Abstract
Double-stranded DNA breaks (DSBs) are highly detrimental DNA lesions, which may be repaired by the homologous recombination-mediated repair pathway. The 5′ to 3′ direction of long-range end resection on one DNA strand, in which 3′-single-stranded DNA overhangs are created from broken DNA ends, is an essential step in this pathway. Dna2 has been demonstrated as an essential nuclease in this event, but the molecular mechanism of how Dna2 is recruited to DNA break sites in vivo has not been elucidated. In this study, a novel recombination factor called Cdc24 was identified in fission yeast. We demonstrated that Cdc24 localizes to DNA break sites during the repair of DNA breaks and is an essential factor in long-range end resection. We also determined that Cdc24 plays a direct role in recruiting Dna2 to DNA break sites through its interaction with Dna2 and replication protein A (RPA). Further, this study revealed that RPA acts as the foundation for assembling the machinery for long-range end resection by its essential role in recruiting Cdc24 and Dna2 to DNA break sites. These results define Cdc24 as an essential factor for long-range end resection in the repair of DSBs, opening the door for further investigations into the enzymes involved in long-range end resection for DSB repair.
Keywords: DNA damage, DNA endonuclease, DNA repair, DNA-protein interaction, homologous recombination
Introduction
Double-stranded DNA breaks (DSBs)2 are one of the most cytotoxic DNA lesions. DSBs can occur naturally inside cells (e.g. meiotic recombination, mating type switch, or V(D)J exchange); they can also be caused by endogenous reactive metabolites and extracellular agents (e.g. UV, IR, and chemical agents such as camptothecin (CPT)) (1). If DSBs are left unrepaired or are repaired inappropriately, it will lead to a variety of mutations such as deletions, insertions, translocation, and genetic fusions, resulting in drastic genomic instability (2, 3).
Eukaryotic cells have two major and conserved pathways to repair DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR) (4). In NHEJ, two broken DNA ends are rejoined by the concerted actions of Ku70/80, DNA-PKcs, DNA ligase IV, and some other proteins and enzymes (5). NHEJ does not need a homologous DNA sequence for the repair of DSBs, so it can occur in any phase of the cell cycles but dominates at G0/G1 (6). Compared with HR, NHEJ is an error-prone pathway because genetic deletion or insertion of a few base pairs is frequently observed at the break-rejoining site (7, 8). HR requires a homologous DNA template that is present in a sister chromatid or an ectopic DNA sequence to fix breaks. Therefore, the HR pathway dominates at the S and G2 phases and is a relatively error-free repair pathway, although genetic mutations can occur at times (9, 10). A central step in the HR pathway is the 5′ to 3′ digestion on one DNA strand from broken DNA ends to generate 3′-ended single-stranded DNA (ssDNA) overhangs, a process termed DNA end resection (11, 12). The HR-mediated repair of DSBs is reliant on this end resection because only ssDNA is able to invade a homologous double-stranded DNA sequence and pair with one of two strands to initiate the subsequent repair process of the DSBs.
DNA end resection is initiated by the Mre11-Rad50-Xrs2 (MRX) complex (or MRE11-RAD50-NBS1 (MRN) in Schizosaccharomyces pombe/metazoans) along with Sae2 (or Ctp1/CtIP in S. pombe/metazoans) in budding yeast (13–15). The initial resection generates 3′-ended ssDNA overhangs of approximately a few dozen to 100 or 200 nucleotides in length. Subsequent extensive resection requires two cooperative nucleases, Exo1 and Dna2 (16–19). After 3′-terminated ssDNA is available, RPA comes to coat the ssDNA, protect DNA ends from degradation, and direct the 5′-3′ resection polarity of Dna2 (16, 17, 20, 21). RPA is subsequently displaced by DNA strand exchange protein Rad51 with help from Rad52. Rad51 forms a nucleoprotein filament on the 3′-ended ssDNA substrate and catalyzes homologous sequence searching and strand invasion on a homologous DNA sequence followed by a series of steps to fix DNA breaks (22, 23). DNA end resection is a critical step in the HR pathway. It commits DSB repair to HR and prevents NHEJ (21, 24). Over the past decade, we have observed significant progress in understanding the end resection event (1, 4, 16, 17, 22). However, the mechanism of how Dna2 and Exo1 are recruited to DNA break sites still remains to be resolved.
In this study, we identified Cdc24 in the fission yeast S. pombe as an essential recombination factor that participates in the repair of DSBs. Cdc24 is an essential protein for cell growth (25). Its function is implicated in DNA replication, but its precise biochemical action has not yet been determined (25–28). We first determined that Cdc24, like Dna2 and Rad52, localizes to DNA break sites during the break-repairing process. Then, Cdc24 was found to be essential for long-range end resection, indicating that Cdc24 participates in the HR pathway. Finally, we demonstrated that Cdc24 is required to recruit Dna2 nuclease to DNA break sites through its interaction with Dna2 and RPA for catalyzing end resection. This finding also may be related to the essential role of Cdc24 in DNA replication and cell growth.
Results
Cdc24 Is Localized to DSB Sites during the Repair of DSBs
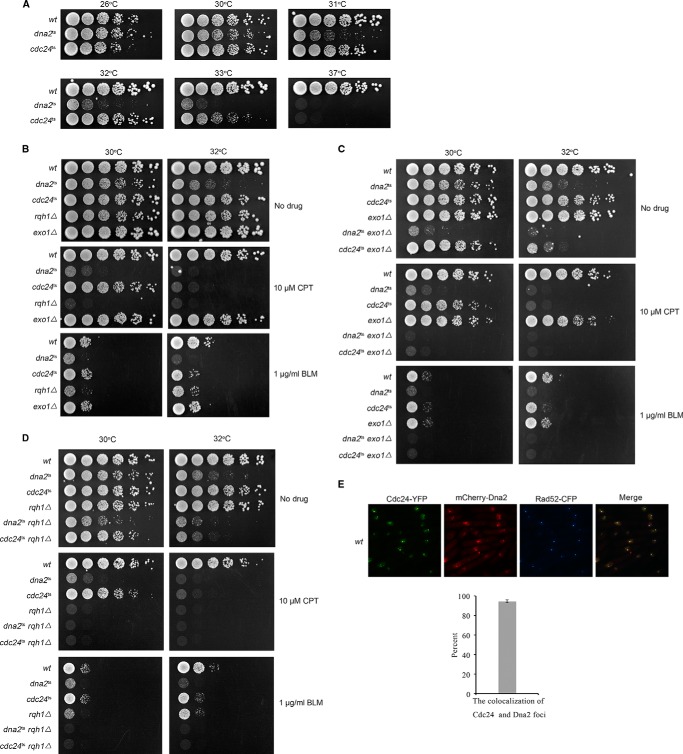
To investigate the mechanism of long-range end resection in fission yeast, we first examined the growth sensitivity of some S. pombe mutant strains to CPT or BLM (bleomycin) (two chemicals that induce DNA breaks), including Cdc24, Dna2, Exo1, and Rqh1 mutant cells. Cdc24 was included in this study because some previous studies suggest that Cdc24 and Dna2 may have a biological interaction with one another (27–29). Cdc24 and Dna2 are essential for cell growth, so their temperature-sensitive mutants were used (25, 30). The results shown in Fig. 1A indicate the extent of growth sensitivity of dna2ts (L1079S mutation) and cdc24ts (C114R mutation) cells at 26–37 °C. As in budding yeast, S. pombe dna2ts, rqh1Δ (the homologue of Sgs1 in fission yeast), and exo1Δ cells showed high sensitivity to CPT/BLM (Fig. 1, B–D). In addition, we found that cdc24ts cells were also highly sensitive to CPT/BLM (Fig. 1, B–D). We further examined dna2ts-rqh1Δ, cdc24ts-rqh1Δ, dna2ts-exo1Δ, and cdc24ts-exo1Δ cells, which showed synthetic sensitivity to CPT and BLM (Fig. 1, C and D), indicating that these proteins have an additive function in DSB repair. We were not able to examine dna2ts-cdc24ts cells because they grew very weakly and died constantly even at 26 °C.
FIGURE 1.
Cdc24 localizes to DSB sites during the DNA break-repairing process. A, growth sensitivity of dna2ts and cdc24ts cells to various temperatures. B–D, a 5-fold dilution assay was conducted. The cells on the plates were incubated at the indicated temperatures and growth conditions for 3 or 4 days, and then the cells were photographed. B, a 5-fold dilution assay showed that the growth of dna2ts, cdc24ts, rqh1Δ, and exo1Δ cells is sensitive to CPT and BLM. C, dna2ts-exo1Δ and cdc24ts-exo1Δ cells are more sensitive to CPT and BLM than dna2ts, cdc24ts, and exo1Δ cells. D, dna2ts-rqh1Δ and cdc24ts-rqh1Δ cells are more sensitive to CPT and BLM than dna2ts, cdc24ts, and rqh1Δ cells. E, Cdc24 and Dna2 localize to DSB sites, similar to Rad52. A HO-induced DSB system was used for this assay. Cdc24, Dna2, and Rad52 were tagged by YFP, mCherry, and CFP, respectively. These cells were incubated in EGRL medium at 30 °C for 18 h for the expression of HO endonuclease and the generation of HO-induced DNA breaks followed by ethanol fixation and a fluorescent focus assay. The percent of co-localized Cdc24 and Dna2 foci is shown in the lower panel.
The high sensitivity of cdc24ts cells to CPT/BLM suggests that Cdc24 may have a direct role in DSB repair. Thus, we first examined whether Cdc24 localizes to DSB sites during break repair. A routine HO-induced DNA break system was used (31). As presented in Fig. 1E, both Cdc24 and Dna2 co-localized with Rad52 based on the overlapping fluorescent foci of these proteins. Co-localization of Cdc24-YFP and mCherry-Dna2 foci was nearly 95%. This result indicates that Cdc24 and Dna2 localize to DSB sites, because Rad52 is a known HR-mediated break repair protein.
Cdc24 Is Required for Long-range DNA End Resection
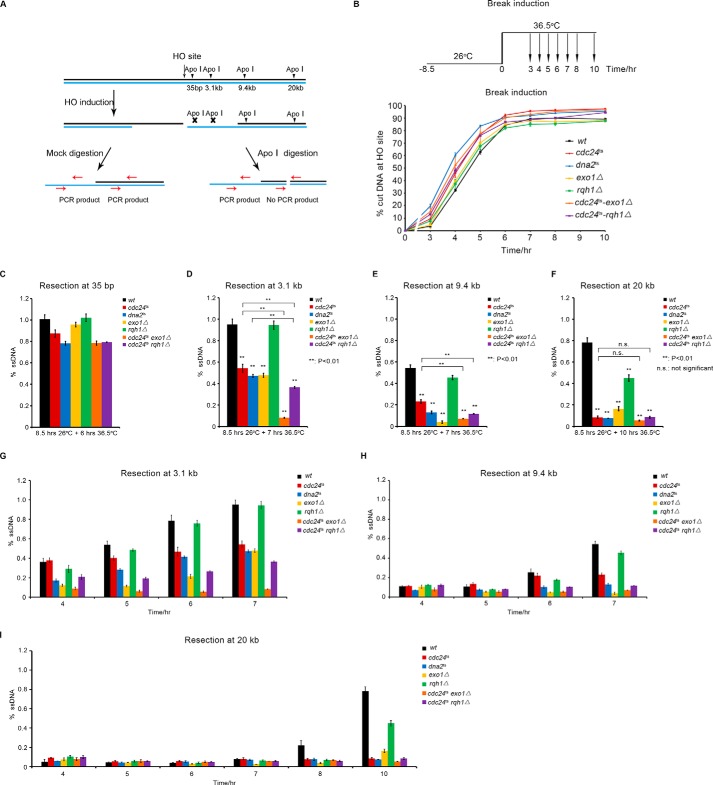
Next, we examined how end resection is affected in cdc24ts cells, as well as in dna2ts, rqh1Δ, exo1Δ, cdc24ts-exo1Δ, and cdc24ts-rqh1Δ (Rqh1 is the homologue of Sgs1 in fission yeast) mutant cells, using a method based on quantitative PCR (qPCR) (31, 32) (Fig. 2A). A scheme for break induction and the percentage of DNA cut at the HO site versus time induction is presented in Fig. 2B. The rate of DNA cut/break at the HO site was similar in the wt and mutant cells (Fig. 2B). However, the long-range end resection was significantly reduced in the cdc24ts, dna2ts, and exo1Δ cells (Fig. 2, C–F). This result indicates that Cdc24 is an essential factor for the repair of DSBs and that it functions in the HR pathway. Consistent with previous findings (16, 17), Dna2 and Exo1 are also required for long-range end resection in fission yeast (Fig. 2, C–F). We also detected a further decrease in end resection in the cdc24ts-rqh1Δ and cdc24ts-exo1Δ cells, indicating that Cdc24, Exo1, and Rqh1 have an additive action in promoting end resection (Fig. 2, C–F), consistent with a greater sensitivity of the cdc24ts-rqh1Δ and cdc24ts-exo1Δ cells to CPT or BLM (Fig. 1, C and D). A previous finding indicates that long-range end resection is not much affected in rqh1Δ cells (32). In this study, we also found that Rqh1 deficiency did not affect end resection as much as in cdc24ts, dna2ts, and exo1Δ cells (Fig. 2, C–F). The high sensitivity of rqh1Δ cells to CPT and BLM suggests that Rqh1 may have an additional function in DSB repair, besides its moderate role in long-range end resection. Fig. 2, G–I, presents the efficiency of long-range end resection at different induction times, which is basically consistent with the results presented in Fig. 2, C–F.
FIGURE 2.
Cdc24 is required for long-range end resection. A, schematic of HO-induced DSB and end resection assay. B, upper panel, the cells were first grown at 26 °C for 8.5 h and then shifted to 36.5 °C for the indicated times. Cell samples were taken at 3, 4, 5, 6, 7, 8, and 10 h to examine the cut rate at the HO site and end resection rate at 35 bp and 3.1, 9.4, and 20 kb from the HO site. Lower panel, a similar percentage of DNA cut at the HO site at the indicated times in the wt and cdc24ts, dna2ts, exo1Δ, rqh1Δ, cdc24ts-exo1Δ, and cdc24ts-rqh1Δ cells. C–F, long-range end resection was significantly reduced in the dna2ts, cdc24ts, exo1Δ, cdc24ts-rqh1Δ, and cdc24ts-exo1Δ cells, but only a moderate reduction was seen in the rqh1Δ cells. The mutant strains that contain the system of HO-induced DSBs were constructed using standard genetic methods. The cells were first incubated in EGRLU medium at 26 °C for 8.5 h and then at 36.5 °C for 6, 7, 7, and 10 h for the examination of end resection at 35 bp and 3.1, 9.4, and 20 kb, respectively. The efficiency of end resection was measured as described under “Experimental Procedures.” G–I, end resection rates at the 3.1-, 9.4-, and 20-kb sites at the indicated induction times in wt and mutant cells. Three independent assays were performed, and the data are presented as means ± S.E. Statistical significance was determined using Student's unpaired t test. **, p < 0.01, statistically significant; n.s., not significant.
To further confirm Cdc24 function in DSB repair, the efficiency of DSB repair was measured according to a method developed by Humphrey and colleagues (33) (Fig. 3A). Consistent with a reduced end resection, the frequency of gene conversion, which reflects the efficiency of DSB repair through the HR pathway, decreased by ∼40% in the cdc24ts and dna2ts cells compared with that in wt cells (Fig. 3B). This assay was conducted at a cell growth temperature of 30 °C, the temperature at which the mutant and wt cells had a similar survival rate. Although the rate of HR was reduced, NHEJ and loss of heterozygosity, as expected, correspondingly increased in the dna2ts and cdc24ts cells (Fig. 3B).
FIGURE 3.
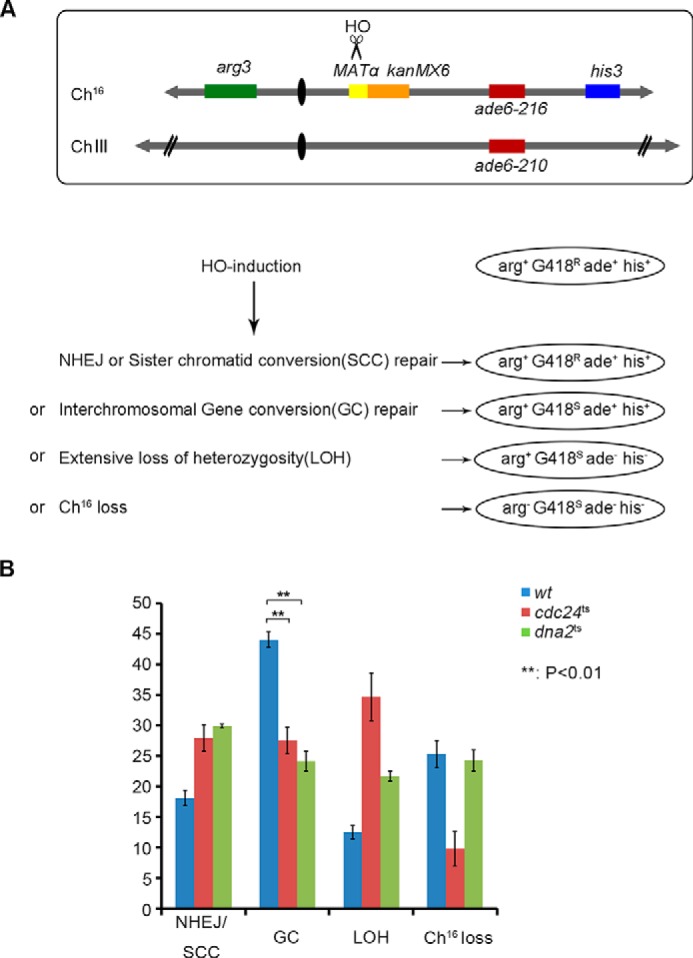
Homologous recombination was reduced in dna2ts and cdc24ts cells. A, schematic of Humphrey's system to examine DSB repair efficiency (33). B, the frequency of HR (gene conversion) and NHEJ was measured using the system developed in T. C. Humphrey's laboratory. The detailed procedure is described under “Experimental Procedures.”
Cdc24 Interacts with Dna2 Inside the Cells
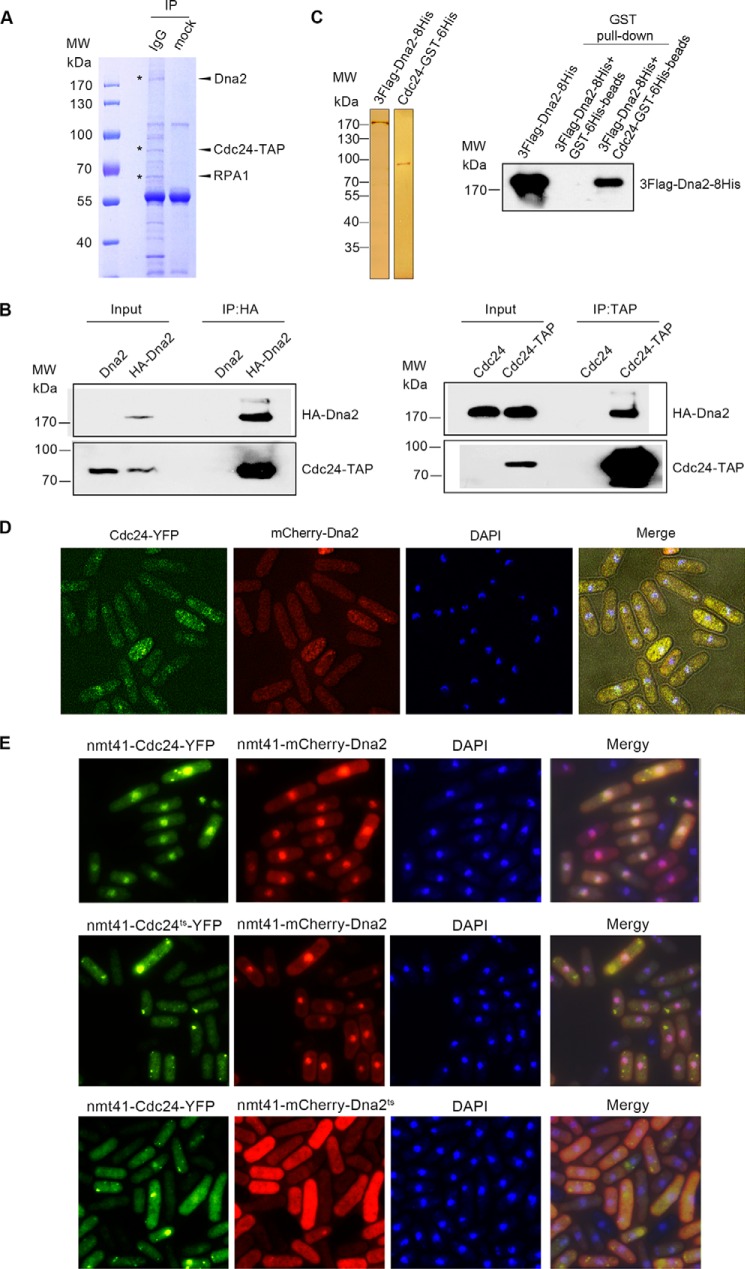
To elucidate the biochemical action of Cdc24 in long-range end resection, we first tried to determine the protein(s) that Cdc24 interacts with in vivo. Thus, an immunoprecipitation (IP) assay was performed using the TAP antibody against Cdc24-TAP in whole cell extracts. A protein of ∼170 kDa was isolated and identified as Dna2 by mass spectroscopy (Fig. 4A). Besides Dna2, we also detected RPA in the pulled-down fraction. Reciprocal IP confirmed the interaction of Dna2 and Cdc24 (Fig. 4B). Further, protein affinity chromatography with purified Cdc24 and Dna2 demonstrated that Dna2 and Cdc24 can interact directly with one another (Fig. 4C). To examine the distribution of Dna2 and Cdc24 inside the cells, Cdc24 was tagged by YFP at its C terminus, and Dna2 was tagged by mCherry at its N terminus. These two tagged genes were integrated into their origin genomic locus under the control of their native promoters as having a physiological level of Cdc24-YFP and mCherry-Dna2 inside the cells. The results shown in Fig. 4D indicate that both Cdc24 and Dna2 are quite evenly distributed in the cytoplasm and nucleoplasm. A portion of Cdc24 and Dna2 showed overlapping fluorescence, indicating again that these two proteins interact with each other to form a complex. Interestingly, when Cdc24-YFP and mCherry-Dna2 were overexpressed, the majority of the Cdc24-YFP and mCherry-Dna2 were found in the nucleus and had a nicely overlapping fluorescence (Fig. 4E, top panel), indicating that an intrinsic interaction exists between Cdc24 and Dna2. mCherry-Dna2 was still enriched in the nucleus when Cdc24ts-YFP and mCherry-Dna2 were overexpressed (Fig. 4E, middle panel). But when mCherry-Dna2ts and Cdc24-YFP were overexpressed, both of them were evenly distributed in the cells and not enriched in the nucleus (Fig. 4, bottom panel), suggesting that the transportation of Cdc24 into the nucleus requires Dna2 function. Here, both in vivo and in vitro assays indicated that Cdc24 is able to interact directly with Dna2.
FIGURE 4.
Cdc24 and Dna2 directly interact with one another. A–C, Dna2 and RPA were isolated in the immunoprecipitation with IgG-beads against Cdc24-TAP (A, Coomassie Blue staining); reciprocal IPs of HA-Dna2 and Cdc24-TAP were carried out using monoclonal anti-HA-agarose beads and IgG beads. (B, Western blotting); and protein affinity chromatography with purified Cdc24 and Dna2 showed that they interact directly (C, Western blotting). D, a portion of Cdc24-YFP and mCherry-Dna2 had overlapping foci. Cdc24-YFP and mCherry-Dna2 were expressed under their native promoters. Unsynchronized cells were subjected to fluorescent analysis. E, overexpressed Cdc24 and Dna2 form a complex in vivo and are located in the nuclei. YFP-tagged Cdc24 or Cdc24ts and mCherry-tagged Dna2 or Dna2ts were overexpressed under the promoter of nmt41. Unsynchronized cells were first incubated at 26 °C for 12 h and at 36.5 °C for 6 h and then subjected to fluorescent analysis. Top panel, both overexpressed Dna2 and Cdc24 were enriched in the nucleus. Middle panel, Dna2, but not Cdc24ts, was enriched in the nucleus. Bottom panel, both Dna2ts and Cdc24 were evenly distributed in the cytoplasm and nucleoplasm and were not enriched in the nucleus.
Cdc24 Is Required for the Formation of Dna2 Foci at DNA Break Sites
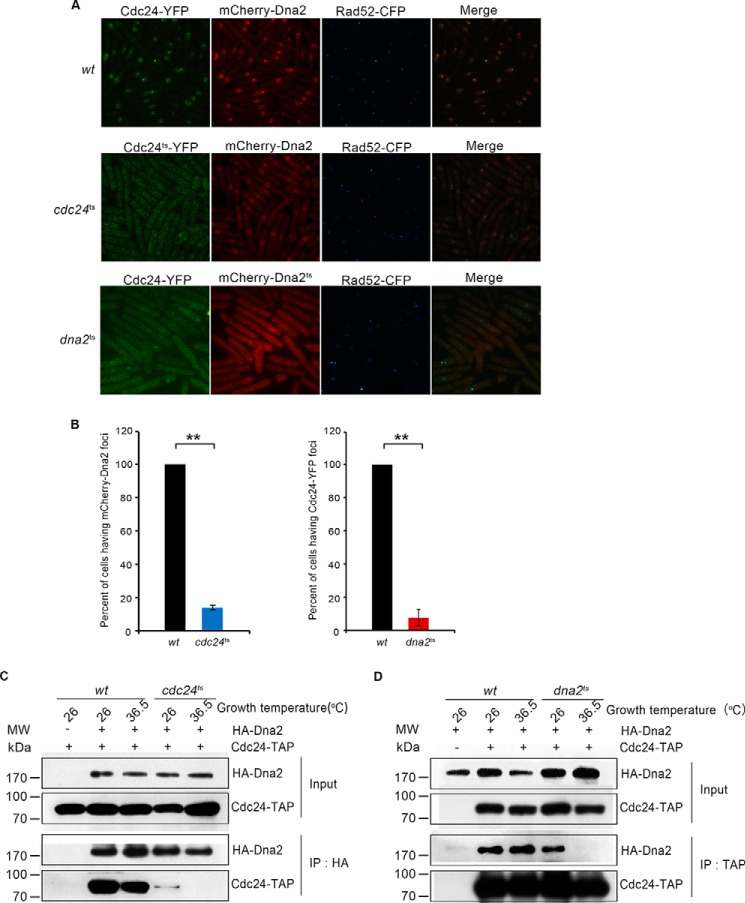
We next examined whether Dna2 could localize to DSB sites when Cdc24 function is defective. In wt cells with normal Cdc24 function, both Cdc24 and Dna2 were enriched in the nuclei after DNA breaks and were localized to DSB sites (Fig. 5A, top panel). However, in the cdc24ts cells, both the number and fluorescent extent of the Dna2 and Cdc24 foci at DSB sites were hardly detectable (Fig. 5A, middle panel). Cdc24ts appeared to be evenly distributed in the cytoplasm and nucleus; and Dna2 existed in the cytoplasm and nucleus, with enrichment in the nuclei (Fig. 5A, middle panel). Similar to the cdc24ts cells, the Dna2 and Cdc24 foci were also significantly decreased in the dna2ts cells (Fig. 5A, bottom panel). Both Cdc24 and Dna2ts were distributed evenly in the cytoplasm and nucleus of the dna2ts cells, but there was no enrichment of either Cdc24 or Dna2ts in the nucleus (Fig. 5A, bottom panel). A statistical measure indicates that the percentage of cells having Dna2 or Cdc24 foci decreased by approximately 7- and 13-fold in the cdc24ts and dna2ts cells, respectively (Fig. 5B). Correspondingly, the intensity of Rad52 florescent foci decreased significantly in cdc24ts and dna2ts cells compared with that in wt cells (Fig. 5A, middle and bottom panels), indicating that end resection was impaired. As expected, even at 26 °C, the interaction between Cdc24ts and Dna2 was markedly reduced; at 36.5 °C, the interaction was undetected (Fig. 5C) based on the IP assay with anti-HA monoclonal antibody against HA-Dna2. The interaction between Dna2ts and Cdc24 was not detectable by IP at 36.5 °C (Fig. 5D). These results indicate that Cdc24 is required to recruit Dna2 to DSB sites. The results shown in Fig. 5A, bottom panel, also suggest that Dna2 has an essential role in the enrichment of Cdc24 in the nucleus or in the transportation of Cdc24 into nucleus.
FIGURE 5.
Cdc24 is required to recruit Dna2 to DSB sites. A, Cdc24 is required for Dna2 to localize to DSB sites. This assay was conducted as described in Fig. 1E except for modifications to the growth temperature. wt, cdc24ts, or dna2ts cells with an A590 of ∼0.05 were each inoculated into an EGRL culture and incubated at 26 °C for 8.5 h. Next, the three cultures were shifted to 36.5 °C for an additional 8-h incubation followed by cell fixation with ethanol and analysis of fluorescent foci. B, a statistical measure shows a significant reduction of cdc24ts cells having mCherry-Dna2 foci or dna2ts cells having Cdc24-YFP foci. **, p < 0.01, statistically significant. C, the interaction between Cdc24ts and Dna2 is significantly impaired at growth temperatures of 26 and 36.5 °C. The cdc24ts and wt cells were grown to an A590 of ∼0.3 at 26 °C and then were shifted to 36.5 °C for a 6-h incubation. Whole cell extracts were prepared and subjected to IP with monoclonal anti-HA-agarose beads against HA-Dna2. The pulled-down proteins were examined using the HA antibody to detect HA-Dna2 and the TAP antibody to detect Cdc24-TAP. D, the interaction between Dna2ts and Cdc24 was undetected at 36.5 °C. The assay was performed as described in C except that IgG beads were used to pull down Cdc24-TAP. HA-Dna2 and Cdc24-TAP were detected by Western blotting using the HA and TAP antibodies.
Through Cdc24, Dna2 and RPA Have an Indirect Interaction Inside the Cells
The above result indicates that Cdc24 is required for Dna2 to localize to DNA break sites. Next, we wanted to know how Cdc24 is recruited to the DSBs. For HR-mediated DSB repair, the first step is MRN-mediated short-range end resection at the broken ends, which creates the 3′-ended ssDNA overhangs of a few dozen to a few hundred nucleotides (4). This ssDNA strand will be coated by RPA. In the IP assay used to search for Cdc24-interacting proteins (Fig. 4A), besides Dna2, RPA was also found in the fraction. Thus, RPA could be the protein that recruits Cdc24 to DNA break sites. Thus, we examined the interaction among Dna2, Cdc24, and RPA. First, an IP assay against Dna2 was carried out in wt and cdc24ts cells. As shown in Fig. 6A, Dna2 interacted with RPA in wt cells at a growth temperature of either 26 or 36.5 °C, because the RPA1 subunit (the large subunit in RPA) was detected in the pulled-down fraction using a monoclonal HA antibody against HA-Dna2. But, the RPA1 subunit was under detectable levels in the cdc24ts cells at either 26 or 36.5 °C in a similar IP assay (Fig. 6A). However, in the wt or dna2ts cells, the interaction between Cdc24 and RPA was detected and was not changed in either the wt cells or dna2ts cells at both the permissive and restrictive temperatures of 26 and 36.5 °C based on the IP assay with IgG beads against Cdc24-TAP (Fig. 6B). Thus, the results shown in Fig. 6, A and B, and Fig. 4, B–D, indicate that Cdc24 can interact with both Dna2 and RPA, but the interaction between Dna2 and RPA requires the presence of Cdc24. In other words, the interaction between Dna2 and RPA occurs through their interactions with Cdc24.
FIGURE 6.
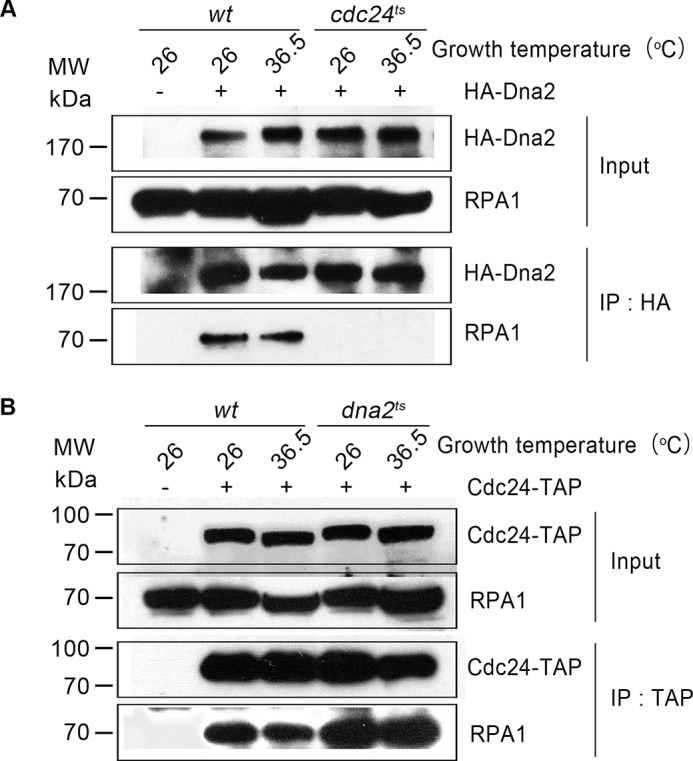
Cdc24 interacts with RPA, and the interaction between Dna2 and RPA is mediated by Cdc24. A, Dna2 does not interact with RPA in the cdc24ts cells. The wt or cdc24ts cells were cultured at the indicated temperatures. IP with anti-HA-agarose beads against HA-Dna2 was carried out with whole cell extracts. Western blotting results are presented. B, Cdc24 interacts with RPA in the wt or dna2ts cells. The assays were carried out as described in A. Western blotting results are presented.
Cdc24 Recruits Dna2 to DNA Break Sites through Its Interaction with RPA and Dna2
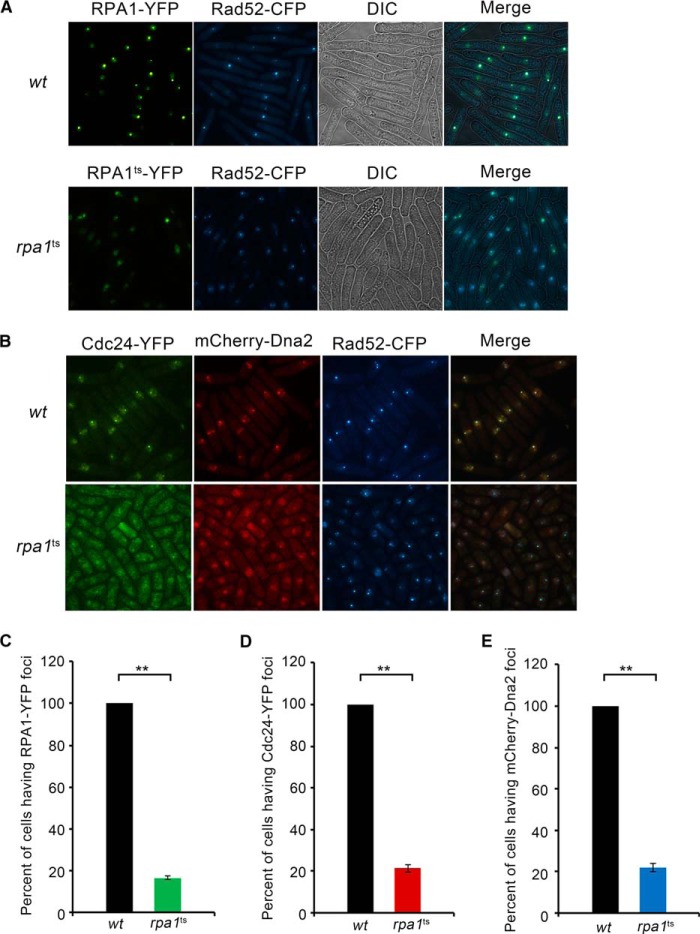
The interactions among Dna2, Cdc24, and RPA suggest the role of RPA in recruiting Cdc24 and Dna2 to DNA break sites. To prove this suggestion, we used a fluorescent assay. First, in rpa1ts cells, both the intensity and number of RPA1 and Rad52 foci significantly decreased compared with that in wt cells at the restrictive temperature of 36.5 °C, confirming the essential function of RPA in the repair of DSB (Fig. 7A). Then, we examined how the recruitment of Cdc24 and Dna2 to break sites is affected when RPA is functionally defective. As shown in Fig. 7B, the foci of Cdc24 and Dna2 were readily detected in wt cells, but they were hardly detectable in the rpa1ts cells. Correspondingly, the intensity and number of Rad52 foci also significantly decreased. A statistically significant reduction of the RPA, Cdc24, and Dna2 foci in rpa1ts cells is shown in Fig. 7, C, D, and E, respectively. These results are consistent with the finding in budding yeast (20) Taking these results together, we conclude that RPA acts as a foundation for recruiting Cdc24 and Dna2 to DNA break sites.
FIGURE 7.
RPA is required to recruit Cdc24 and Dna2 to DNA break sites. A, the intensity and number of RPA foci decrease in the rpa1ts cells. The RPA1-YFP gene was integrated into its original genomic locus under the control of the native promoter. The cultures of wt and rpa1ts cells were first grown to an A590 of ∼0.3 at 26 °C and then shifted to 36.5 °C for an additional 6-h incubation. The cells were fixed and subjected to fluorescent analysis. B, Cdc24 and Dna2 are not localized to DNA break sites in the rpa1ts cells. The assays were conducted as described in Fig. 6A. The foci of Cdc24-YFP, mCherry-Dna2, and Rad52-CFP are presented. C–E, quantification of RPA (C), Cdc24 (D), and Dna2 (E) foci in the wt and rpa1ts cells. The percentage of cells having RPA, Cdc24, or Dna2 foci is arbitrarily assumed to be 100% in the wt cells.
Cdc24 Is Required for Dna2 to Digest RPA-coated Fork-structured DNA
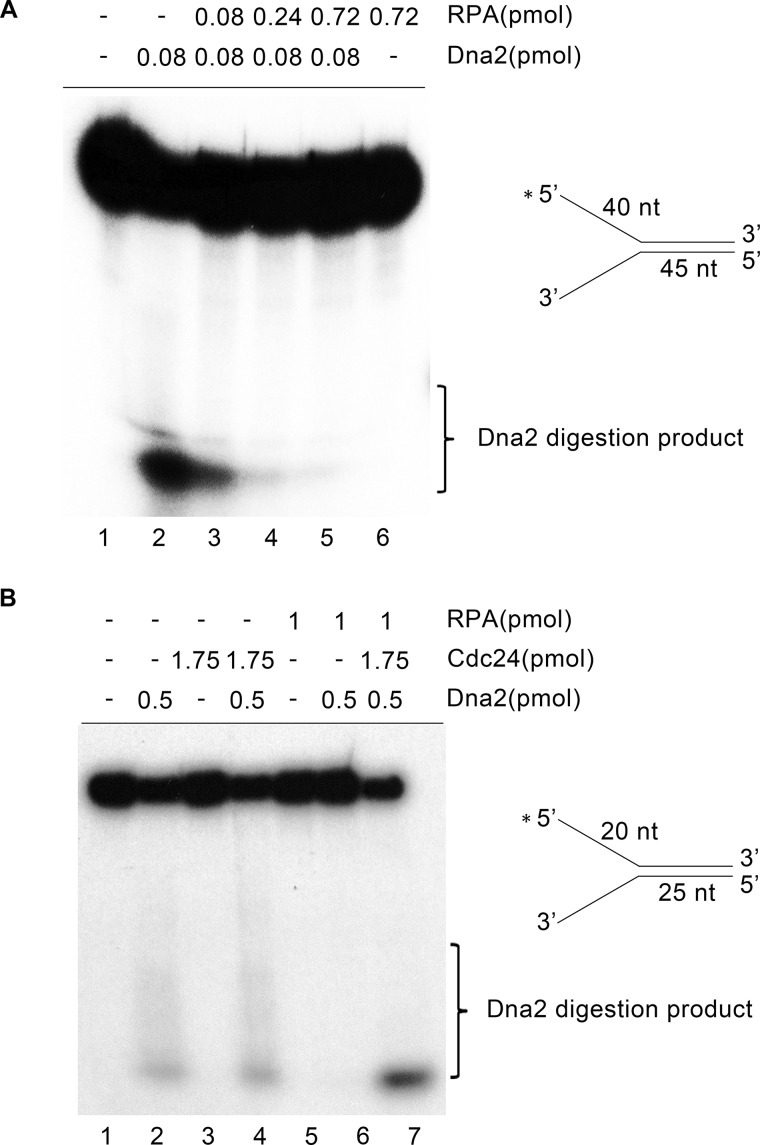
To further demonstrate that Cdc24 is responsible for recruiting Dna2 to RPA-coated DNA, an in vitro DNA digestion assay was performed. As shown in Fig. 8A, lane 2, Dna2 alone was able to digest a naked DNA fork, consistent with previous findings that Dna2 is able to access a naked ssDNA (30, 34). However, when an increasing amount of RPA was added to the reactions, DNA digestion was reduced and eventually blocked (Fig. 8A, lanes 3–5). This result indicates that S. pombe Dna2 is unable to access RPA-coated ssDNA. When Cdc24 was further added to the reactions, DNA digestion was restored (Fig. 8B, lane 7). Moreover, when both RPA and Cdc24 were present, the DNA digestion of Dna2 was stimulated (Fig. 8B, lane 7).
FIGURE 8.
Cdc24 is required for Dna2 to digest RPA-coated DNA substrate. A 32P-labeled fork-structured DNA was used to examine Dna2-mediated digestion in the presence or absence of RPA and Cdc24. A, RPA blocks Dna2 digestion. Increasing amounts of RPA as indicated were added to the reactions. 15 fmol of forked DNA molecule was used, and the reactions were incubated at 30 °C for 30 min. Dna2 digestion products were detected by denatured PAGE. B, Cdc24 restores Dna2 digestion of RPA-coated fork-structured DNA. The amounts of Cdc24, Dna2, and RPA in the reactions were indicated. 50 fmol of forked DNA molecule was used, and the reactions were incubated at 30 °C for 30 min.
Discussion
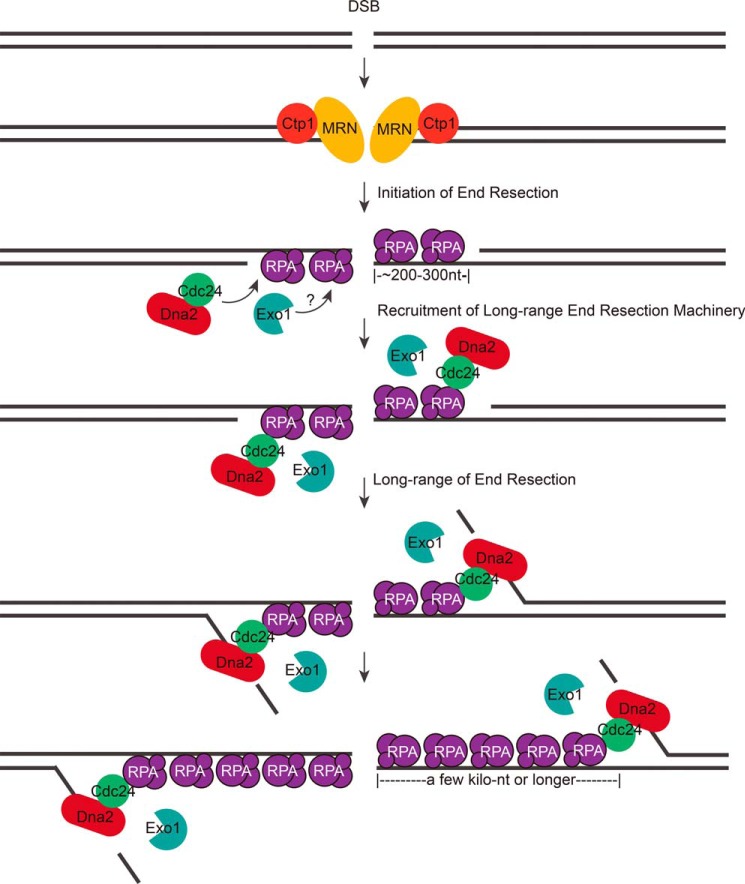
In this study, a new recombination and DSB repair factor, called Cdc24 in the fission yeast S. pombe, was identified. Cdc24 was found to be an essential factor for long-range end resection in the repair of DSBs (Fig. 2). The molecular mechanism of Cdc24 action in DSB repair is to recruit Dna2 to DNA break sites through its interaction with both Dna2 and RPA for subsequent Dna2-catalyzing end resection (Figs. 4–8). RPA was demonstrated to act as a foundation in recruiting Dna2 and Cdc24 at break sites (Figs. 6–8). Thus, this study answers the question of how Dna2, a nuclease critical for long-range end resection, is recruited to DNA break sites (Fig. 9).
FIGURE 9.
A model of the long-range end resection in fission yeast. After DNA breaks, the MRN complex and Ctp1 first bind to DNA break ends and carry out a limited DNA end resection, which generates a 3′-ssDNA overhang of a few dozen to a few hundred nucleotides in length. Then, RPA comes to bind the ssDNA. Subsequently, RPA recruits the Cdc24-Dna2 complex to DNA break sites through the interaction between RPA and Cdc24. Exo1 is also recruited to the break sites. Then, the Cdc24-Dna2 complex and Exo1 coordinate to conduct long-range end resection, generating an ssDNA of a few kilobases.
Although Cdc24 is required to recruit Dna2 to DSB sites or to RPA-coated ssDNA (Fig. 5A, 8), Dna2 also appears to play a critical role in the transportation of Cdc24 into nucleus. When both Dna2 and Cdc24 were overexpressed, they were enriched in the nucleus (Fig. 4E, top panel). But Cdc24 was not enriched in the nucleus when Cdc24 and Dna2ts were co-overexpressed (Fig. 4E, bottom panel). In contrast, Dna2 was enriched in the nucleus when Dna2 and Cdc24ts were overexpressed (Fig. 4E, middle panel). These results suggest that the nuclear localization of overexpressed Cdc24 requires Dna2 function but that overexpressed Dna2 can localize to the nucleus in the absence of Cdc24 function. Further, under DNA breaks, both Cdc24 and Dna2, having amounts at a physiological level, were enriched in the nucleus and formed foci at DSB sites (Fig. 5A, top panel). However, when Dna2 was functionally defective (Dna2ts), Cdc24 was not enriched in the nucleus and did not form foci at DSB sites (Fig. 5A, bottom panel). These results taken together suggest that Dna2 is required for transporting Cdc24 into the nucleus, probably through the formation a Cdc24-Dna2 complex.
Cdc24 is required for cell growth, and its essential function appears to be related to DNA replication (25, 27). However, its exact biochemical function in replication has not been determined. We did not find Cdc24 to have any nuclease activity (Fig. 8B). Neither did we detect any DNA binding activity in Cdc24.3 Besides Dna2 and RPA, there were no other replication-related proteins in the Cdc24 pulled-down fraction (Fig. 4A). Both RPA and Dna2 are essential factors for DNA replication. RPA, a single-stranded DNA-binding protein in eukaryotes, binds to ssDNA regions at replication forks. Besides protecting ssDNA regions, RPA coordinates biochemical reactions at forks (35). Dna2 is required for processing Okazaki fragments for the synthesis of lagging strand DNA (36). Therefore, based on the above findings, we suggest that the essential function of Cdc24 in replication is possibly similar to its role in long-range end resection and repair of DSBs: recruiting Dna2 to replication forks through its interaction with RPA and Dna2.
As in budding yeast (16, 17), both Dna2 and Exo1 nucleases are required for long-range end resection in fission yeast (Fig. 2), suggesting that the basic mechanism of this event should be conserved. But different from budding yeast, Rqh1, the homologue of Sgs1 in fission yeast, does not appear to have an important role in end resection in fission yeast, although a moderate decrease in long-range end resection was detected in the rqh1Δ cells (Fig. 2, E and F). A moderate role for Rqh1 in end resection is also reported in a previous study (32). But Rqh1 has been shown to have an important role in end resection and homologous recombination in other studies (37, 38). The biochemical properties of Dna2 have been thoroughly characterized in vitro; it preferentially cleaves a flap DNA structure (39). If the basic enzymatic activity of S. pombe Dna2 is not changed in vivo, a flap structure should be generated during long-range end resection. In this case, Rqh1 does function in long-range end resection (Fig. 2, E and F). In support of this, a further decrease in end resection was also observed in the cdc24ts-rqh1Δ double mutant compared with that in the cdc24ts cells (Fig. 2, D and E). However, considering a moderate role for Rqh1 in long-range end resection, the question remained as to what extent flap DNA structures are generated in vivo during long-range end resection in fission yeast. It is also unknown at present whether another mechanism generates flap structures besides DNA helicase action.
The identification of Dna2 and Exo1 in long-range end resection is a big step in elucidating the molecular mechanism of long-range end resection (16, 17). In this study, we solved the question how Dna2 is recruited to DNA break sites (Fig. 9). However, in this end resection event, a few obvious questions still remain to be answered. For example, how is Exo1 recruited to DNA break sites? Considering that Exo1 is an exonuclease and Dna2 is a flap endonuclease, a question arises as to how these two nucleases coordinate to catalyze long-range end resection. It is also not clear how MRN- and Ctp1-initiated end resection is transitioned to long-range end resection. In the future, these questions will require elucidation in order to gain a better understanding of the end resection event.
Experimental Procedures
Dilution Assays
Routinely, a single colony was picked and incubated in 5 ml of YES medium (0.5% yeast extract, 3% glucose, 225 mg/liter adenine, histidine, leucine, uracil, and arginine) at 26 °C to an A590 = 0.5–1.0. Each culture was measured for its A590 value and was diluted to A590 = 0.5; 5-fold serial dilutions of each culture were spotted on the indicated plates, allowed to grow for 3–4 days, and then photographed.
Localization Assay of Cdc24 and Dna2 to DNA Break Sites
Originally developed by Dr. James Haber (40), the HO-induced system was used to determine whether Cdc24 and Dna2, under the control of their native promoters, were localized to DNA break sites during the repair process (31). Cdc24, Dna2, and Rad52 were tagged with YFP, mCherry, and CFP, respectively. Cells were first incubated until log growth phase in YES medium and were then collected, washed with ddH2O, and inoculated in EGRL medium (EMM medium with a replacement of NH4Cl with 3.7 g/l sodium glutamate, and supplemented of 225 mg/liter arginine and leucine) with an initial A590 = 0.05. After incubation at 30 °C for 18 h for the formation of the HO-induced break site, the cells were fixed with 70% ethanol and examined by DeltaVision fluorescent microscopy to determine the localization of Cdc24 and Dna2 inside the cells.
To examine the localization of Dna2 in Cdc24-defective cdc24ts cells, a culture with an initial A590 = 0.05 was incubated in EGRL medium at 26 °C for 8.5 h and then shifted to 36.5 °C for an additional 8 h. The cells were fixed with ethanol and subsequently subjected to a fluorescence assay.
End Resection
Using the HO system (31), the efficiency of end resection was measured in the strains with a genetic background of dna2ts, cdc24ts, exo1Δ, rqh1Δ, dna2ts-rqh1Δ, cdc24ts-rqh1Δ, dna2ts-exo1Δ, or cdc24ts-exo1Δ (Table 1). These cells were first incubated in YES medium at 26 °C to log growth phase. Next, the cells were transferred to EGRLU medium (EMM medium with a replacement of NH4Cl with 3.7 g/l sodium glutamate, and supplemented of 225 mg/liter arginine, leucine and uracil) and incubated at 26 °C for 8.5 h and at 36.5 °C for the indicated time, for HO-induced break formation and subsequent end resection. Genomic DNA was prepared from these cells and subjected to the examination of end resection efficiency using qPCR as described previously (31, 32).
TABLE 1.
Strains used in this study
h+ and h− are two fission yeast mating types.
| Strain | Mating type | Genotype | Source |
|---|---|---|---|
| LD330 | h− | ura4-D18 | Li-Lin Du |
| cdc24ts | h? | cdc24C114R | M. Yanagida |
| dna2ts | h? | dna2L1079S | Laboratory stock |
| YHMZ1 | h− | ura4-D18 cdc24-TAP::hphMX His-HA-dna2::ura4 | This study |
| YHMZ2 | h− | ura4-D18 cdc24-TAP::hphMX His-HA-dna2ts::ura4 | This study |
| YHMZ3 | h− | ura4-D18 cdc24ts-TAP::hphMX His-HA-dna2::ura4 | This study |
| YHMZ4 | h− | ura4-D18 His-HA-dna2::ura4 | This study |
| YHMZ5 | h− | ura4-D18 cdc24-TAP::hphMX | This study |
| TK8 | h+ | leu1–32 ura4-D18 ade-M216 | Laboratory stock |
| YHMZ10 | h+ | leu1–32 ura4-D18 ade-M216 cdc24ts::kanMX | This study |
| YHMZ14 | h+ | leu1–32 ura4-D18 ade-M216 dna2ts::kanMX | This study |
| YHMZ15 | h+ | leu1–32 ura4-D18 ade-M216 exo1Δ::ura4 | This study |
| YHMZ16 | h+ | leu1–32 ura4-D18 ade-M216 rqh1Δ::ura4 | This study |
| YHMZ17 | h+ | leu1–32 ura4-D18 ade-M216 exo1Δ::ura4 cdc24ts::kanMX | This study |
| YHMZ18 | h+ | leu1–32 ura4-D18 ade-M216 rqh1Δ::ura4 cdc24ts::kanMX | This study |
| YHMZ53 | h+ | leu1–32 ura4-D18 ade-M216 rqh1Δ::ura4 dna2ts::kanMX | This study |
| YHMZ54 | h+ | leu1–32 ura4-D18 ade-M216 exo1Δ::ura4 dna2ts::kanMX | This study |
| YHMZ55 | h+ | leu1–32 ura4-D18 ade-M216 mcherry-3Flag-dna2::ura4 cdc24-YFP::kanMX | This study |
| DY49 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) | Li-Lin Du |
| YHMZ23 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX | This study |
| YHMZ24 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg−) ars1::[pJR1–41XH + HO](his3+) rad22-2xCFP::hphMX ura4C721A | This study |
| YHMZ25 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A mcherry-3Flag-dna2::ura4 | This study |
| YHMZ26 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) ura4C721A rad22–2xCFP::hphMX mcherry-3Flag-dna2::ura4 cdc24-YFP::kanMX | This study |
| YHMZ27 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A cdc24ts-YFP::kanMX | This study |
| YHMZ28 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) ura4C721A rad22–2xCFP::hphMX cdc24ts-YFP::kanMX mcherry-3Flag-dna2::ura4 | This study |
| YHMZ29 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A cdc24ts::kanMX | This study |
| YHMZ30 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A dna2ts::ura4 | This study |
| YHMZ31 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A exo1Δ::ura4 | This study |
| YHMZ32 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A rqh1Δ::ura4 | This study |
| YHMZ33 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A exo1Δ::ura4 cdc24ts::kanMX | This study |
| YHMZ34 | h+ | leu1–32 his3-D1 arg3Δ::Hosite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A rqh1Δ::ura4 cdc24ts::kanMX | This study |
| TH2125 | h+ | leu1–32 arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH | Tim Humphrey |
| KHY2125 | h+ | arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41-HO(leu1+) | This study |
| YHMZ36 | h+ | leu1–32 arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41-HO(leu1+) cdc24ts-TAP::hphMX | This study |
| YHMZ37 | h+ | leu1–32 arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41-HO(leu1+) His-HA-dna2ts::ura4 | This study |
| KHY2126 | h+ | arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41leu1+) | This study |
| YHMZ40 | h+ | leu1–32 arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41(leu1+) cdc24ts-TAP::hphMX | This study |
| YHMZ41 | h+ | leu1–32 arg3-D4 ade6-M210 ura4-D18 his3-D1 Ch16-RMGAH leu1–32::nmt41(leu1+) His-HA-dna2ts::ura4 | This study |
| YHMZ56 | h+ | leu1–32 his3-D1 arg3Δ::HOsite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A rpa1-YFP-Flag-His6::leu1+ | This study |
| YHMZ57 | h+ | leu1–32 his3-D1 arg3Δ::HOsite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) rad22–2xCFP::hphMX ura4C721A rpa1ts-YFP-Flag-His6::leu1+ | This study |
| YHMZ58 | h+ | leu1–32 his3-D1 arg3Δ::HOsite-natMX(arg3−) ars1::[pJR1–41XH + HO](his3+) ura4C721A rad22–2xCFP::hphMX mcherry-3Flag-dna2::ura4 cdc24-YFP::kanMX rpa1ts-Flag::leu1+ | This study |
HR Frequency
The effect of defective Cdc24 (cdc24ts) or Dna2 (dna2ts) on homologous recombination was measured according to the method developed by Humphrey and his colleagues (33). To obtain accuracy of the measured recombination frequency with this assay, it was necessary to verify that each cdc24ts or dna2ts cell was viable. Thus, the recombination frequencies in the cdc24ts or dna2ts cells were measured at a growth temperature of 30 °C.
Overexpression and Purification of Dna2 and Cdc24 Protein
Cdc24 and Dna2 were overexpressed and purified from S. pombe cells. To facilitate purification, Cdc24 was tagged with GST and His6 at its C terminus; Dna2 was tagged with 3FLAG at the N terminus and His8 at the C terminus. Both genes were under the control of the nmt1 promoter. Four liters of Edinburgh minimal medium (5 g/liter NH4Cl; Ref. 41) were inoculated with cells containing the Cdc24-GST-His6 or 3FLAG-Dna2-His8 overexpression vectors to an A590 = 0.05 and incubated at 26 °C for 19 h. The following procedure was performed at 4 °C. To purify Cdc24, cells were collected, resuspended in 30–40 ml of buffer A (25 mm Tris (pH 7.5), 100 mm NaCl, 5 mm magnesium acetate, 8 mm β-mercaptoethanol, and 10% glycerol), and lysed with glass beads. Cell debris was removed by centrifugation at 18,000 rpm for 30 min. The supernatant was further filtered through a 0.45-μm membrane. Next, 1 ml of Ni-NTA (nickel-nitrilotriacetic acid) beads and 10 mm imidazole were added and mixed by gentle rotation for 1 h. Cdc24-GST-His6-bound Ni-NTA (nickel-nitrilotriacetic acid) beads were washed with 500 mm NaCl and 10 mm imidazole a few times to remove nonspecific bound proteins. Cdc24-GST-His6 was released from the beads with 250 mm imidazole and then the mixture with Cdc24 subjected to GST affinity chromatography with 800 μl of GST beads. After mixing for 2 h, the beads were washed with buffer B (25 mm Tris (pH 7.5), 100 mm NaCl, 5 mm magnesium acetate, 5 mm DTT, 10% glycerol, and 1% Triton X-100), and Cdc24 was released with glutathione. Cdc24-GST-His6 was further purified by gel filtration using an S200 column.
To purify 3FLAG-Dna2-His8, the cells containing overexpressed Dna2 were collected, resuspended in 30–40 ml of D100 (25 mm Tris (pH 7.5), 100 mm NaCl, and 10% glycerol), and lysed with glass beads. After the cell debris was removed following centrifugation at 18,000 rpm for 30 min, the supernatant was filtered through a 0.45-μm membrane. Nickel-bead affinity chromatography was performed as described above. 3FLAG-Dna2-His8 was then further purified with 500 μl of FLAG-beads and S200 gel filtration. The purified Cdc24 and Dna2 were stored at −80 °C. During the above procedures of protein purification, proteinase inhibitors were added.
Author Contributions
D. K. and H. Z. designed this study, and H. Z. performed all experiments. R. L. helped with DNA end resection and protein purification experiments and Y. H. helped with the end resection assays and the analysis of the data during the course of this study.
Acknowledgments
We thank Li-Lin Du and T. C. Humphrey for gifting us with the strains used in this study, M. Yanagida for the cdc24ts mutant, X. Li for help with the fluorescent assays, Y. Yu and J. Zhang for experimental advice, and the members of the Kong laboratory for their support during the course of this study.
This work was supported by Grant 31230021 from the National Natural Science Foundation of China and Grant 2013CB911000 from the Ministry of Science and Technology of China. This work also was supported by grants from the Peking-Tsinghua Center for Life Sciences and the National Key Laboratory of Protein and Plant Gene Research. The authors declare that they have no conflicts of interest with the contents of this article.
H. Zhang, Y. Hua, R. Li, and D. Kong, unpublished data.
- DSB
- double-stranded DNA break
- CPT
- camptothecin
- BLM
- bleomycin
- NHEJ
- non-homologous end joining
- HR
- homologous recombination
- ssDNA
- single-stranded DNA
- RPA
- replication protein A
- IP
- immunoprecipitation
- MRN
- MRE11-RAD50-NBS1
- CFP
- cyan fluorescent protein
- TAP
- tandem affinity purification
- GC
- gene conversion
- LOH
- loss of heterozygosity
- SCC
- sister chromatin conversion.
References
- 1. Mehta A., and Haber J. E. (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6, a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malkova A., and Haber J. E. (2012) Mutations arising during repair of chromosome breaks. Annu. Rev. Genet. 46, 455–473 [DOI] [PubMed] [Google Scholar]
- 3. Daley J. M., Niu H., Miller A. S., and Sung P. (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst.) 32, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Symington L. S., and Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 [DOI] [PubMed] [Google Scholar]
- 5. Chiruvella K. K., Liang Z., and Wilson T. E. (2013) Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5, a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karanam K., Kafri R., Loewer A., and Lahav G. (2012) Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid-S phase. Mol. Cell 47, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daley J. M., Palmbos P. L., Wu D., and Wilson T. E. (2005) Nonhomologous end joining in yeast. Annu. Rev. Genet. 39, 431–451 [DOI] [PubMed] [Google Scholar]
- 8. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strathern J. N., Shafer B. K., and McGill C. B. (1995) DNA synthesis errors associated with double-strand-break repair. Genetics 140, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hicks W. M., Kim M., and Haber J. E. (2010) Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329, 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Symington L. S. (2014) End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6, a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blackwood J. K., Rzechorzek N. J., Bray S. M., Maman J. D., Pellegrini L., and Robinson N. P. (2013) End-resection at DNA double-strand breaks in the three domains of life. Biochem. Soc. Trans. 41, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cannavo E., and Cejka P. (2014) Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 [DOI] [PubMed] [Google Scholar]
- 14. Andres S. N., Appel C. D., Westmoreland J. W., Williams J. S., Nguyen Y., Robertson P. D., Resnick M. A., and Williams R. S. (2015) Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 22, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies O. R., Forment J. V., Sun M., Belotserkovskaya R., Coates J., Galanty Y., Demir M., Morton C. R., Rzechorzek N. J., Jackson S. P., and Pellegrini L. (2015) CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., and Ira G. (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mimitou E. P., and Symington L. S. (2008) Sae2, Exo1, and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., and Kowalczykowski S. C. (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., Ira G., and Sung P. (2010) Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H., Lisby M., and Symington L. S. (2013) RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cannavo E., Cejka P., and Kowalczykowski S. C. (2013) Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 110, E1661–E1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 23. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 24. Cejka P. (2015) DNA End resection: nucleases team up with the right Partners to initiate homologous recombination. J. Biol. Chem. 290, 22931–22938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gould K. L., Burns C. G., Feoktistova A., Hu C. P., Pasion S. G., and Forsburg S. L. (1998) Fission yeast cdc24(+) encodes a novel replication factor required for chromosome integrity. Genetics 149, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nasmyth K., and Nurse P. (1981) Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 182, 119–124 [DOI] [PubMed] [Google Scholar]
- 27. Tanaka H., Tanaka K., Murakami H., and Okayama H. (1999) Fission yeast cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol. 19, 1038–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanaka H., Ryu G. H., Seo Y. S., and MacNeill S. A. (2004) Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2-Cdc24 complex to DNA polymerase delta. Nucleic Acids Res. 32, 6367–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J. M., Liu X. M., Ding Y. H., Xiong L. Y., Ren J. Y., Zhou Z. X., Wang H. T., Zhang M. J., Yu Y., Dong M. Q., and Du L. L. (2014) Fission yeast Pxd1 promotes proper DNA repair by activating Rad16XPF and inhibiting Dna2. PLoS Biol. 12, e1001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu J., Sun L., Shen F., Chen Y., Hua Y., Liu Y., Zhang M., Hu Y., Wang Q., Xu W., Sun F., Ji J., Murray J. M., Carr A. M., and Kong D. (2012) The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149, 1221–1232 [DOI] [PubMed] [Google Scholar]
- 31. Yu Y., Ren J. Y., Zhang J. M., Suo F., Fang X. F., Wu F., and Du L. L. (2013) A proteome-wide visual screen identifies fission yeast proteins localizing to DNA double-strand breaks. DNA Repair (Amst.) 12, 433–443 [DOI] [PubMed] [Google Scholar]
- 32. Langerak P., Mejia-Ramirez E., Limbo O., and Russell P. (2011) Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tinline-Purvis H., Savory A. P., Cullen J. K., Davé A., Moss J., Bridge W. L., Marguerat S., Bähler J., Ragoussis J., Mott R., Walker C. A., and Humphrey T. C. (2009) Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast. EMBO J. 28, 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kao H. I., Campbell J. L., and Bambara R. A. (2004) Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 35. Prakash A., and Borgstahl G. E. (2012) The structure and function of replication protein A in DNA replication. Subcell. Biochem. 62, 171–196 [DOI] [PubMed] [Google Scholar]
- 36. Zheng L., and Shen B. (2011) Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 3, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nanbu T., Nguyen L. C., Habib A. G., Hirata N., Ukimori S., Tanaka D., Masuda K., Takahashi K., Yukawa M., Tsuchiya E., and Ueno M. (2015) Fission yeast Exo1 and Rqh1-Dna2 redundantly contribute to resection of uncapped telomeres. PloS One 10, e0140456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osman F., Ahn J. S., Lorenz A., and Whitby M. C. (2016) The RecQ DNA helicase Rqh1 constrains Exonuclease 1-dependent recombination at stalled replication forks. Sci. Rep. 6, 22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stewart J. A., Campbell J. L., and Bambara R. A. (2010) Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 38, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rudin N., and Haber J. E. (1988) Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 8, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moreno S., Klar A., and Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]