Abstract
Nucleosomes affect Cas9 binding and activity at on-target sites, but their impact at off-target sites is unknown. To investigate how nucleosomes affect Cas9 cleavage at off-target sites in vitro, we used a single guide RNA (sgRNA) that has been previously shown to efficiently direct Cas9 cleavage at the edge of the strongly positioned 601 nucleosome. Our data indicate that single mismatches between the sgRNA and DNA target have relatively little effect on Cas9 cleavage of naked DNA substrates, but strongly inhibit cleavage of nucleosome substrates, particularly when the mismatch is in the sgRNA “seed” region. These findings indicate that nucleosomes may enhance Cas9 specificity by inhibiting cleavage of off-target sites at the nucleosome edge.
Keywords: chromatin, CRISPR/Cas, endonuclease, histone, nucleosome, PAM, mismatch, off-target
Introduction
The RNA-guided Cas9 endonuclease is a promising tool for genome editing, due to its ease and efficiency at targeting DNA double-strand breaks (1, 2). However, a key limitation of Cas9 genome editing is its propensity to induce DNA breaks at off-target sites, particularly in large eukaryotic genomes (3–8). Such breaks can lead to mutagenesis at off-target sites, which is a significant liability for all genome editing applications, especially those potentially involving human patients.
Cas9 searches for target sites by initially binding DNA sequences containing a proto-spacer adjacent motif (PAM)2 (9), which is 5′-NGG (where N indicates any nucleotide) for Cas9 from Streptococcus pyogenes (10, 11). Following PAM recognition, Cas9 helicase activity promotes target DNA unwinding and the progressive invasion of the 20-nucleotide guide segment of the Cas9-bound sgRNA, beginning with the sgRNA “seed” region at the 3′ end of the guide segment. DNA cleavage by Cas9 requires base pairing of the entire guide segment with the DNA target, which only then triggers the Cas9 protein to adopt an endonuclease-active conformation (12, 13).
Despite this “multilayered” regulation of Cas9 endonuclease activity (14), in vivo and in vitro studies indicate that Cas9 frequently cleaves off-target sites, in some cases as efficiently as the on-target site (7, 8, 11). Off-target sites can contain up to 4–5 mismatches between the guide RNA and the DNA target, and even 1-bp insertions or deletions. Although multiple studies have suggested that mismatches within the PAM-proximal seed region are more detrimental to Cas9 activity, efforts to use such data to computationally predict off-target sites for guide RNA sequences often perform poorly in practice (7, 15).
We, and others, have recently shown that Cas9 cleavage of on-target sites is significantly inhibited in vitro by strongly positioned nucleosomes (16–18). These studies indicate that nucleosomes primarily inhibit the initial binding of Cas9 to the PAM. Cas9 was unable to bind PAM sites located within strongly positioned nucleosomes (17, 18), but was able to efficiently cleave target sites when the PAM was located in adjacent linker DNA (16). Moreover, Cas9 can efficiently cleave a target within a nucleosome if the corresponding PAM sequence is located in accessible linker DNA, indicating that Cas9 can efficiently catalyze guide RNA strand invasion within nucleosomes at on-target sites (16).
However, it is not known how nucleosomes influence Cas9 activity at potential off-target sites. To address this question, we investigated how single (or multiple) mismatches between the sgRNA and the DNA target affect Cas9 activity on a nucleosome substrate in vitro.
Results
sgRNA Mismatches Specifically Impact Cas9 Activity in Nucleosomes
In our previous study, we demonstrated that Cas9 activity in vitro is almost completely inhibited at target sites within the strongly positioned 601 nucleosome (16). However, a guide RNA targeting the nucleosome edge (sgRNA3 in Ref. 16) could efficiently direct Cas9 to cleave 601 nucleosomes, because the PAM site was located in accessible linker DNA (16). We exploited the high activity of this sgRNA (labeled “wt-sgRNA” in this study) to investigate how cleavage of off-target sites is impacted by nucleosomes.
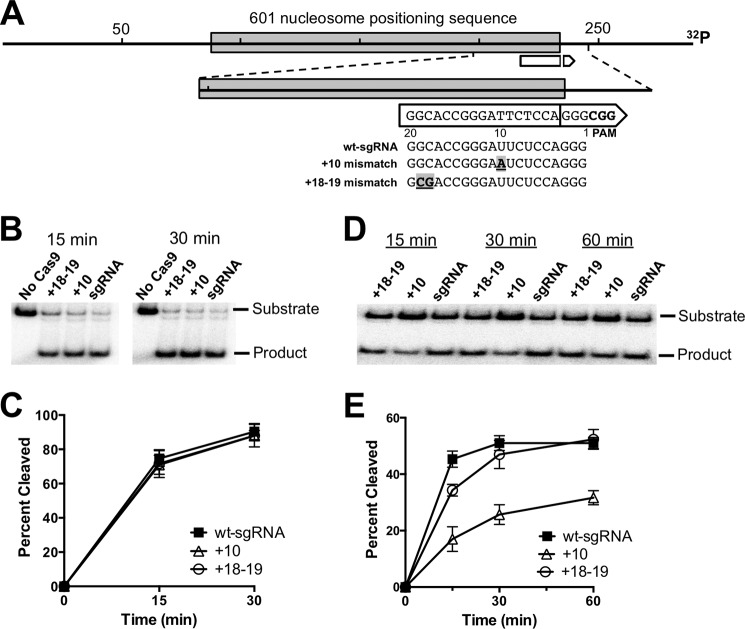
We initially tested mismatches at two locations in the sgRNA-DNA interface: a single mismatch at a position 10 nucleotides from the PAM (+10) and a double mismatch at positions 18 and 19 nucleotides from the PAM (+18–19; Fig. 1A). We prepared sgRNAs containing a U to A mutation at position +10 or a 5′-GC to 5′-CG mutation at position +18–19, and then measured their activity on naked DNA and nucleosome substrates, consisting of a radiolabeled 289-bp DNA substrate containing the 601 nucleosome positioning sequence. Analysis of Cas9 cleavage of the naked DNA substrate over a 30-min time course revealed no significant difference in Cas9 activity between the wild-type sgRNA and the two mismatch sgRNAs (Fig. 1, B and C). This finding is in accordance with previous results, indicating that single or double mismatches at locations more distal from the PAM often have relatively little impact on Cas9 activity on naked DNA (3, 11).
FIGURE 1.
Time course of Cas9 cleavage on naked DNA and nucleosome substrates containing sgRNA-target DNA mismatches. A, diagram showing the location of mismatches within the sgRNA relative to the DNA target (white arrow) in the 289-bp substrate containing the 601 nucleosome positioning sequence. The break in the arrow denotes the Cas9 cleavage site, and the point of the arrow corresponds to the PAM site. The locations and sequences of the sgRNA mismatches are indicated, with the mismatch location (e.g. +10) denoting nucleotide distance from the PAM. B, representative polyacrylamide gels showing the full-length substrate and Cas9 cleavage product of the naked DNA substrate for each sgRNA at different time points. No Cas9 represents control samples with only the wt-sgRNA. C, graph showing the percentage of cleavage of the naked DNA substrate as a function of time. Data points represent the average of three independent experiments, and error bars represent standard deviations. D, same as in panel B, except showing the time course of Cas9 cleavage of nucleosome substrate. E, summary of Cas9 cleavage of nucleosome substrates for the indicated sgRNAs. Data points represent the average of three independent experiments, and error bars represent standard deviations.
We repeated these experiments on substrates that had been reconstituted into nucleosomes. Relatively high levels of Cas9 cleavage in the nucleosome were detected with the wt-sgRNA (Fig. 1D), although to a lesser extent than with naked DNA (compare Fig. 1, C and E), in accordance with our previous study (16). However, Cas9 cleavage of the nucleosome substrate was diminished with both mismatch sgRNAs relative to wt-sgRNA (Fig. 1, D and E). This was particularly apparent for the +10 mismatch, which had significantly reduced levels of cleavage product even after a 1-h incubation (Fig. 1E). These results suggest that the effect of sgRNA mismatches on Cas9 activity in our in vitro system is significantly modulated by the packaging of the DNA target into a nucleosome.
sgRNA Complementarity in the Seed Region Is Critical for Cas9 Cleavage of Nucleosomes
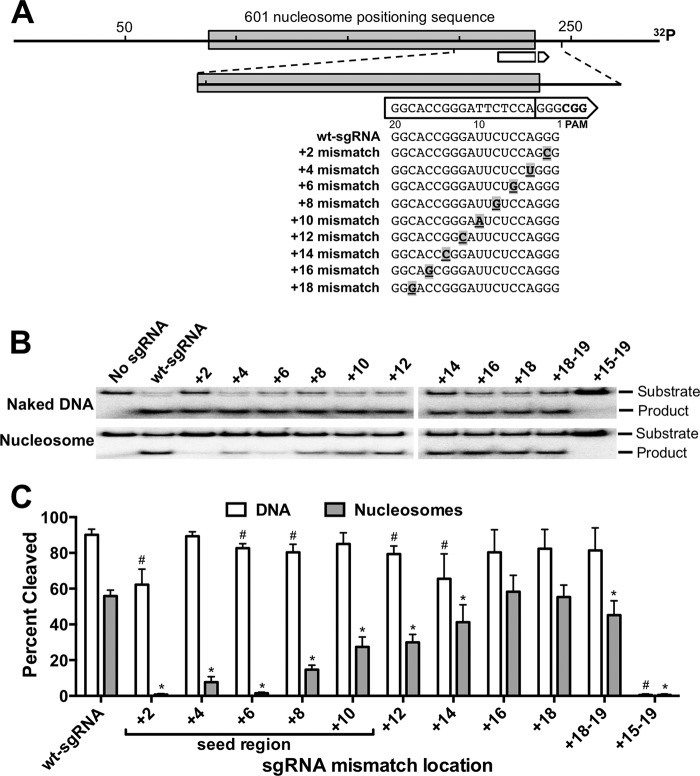
To systematically screen how sgRNA mismatches impact Cas9 activity on nucleosomes, we generated a panel of sgRNA mutants containing single mismatches at locations throughout the sgRNA guide segment (Fig. 2A). Each mutant sgRNA was tested for its activity in directing Cas9 cleavage of naked DNA or nucleosome substrates, following a 30-min incubation. A number of single sgRNA mismatches had significant effects on cleavage of the naked DNA substrate, particularly mismatches in the sgRNA seed region (Fig. 2, B and C), consistent with previous studies (3, 11). This effect was most pronounced for single mismatches at positions +2 and +14 nucleotides from the PAM, which had as much as ∼30% lower product yield (Fig. 2, B and C). Five simultaneous mismatches at positions +15–19 from the PAM completely eliminated Cas9 cleavage of the naked DNA substrate (Fig. 2B). This is in accordance with previous studies, which have shown that four or more mismatches in the distal end of the guide segment largely abolish Cas9 activity in vitro, due to the importance of this region in triggering Cas9 to adopt a nuclease-active conformation (12).
FIGURE 2.
sgRNA mismatches primarily affect Cas9 activity on nucleosome substrates. A, diagram showing the location of sgRNA mismatches relative to the DNA target in the 601 nucleosome. See the legend for Fig. 1 for more details. B, gel electrophoresis of Cas9 cleavage products following a 30-min incubation with naked DNA (top panel) and nucleosome substrate (lower panel), targeted by the indicated wt or mismatch sgRNAs. No sgRNA represents control samples with only the Cas9 enzyme (1 pmol) without the addition of an sgRNA. C, summary of the percentage of substrate cleavage by Cas9 after 30 min, targeted by sgRNAs with mismatches at different distances from the PAM site. Data points represent the average of at least three independent experiments, and error bars represent standard deviations. # indicates that Cas9 cleavage of naked DNA is significantly lower with mismatch sgRNA relative to wt-sgRNA (p < 0.05). * indicates that Cas9 cleavage of nucleosome is significantly lower with mismatch sgRNA relative to wt-sgRNA (p < 0.05).
The inhibitory effect of single sgRNA mismatches on Cas9 activity was greatly exacerbated on the nucleosome substrate (Fig. 2, B and C). This was particularly apparent for sgRNA mismatches located in the seed region of the guide segment (i.e. proximal to the PAM). For example, mismatches at positions +2 and +6 from the PAM almost completely abolished cleavage of the nucleosome (Fig. 2, B and C), whereas mismatches at positions +4 and +8 from the PAM reduced Cas9 activity on nucleosomes as much as 7-fold. Mismatches adjacent to the seed region (i.e. positions +12 and +14) also decreased Cas9 activity on the nucleosome substrate, but to a lesser extent than mismatches within the seed region (Fig. 2C). In contrast, single mismatches in the distal end of the guide segment (i.e. positions +16 and +18) did not significantly affect Cas9 activity on the nucleosome or naked DNA relative to wt-sgRNA.
The decrease in nucleosome cleavage observed for the +14 mismatch sgRNA could be explained in part by a general decrease in Cas9 activity with this sgRNA, seen even on naked DNA (Fig. 2C). However, for other sgRNA mismatches, differences in Cas9 activity on naked DNA cannot explain the differences observed for the nucleosome substrate. For example, Cas9 activity on naked DNA was >25% higher with the +4 mismatch than the +14 mismatch sgRNA; however, Cas9 activity on the nucleosome substrate was ∼5-fold lower with the +4 mismatch than the +14 mismatch sgRNA (Fig. 2C). Hence, in many instances, the effect of mismatches on Cas9 activity on nucleosomes differed greatly from the effect seen on naked DNA.
As a further test that the sgRNA mismatch constructs did not nonspecifically affect Cas9 activity, we constructed a nucleosome substrate (601-m6) containing a complementary mutation for the +6 mismatch sgRNA (Fig. 3A). The 601-m6 substrate was able to reconstitute into well positioned nucleosomes in vitro, similar to the wild-type 601 sequence (Fig. 3B). Cas9 cleaved the 601-m6 naked DNA when directed by either the wt or +6 mismatch sgRNAs (Fig. 3C). However, other mismatch sgRNAs (i.e. +2 or +10 mismatch) did not direct Cas9 to cleave the 601-m6 DNA (Fig. 3C), likely because there are two mismatches in the seed region for each of these sgRNAs (e.g. mismatches at positions +2 and +6 for the +2 mismatch sgRNA; Fig. 3A). The more distal +18 mismatch sgRNA did direct Cas9 to cleave the 601-m6 DNA, although less efficiently than either the wt or +6 mismatch sgRNAs (Fig. 3, C and D).
FIGURE 3.
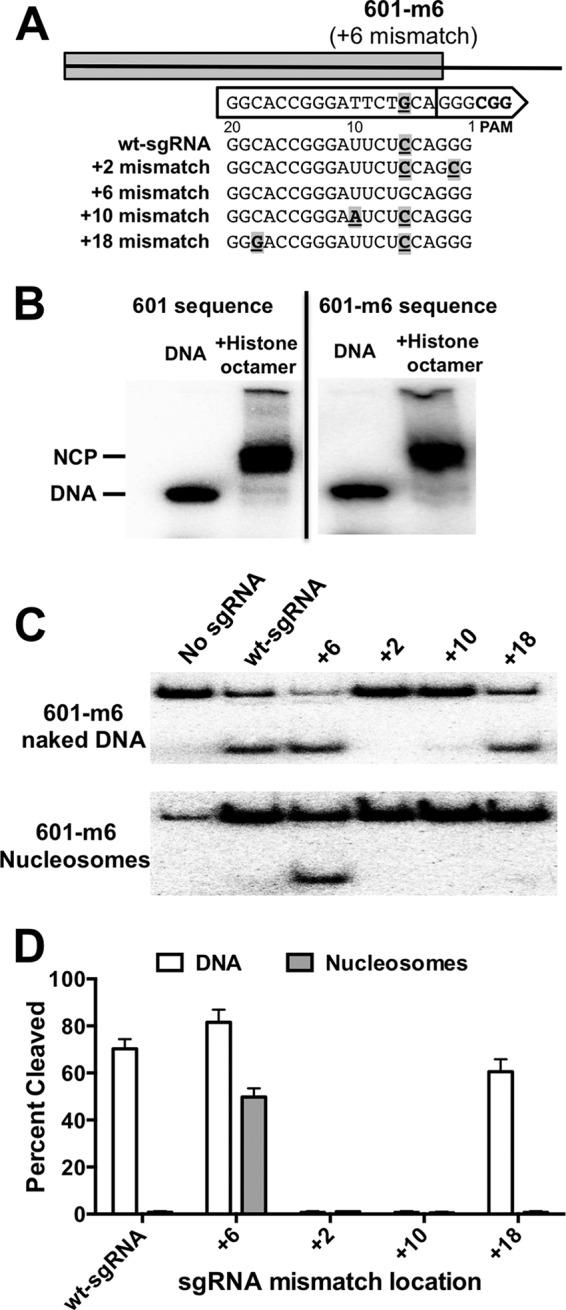
Cas9 activity with a mismatch sgRNA is restored with complementary mutation in 601 nucleosome substrate. A, diagram showing mismatch sgRNAs and complementary mutation in the 601 nucleosome substrate at position +6 nucleotides from the PAM site. B, representative reconstitutions of the wild-type 601 sequence and mutant DNA substrate (601-m6, with sequence change to complement the sgRNA +6 mismatch) into mononucleosomes analyzed by native polyacrylamide gel electrophoresis. NCP indicates the band corresponding to reconstituted nucleosome core particles. C, representative polyacrylamide gels showing cleavage of the 601-m6 naked DNA (top panel) and nucleosome (bottom panel) substrates after 30 min by Cas9 targeted by different sgRNAs used in the study. No sgRNA represents control samples with only the Cas9 enzyme (1 pmol) without the addition of an sgRNA. D, graph showing the percentage of cleavage of the 601-m6 DNA and nucleosome substrates by Cas9. Note that the +2, +10, and +18 sgRNAs have two mismatches with the 601-m6 substrate, because all have the equivalent of an additional +6 mismatch. Data points represent the average of at least three independent experiments, and error bars represent standard deviations.
Cas9 efficiently cleaved the 601-m6 nucleosome when guided by the complementary +6 mismatch sgRNA, but no cleavage activity was detected with wt-sgRNA (Fig. 3, C and D), presumably due to the mismatch at position +6 between the wt-sgRNA and the 601-m6 sequence (Fig. 3A). These results confirm that the +6 mismatch sgRNA can direct efficient Cas9 cleavage of a nucleosome containing a complementary target sequence (i.e. 601-m6), but not for the nucleosome containing a +6 mismatch in the seed region of the guide segment (i.e. 601 wild type). Similarly, the wt-sgRNA directed efficient cleavage of the wild-type 601 nucleosome, but not the mutant 601-m6 nucleosome, due to the mismatch at the +6 position.
Discussion
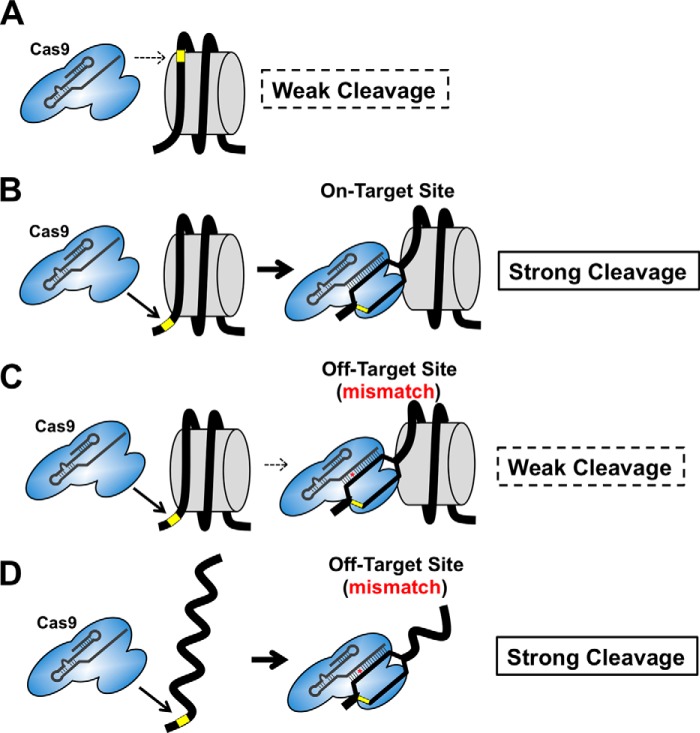
In this study, we used a model guide RNA targeting the edge of the strongly positioned 601 nucleosome sequence to investigate how nucleosomes impact Cas9 cleavage of mismatch-containing off-target sites. These data indicate that Cas9 endonuclease activity on nucleosomes in vitro is particularly sensitive to mismatches between the sgRNA and target DNA sequences. Mismatches within the PAM-proximal seed region were most prone to inhibition by nucleosomes, whereas mismatches in the PAM-distal end of the guide segment had little affect on Cas9 activity in nucleosomes. We conclude that nucleosomes not only inhibit the initial binding and recognition of PAM sequences by Cas9 (16), but may also impact subsequent DNA unwinding and sgRNA invasion of the target DNA (Fig. 4). Although this process can occur relatively efficiently at on-target sites, even in strongly positioned nucleosomes, the energetic penalty of sgRNA-target DNA mismatches is apparently magnified in the context of a nucleosome. Our conclusion that nucleosomes can oppose sgRNA invasion of the target DNA is consistent with previous studies, which showed that generation of RNA:DNA hybrids is incompatible with the formation of stable nucleosomes (19, 20).
FIGURE 4.
Model of how nucleosomes impact Cas9 activity at off-target sites containing mismatches. A, Cas9 cannot efficiently cleave target sites in strongly positioned nucleosomes if the PAM site (yellow rectangle) is located within the nucleosome. B, Cas9 can efficiently cleave target sites in the nucleosome if the PAM site (yellow rectangle) is located in the accessible linker DNA. C, a single mismatch between the sgRNA and DNA target (red asterisk) inhibits Cas9 cleavage of nucleosome substrates, particularly if the mismatch occurs in the PAM-proximal seed region. D, Cas9 can efficiently cleave naked DNA substrates containing single mismatches (red asterisk).
Our study utilized an sgRNA target site in which the PAM motif was located in accessible linker DNA, but much of the target sequence overlapped with the 601 nucleosome (Fig. 1A). This specific arrangement was necessary to obtain high levels of on-target cleavage within the 601 nucleosome, which otherwise strongly inhibits Cas9 activity, even at on-target sites, when the PAM motif is occluded within the nucleosome (16–18). However, bioinformatics analysis (see “Experimental Procedures”) indicates that as many as 11.5% of potential Cas9 target sites (on- and off-target) in the yeast genome have a similar arrangement (i.e. PAM located outside the nucleosome, whereas the remainder of the guide target overlaps with the nucleosome), indicating that this sort of target site arrangement is commonly encountered in Cas9 genome editing in vivo. It will be important to determine whether nucleosomes have a similar effect on Cas9 off-target activity for other sgRNA targets that have a similar arrangement relative to nucleosomes. It has recently been shown that more weakly positioned nucleosomes are more permissive to Cas9 cleavage, even when the PAM motif is located within the nucleosome (18). It is not clear to what extent such nucleosomes may impact Cas9 cleavage of off-target sites located entirely within the nucleosome boundaries.
In summary, our results indicate that chromatin can exacerbate the impact of sgRNA-target DNA mismatches on Cas9 activity and thus potentially limit Cas9 off-target activity in eukaryotic cells. These findings have important ramifications for off-target mutagenesis in CRISPR-Cas9 genome editing. First, inclusion of chromatin data may significantly improve the computational prediction of off-target sites for guide RNA sequences. This conclusion is supported by a recent bioinformatics analysis of in vivo Cas9 genome editing data (21). Second, cellular enzymes that enhance DNA accessibility in nucleosomes may increase off-target mutagenesis during Cas9 genome editing. It will be important to determine to what extent our observations can be generalized to other off-target sites within nucleosomes, and whether Cas9 off-target activity in chromatin may actually be enhanced by ATP-dependent chromatin remodelers or other histone-modifying enzymes.
Experimental Procedures
DNA and Nucleosome Substrates
The 147-bp Widom 601 nucleosomal positioning sequence (22), flanked by 87 and 55 bp of linker DNA (16), was used as the DNA substrate for all experiments. The DNA substrate was generated by PCR amplification of the template pGEM3Z.601 (a gift from the late Dr. Jonathon Widom, Northwestern University) and radiolabeled at one end (Fig. 1A), following our previously published procedures (16). The pGEM3Z.601 template was mutated using a modified version of the QuikChange method (23) with primers OWY359 (CGGCACCGGGATTCTGCAGGGCGGCCG) and OWY360 (CGGCCGCCCTGCAGAATCCCGGTGCCG) to generate plasmid pML123, containing a C-to-G mutation complementary to the +6 mismatch sgRNA. This plasmid (i.e. pML123) was used to PCR-amplify the 601-m6 substrate.
Nucleosomes were prepared by salt dialysis as described previously (16, 24). Briefly, 7 pmol of purified recombinant Xenopus laevis histone octamers were mixed with 7 pmol of the radiolabeled 289-bp DNA substrate (a 1:1 molar ratio of DNA to octamer). The mixture was transferred to dialysis tubes (3500-dalton molecular weight cut-off; Thermo Scientific) and reconstituted by sequential salt dialysis (from 4 m NaCl to 50 mm NaCl over the course of ∼5 h). Nucleosomes were verified by polyacrylamide gel electrophoresis on 6% gels.
Mismatch sgRNA Preparation
DNA templates for single guide RNAs were prepared by PCR amplification using long oligonucleotide templates. The resulting purified PCR products were used as the template for in vitro transcription using the MEGAscript T7 transcription kit (Ambion) and then purified using an RNA purification kit (Qiagen). Oligonucleotide sequences are available upon request.
Cas9 Endonuclease Assays
Cas9 endonuclease assays were performed in 20-μl reactions at 37 °C for various times: 0–30 min for the DNA time course; 0–60 min for the nucleosome time course; and 30 min for all other measurements. Cas9 enzyme (recombinant S. pyogenes Cas9, New England Biolabs) was preincubated with the indicated sgRNA at a 1:1.6 ratio of Cas9 to sgRNA for 30 min at 37 °C (9) in 1× Cas9 reaction buffer (20 mm HEPES, 100 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA; New England Biolabs) prior to the addition of radiolabeled DNA or nucleosome substrates to the reactions. Reactions were terminated by adding phenol:chloroform:isoamyl alcohol (20:19:1), and the resulting cleavage products were analyzed by electrophoresis on 10% native polyacrylamide gels.
To determine the relative efficiencies of the different mismatched sgRNAs to promote Cas9 nuclease activity on naked DNA and nucleosomes, 1 pmol of Cas9 was preincubated with 1.6 pmol of each sgRNA prior to the addition of 0.04 pmol of DNA or nucleosome substrates. An equal amount (0.04 pmol) of recombinant histone octamer was present in naked DNA reactions to control for the presence of histones in the nucleosome samples. The “No sgRNA” control samples have only the Cas9 enzyme (1 pmol) without the addition of an sgRNA, and the “No Cas9” control samples have only the wt-sgRNA RNA (1.6 pmol). Each experimental measurement was independently performed at least three times, and statistical significance was determined using Student's t test.
Bioinformatics Analysis of Potential Cas9 Target Sites in the Yeast Genome
Custom Perl scripts were used to identify potential Cas9 target sites in the yeast genome (SacCer3), defined as 23-nucleotide-long sequences ending with an NGG PAM sequence. A high resolution map of nucleosome positions (25) was used to identify potential Cas9 target sites in which the GG sequence of the PAM motif was located outside the nucleosome boundary, but the remainder of the target site partially overlapped with the nucleosome. This analysis revealed that 11.5% of the potential Cas9 target sites have this arrangement relative to nucleosomes. Even when potential target sites that overlapped with neighboring nucleosomes were excluded from the analysis, we still found that 6.9% of potential Cas9 target sites had this arrangement in chromatin.
Author Contributions
J. M. H. coordinated the study, performed the experiments shown, and co-wrote the paper. M. F. L. provided technical assistance, prepared all sgRNAs, and edited the paper. J. J. W. conceived the study, helped oversee the experiments, and co-wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Amelia Hodges, Dr. Peng Mao, and Dr. Michael Smerdon for helpful comments on the manuscript. We thank Dr. Mingrui Duan for providing reagents.
This work was supported by National Institutes of Health Grant ES002614 (to J. J. W.) from the NIEHS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PAM
- proto-spacer adjacent motif
- CRISPR
- clustered regularly interspaced short palindromic repeats
- sgRNA
- single guide RNA.
References
- 1. Doudna J. A., and Charpentier E. (2014) Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- 2. Hsu P. D., Lander E. S., and Zhang F. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., and Liu D. R. (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., and Sander J. D. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O., Cradick T. J., Marraffini L. A., Bao G., and Zhang F. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., Kosuri S., Yang L., and Church G. M. (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai S. Q., and Joung J. K. (2016) Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genetics 17, 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tycko J., Myer V. E., and Hsu P. D. (2016) Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell 63, 355–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sternberg S. H., Redding S., Jinek M., Greene E. C., and Doudna J. A. (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., Eckert M. R., Vogel J., and Charpentier E. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., and Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sternberg S. H., LaFrance B., Kaplan M., and Doudna J. A. (2015) Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang F., Taylor D. W., Chen J. S., Kornfeld J. E., Zhou K., Thompson A. J., Nogales E., and Doudna J. A. (2016) Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang F., Zhou K., Ma L., Gressel S., and Doudna J. A. (2015) A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 [DOI] [PubMed] [Google Scholar]
- 15. Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., Wyvekens N., Khayter C., Iafrate A. J., Le L. P., Aryee M. J., and Joung J. K. (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinz J. M., Laughery M. F., and Wyrick J. J. (2015) Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry 54, 7063–7066 [DOI] [PubMed] [Google Scholar]
- 17. Horlbeck M. A., Witkowsky L. B., Guglielmi B., Replogle J. M., Gilbert L. A., Villalta J. E., Torigoe S. E., Tjian R., and Weissman J. S. (2016) Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife 5, e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isaac R. S., Jiang F., Doudna J. A., Lim W. A., Narlikar G. J., and Almeida R. (2016) Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife 5, e13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunn K., and Griffith J. D. (1980) The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 8, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hovatter K. R., and Martinson H. G. (1987) Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc. Natl. Acad. Sci. U.S.A. 84, 1162–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh R., Kuscu C., Quinlan A., Qi Y., and Adli M. (2015) Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 43, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez A. G., and Anderson J. N. (2007) Nucleosome positioning determinants. J. Mol. Biol. 371, 649–668 [DOI] [PubMed] [Google Scholar]
- 23. Hodges A. J., Gallegos I. J., Laughery M. F., Meas R., Tran L., and Wyrick J. J. (2015) Histone sprocket arginine residues are important for gene expression, DNA repair, and cell viability in Saccharomyces cerevisiae. Genetics 200, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luger K., Rechsteiner T. J., and Richmond T. J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 25. Brogaard K., Xi L., Wang J. P., and Widom J. (2012) A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]