introduction, there is considerable evidence that NM2s, in addition to their association with the actin cytoskeleton, are bound to diverse membranes in many cell types. Also, full-length NM2s (22, 32) and the C-terminal half of LMM (32, 33) have been shown to bind to acidic liposomes, but there is no evidence for the molecular basis of these interactions. The data in this paper show that both filamentous full-length recombinant NM2A, NM2B, and NM2C and monomeric recombinant HMM bind to liposomes containing acidic phospholipids (PS, PIP2, or PIP3) principally, if not exclusively, through the regulatory light chain binding site of the heavy chain with concomitant displacement of the RLC. Also, endogenous and expressed NM2A remained associated with the plasma membrane of live HeLa and HT1080 fibrosarcoma cells in which actin filaments were removed by latrunculin A. The relatively weak binding to PS-liposomes of recombinant HMM minus the RLC-binding site and of recombinant LMM may result from structural differences between these molecules and native myosin. For example, LMM associates into paracrystals with a very different structure than bipolar myosin filaments.
The RLC-binding sites (IQ2 domains) of NMIIA (AYLKLRNWQWWRLFTKV), NMIIB (AYLKLRHWQWWRVFTKV), and NMIIC (AYLKLRNWQWWRLFIKV) have a high percentage of basic amino acids (∼24%) and hydrophobic amino acids (∼55%). Similar basic/hydrophobic sites in the non-helical tails of Acanthamoeba and Dictyostelium class I myosins have been shown to be the sites by which these myosins bind to acidic phospholipid vesicles in vitro and to PIP2/PIP3-enriched regions of the plasma membrane in vivo (34, 35). Also, Ca2+-stimulated binding of chicken brush border myosin I to phosphatidylserine vesicles occurs with concomitant dissociation of calmodulin from sites analogous to the light chain binding sites of NM2s (36), and Ca2+-enhanced binding of mammalian myosin IC to acidic phospholipids (37) occurs at a Ca2+ concentration at which calmodulin dissociates from the HC (38). These in vitro observations are directly relevant to in vivo studies showing that myosin IC interacts with hair cell receptors via its calmodulin binding domains (39) and that PIP2 is required for hair cell activities that involve myosin IC (40).
The present paper is the first in which observations similar to those summarized above for class I myosins have been made for class II myosins. Breckenridge et al. (41) did observe, however, that contrary to endogenous NM2A, expressed NM2A with the RLC-binding site deleted did not relocalize from the cytoplasm to the cell periphery during spreading of cultured human cervical cancer cells. This observation was interpreted as demonstrating that HC-bound RLC is required for NM2A to form a folded monomer that could diffuse through the cytoplasm to the periphery. However, it is possible that the RLC-binding site was required in those experiments, not for binding of RLC to NM2A but for binding of NM2A to the plasma membrane.
Although an NM2 monomer bound to membranes by its RLC-binding site would probably not be accessible to F-actin, a membrane-bound monomer might initiate polymerization of myosin filaments, or myosin filaments could bind directly to membranes as they do to liposomes. Membrane-bound filamentous myosin would interact with F-actin, and if the myosin filaments contained RLC-phosphorylated subunits (which is likely because phosphorylated RLC is more resistant than unphosphorylated RLC to displacement by lipids) the membrane-bound myosin filaments would have actin-activated ATPase activity. Whether ATP hydrolysis by membrane-bound actomyosin could generate a useful powerstroke requires further investigation.
Experimental Procedures
Cloning of DNAs
Full-length cDNAs of the HCs of NM2A (Homo sapiens myosin heavy chain 9), NM2B (H. sapiens myosin heavy chain 10 transcript variant 2), and mouse NM2C (Mus musculus myosin heavy chain 14 transcript variant 1) were cloned onto pFastBac1, the Bac-to-Bac plasmid for expression in SF-9 cells (Invitrogen). A FLAG tag (DYKDDDDK) was added to the N termini of the HCs for purification of the myosins by affinity chromatography. The cDNAs for the HCs of NM2A-HMM (terminating at Leu-1355), NM2A-LMM (beginning at Met-1565), and deletion of the RLC-binding site (Gln-804–Val-834) from HMM (HMMΔIQ2) were prepared according to standard molecular biology protocols. For expression in SF-9 cells, the cDNAs of human NM2 RLC and ELC were synthesized and cloned onto pFastBac1. For their expression in Escherichia coli, both light chain cDNAs with a FLAG-tag sequence at the N terminus were cloned onto pETDuet-1. All sequences obtained through PCR were confirmed by DNA-sequencing by GENEWIZ (South Plainfield, NJ).
Expression and Purification of Proteins
Full-length NM2A, NM2B, NM2C, and HMM heavy chain viruses were individually co-expressed with RLC and ELC viruses in SF-9 cells. The recombinant wild-type and mutant NM2s and recombinant HMMs were purified from cell lysates by affinity chromatography on anti-FLAG-resin (Sigma) and eluted with FLAG peptide, as described (42). LMM, ELC, and RLC with N-terminal FLAG tags were individually expressed in E. coli. Bacteria were lysed by the same procedure as for SF-9 cells (42). Purified proteins were dialyzed against buffer (20 mm Tris, pH 7.0, 600 mm NaCl) to remove the FLAG peptides used to elute the proteins from the affinity columns, and the proteins were stored in liquid nitrogen until use.
Preparation of Unilamellar Phospholipid Liposomes
All phospholipids were from Avanti Polar Lipids: dioleoyl-PC (catalog #850375C), dioleoyl-PS (catalog #840035C), dioleoyl-PE (catalog #850725C), dioleoyl-PI (catalog #850149P0), TopFluor PC (catalog #810281C), brain PIP2 (catalog #840046x), brain PIP3 (catalog #850156x), and cholesterol (catalog #700000P). Phospholipid liposomes were prepared as described (32) with modifications. Phospholipids in chloroform and/or methanol were mixed in appropriate proportions and dried sequentially for 5 min each under gentle and strong streams of N2, 30 min under vacuum, and 30 min under N2. Dried lipids were resuspended in 10 mm imidazole, pH 7.0, and 180 mm sucrose to a final concentration of total lipid of 5 mm. The mixture was vortexed for 1 min and sonicated in a bath sonicator 5 times for 5 min each. Vesicles were frozen and thawed in liquid N2 five times and then manually passed 30 times through an Avanti Mini-Extruder fitted with two 100-nm membranes. The final liposomes contained either 100% PC, 100% PS, or PC with varied percentages of PS, PE, PI, PIP2, PIP3, and cholesterol. The pH of the final liposome preparations varied between 6.9 and 7.2 with no correlation with their lipid compositions. All liposomes contained 0.25% fluorescent TopFluor PC, and liposome concentrations were determined by fluorescence measurements in a LS55 luminescence spectrometer with an excitation wavelength of 495 nm, an emission wavelength of 503 nm, and a slit width of 4 nm.
Phosphorylation of Myosins
Phosphorylation was done as described (3) in a reaction mixture containing 10 mm MOPS, pH 7.0, 150 mm NaCl, 2 mm MgCl2, 1 mm ATP, 0.2 mm CaCl2, 1 mm DTT, 100 nm calmodulin, and ∼10 nm myosin light chain kinase for 1 h at room temperature.
Binding Assays
Myosins (100–200 nm) and different concentrations of liposomes (0–125 μm) were mixed together in 20 mm imidazole, pH 7.0, 150 mm NaCl, and 1 mm EGTA containing 0.5 mg/ml BSA to block nonspecific binding of myosins to liposomes. The mixture, 100 μl, was incubated at room temperature for 1 h. To separate liposome-bound myosin from unbound myosin, mixtures of liposomes and filaments of full-length myosins or LMM were centrifuged in a Beckman TL-100 for 20 min at 13,000 × g. Under these conditions, liposomes with either bound myosin or bound LMM pellet but neither myosin filaments, LMM, nor liposomes alone, did. To separate liposome-bound HMM from unbound non-filamentous HMM, mixtures were centrifuged for 30 min at 189,000 × g, conditions under which liposomes and liposome bound the HMM pellet but not unbound HMM. For NM2A binding to F-actin, 200 nm NM2A was incubated with 1 μm muscle F-actin for 1 h at room temperature, and actin-bound myosin separated from unbound myosin by centrifugation for 20 min at 13,000 × g. After centrifugation, 100-μl aliquots of the supernatants were added to 15 μl of 2× SDS sample buffer, and the pellets were suspended in 115 μl of the SDS buffer with the same composition. After SDS-PAGE, the bound (P) and unbound (S) proteins were quantified by Odyssey infrared imaging (LI-COR Biosciences, Lincoln, NE) of the protein bands.
Preparation of Actin, Calmodulin, and Myosin Light Chain Kinase
Actin was purified from rabbit muscle acetone powder (PelFreez, Rogers, AZ) as described by Spudich and Watt (43). Rat calmodulin was expressed and purified from E. coli. FLAG-tagged rabbit smooth muscle myosin light chain kinase (NP_001075775) was expressed using Baculovirus and purified from SF-9 cells.
Electrophoresis
SDS-PAGE was performed by standard procedures on 10% NUPAGE gels (Invitrogen). Urea-glycerol PAGE was performed by the method described by Ruppel et al. (44). Gels were stained by Instant Blue (Expedeon, San Diego, CA).
Fluorescence Microscopy
HeLa and HT1080 human fibrosarcoma cells were maintained in DMEM or EMEM (Thermo Fisher Scientific), respectively, supplemented with 10% FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2.
For immunolocalization, HeLa cells were seeded on coverslips for 24 h and then treated at 37 °C with either 4 μm latrunculin A, to depolymerize F-actin, or DMSO for 15 min before fixation with 2% paraformaldehyde in PBS at room temperature for 10 min. After fixation, cells were washed with PBS supplemented with 10% FBS for 1 h. Cells were then incubated with primary antibodies diluted in PBS/FBS containing 0.2% saponin for 1 h. Cells were washed 3 times in PBS and incubated with the appropriate secondary antibodies in PBS/FBS containing 0.2% saponin for 1 h. Cells were then washed 3 times in PBS and mounted on glass slides. Rabbit polyclonal NM2A-specific antibody (Sigma, M8064) was diluted 1:2000, and anti-PIP2 mouse monoclonal IgG antibody (Abcam, Cambridge, MA), which also recognizes PIP3, was diluted 1:100. Fluorescently labeled secondary goat anti-rabbit and goat anti-mouse IgG antibodies (Alexa-Fluoro 488 and Alexa-Fluoro 594) from Molecular Probes, Inc. (Eugene, OR) were diluted 1:750. F-actin was detected by staining with rhodamine-labeled phalloidin. Images were acquired with an LSM-780 laser scanning confocal microscope (Carl Zeiss) equipped with a PLANapo 63× oil objective (NA 1.4). Pixel dimensions for Fig. 10A were 0.26 μm × 0.26 μm (top panel) and 0.10 μm x 0.10 μm (bottom panel).
Live-cell imaging experiments were carried out 18–24 h after transfection by transient electroporation of NM2A-mTomato and PLCδ1-PH-GFP in X-tremeGENE by Amaxa Nucleofection kit according to the manufacturer's instructions (Lonza, Allendale, NJ). Cells were treated with 4 μm latrunculin A for 15 min to depolymerize F-actin, and images were acquired every 8 s for 30 min with an LSM-780 or LSM-880 laser scanning confocal microscope (Carl Zeiss) equipped with a PLAN apo63× oil objective (NA = 1.4). Pixel dimensions were 0.081 μm × 0.081 μm (Fig. 10C), and 0.87 μm × 0.87 μm (Fig. 11).
Negative Staining Electron Microscopy of Proteins Bound to Liposomes
Proteins and liposomes were diluted in buffer with final concentrations of 150 mm NaCl, 10 mm MOPS, pH 7.0, 0.1 mm EGTA, 1 mm MgCl2, and either 100 nm myosin or 125 μm liposomes. In the case of mixtures, myosins and liposomes were first diluted separately to concentrations of 200 nm each. Equal volumes of protein and liposomes were then mixed and vortexed to avoid excessive aggregation, and 3-μl samples were applied immediately to UV-treated carbon-coated copper grids and negatively stained with 1% uranyl acetate. Images were recorded on an AMT XR-60 CCD camera using a JEOL 1200EX II microscope operating at 80 kV. Catalase crystals were used as a size calibration standard.
Author Contributions
X. L. and S. S. designed, conducted, and interpreted the results of most of the experiments. N. B. conducted and interpreted the experiments in Figs. 3 and 5C. S. Y. conducted the experiments. H. B. contributed to the design and preparation of liposomes and interpretation of the binding data. C. D. W. designed, performed, and interpreted the experiment described in Figs. 10C and 11. J. G. D. contributed to the experimental approach and analysis of Figs. 10C and 11. J. R. S. contributed to the interpretation of the experimental results. E. D. K. conceived the project, assisted in the design of the experiments and the interpretation of the experimental results, and wrote the paper. All authors read and contributed to the paper, and approved its submission.
Acknowledgments
We acknowledge the support of the Electron and Light Microscopy Cores of the NHLBI, National Institutes of Health. We thank Dr. John Hammer 3rd (NHLBI) for NM2A-mTomato and Dr. Fang Zhang (NHLBI) for purified myosin light chain kinase.
This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NM2
- nonmuscle myosin II
- ELC
- essential light chain
- HC
- heavy chain
- HMM
- heavy meromyosin
- LMM
- light meromyosin
- RLC
- regulatory light chain
- pRLC
- phosphorylated RLC
- PC
- phosphatidylcholine
- PS
- phosphatidylserine
- PIP2 and PIP3
- phosphatidylinositol di- and triphosphate.
References
- 1. Sellers J. R. (2000) Myosins: a diverse superfamily. Biochim. Biophys. Acta 1496, 3–22 [DOI] [PubMed] [Google Scholar]
- 2. Dulyaninova N. G., and Bresnick A. R. (2013) The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture 3, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Billington N., Wang A., Mao J., Adelstein R. S., and Sellers J. R. (2013) Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem. 288, 33398–33410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suzuki H., Kamata T., Onishi H., and Watanabe S. (1982) Adenosine triphosphate-induced reversible change in the conformation of chicken gizzard myosin and heavy meromyoisin. J. Biochem. 91, 1699–1705 [DOI] [PubMed] [Google Scholar]
- 5. Trybus K. M., Huiatt T. W., and Lowey S. (1982) A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. U.S.A. 79, 6151–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Craig R., Smith R., and Kendrick-Jones J. (1983) Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature 302, 436–439 [DOI] [PubMed] [Google Scholar]
- 7. Ikebe M., Koretz J., and Hartshorne D. J. (1988) Effects of phosphorylation of light chain residues threonine 18 and serine 19 on the properties and conformation of smooth muscle myosin. J. Biol. Chem. 263, 6432–6437 [PubMed] [Google Scholar]
- 8. Adelstein R. S., and Conti M. A. (1975) Phosphorylation of platelet myosin increases actin-activated ATPase activity. Nature 256, 597–598 [DOI] [PubMed] [Google Scholar]
- 9. Lebowitz E. A., and Cooke R. (1978) Contractile properties of actomyosin from human blood platelets. J. Biol. Chem. 253, 5443–5447 [PubMed] [Google Scholar]
- 10. Trotter J. A., and Adelstein R. S. (1979) Macrophage myosin: regulation of actin-activated ATPase activity by phosphorylation of the 20,000-dalton light chain. J. Biol. Chem. 254, 8781–8785 [PubMed] [Google Scholar]
- 11. Fechheimer M., and Cebra J. J. (1982) Phosphorylation of lymphocyte myosin catalyzed in vitro and in intact cells. J. Cell Biol. 93, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conti M. A., and Adelstein R. S. (2008) Nonmuscle myosin II moves in new directions. J. Cell Sci. 121, 11–18 [DOI] [PubMed] [Google Scholar]
- 13. Vicente-Manzanares M., Ma X., Adelstein R. S., and Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DePina A. S., Wöllert T., and Langford G. M. (2007) Membrane associated nonmuscle myosin II functions as a motor for actin-based vesicle transport in clam oocyte extracts. Cell Motil. Cytoskeleton 64, 739–755 [DOI] [PubMed] [Google Scholar]
- 15. Ikonen E., de Almeid J. B., Fath K. R., Burgess D. R., Ashman K., Simons K., and Stow J. L. (1997) Myosin II is associated with Golgi membranes: identification of p200 as nonmuscle myosin II on Golgi-derived vesicles. J. Cell Sci. 110, 2155–2164 [DOI] [PubMed] [Google Scholar]
- 16. Müsch A., Cohen D., and Rodriguez-Boulan E. (1997) Myosin II is involved in the production of constitutive transport vesicles from the TGN. J. Cell Biol. 138, 291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stow J. L., and Heimann K. (1998) Vesicle budding on Golgi membrane: regulation by G proteins and myosin motors. Biochim. Biophys. Acta 1404, 161–171 [DOI] [PubMed] [Google Scholar]
- 18. Durán J. M., Valderrama F., Castel S., Magdalena J., Tomás M., Hosoya H., Renau-Piqueras J., Malhotra V., and Egea G. (2003) Myosin motors and not actin comets are mediators of the actin-based Golgi-to-endoplasmic reticulum protein transport. Mol. Biol. Cell 14, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fath K. R. (2005) Characterization of myosin-II binding to Golgi stacks in vitro. Cell Motil. Cytoskeleton 60, 222–235 [DOI] [PubMed] [Google Scholar]
- 20. Miserey-Lenkei S., Chalancon G., Bardin S., Formstecher E., Goud B., and Echard A. (2010) Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 12, 645–654 [DOI] [PubMed] [Google Scholar]
- 21. DeGiorgis J. A., Reese T. S., and Bearer E. L. (2002) Association of a nonmuscle myosin II with axoplasmic organelles. Mol. Biol. Cell 13, 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li D., Miller M., and Chantler P. D. (1994) Association of a cellular myosin II with anionic phospholipids and the neuronal plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 91, 853–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishmael J. E., Safic M., Amparan D., Vogel W. K., Pham T., Marley K., Filtz T. M., and Maier C. S. (2007) Nonmuscle myosin II-B and Va are components of detergent-resistant membrane cytoskeletons derived from mouse forebrain. Brain Res. 1143, 46–59 [DOI] [PubMed] [Google Scholar]
- 24. Ecay T. W., Conner T. D., and Decker E. R. (1997) Nonmuscle myosin IIA copurifies with chloride channel-enriched membranes from epithelia. Biochem. Biophys. Res. Commun. 231, 369–372 [DOI] [PubMed] [Google Scholar]
- 25. Linz-McGillem L. A., and Alliegro M. C. (2003) Myosin II in retinal pigmented epithelial cells: evidence for an association with membranous vesicles. Exp. Eye Res. 76, 543–552 [DOI] [PubMed] [Google Scholar]
- 26. Sanborn K. B., Mace E. M., Rak G. D., Difeo A., Martignetti J. A., Pecci A., Bussel J. B., Favier R., and Orange J. S. (2011) Phosphorylation of the myosin IIA tailpiece regulates myosin IIA molecule association with lytic granules to promote NK-cell cytotoxicity. Blood 118, 5862–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nebl T., Pestonjamasp K. N., Leszyk J. D., Crowley J. L., Oh S. W., and Luna E. J. (2002) Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem. 277, 43399–43409 [DOI] [PubMed] [Google Scholar]
- 28. Morimura S., Suzuki K., and Takahashi K. (2011) Nonmuscle myosin IIA is required for lamellipodia formation through binding to WAVE2 and phosphatidyl-3,4,5-triphosphate. Biochem. Biophys. Res. Commun. 404, 834–840 [DOI] [PubMed] [Google Scholar]
- 29. Nie Z., Hirsch D. S., Luo R., Jian X., Stauffer S., Cremesti A., Andrade J., Lebowitz J., Marino M., Ahvazi B., Hinshaw J. E., and Randazzo P. A. (2006) A BAR domain in the N-terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr. Biol. 16, 130–139 [DOI] [PubMed] [Google Scholar]
- 30. Jian X., Brown P., Schuck P., Gruschus J. M., Balbo A., Hinshaw J. E., and Randazzo P. A. (2009) Autoinhibition of Arf GTPase-activating protein activity by the BAR domain of ASAP1. J. Biol. Chem. 284, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown F. D., Rozelle A. L., Yin H. L., Balla T., and Donaldson J. G. (2001) Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murakami N., Elzinga M., Singh S. S., and Chauhan V. P. (1994) Direct binding of myosin II to phospholipid vesicles via tail regions and phosphorylation of the heavy chains by protein kinase C. J. Biol. Chem. 269, 16082–16090 [PubMed] [Google Scholar]
- 33. Murakami N., Singh S. S., Chauhan V. P., and Elzinga M. (1995) Phospholipid binding, phosphorylation by protein kinase C, and filament assembly of the COOH-terminal heavy chain fragments of nonmuscle myosin II isoforms MIIA and MIIB. Biochemistry 34, 16046–16055 [DOI] [PubMed] [Google Scholar]
- 34. Brzeska H., Hwang K. J., and Korn E. D. (2008) Acanthamoebba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J. Biol. Chem. 283, 32014–32023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brzeska H., Guag J., Preston G. M., Titus M. A., and Korn E. D. (2012) Molecular basis of dynamic relocalization of Dictyostelium myosin IB. J. Biol. Chem. 287, 14923–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swanljung-Collins H., and Collins J. H. (1992) Phosphorylation of brush border myosin I by protein kinase c is regulated by Ca2+-stimulated binding of myosin I to phosphatidylserine concerted with calmodulin dissociation. J. Biol. Chem. 267, 3445–3454 [PubMed] [Google Scholar]
- 37. Tang N., Lin T., and Ostap E. M. (2002) Dynamics of myo1c (myosin-Iβ) lipid binding and dissociation. J. Biol. Chem. 277, 42763–42768 [DOI] [PubMed] [Google Scholar]
- 38. Barylko B., Jung G., and Albanesi J. (2005) Structure, function, and regulation of myosin1C. Acta Biochim. Pol. 52, 373–380 [PubMed] [Google Scholar]
- 39. Cyr J. L., Dumont R. A., and Gillespie P. G. (2002) Myosin-1c interacts with hair-cell receptors through its calmodulin-binding IQ domains. J. Neurosci. 22, 2487–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirono M., Denis C. S., Richardson G. P., and Gillespie P. G. (2004) Hair cells require phosphatidylinositol 4,5-bisphospohate for mechanical transduction and adaptation. Neuron 44, 309–320 [DOI] [PubMed] [Google Scholar]
- 41. Breckenridge M. T., Dulyaninova N. G., and Egelhoff T. T. (2009) Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol. Biol. Cell 20, 338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu X., Hong M. S., Shu S., Yu S., and Korn E. D. (2013) Regulation of the filament structure and assembly of Acanthamoeba myosin II by phosphorylation of serines in the heavy-chain nonhelical tailpiece. Proc. Natl. Acad. Sci. U.S.A. 110, E33–E40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spudich J. A., and Watt S. (1971) The regulation of rabbit skeletal muscle contraction. 1. Biochemical studies of the interaction of tropomyosin-troponin complex with actin and proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 44. Ruppel K. M., Uyeda T. Q., and Spudich J. A. (1994) Role of highly conserved lysine 130 of myosin motor domain: in vivo and in vitro characterization of site specifically mutated myosin. J. Biol. Chem. 269, 18773–18780 [PubMed] [Google Scholar]