Abstract
Desmosomes are prominent adhesive junctions present between many epithelial cells as well as cardiomyocytes. The mechanisms controlling desmosome assembly and remodeling in epithelial and cardiac tissue are poorly understood. We recently identified protein palmitoylation as a mechanism regulating desmosome dynamics. In this study, we have focused on the palmitoylation of the desmosomal cadherin desmoglein-2 (Dsg2) and characterized the role that palmitoylation of Dsg2 plays in its localization and stability in cultured cells. We identified two cysteine residues in the juxtamembrane (intracellular anchor) domain of Dsg2 that, when mutated, eliminate its palmitoylation. These cysteine residues are conserved in all four desmoglein family members. Although mutant Dsg2 localizes to endogenous desmosomes, there is a significant delay in its incorporation into junctions, and the mutant is also present in a cytoplasmic pool. Triton X-100 solubility assays demonstrate that mutant Dsg2 is more soluble than wild-type protein. Interestingly, trafficking of the mutant Dsg2 to the cell surface was delayed, and a pool of the non-palmitoylated Dsg2 co-localized with lysosomal markers. Taken together, these data suggest that palmitoylation of Dsg2 regulates protein transport to the plasma membrane. Modulation of the palmitoylation status of desmosomal cadherins can affect desmosome dynamics.
Keywords: cadherin, desmoglein, desmosome, lipid raft, protein palmitoylation
Introduction
Desmosomes are multiprotein complexes that serve as points of contact for the intermediate filament cytoskeleton of neighboring cells of a given tissue. Desmosomes are particularly prominent in skin and cardiac tissue, and disruption of desmosomal adhesion results in a variety of epithelial blister phenotypes and ventricular cardiomyopathy, respectively (1). The transmembrane protein core of the desmosome is comprised of the desmosomal cadherins, desmogleins, and desmocollins (2, 3). These single-pass transmembrane adhesion proteins interact extracellularly to mediate cell-cell adhesion (4). The intracellular domains of the desmosomal cadherins recruit desmosomal plaque proteins (plakophilins and plakoglobin) and recruit the intermediate filament cytoskeleton via interactions with plakin family members (i.e. desmoplakin). In humans, there are four desmoglein genes (Dsg1–4). Dsg1, Dsg3, and Dsg4 are expressed in complex stratified epithelial tissues, whereas Dsg2 is widely expressed in a variety of epithelial tissues as well as in cardiomyocytes (2, 5, 6). Disruption of desmosomal adhesion through inactivation of desmoglein adhesive activity results in a variety of cardiocutaneous syndromes (7), underlining the importance of desmogleins in the maintenance of strong cell-cell adhesion.
Protein palmitoylation is a reversible posttranslational modification whereby a 16-carbon fatty acid (palmitate) is linked to specific cysteine residues via a labile thioester linkage (8, 9). Palmitoylation of cellular proteins is thought to influence protein function by increasing their association with cellular membranes or membrane microdomains and thereby regulating diverse protein activities, including protein localization, trafficking, activity, and stability (10). Unlike other lipid moieties added to cellular targets, palmitoylation of cysteine residues has been shown to be a reversible posttranslational modification. The best studied example of reversible protein palmitoylation is that of H-RAS. This acylation-deacylation cycle is important for the proper trafficking of H-RAS between the Golgi apparatus and the plasma membrane. Palmitoylation of both H-RAS and N-RAS occurs on membranes of the Golgi apparatus and increases their affinity for cellular membranes and promotes trafficking to the plasma membrane, where deacylation occurs, leading to the return of the deacylated proteins to the Golgi apparatus (11, 12).
Although the composition of the desmosome has been extensively studied, relatively little is known regarding the mechanisms controlling the assembly and remodeling of this junction. We recently demonstrated that several desmosomal components are palmitoylated in cultured cells and that preventing the palmitoylation of plakophilin-2 and 3 resulted in disruption of desmosomal adhesion through a dominant-negative mechanism (13). These findings suggest that palmitoylation plays an important regulatory role in desmosome assembly, stability, or adhesive strength. In this study, we characterized the role of palmitoylation on the localization of Dsg2. We identified two cysteine residues in the cytoplasmic tail of Dsg2 as palmitoylated residues and determined that palmitoylation affects the trafficking of Dsg2 to the plasma membrane as well as the stability of the protein.
Results
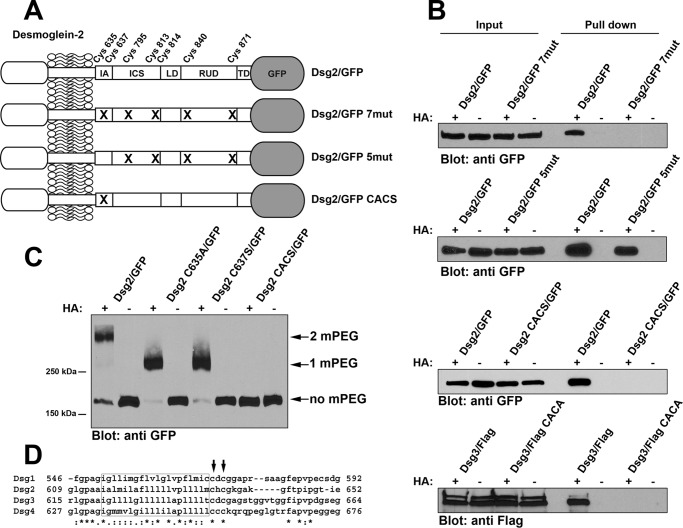
Previous work from our laboratory demonstrated that several desmosomal components were palmitoylated in cultured cells, including the desmosomal cadherins (13). We chose to more closely examine the effects of palmitoylation on the localization and dynamics of Dsg2. We generated Dsg2 fused to monomeric enhanced green fluorescent protein (Dsg2/GFP) as well as Dsg2/GFP mutants in which the cysteine residues present in the cytoplasmic domain were mutated (Fig. 1A). When stably expressed in A431 cells, wild-type Dsg2/GFP was palmitoylated whereas mutant desmoglein 2/GFP 7mut with all seven cytoplasmic cysteine residues mutated was not (Fig. 1B). Furthermore, restoring two cysteine residues (Cys-635 and Cys-637) in the mutant construct Dsg2/GFP 5mut resulted in the restoration of palmitoylation. Mutating only cysteine 635 and cysteine 637 (Dsg2/GFP CACS), leaving the other five cysteine residues intact, resulted in abrogation of palmitoylation (Fig. 1B).
FIGURE 1.
Cysteine residues in the IA domain of Dsg2 and Dsg3 are palmitoylated. A, the relative location of the seven cysteine residues present in the cytoplasmic tail of Dsg2. The extracellular domain of the Dsg2 is not shown in detail. Dsg2/GFP is wild-type full-length Dsg2 with monomeric GFP fused to the carboxyl terminus. Dsg2/GFP 7mut has all seven cysteine residues mutated. Dsg2/GFP 5mut retains Cys-635 and Cys-637 but has the five distal cysteines mutated. Dsg2/GFP CACS contains only the mutations C635A and C637S. IA, intracellular anchor; LD, linker domain; RUD, repeat unit domain; TD, terminal domain. B, A431 cells stably expressing wild-type Dsg2/GFP and the various mutant Dsg2/GFP were analyzed using the acyl biotin exchange assay. 10% of the input lysate used for each pulldown was loaded (left). Streptavidin-agarose pulldowns of biotin-labeled proteins were also loaded (right). Immunoblot analysis was performed using anti-GFP (JL-8) to detect Dsg2/GFP fusion proteins and anti-FLAG to detect FLAG-tagged Dsg3. C, A431 cells stably expressing wild-type Dsg2/GFP or various point mutants (Dsg2 C635A/GFP, Dsg2 C637S/GFP, and Dsg2/GFP CACS) were analyzed by mass tag labeling. Immunoblot analysis was performed using anti-GFP (JL-8) to detect Dsg2/GFP fusion proteins. The arrows indicate the bands migrating at the expected molecular weight for the incorporation of two mPEG, one mPEG, or no mPEG moieties. Acyl biotin exchange assays and mass tag labeling assays were repeated three times using independent cell cultures for each experiment. D, amino acid sequence alignment of the four human Dsg proteins. The predicted transmembrane segments are boxed, and the cysteine residues corresponding to Cys-635 and Cys-637 of Dsg2 are indicated by arrows.
We further investigated the palmitoylation of cysteine 635 and 637 by generating single point mutants (Dsg2 C635A/GFP and Dsg2 C637S/GFP) fused to GFP and expressed these fusion proteins in A431 cells. We performed mass tag labeling as described recently by Percher et al. (14) (Fig. 1C). A significant pool of wild-type Dsg2/GFP migrated at a molecular weight consistent with the incorporation of two mPEG moieties, and a faint band could be detected at the molecular weight corresponding to the addition of one mPEG.2 Dsg2 C635A/GFP and Dsg2 C637S/GFP were found to migrate at the molecular weight corresponding to addition of a single mPEG moiety. This analysis suggests that each palmitoylated cysteine is modified independently of the other and that each does not depend on previous modification of the adjacent cysteine. In addition, the results show that the majority of Dsg2 is palmitoylated on both cysteine 635 and cysteine 637. Dsg2/GFP CACS did not incorporate mPEG, consistent with the acyl-biotin exchange results shown in Fig. 1B. Comparing the sequences of the human desmoglein proteins (Dsg1, 2, 3, and 4) revealed that all four proteins have cysteines in the positions that correspond to Cys-635 and Cys-637 in Dsg2 (Fig. 1D). Mutating murine Dsg3 at the corresponding cysteine residues (i.e. Cys-640 and Cys-642) also resulted in abrogation of palmitoylation (Fig. 1B). These data demonstrate that palmitoylation of the Dsg proteins occurs at two sites corresponding to conserved cysteine residues in the intracellular anchor (IA) domain.
Next we determined the subcellular localization of wild-type and mutant Dsg2/GFP proteins that were stably expressed in A431 cells (Fig. 2). Wild-type Dsg2/GFP was localized in a punctate staining pattern at cell-cell borders and co-localized with the desmosomal plaque protein desmoplakin (Fig. 2, A–C). Similarly, Dsg2/GFP 5mut, the construct with five sites mutated that was still palmitoylated (Fig. 1B), co-localized with desmoplakin at cell-cell borders in a punctate pattern (Fig. 2, G–I). In contrast, Dsg2/GFP 7mut (Fig. 2, D–F) and Dsg2/GFP CACS (Fig. 2, J–L), two Dsg2/GFP fusion proteins that were palmitoylation-deficient, were largely localized at cell-cell borders in a linear pattern. A pool of these non-palmitoylated Dsg2/GFP proteins was also localized in a perinuclear region.
FIGURE 2.
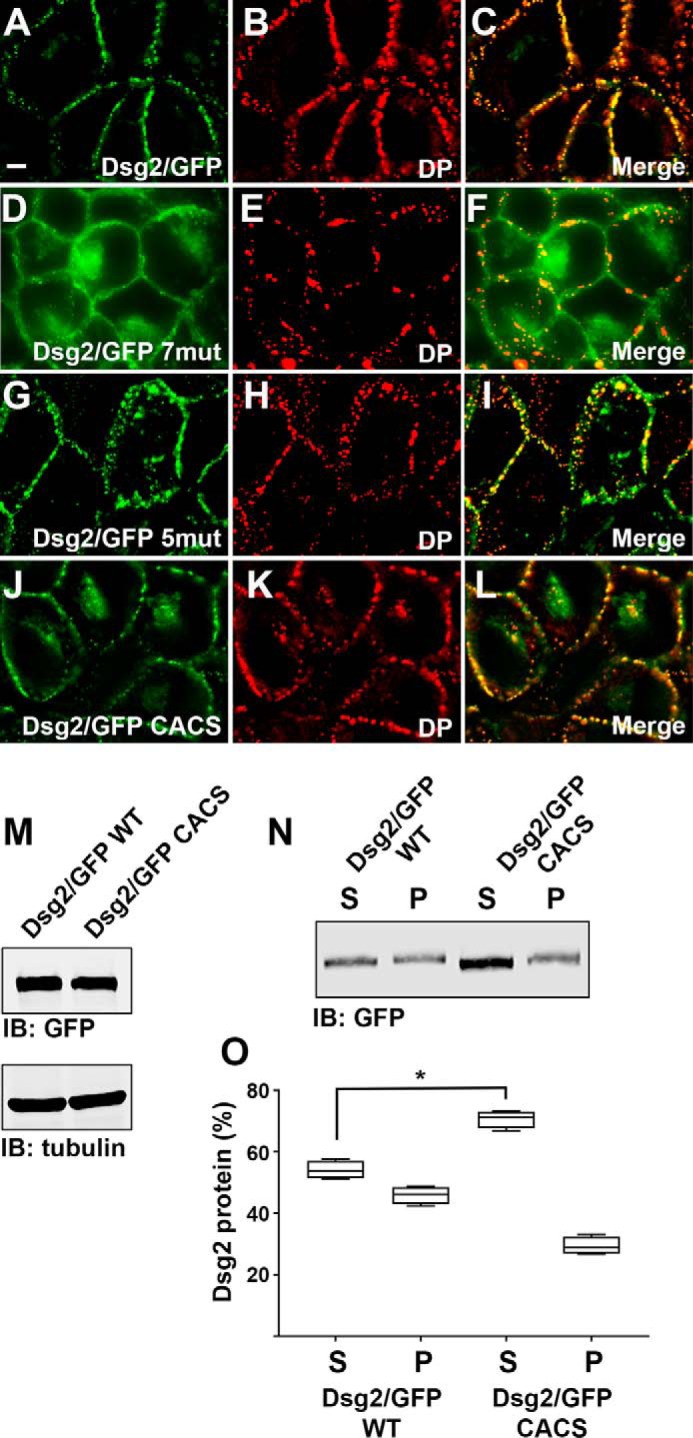
Palmitoylation-deficient Dsg2 localizes to a perinuclear subcellular compartment. A–L, A431 cells stably expressing wild-type Dsg2/GFP (A–C), Dsg2/GFP 7mut (D–F), Dsg2/GFP 5mut (G–I), and Dsg2/GFP CACS (J–L) were immunostained using anti-desmoplakin antibody, and co-localization of GFP with desmoplakin is shown. The localization of Dsg2/GFP fusion proteins was determined in at least three independent cultures grown on individual coverslips. Scale bar = 10 μm. M, whole cell lysates were prepared from A431 cells expressing wild-type Dsg2/GFP and Dsg2/GFP CACS. Lysates were subjected to immunoblot (IB) analysis. Immunoblot analysis was performed using cell lysates prepared from three different cultures. These data demonstrate equal expression of the GFP fusion proteins. N, A431 cells expressing Dsg2/GFP and Dsg2/GFP CACS were separated into Triton X-100-insoluble (P) and Triton X-100-soluble (S) fractions and subjected to immunoblot analysis using anti-GFP. O, quantitation of soluble and insoluble fractions shown in N. The lysates were prepared from four independent cultures, and immunoblot analysis band intensities were collected using LiCor Odyssey infrared scanning. Student's t test was performed to determine differences in solubility (*, p < 0.05). DP, desmoplakin.
Immunoblot analysis of cell lysates prepared from A431 cells expressing Dsg2/GFP and Dsg2/GFP CACS demonstrated that each fusion protein is expressed at similar levels (Fig. 2M). Comparing the Triton X-100 solubility of wild-type Dsg2/GFP with Dsg2/GFP CACS revealed an increase in the amount of the CACS mutant present in the Triton X-100 soluble fraction (70.6% of total ± 1.4%) (Fig. 2, N and O) compared with wild-type Dsg2/GFP (54.1% of total ± 1.5%) and a decrease in the amount of the insoluble pellet fraction.
Palmitoylation-deficient Desmoglein 2 Displays Altered Desmosome Assembly Dynamics
Using a calcium switch assay, we tested the ability of Dsg2/GFP fusion proteins to incorporate into desmosomes. We generated HaCaT keratinocytes stably expressing wild-type Dsg2/GFP and the palmitoylation-deficient mutant Dsg2/GFP CACS. Dsg2/GFP fusion proteins in HaCaT cells grown in medium containing low extracellular calcium (50 μm Ca2+) localized in a diffuse cytoplasmic pattern (Fig. 3, A and E). Upon elevating the calcium concentration to 1.8 mm (high calcium), within 30 min both Dsg2/GFP and Dsg2/GFP CACS partially localized to cell-cell borders. After 3 h in high calcium-containing medium, wild-type Dsg2/GFP was almost entirely found at cell-cell borders (Fig. 3C) and was identical to the localization seen after 18 h in high calcium medium (Fig. 3D). In contrast, a significant pool of Dsg2/GFP CACS was still present in the cytoplasm at the 3-h time point, and this cytoplasmic localization was still seen after 18 h in high calcium-containing medium (Fig. 3, F–H). Quantification of fluorescence intensity (Fig. 3I) revealed that wild-type Dsg2/GFP fluorescence intensity at the plasma membrane increased following addition of calcium to the culture medium with a corresponding decrease in the cytoplasmic fluorescence intensity. In contrast, Dsg2/GFP CACS fluorescence intensity at cell-cell borders and the cytoplasm did not change dramatically over the 18-h time course.
FIGURE 3.

Palmitoylation-deficient Dsg2/GFP CACS shows delayed incorporation into desmosomes. A–H, HaCaT keratinocytes stably expressing wild-type Dsg2/GFP (A–D) and Dsg2/GFP CACS (E–H) were subjected to a calcium switch assay. Calcium switch assays were performed three times using different cell cultures on three different days. Subcellular localization of the GFP fusion protein was determined at the indicated times following elevation of the extracellular calcium concentration to 1.8 mm. Scale bar = 10 μm. I, the fluorescence intensity was measured at cell-cell borders and the cytoplasm, total signal intensity was determined, and the percentage of signal in the cytosol and membrane was determined. A minimum of 20 individual cells at each time point were examined, individual data points are shown, and the standard deviation is depicted by error bars. Note the continued presence of cytoplasmic Dsg2/GFP CACS. An unpaired Student's t test was used to determine differences in the percentage of cytoplasmic signal after overnight addition of calcium (*, p < 0.0001).
Palmitoylation-deficient Desmogleins Partitions with Lipid Raft Components
Palmitoylation is widely believed to increase the association of proteins with cellular membranes and lipid raft microdomains in particular (15). Proteomic analysis of isolated lipid raft microdomains revealed an enrichment of proteins known to be palmitoylated (16). Recent evidence has demonstrated that numerous desmosomal components are also associated with lipid rafts, including desmogleins (17–19). In addition, mutation of the cysteine palmitoylated in plakophilin-3 decreased plakophilin-3 association with lipid rafts (13).
We examined the ability of Dsg2/GFP, Dsg2/GFP CACS Dsg3/FLAG, and Dsg3/FLAG CACA to associate with lipid rafts by sucrose gradient centrifugation. Cell lysates were prepared from A431 cells expressing wild-type desmoglein or palmitoylation-deficient desmoglein mutants fused to GFP or FLAG, and cell lysates were separated by sucrose gradient centrifugation. Co-sedimentation of the GFP-tagged Dsg2 or FLAG-tagged Dsg3 with the lipid raft component Caveolin1 was examined by immunoblot analysis (Fig. 4). Wild-type Dsg2 (Fig. 4A) and Dsg3 (Fig. 4B) partition with the lipid raft component caveolin-1 in fractions 4 and 5, indicating an association with lipid raft components. Palmitoylation-deficient Dsg2 and Dsg3 also partition to fractions containing caveolin-1, demonstrating that these mutant are also capable of associating with lipid raft microdomains. These data suggest that palmitoylation of the desmogleins is not the primary mechanism directing these transmembrane proteins to lipid raft microdomains and that other, still unidentified mechanism are likely responsible for lipid raft targeting of these desmosomal cadherins.
FIGURE 4.
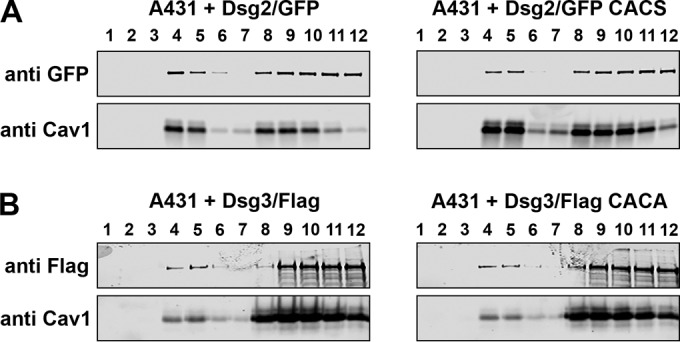
Palmitoylation-deficient desmogleins associate with lipid raft microdomains. A and B, cell lysates were prepared from A431 cells expressing Dsg2/GFP or Dsg2/GFP CACS (A) and A431 cells expressing Dsg3/FLAG or Dsg3/FLAG CACA (B). Lysates were separated by sucrose gradient centrifugation, and fractions were analyzed by immunoblot analysis and anti-GFP and anti-FLAG. The presence of caveolin-1 (Cav1) in fractions 4 and 5 identify lipid raft-containing fractions.
Palmitoylation-deficient Desmoglein-2 Partially Co-localizes with Golgi Compartment and Lysosomal Markers
We sought to identify the perinuclear subcellular compartment to which the Dsg2/GFP CACS fusion protein localized. We compared the localization of wild-type Dsg2/GFP and Dsg2/GFP CACS with that of the Golgi network using antibodies specific for the trans Golgi network marker protein Golgin-97. Golgin-97 partially co-localized with the GFP signal in A431 cells expressing Dsg2/GFP CACS (Fig. 5, E–H), whereas there was no discernible co-localization with the wild-type Dsg2/GFP fusion protein (Fig. 5, A–D). Additionally, we examined the ability of the Dsg2/GFP fusion proteins to co-localize with the lysosomal marker Lamp-1. Lamp-1 partially co-localized with Dsg2/GFP CACS, suggesting that the pool of Dsg2/GFP CACS associated with the Golgi compartment is being degraded by a lysosomal pathway (Fig. 6, E–H). No co-localization was observed between wild-type Dsg2/GFP and Lamp-1 (Fig. 6, A–D).
FIGURE 5.
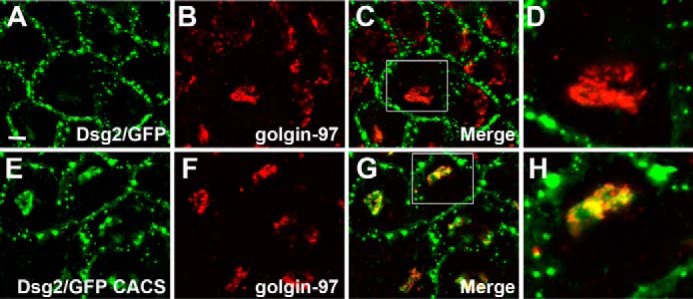
Dsg2/GFP CACS co-localizes with the Golgi network. A–H, A431 cells expressing wild-type Dsg2/GFP (A–D) and Dsg2/GFP CACS (E–H) were stained using anti-Golgin-97. Co-localization of Golgin-97 with the GFP fusion proteins is shown. The boxed areas in C and G are shown at higher magnification in D and H, respectively. Immunofluorescence microscopy demonstrating co-localization was repeated three times. Scale bar = 10 μm.
FIGURE 6.
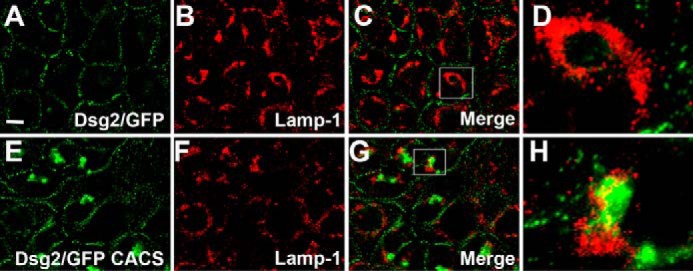
Dsg2/GFP CACS partially co-localizes with lysosomes. A–H, A431 cells expressing wild-type Dsg2/GFP (A–D) and Dsg2/GFP CACS (E–H) were stained using anti-LAMP-1. Co-localization of Lamp-1 with the GFP fusion proteins is shown. The boxed areas in C and G are shown at higher magnification in D and H, respectively. Immunofluorescence microscopy demonstrating co-localization was repeated three times. Scale bar = 20 μm.
Palmitoylation-deficient Dsg2 Is Degraded More Rapidly Than Wild-type Dsg2
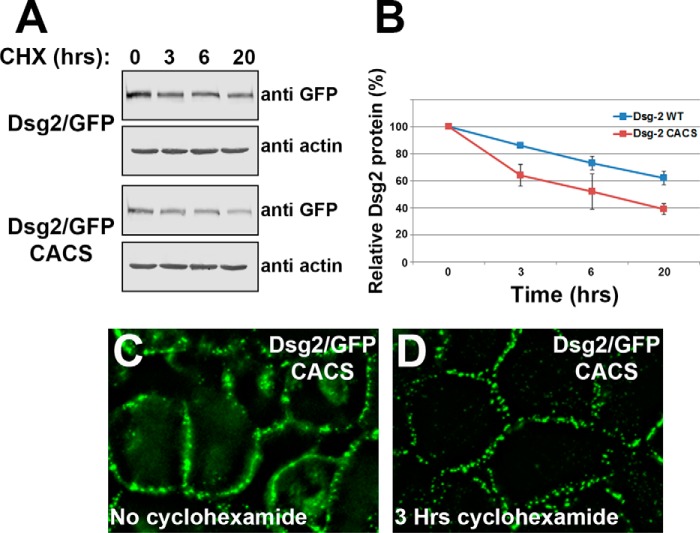
We compared the stability of wild-type Dsg2/GFP and Dsg2/GFP CACS by treating cells with the protein synthesis inhibitor cycloheximide. Cell lysates were prepared after various times of growth in medium containing cycloheximide, and Dsg2/GFP levels were compared by immunoblot analysis (Fig. 7A). After 20 h in medium containing cycloheximide, wild-type Dsg2/GFP was relatively stable, and ∼60% of the fusion protein remained. In contrast, only 40% of Dsg2/GFP CACS remained after 20 h (Fig. 7B). As a loading control, cell lysates were probed with anti-actin antibody. A difference in degradation of the Dsg2/GFP CACS from that of wild-type Dsg2/GFP was observed as early as 3 h. We examined the subcellular localization of Dsg2/GFP CACS after 3 h of growth in medium containing cycloheximide and observed a distinct clearing of the perinuclear compartment compared with untreated cells (Fig. 7, C and D). Fluorescence recovery after photobleaching analysis and cell surface biotinylation assays revealed no difference in the internalization rates or in the mobile fraction of Dsg2/GFP CACS compared with wild-type Dsg2/GFP (data not shown). Taken together, these data suggest that the perinuclear pool of Dsg2/GFP CACS is being degraded rather than being transported to the plasma membrane, whereas the Dsg2/GFP CACS residing in the desmosome is relatively stable despite not being palmitoylated. Thus, palmitoylation of Dsg2 affects transport to the plasma membrane but not its stability at the desmosome.
FIGURE 7.
Palmitoylation-deficient Dsg2/GFP CACS is degraded more rapidly than wild-type Dsg2/GFP. A, A431 cells expressing wild-type Dsg2/GFP or Dsg2/GFP CACS were grown in medium containing 30 μg/ml cycloheximide (CHX) for the indicated times. Cell lysates were prepared, and equal protein was loaded for immunoblot analysis (cycloheximide treatments were performed in triplicate, and representative blots are shown). B, band intensities were collected using Odyssey infrared scanning of three independent experiments. Error bars represent the standard deviation. C and D, subcellular localization of Dsg2/GFP CACS stably expressed in A431 cells that were grown in control medium and in medium containing 30 μg/ml cycloheximide for 3 h.
Discussion
Desmosomes are prominent adhesive junctions in a variety of epithelial tissues, and disruption of desmosomal adhesion has severe effects on tissue homeostasis (20–22) and important consequences for the integrity of epithelial and cardiac tissues (23, 24). In this study, we demonstrate that palmitoylation of the cytoplasmic domain affects the transport of Dsg2 to the plasma membrane, where it is incorporated into junctional complexes. Interestingly, when incorporated into the desmosomal plaque, palmitoylation-deficient desmoglein was retained at the junction in a manner indistinguishable from the wild type. Failure to properly traffic to the plasma membrane resulted in accelerated degradation of palmitoylation-deficient Dsg2 through a lysosomal pathway.
Palmitoylation of cellular proteins has been shown to affect protein localization and targeting to specific membrane compartments (25, 26). For example, reversible palmitoylation regulates Ras localization and activation through an acylation/deacylation cycle (11). Previous studies from our laboratory have demonstrated that palmitoylation-deficient plakophilins fail to efficiently localize to the plasma membrane and act in a dominant-negative manner to disrupt adhesion (13). Mechanisms regulating the transport to and stability of desmogleins at the plasma membrane are beginning to be elucidated (27). Here we have shown that preventing the palmitoylation of Dsg2 has less pronounced effects on desmosome dynamics than when plakophilin palmitoylation is disrupted. In the case of Dsg2, mutation of the two cysteines in the IA domain that can be palmitoylated results in defects in Dsg2 trafficking and degradation. In contrast to the altered Dsg2 protein turnover shown here, plakophilin-3 stability was not noticeably affected by preventing its palmitoylation (data not shown).
For our experiments, we chose to examine the behavior of Dsg2 in A431 cells because this cell line assembles numerous desmosomes, and the composition of the junctions in this cell line are well documented (28–30). A confounding issue with our cell culture approach is the endogenous expression of other desmosomal cadherins (Dsg3, Dsc2, and Dsc3) as well as endogenously expressed Dsg2. Our experiments demonstrate that A431 cells were capable of processing and transporting wild-type Dsg2/GFP to sites of cell-cell contact where it co-localized with endogenously expressed desmoplakin. Dsg2 palmitoylation-deficient mutants (Dsg2/GFP CACS and Dsg2/GFP 7mut) also partially localized with desmoplakin but, in contrast to the wild-type protein, also localized in a non-punctate distribution at the plasma membrane and to a perinuclear compartment. We suspect that Dsg2/GFP palmitoylation mutants were able to associate with endogenous desmosomal components, and when they were incorporated into desmosomes, they were resistant to degradation (Fig. 7). The nature of these interactions is currently unclear, but it is possible that desmosomal cadherins interact through lateral interactions at the plasma membrane. Palmitoylation of transmembrane proteins has been proposed to affect local packing of the protein in the context of the plasma membrane (26, 31). Thus, packing of the transmembrane components of the desmosome in the plane of the plasma membrane to assemble the desmosomal plaque could be facilitated by palmitoylation of the desmosomal cadherins. Alternatively, desmosomal plaque proteins (plakoglobin and plakophilins) may act to cluster the desmosomal cadherins at the plasma membrane, and these interactions may be independent of desmoglein palmitoylation. However, based on our findings, we predict that palmitoylation of the desmosomal cadherins promotes proper membrane targeting and incorporation into stable desmosomal plaques.
Palmitoylation has been shown to affect protein association with membranes in a number of divergent systems (25). One consequence of disrupted Dsg2 palmitoylation could be altered retention time at the plasma membrane. We tested the effect of protein palmitoylation on the ability of Dsg2/GFP to maintain plasma membrane association by fluorescence recovery after photobleaching analysis. Analysis of the Dsg2/GFP localized at cell-cell contacts revealed no difference in fluorescence recovery between wild-type Dsg2/GFP and Dsg2/GFP CACS. Additionally, we performed cell surface biotinylation assays and again observed no significant difference in the rates of internalization of the Dsg2/GFP fusion proteins (data not shown). These results show that the stability of Dsg2 at the cell surface is not affected by loss of palmitoylation.
Palmitate is added to specific cysteine residues through the action of palmitoyl acyltransferase activity. Recent studies have identified the DHHC family of proteins as palmitoyl acyltransferases (32). There are 23 evolutionarily conserved ZDHHC genes in mammals; recent studies have demonstrated that deregulation of several ZDHHC proteins is linked to carcinogenesis (9) and skin differentiation (33–35). The substrate specificity of the different ZDHHCs is currently unknown. We hypothesize that a limited number of ZDHHCs present in the Golgi compartment are responsible for palmitoylation of the desmogleins. Identification of these palmitoyl acyltransferases is a source of ongoing investigation, and targeted inhibition of the relevant ZDHHC is likely to have effects on desmosome assembly and dynamics.
At the beginning of our study, we examined the conservation of cysteine residues present in the cytoplasmic domain of desmogleins, and it was interesting to note that, in addition to the membrane-proximal cysteines in Dsg2, there were several other cysteines well conserved across different species. In particular, the two cysteines present near the plakoglobin interaction domain (human Dsg2 cysteines 813 and 814) in the ICS domain are conserved in all members of the desmoglein family. These cysteines are also present in the ICS domain of the desmocollins. A previous study had shown that cysteine 813 in human Dsg2 was mutated in arrhythmogenic right ventricular cardiomyopathy patients (36). When cysteine 813 was mutated to arginine, the authors noted abnormal migration of the protein in SDS-PAGE and speculated that this cysteine may be posttranslationally modified. Although adjacent Cys residues are candidates for palmitoylation, we found no evidence that these cysteines are palmitoylated in either Dsg2 or Dsg3 (Fig. 1). Preliminary studies suggest that this motif in Dsc2a is also not palmitoylated.3 However, our experiments do not exclude the possibility that these residues in the desmosomal cadherins are modified in some other manner, as Cys residues can be modified in many ways (37, 38).
In the future, it will be beneficial to determine the mechanisms regulating desmosomal cadherin palmitoylation and the effect this posttranslational modification has on desmosome assembly and dynamics. Our study focused on Dsg2 palmitoylation in epithelial cells, but the effect of Dsg2 palmitoylation in cardiomyocytes has important implications for the understanding of Dsg2 mutations in the context of arrhythmogenic right ventricular cardiomyopathy. Identification of the palmitoyl acyltransferases involved in the epithelium and cardiac tissue will be an important component of this analysis.
Experimental Procedures
Cell Culture
The A431 cervical squamous cell carcinoma cell line was obtained from the ATCC (Manassas, VA). A431 and HaCaT keratinocytes were routinely grown in DMEM (Sigma) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT). Generation of retroviral particles and retroviral infection have been described previously (13, 39, 40). Retrovirally infected cell populations were routinely grown in DMEM containing 500 μg/ml G418 (Mediatech Inc., Herndon, VA.). For calcium switch experiments, HaCaT cells were grown in DMEM containing 50 μm calcium supplemented with 10% dialyzed fetal bovine serum for at least 48 h prior to the addition of calcium to a final concentration of 1.8 mm CaCl2. Fluorescence images were collected at the indicated times following addition of calcium.
Antibodies
Anti-desmoplakin (20B6) was described previously (41). Commercially available antibodies were as follows. Anti-GFP (clone JL-8 used at 1:1000 dilution for immunoblot analysis) was purchased from Clontech. Anti-FLAG epitope antibody (DYKDDDDK tag) was purchased from Cell Signaling Technology (used at 1:1000, Boston, MA). Anti-Golgin-97 (clone CDF4) was purchased from Thermo Scientific (Rockford, IL) and diluted 1:100 for immunofluorescence microscopy. Anti-Lamp-1 hybridoma (clone H4A3), anti-β-tubulin (clone E7), and anti-actin (clone JLA20) were purchased from the Developmental Studies Hybridoma Bank at the University of Iowa. Hybridoma conditioned medium was prepared, stored at −80 °C, and used at 1:10 dilution for immunoblots and undiluted for immunofluorescence microscopy.
Detergent Extraction
Cell lysates were prepared as described previously (41, 42). Triton X-100 soluble (s) and insoluble (p) fractions were isolated by first washing cell monolayers with 1× phosphate-buffered saline. Cells grown in a T25 flask were scraped into 1 ml of Triton X-100-containing buffer (10 mm Tris-HCl (pH 7.0), 0.5% Triton X-100 (Sigma), 5 mm EDTA, 2 mm EGTA, 30 mm sodium fluoride, 40 mm β-glycerophosphate, 10 mm sodium pyrophosphate, 2 mm sodium orthovanadate, and 2 mm phenylmethylsulfonyl fluoride) and incubated on ice while shaking for 15 min. The insoluble material was collected by centrifugation for 15 min at 14,000 × g. The insoluble pellet was washed once with Triton X-100 lysis buffer prior to resuspension of the pellet in 1 ml of SDS containing buffer (10 mm Tris HCl (pH 8.0), 2% SDS, and 2 mm EDTA). Lysates were prepared in Laemmli sample buffer, and proteins were resolved by SDS-PAGE.
Lipid Raft Analysis
Cell lysates were prepared from A431 cell lines expressing wild-type or mutant desmogleins and analyzed by sucrose gradient centrifugation as described previously (43).
Generation of cDNA Constructs
Human Dsg2 cDNA (GenBank no. BC099655) was purchased from the ATCC. The β globin 5′ untranslated sequence from pSPUTK (44) (Stratagene/Agilent Technologies, Santa Clara, CA) was added upstream of the open reading frame. PCR was used to remove the stop codon and add sequences encoding monomeric (45) GFP. To generate monomeric GFP, we used PCR to mutate alanine 206 to lysine (A206K) using pEGFP-N3 (Clontech) as a template for PCR. Monomeric GFP was added to the 3′ end of the Dsg2 open reading frame. The fully assembled cDNA was ligated to the retroviral expression vector LZBob neo (13, 46, 47) as an AfeI/NotI fragment. LZBob neo is a modified version of LZRS ms neo (46) containing a modified multiple cloning site to facilitate cloning of cDNA fragments. Full-length murine desmoglein 3 with a carboxyl-terminal FLAG epitope has been described previously (48). The Dsg3/FLAG cDNA was ligated to LZBob neo as described for Dsg2/GFP. Dsg2 and Dsg3 point mutations were generated using the QuikChange site directed mutagenesis kit (Stratagene/Agilent Technologies). The modified cDNAs were completely sequenced and shown to have no unintended changes.
Acyl Biotin Exchange and Mass Tag Labeling
For acyl biotin exchange, A431 cells expressing wild-type and mutant Dsg2/GFP were grown to 80–90% confluence, and lysates were prepared as described previously (49). Cells were harvested in lysis buffer (150 mm NaCl, 50 mm Tris HCl (pH 7.4), 1% Triton X-100, and 5 mm EDTA) containing 10 mm N-ethylmaleimide (NEM, Sigma), and lysates were collected by scraping on ice and passed through a 25-gauge needle. Samples were chloroform-methanol-precipitated, and the pellet was allowed to air-dry for 2–3 min. The pellet was resuspended in 300 μl of 4% SDS buffer (4% sodium dodecyl sulfate, 50 mm Tris HCl (pH 7.4), and 5 mm EDTA) and diluted 4-fold in lysis buffer containing 10 mm NEM. The samples were incubated at 4 °C overnight with gentle agitation. NEM was removed by performing three sequential chloroform-methanol precipitations, and following the last precipitation, the pellet was resuspended in 100 μl of 4% SDS buffer. The sample was divided in two, and one half was diluted 5-fold with lysis buffer containing 0.7 m hydroxylamine (+HA) (0.7 m hydroxylamine, 1 mm HPDP-biotin, 0.2% Triton X-100, 1 mm PMSF, and 1× protease inhibitor mixture (Sigma), and the other sample was diluted 5-fold with buffer lacking hydroxylamine (−HA). Samples were incubated at room temperature with gentle rocking for 1 h. Samples were chloroform-methanol-precipitated three times, and the final pellets were resuspended in 240 μl of 4% SDS buffer and diluted with 960 μl of low-HPDP-biotin buffer (150 mm NaCl, 50 mm Tris HCl (pH 7.4), 5 mm EDTA, 0.2 mm HPDP-biotin, 0.2% Triton X-100, 1 mm PMSF, and 1× protease inhibitor mixture). Samples were incubated at room temperature with gentle agitation for 1 h prior to three sequential chloroform-methanol precipitations. The final pellet was resuspended in 75 μl of 2% SDS buffer (2% SDS, 50 mm Tris HCl (pH 7.4), and 5 mm EDTA), and samples were diluted to 0.1% SDS in lysis buffer containing 0.2% Triton X-100, 1× protease inhibitor mixture, and 1 mm PMSF. Protein concentrations were determined, and equal amounts of protein were added to streptavidin-agarose. Biotin-labeled proteins were captured on streptavidin-agarose, and non-specifically bound proteins were removed by washing the beads in lysis buffer containing 0.1% SDS and 0.2% Triton X-100. Captured proteins were boiled in 2× Laemmli sample buffer and resolved by SDS-PAGE.
For mass tag labeling, we followed the procedure described by Percher et al. (14). Cell lysates were prepared in TEA buffer (50 mm triethanolamine (pH 7.3), 150 mm NaCl, and 5 mm EDTA) containing 4% SDS. 200 μg of total cellular protein was treated with a final concentration of 10 mm neutralized tris(2-carboxyethyl)phosphine for 30 min with end-over-end rotation. NEM was added to a final concentration of 25 mm, and rocking continued for 2 h. NEM was removed by three rounds of chloroform/methanol/H2O precipitation. The final pellet was resuspended in TEA buffer containing 0.2% Triton X-100. Samples were treated with 0.75 m NH2OH (+HA) or without hydroxylamine (−HA) and incubated at room temperature for 1 h. Excess hydroxylamine was removed with one round of chloroform/methanol/H2O precipitation, and the pellet was resuspended in TEA buffer containing 0.2% Triton X-100 supplemented with 1 mm mPEG (10 kDa, Sigma). Samples were incubated with rocking for 2 h, and reactions were terminated by one round of chloroform/methanol/H2O precipitation. The final pellet was suspended in 1× Laemmli sample buffer and resolved by SDS-PAGE.
Immunofluorescence Microscopy
Cells grown on glass coverslips were washed briefly in Hepes-buffered Hanks' balanced salt solution and fixed in 1% formaldehyde prepared from paraformaldehyde in Hepes-buffered Hanks' balanced salt solution. Antibody staining was performed as described previously (13, 40, 41). GFP fluorescence at cell-cell borders was measured using a Marianas live cell microscopy system consisting of a Zeiss Axiovert 200M microscope and SlideBook 6.0 imaging software (Intelligent Imaging Innovations Inc., Denver, CO) and equipped with a Stanford Research Systems laser ablation system (model NL100). Images were collected using a Plan-Apochromat ×63 (1.4 numerical aperture) objective. Z stack images were collected. Raw images were deconvolved and collapsed into a projection image prior to determination of fluorescence intensity.
Statistical Analysis
Comparisons between two groups used unpaired Student's t test. For immunoblot analysis, samples were prepared and resolved in triplicate, and band intensities were collected using LiCor Odyssey infrared scanning.
Author Contributions
J. K. W. conceived and coordinated the study and wrote the manuscript. B. J. R. designed and performed the experiments in Figs. 1–3 and 5–7. R. A. S. and K. R. J. designed and constructed the expression vectors and point mutants used in the generation of all data presented in Figs. 1–8. A. M. O. and M. G. M. performed the experiments and data analysis presented in Fig. 4. J. D. L. and A. P. K. generated the Dsg3 constructs analyzed in Fig. 1. All authors contributed intellectually to the design of experiments and had input into analyses throughout the study.
Acknowledgments
Microscopy images were acquired and processed at the University of Nebraska Medical Center Advanced Microscopy Core Facility supported by the Nebraska Research Initiative and the Eppley Cancer Center (P30CA036727) and Nebraska Center for Cellular Signaling CoBRE (National Institutes of Health P30GM106397). The anti-Lamp-1 hybridoma supernatant developed by J. T. August and J. E. K. Hildreth was obtained from the Developmental Studies Hybridoma Bank, created by NICHD, National Institutes of Health and maintained at the Department of Biology, University of Iowa, Iowa City, IA 52242.
The research reported in this publication was supported by an Institutional Development Award (Grant 5P30GM106397 to K. R. J.), by NIAMS, National Institutes of Health Grant R15AR065074 (to J. K. W.), and by National Institutes of Health Grants R01AR048266 (to A. P. K.) and F31AR066476 (to J. D. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
B. J. Roberts, R. A. Svoboda, K. R. Johnson, and J. K. Wahl, manuscript in preparation.
- mPEG
- methoxy-PEG-maleimide
- ARVC
- arrhythmogenic right ventricular cardiomyopathy
- IA
- intracellular anchor
- NEM
- N-ethylmaleimide
- HA
- hydroxylamine
- HPDP
- N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide.
References
- 1. Nitoiu D., Etheridge S. L., and Kelsell D. P. (2014) Insights into desmosome biology from inherited human skin disease and cardiocutaneous syndromes. Cell Commun. Adhes. 21, 129–140 [DOI] [PubMed] [Google Scholar]
- 2. Schäfer S., Koch P. J., and Franke W. W. (1994) Identification of the ubiquitous human desmoglein, Dsg2, and the expression catalogue of the desmoglein subfamily of desmosomal cadherins. Exp. Cell Res. 211, 391–399 [DOI] [PubMed] [Google Scholar]
- 3. Nuber U. A., Schäfer S., Schmidt A., Koch P. J., and Franke W. W. (1995) The widespread human desmocollin Dsc2 and tissue-specific patterns of synthesis of various desmocollin subtypes. Eur. J. Cell Biol. 66, 69–74 [PubMed] [Google Scholar]
- 4. Harmon R. M., and Green K. J. (2013) Structural and functional diversity of desmosomes. Cell Commun. Adhes. 20, 171–187 [DOI] [PubMed] [Google Scholar]
- 5. Schmelz M., Duden R., Cowin P., and Franke W. W. (1986) A constitutive transmembrane glycoprotein of Mr 165,000 (desmoglein) in epidermal and non-epidermal desmosomes: I: biochemical identification of the polypeptide. Eur. J. Cell Biol. 42, 177–183 [PubMed] [Google Scholar]
- 6. Franke W. W., Schumacher H., Borrmann C. M., Grund C., Winter-Simanowski S., Schlechter T., Pieperhoff S., and Hofmann I. (2007) The area composita of adhering junctions connecting heart muscle cells of vertebrates: III: assembly and disintegration of intercalated disks in rat cardiomyocytes growing in culture. Eur. J. Cell Biol. 86, 127–142 [DOI] [PubMed] [Google Scholar]
- 7. Broussard J. A., Getsios S., and Green K. J. (2015) Desmosome regulation and signaling in disease. Cell Tissue Res. 360, 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aicart-Ramos C., Valero R. A., and Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808, 2981–2994 [DOI] [PubMed] [Google Scholar]
- 9. Greaves J., and Chamberlain L. H. (2014) New links between S-acylation and cancer. J. Pathol. 233, 4–6 [DOI] [PubMed] [Google Scholar]
- 10. Greaves J., and Chamberlain L. H. (2011) DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 11. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., and Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 12. Goodwin J. S., Drake K. R., Rogers C., Wright L., Lippincott-Schwartz J., Philips M. R., and Kenworthy A. K. (2005) Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts B. J., Johnson K. E., McGuinn K. P., Saowapa J., Svoboda R. A., Mahoney M. G., Johnson K. R., and Wahl J. K. 3rd. (2014) Palmitoylation of plakophilin is required for desmosome assembly. J. Cell Sci. 127, 3782–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Percher A., Ramakrishnan S., Thinon E., Yuan X., Yount J. S., and Hang H. C. (2016) Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl. Acad. Sci. U.S.A. 113, 4302–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levental I., Grzybek M., and Simons K. (2010) Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49, 6305–6316 [DOI] [PubMed] [Google Scholar]
- 16. Yang W., Di Vizio D., Kirchner M., Steen H., and Freeman M. R. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brennan D., Peltonen S., Dowling A., Medhat W., Green K. J., Wahl J. K. 3rd, Del Galdo F., and Mahoney M. G. (2012) A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene 31, 1636–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Resnik N., Sepcic K., Plemenitas A., Windoffer R., Leube R., and Veranic P. (2011) Desmosome assembly and cell-cell adhesion are membrane raft-dependent processes. J. Biol. Chem. 286, 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stahley S. N., Saito M., Faundez V., Koval M., Mattheyses A. L., and Kowalczyk A. P. (2014) Desmosome assembly and disassembly are membrane raft-dependent. PLoS ONE 9, e87809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allen E., Yu Q. C., and Fuchs E. (1996) Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J. Cell Biol. 133, 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koch P. J., Mahoney M. G., Ishikawa H., Pulkkinen L., Uitto J., Shultz L., Murphy G. F., Whitaker-Menezes D., and Stanley J. R. (1997) Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J. Cell Biol. 137, 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samuelov L., Sarig O., Harmon R. M., Rapaport D., Ishida-Yamamoto A., Isakov O., Koetsier J. L., Gat A., Goldberg I., Bergman R., Spiegel R., Eytan O., Geller S., Peleg S., Shomron N., et al. (2013) Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat. Genet. 45, 1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stahley S. N., and Kowalczyk A. P. (2015) Desmosomes in acquired disease. Cell Tissue Res. 360, 439–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bezzina C. R., Lahrouchi N., and Priori S. G. (2015) Genetics of sudden cardiac death. Circ. Res. 116, 1919–1936 [DOI] [PubMed] [Google Scholar]
- 25. Linder M. E., and Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 26. Blaskovic S., Blanc M., and van der Goot F. G. (2013) What does S-palmitoylation do to membrane proteins? FEBS J. 280, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 27. Chen J., Nekrasova O. E., Patel D. M., Klessner J. L., Godsel L. M., Koetsier J. L., Amargo E. V., Desai B. V., and Green K. J. (2012) The C-terminal unique region of desmoglein 2 inhibits its internalization via tail-tail interactions. J. Cell Biol. 199, 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewis J. E., Wahl J. K. 3rd, Sass K. M., Jensen P. J., Johnson K. R., and Wheelock M. J. (1997) Cross-talk between adherens junctions and desmosomes depends on plakoglobin. J. Cell Biol. 136, 919–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Setzer S. V., Calkins C. C., Garner J., Summers S., Green K. J., and Kowalczyk A. P. (2004) Comparative analysis of armadillo family proteins in the regulation of a431 epithelial cell junction assembly, adhesion and migration. J. Invest. Dermatol. 123, 426–433 [DOI] [PubMed] [Google Scholar]
- 30. Bornslaeger E. A., Corcoran C. M., Stappenbeck T. S., and Green K. J. (1996) Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134, 985–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chum T., Glatzová D., Kvíčalová Z., Malínský J., Brdička T., and Cebecauer M. (2016) The role of palmitoylation and transmembrane domain in sorting of transmembrane adaptor proteins. J. Cell Sci. 129, 95–107 [DOI] [PubMed] [Google Scholar]
- 32. Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., and Kihara A. (2012) Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23, 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saleem A. N., Chen Y. H., Baek H. J., Hsiao Y. W., Huang H. W., Kao H. J., Liu K. M., Shen L. F., Song I. W., Tu C. P., Wu J. Y., Kikuchi T., Justice M. J., Yen J. J., and Chen Y. T. (2010) Mice with alopecia, osteoporosis, and systemic amyloidosis due to mutation in Zdhhc13, a gene coding for palmitoyl acyltransferase. PLoS Genet. 6, e1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu K. M., Chen Y. J., Shen L. F., Haddad A. N., Song I. W., Chen L. Y., Chen Y. J., Wu J. Y., Yen J. J., and Chen Y. T. (2015) Cyclic alopecia and abnormal epidermal cornification in Zdhhc13-deficient mice reveal the importance of palmitoylation in hair and skin differentiation. J. Invest. Dermatol. 135, 2603–2610 [DOI] [PubMed] [Google Scholar]
- 35. Mill P., Lee A. W., Fukata Y., Tsutsumi R., Fukata M., Keighren M., Porter R. M., McKie L., Smyth I., and Jackson I. J. (2009) Palmitoylation regulates epidermal homeostasis and hair follicle differentiation. PLoS Genet. 5, e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gehmlich K., Asimaki A., Cahill T. J., Ehler E., Syrris P., Zachara E., Re F., Avella A., Monserrat L., Saffitz J. E., and McKenna W. J. (2010) Novel missense mutations in exon 15 of desmoglein-2: role of the intracellular cadherin segment in arrhythmogenic right ventricular cardiomyopathy? Heart Rhythm. 7, 1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chalker J. M., Bernardes G. J., Lin Y. A., and Davis B. G. (2009) Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem. Asian J. 4, 630–640 [DOI] [PubMed] [Google Scholar]
- 38. Jeong J., Jung Y., Na S., Lee E., Kim M. S., Choi S., Shin D. H., Paek E., Lee H. Y., and Lee K. J. (2011) Novel oxidative modifications in redox-active cysteine residues. Mol. Cell Proteomics 10, M110.000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts B. J., Pashaj A., Johnson K. R., and Wahl J. K. 3rd. (2011) Desmosome dynamics in migrating epithelial cells requires the actin cytoskeleton. Exp. Cell Res. 317, 2814–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sobolik-Delmaire T., Katafiasz D., Keim S. A., Mahoney M. G., and Wahl J. K. 3rd. (2007) Decreased plakophilin-1 expression promotes increased motility in head and neck squamous cell carcinoma cells. Cell Commun. Adhes. 14, 99–109 [DOI] [PubMed] [Google Scholar]
- 41. Sobolik-Delmaire T., Katafiasz D., and Wahl J. K. 3rd. (2006) Carboxyl terminus of Plakophilin-1 recruits it to plasma membrane, whereas amino terminus recruits desmoplakin and promotes desmosome assembly. J. Biol. Chem. 281, 16962–16970 [DOI] [PubMed] [Google Scholar]
- 42. Wahl J. K. 3rd, Nieset J. E., Sacco-Bubulya P. A., Sadler T. M., Johnson K. R., and Wheelock M. J. (2000) The amino- and carboxyl-terminal tails of β-catenin reduce its affinity for desmoglein 2. J. Cell Sci. 113, 1737–1745 [DOI] [PubMed] [Google Scholar]
- 43. McGuinn K. P., and Mahoney M. G. (2014) Lipid rafts and detergent-resistant membranes in epithelial keratinocytes. Methods Mol. Biol. 1195, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falcone D., and Andrews D. W. (1991) Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell Biol. 11, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zacharias D. A., Violin J. D., Newton A. C., and Tsien R. Y. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 46. Ireton R. C., Davis M. A., van Hengel J., Mariner D. J., Barnes K., Thoreson M. A., Anastasiadis P. Z., Matrisian L., Bundy L. M., Sealy L., Gilbert B., van Roy F., and Reynolds A. B. (2002) A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fukumoto Y., Shintani Y., Reynolds A. B., Johnson K. R., and Wheelock M. J. (2008) The regulatory or phosphorylation domain of p120 catenin controls E-cadherin dynamics at the plasma membrane. Exp. Cell Res. 314, 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delva E., Jennings J. M., Calkins C. C., Kottke M. D., Faundez V., and Kowalczyk A. P. (2008) Pemphigus vulgaris IgG-induced desmoglein-3 endocytosis and desmosomal disassembly are mediated by a clathrin- and dynamin-independent mechanism. J. Biol. Chem. 283, 18303–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wan J., Roth A. F., Bailey A. O., and Davis N. G. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]