Abstract
We recently reported that transglutaminase (TG) suppresses immune deficiency pathway-controlled antimicrobial peptides (IMD-AMPs), thereby conferring immune tolerance to gut microbes, and that RNAi of the TG gene in flies decreases the lifespan compared with non-TG-RNAi flies. Here, analysis of the bacterial composition of the Drosophila gut by next-generation sequencing revealed that gut microbiota comprising one dominant genus of Acetobacter in non-TG-RNAi flies was shifted to that comprising two dominant genera of Acetobacter and Providencia in TG-RNAi flies. Four bacterial strains, including Acetobacter persici SK1 and Acetobacter indonesiensis SK2, Lactobacillus pentosus SK3, and Providencia rettgeri SK4, were isolated from the midgut of TG-RNAi flies. SK1 exhibited the highest resistance to the IMD-AMPs Cecropin A1 and Diptericin among the isolated bacteria. In contrast, SK4 exhibited considerably lower resistance against Cecropin A1, whereas SK4 exhibited high resistance to hypochlorous acid. The resistance of strains SK1–4 against IMD-AMPs in in vitro assays could not explain the shift of the microbiota in the gut of TG-RNAi flies. The lifespan was reduced in gnotobiotic flies that ingested both SK4 and SK1, concomitant with the production of reactive oxygen species and apoptosis in the midgut, whereas the survival rate was not altered in gnotobiotic flies that mono-ingested either SK4 or SK1. Interestingly, significant amounts of reactive oxygen species were detected in the midgut of gnotobiotic flies that ingested SK4 and SK2, concomitant with no significant apoptosis in the midgut. In gnotobiotic flies that co-ingested SK4 and SK1, an additional unknown factor(s) may be required to cause midgut apoptosis.
Keywords: antimicrobial peptide (AMP), Drosophila, reactive oxygen species (ROS), RNA interference (RNAi), transglutaminase, dysbiosis, gnotobiotic fly, microbiota
Introduction
Foreign substances, including foods, minerals, and microbes, continually pass through the intestinal tract and attach to the gut epithelium. Development of physical and immunologic barriers against foreign substances is thus essential to protect the gut epithelia. Healthy and balanced gut microbiota provide essential nutrients for their host and help to maintain gut immune homeostasis, whereas disruption of the balance, called dysbiosis, is associated with various animal diseases (1, 2). The fruit fly, Drosophila melanogaster, is a useful model for investigating the close relationship between bacteria and bacteria or host and bacteria interactions (3, 4), and the gut microbial community of Drosophila comprises ∼105 microbes of ∼20 species (5–7). The Drosophila gut is functionally analogous to the mammalian intestinal tract (8, 9), producing antimicrobial peptides (AMPs)2 and reactive oxygen species (ROS) (10, 11). Microbes or microbe-derived immune elicitors in the fly gut can initiate several immune signaling pathways, such as the immune deficiency (IMD) pathway activated by peptidoglycans to produce AMPs and the dual oxidase (DUOX) signaling pathway activated by uracils to produce ROS (11–14). In addition to pathogenic bacteria, commensal microbe-derived peptidoglycans constitutively activate the IMD pathway in the gut through peptidoglycan recognition protein long transcript C and peptidoglycan recognition protein long transcript E, and several types of suppression of the IMD pathway have been reported (10, 15). On the other hand, non-commensal bacteria, such as the opportunistic insect pathogen Ecc15 and the insect pathogens Pseudomonas entomophila and Gluconobacter morbifer G707T, secrete uracil to trigger the DUOX-dependent ROS production (16). The IMD pathway-controlled antimicrobial peptides (IMD-AMPs) exhibit a microbicidal effect on a narrow range of virulent bacteria, whereas ROS-based immunity appears to be a more effective antimicrobial system against gut infection (11–14).
Transglutaminase (TG) catalyzes the isopeptide bond formation between Lys and Gln residues in a Ca2+-dependent manner (17). In mammals, eight TG isozymes are involved in various biologic processes, such as blood coagulation, extracellular matrix formation, and apoptosis (18). In invertebrates, such as horseshoe crabs, crayfish, and Drosophila, TG-mediated cross-linking of specific proteins is involved in hemolymph coagulation and cuticle formation (19–22). Drosophila carries a single TG gene, which cross-links the nuclear factor-κB-like transcription factor Relish in the IMD pathway in midgut cells to suppress the expression of antimicrobial peptides (23). RNAi of the TG gene enhances the expression of IMD-AMPs, including Cecropin A1 and Diptericin, which decreases the lifespan concomitant with apoptosis of the midgut cells, compared with non-TG-RNAi flies (23). Interestingly, TG-RNAi does not decrease the lifespan of germ-free flies, whereas non-TG-RNAi flies that ingest gut lysates prepared from conventionally reared TG-RNAi flies have a short lifespan (23). We hypothesized that TG-RNAi causes dysbiosis of the gut microbiota, leading to a short lifespan. Here we identified the bacterial composition in the gut of TG-RNAi and non-TG-RNAi flies by next-generation sequencing and characterized the microbes isolated from the gut.
Results
Effects of TG-RNAi on Gut Flora
The survival rates of conventionally reared TG-RNAi (Da>TG IR) and non-TG-RNAi (Da>+) flies as control flies are shown in Fig. 1, which confirmed that RNAi directed against the TG gene reduces the lifespan of flies under conventional non-sterile conditions (23). To investigate the effects of TG-RNAi on the gut microbiota, gut microbes of 0.5-day-old and 10-day-old non-TG-RNAi or TG-RNAi flies were analyzed using next-generation sequencing of 16S rRNA gene (rDNA) clones. Four genera of bacteria were identified in both non-TG-RNAi and TG-RNAi flies, including Acetobacter, Lactobacillus, Providencia, and Wolbachia (Table 1). These genera were previously reported as bacterial communities in D. melanogaster (24–30). Bacteria that highly matched the sequences of the V4 region of the 16S rDNA clones are shown in Table 2. In this study, Wolbachia is not discussed because it is an endosymbiont bacteria, not a gut microbe (31). The effective number of bacterial cells was estimated as described under “Experimental Procedures.” In 0.5-day-old non-TG-RNAi flies, the most dominant genus was Acetobacter, followed by Providencia (Fig. 2A, left). In 0.5-day-old TG-RNAi flies, Providencia was the most dominant, followed by Acetobacter (Fig. 2A, right). Lactobacillus was a recessive genus in both cases in 0.5-day old flies (Fig. 2A, right and left). Acetobacter became more dominant, whereas Providencia and Lactobacillus were not identified in 10-day-old non-TG-RNAi flies in the sequencing analysis (Fig. 2B, left). Interestingly, microbiota in 10-day-old TG-RNAi flies contained two bacterial dominant microbiota, Acetobacter and Providencia, at a ∼1:1 ratio in the gut (Fig. 2B, right). To determine whether the total number of gut bacteria was changed by TG-RNAi, the copy number of the 16S rDNA was analyzed by quantitative PCR (qPCR) after considering the 16S rRNA copy number of each genus (Fig. 2C). The total number of gut bacteria did not differ significantly between 0.5-day-old non-TG-RNAi and 0.5-day-old TG-RNAi flies, whereas the bacterial load was increased 10-fold in 10-day-old TG-RNAi flies compared with 10-day-old non-TG-RNAi flies, suggesting that dysbiosis of the gut microbiota was induced by TG-RNAi to increase the number of gut microbes compared with that in non-TG-RNAi flies. We previously reported that TG-RNAi enhances the expression of IMD-AMP genes, such as Cecropin A1 and Diptericin (23). To confirm the induction of IMD-AMPs by TG-RNAi, the expression of Cecropin A1 was measured by qPCR. No difference in the expression of Cecropin A1 was observed in the guts of 0.5-day-old flies between non-TG-RNAi and TG-RNAi flies, whereas in 10-day-old flies, the expression of Cecropin A1 was significantly increased in TG-RNAi flies, but not in non-TG-RNAi flies (Fig. 2D).
FIGURE 1.
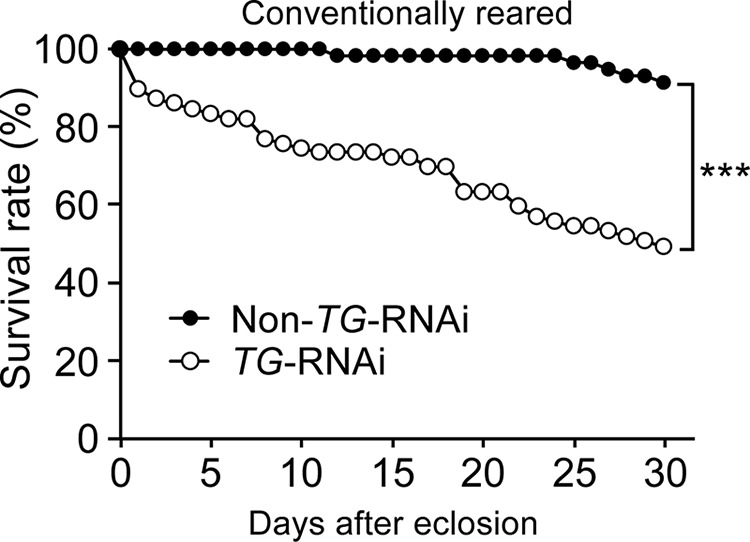
Survival rates of non-TG-RNAi flies and TG-RNAi flies. The survival rates of conventionally reared non-TG-RNAi (Da>+, n = 59; closed circles) and TG-RNAi (Da>TG IR, n = 80; open circles) flies are shown. p values were calculated by the log-rank test. ***, p < 0.005.
TABLE 1.
Sequences of the V4 region of 16S rDNA clones obtained from the gut of non-TG-RNAi or TG-RNAi flies
| Genera identified | Sequences of the V4 region of the 16S rDNA clones |
|---|---|
| Acetobacter | 5′-tacgaagggggctagcgttgctcggaatgactgggcgtaaagggcgtgtaggcggtttgcacagttagatgtgaaatccccgggcttaacctgggagctgcatttaagacgtgcagactagagtgtgagagagggttgtggaattcccagtgtagaggtgaaattcgtagatattgggaagaacaccggtggcgaaggcggcaacctggctcatgactgacgctgaggcgcgaaagcgtggggagcaaacagg-3′ |
| Lactobacillus | 5′-tacgtaggtggcaagcgttgtccggatttattgggcgtaaagcgagcgcaggcggttttttaagtctgatgtgaaagccttcggctcaaccgaagaagtgcatcggaaactgggaaacttgagtgcagaagaggacagtggaactccatgtgtagcggtgaaatgcgtagatatatggaagaacaccagtggcgaaggcggctgtctggtctgtaactgacgctgaggctcgaaagtatgggtagcaaacagg-3′ |
| Providencia | 5′-tacggagggtgcaagcgttaatcggaattactgggcgtaaagcgcacgcaggcggttgattaagttagatgtgaaatccccgggcttaacctgggaatggcatctaagactggtcagctagagtcttgtagaggggggtagaattccatgtgtagcggtgaaatgcgtagagatgtggaggaataccggtggcgaaggcggccccctggacaaagactgacgctcaggtgcgaaagcgtggggagcaaacagg-3′ |
| Wolbachia | 5′-tacggagagggctagcgttattcggaattattgggcgtaaagggcgcgtaggcggattagtaagttaaaagtgaaatcccaaggctcaaccttggaattgcttttaaaactgctaatctagagattgaaagaggatagaggaattcctagtgtagaggtgaaattcgtaaatattaggaggaacaccagtggcgaaggcgtctatctggttcaaatctgacgctgaggcgcgaaggcgtggggagcaaacagg-3′ |
TABLE 2.
Bacteria highly matched with the sequences of the V4 region of the 16S rDNA clones
| Genera identified | Highly matched bacterial strains | Identity |
|---|---|---|
| % | ||
| Acetobacter | A. persici T-120T | 99.6 |
| A. farinalis G360–1T | 99.2 | |
| A. cerevisiae LMG 1625T | 98.8 | |
| A. indonesiensis NRIC 0313T | 98.0 | |
| Lactobacillus | L. pentosus JCM 1558T | 100 |
| L. fabifermentans DSM 21115T | 100 | |
| L. paraplantarum DSM 10667T | 100 | |
| L. plantarum subsp. plantarum ATCC 14917T | 100 | |
| Providencia | P. rettgeri Dmel1a | 100 |
| P. alcalifaciens DSM 30120T | 100 | |
| P. rustigianii DSM 4541T | 100 | |
| P. vermicola OP1T | 100 | |
| P. sneebia DSM 19967T | 100 | |
| Wolbachia | W. pipientis ωPip | 95.7 |
a Ref. 38.
FIGURE 2.
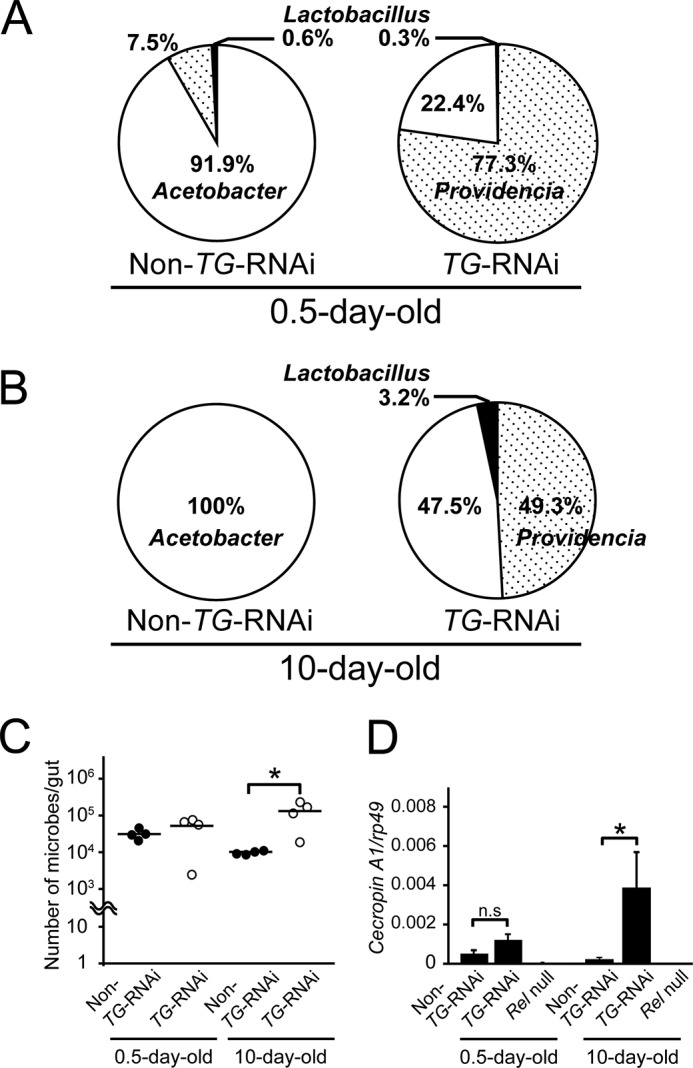
Gut flora of non-TG-RNAi flies and TG-RNAi flies. A, gut microbiota in 0.5-day-old flies. For non-TG-RNAi and TG-RNAi flies, 160 and 162 flies were analyzed, respectively. B, gut microbiota in 10-day-old flies. For non-TG-RNAi and TG-RNAi flies, 200 and 126 flies were analyzed, respectively. In all cases, the male/female ratio was 1.0. C, four midguts of 0.5- or 10-day-old flies (male/female ratio was 1.0) were mixed into one tube, and extracted 16S rDNA was analyzed by quantitative PCR. The bacterial loads are shown as 16S rDNA copies per midgut. The analysis was repeated four times. Closed circles, non-TG-RNAi; open circles, TG-RNAi. Bars, means of four independent experiments. p values were calculated using Student's t test. *, p < 0.05. D, mRNA expression levels of Cecropin A1 in the gut of 0.5- and 10-day-old non-TG-RNAi, TG-RNAi, and Relish null (RelE20) flies. Results are shown as the Cecropin A1/rp49 ratio. Values shown are means ± S.E. (error bars) (n = 2). p values were calculated by one-way ANOVA followed by the Bonferroni post hoc test. *, p < 0.05; n.s, not significant.
Antimicrobial Peptide Resistance of the Isolated Bacteria
IMD-AMPs in the Drosophila gut act as a selective pressure, and overexpression of IMD-AMPs induces dysbiosis, although the importance of IMD-AMPs in immune responses in the gut is unclear (12, 24). We speculate that TG-RNAi triggers a shift in the gut microbiota from the single bacterial dominant microbiota of Acetobacter to the two bacterial dominant microbiota of Acetobacter and Providencia through regulation of the production of IMD-AMPs. We isolated gut bacteria from 10–12-day-old non-TG-RNAi or TG-RNAi flies by plating and incubating gut homogenates on Hestrin-Schramm medium agar plates and nutrient agar medium plates. Four bacterial strains were isolated from the midgut of the flies; the SK1 and SK3 strains were isolated from both non-TG-RNAi and TG-RNAi flies, and the SK2 and SK4 strains were isolated from TG-RNAi flies, but not from non-TG-RNAi flies. Sequence analyses of the entire 16S rDNAs identified these isolated strains (SK1, SK2, SK3, and SK4) as Acetobacter persici, Acetobacter indonesiensis, Lactobacillus pentosus, and Providencia rettgeri, respectively (Table 3).
TABLE 3.
The closest bacterium matching the 16S rDNA sequence of the bacterium isolated from the gut of non-TG-RNAi or TG-RNAi flies
GenBankTM accession numbers are shown in parentheses.
To evaluate the colonization ability of strains SK1–4, bacterial loads in the midguts of germ-free TG-RNAi or germ-free non-TG-RNAi flies that transiently ingested each bacterial strain were examined (Fig. 3). SK1 and SK4 colonized equally in the midguts of both non-TG-RNAi and TG-RNAi flies. In contrast, little or no bacterial load of SK2 and SK3 was found in non-TG-RNAi flies, especially the middle and posterior midgut regions, but sufficient bacterial loads of SK2 and SK3 were counted in the midguts of TG-RNAi flies. These findings suggest that a better intestinal habitat is provided for specific bacterial strains under the conditions of TG-RNAi despite the increasing expression of IMD-AMPs in the fly gut. To investigate the resistance of strains SK1–4 to IMD-AMPs, the antimicrobial activities of Cecropin A1 and Diptericin were assayed (Figs. 4 and 5 and Table 4). SK1 exhibited the strongest resistance to Cecropin A1, with an IC50 value of >10 μg/ml in isolated and established bacteria (Fig. 4 and Table 4). The IC50 value of SK2 and SK3 to Cecropin A1 was an order of magnitude higher (0.22–0.35 μg/ml) than the IC50 values of established bacteria (0.05–0.06 μg/ml; Fig. 4 and Table 4). In contrast, SK4 exhibited considerably lower resistance against Cecropin A1, with an IC50 of 0.07 μg/ml, similar to that of established bacteria (Fig. 4 and Table 4). Notably, Diptericin had no or little inhibitory effect on the growth of SK1 or SK3 at a concentration of 20 μg/ml (Fig. 5 and Table 4). SK4 also had significantly higher resistance to Diptericin than the established bacteria, including Erwinia carotovora carotovora 15 2141, Pseudomonas azotoformans IAM 1603, and Escherichia coli K12 (Fig. 5 and Table 4). Providencia alcalifaciens JCM1673T, belonging to the same genus of SK4, exhibited rather high resistance to Diptericin (Fig. 5 and Table 4).
FIGURE 3.
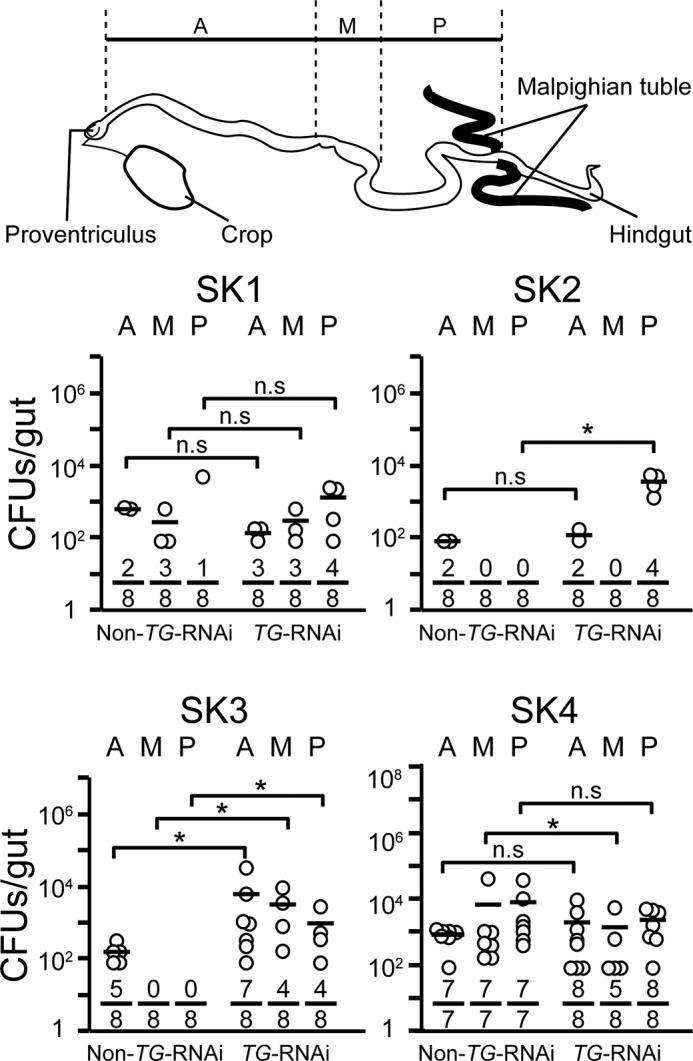
Bacterial colonization ability in fly midguts after transient ingestion of strains SK1–4. The bacterial strains were ingested by 3–5-day-old adult non-TG-RNAi and TG-RNAi flies for 24 h, and then the flies were transferred into sterile vials containing sterile food. After a 5-h incubation, midguts were removed and dissected into three regions: the anterior midgut (A), middle midgut (copper cell region) (M), and posterior midgut (P). The three dissected parts were homogenized and spotted onto the strain-specific agar plates, and the resulting bacterial colonies were counted as described under “Experimental Procedures.” Top, schematic image of the Drosophila midgut. Horizontal bars represent the median cfu/fly for each ingestion group. Each denominator and numerator of the fractions shows the number of all guts used for the experiments and the number of guts with bacteria growing on agarose plates, respectively. p values were calculated by the non-parametric Mann-Whitney U test. *, p < 0.05; n.s, not significant.
FIGURE 4.
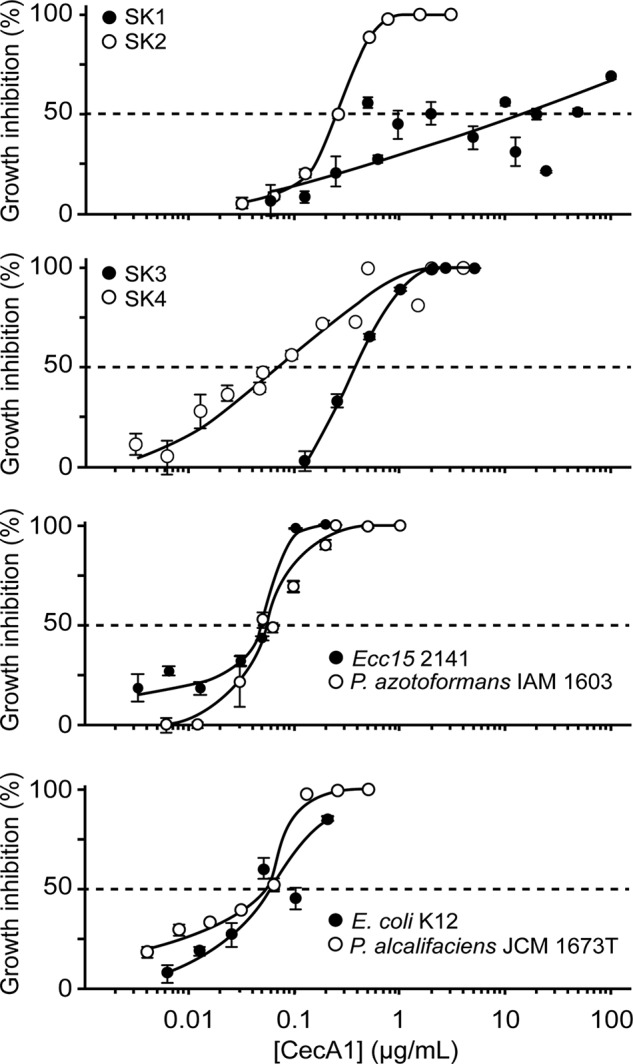
Antimicrobial resistance of strains SK1–4 and established bacteria to Cecropin A1. Inhibitory activity was measured after incubating bacteria in the presence of different concentrations of Cecropin A1 against strains SK1–4 and established bacteria. Values shown are means ± S.E. (error bars) (n = 3). The dashed line represents the half-maximal inhibitory concentration (IC50) of Cecropin A1.
FIGURE 5.
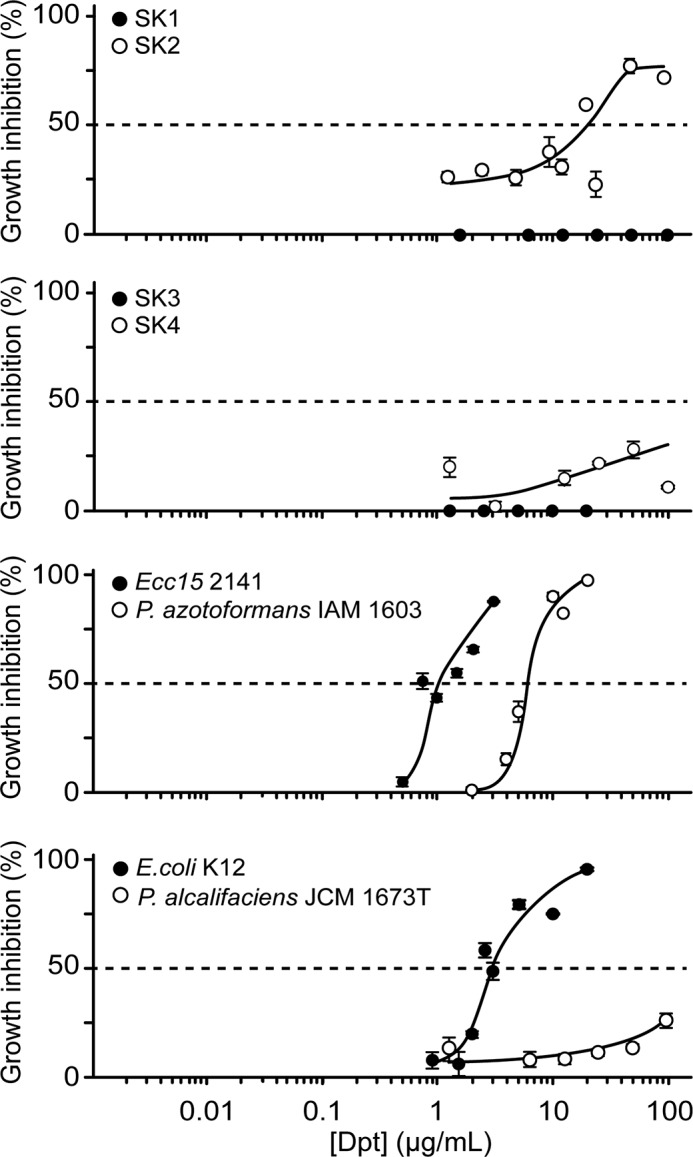
Antimicrobial resistance of strains SK1–4 and established bacteria to Diptericin. Inhibitory activity was measured after incubating bacteria in the presence of different concentrations of Diptericin (Dpt) against strains SK1–4 and established bacteria. Values are shown as means ± S.E. (error bars) (n = 3). The dashed line represents the half-maximal inhibitory concentration (IC50) of Diptericin.
TABLE 4.
IC50 of Cecropin A1 and Diptericin to strains SK1–4 and established bacteria
| Bacterial strains | IC50 |
|
|---|---|---|
| Cecropin A1 | Diptericin | |
| μg/ml | ||
| A. persici SK1 | >10 | No inhibition |
| A. indonesiensis SK2 | 0.22 | >20 |
| L. pentosus SK3 | 0.35 | No inhibition |
| P. rettgeri SK4 | 0.07 | >100 |
| Ecc15 2141 | 0.05 | 1.0 |
| P. azotoformans IAM1603 | 0.06 | 6.0 |
| E. coli K12 | 0.06 | 3.0 |
| P. alcalifaciens JCM1673T | 0.05 | >100 |
No experimental conditions to culture all microbial communities in the fly gut have been established, and strains SK1–4 may not reflect the complete microbiome of TG-RNAi flies. Gluconobacter morbifer G707T is reported to be a microbe with pathologic consequences in the Drosophila gut (24). G. morbifer G707T and reported intestinal strains, including Commensalibacter intestini A911T, Acetobacter pomorum, Lactobacillus brevis EW, and Lactobacillus plantarum WJL (24), could be cultured under the same conditions used for SK1, and their antimicrobial resistance was examined (Table 5). G. morbifer G707T exhibited significantly lower resistance against Cecropin A1 at 1.0 μg/ml, compared with the other commensal stains, whereas Diptericin at 10 μg/ml had little effect on the growth of G. morbifer G707T. On the other hand, in this study, the genera Gluconobacter and Commensalibacter were not identified in the midguts of non-TG-RNAi and TG-RNAi flies by next-generation sequencing of 16S rDNA. Therefore, we used strains SK1–4 as representative microbiota of TG-RNAi flies.
TABLE 5.
Antimicrobial resistance of the gut strains isolated by Ryu et al. (24)
Cecropin A1 and Diptericin resistance activities are expressed as relative CFUs on plates with the number of colony-forming units in the untreated bacteria arbitrarily set to 100%. Data represent means ± S.E. (n = 3).
| Bacterial strains | Growth inhibition |
|
|---|---|---|
| Cecropin A1 at 1 μg/ml | Diptericin at 10 μg/ml | |
| % | ||
| G. morbifer G707T | 80 ± 0.6 | 20 ± 5.9 |
| C. intestini A911T | 8.9 ± 8.0 | 11 ± 8.2 |
| A. pomorum | 7.8 ± 0.4 | 3.5 ± 1.8 |
| L. brevis EW | No inhibition | 5.4 ± 2.4 |
| L. plantarum WJL | 8.0 ± 4.3 | 8.9 ± 2.3 |
Anti-ROS Activities of the Isolated Bacteria
In Drosophila, midgut cells may be tolerant toward commensal bacteria, and at the same time, they should be protected against the invasion of virulent pathogens. The Drosophila genome contains one DUOX gene, which is indispensable for gut immunity, and an extracellular peroxidase homology domain of DUOX has the enzymatic activity to produce highly microbicidal hypochlorous acid (HOCl) from H2O2 in the presence of chlorine (32). On the other hand, severe gut infection by pathogenic bacteria induces excessive production of DUOX-dependent ROS to damage host tissues and cells and thus decreases the survival rate (11, 16). Strains SK1–4 were incubated in the presence of HOCl containing 15 μg/liter or 50 μg/liter available chlorine to determine the resistance to ROS (Table 6). Treatment with 15 μg/liter HOCl had little or no effect on strains SK1–4. Furthermore, SK1 and SK4 were quite resistant to HOCl at a higher concentration of 50 μg/liter. On the other hand, the growth of SK3 was strongly suppressed at 50 μg/liter HOCl. These findings suggest that SK1 and SK4 acquire high resistance against ROS in the Drosophila gut. Moreover, the intestinal strains of A. pomorum and L. brevis EW and the insect opportunistic pathogen Ecc15 2141 exhibited very high anti-ROS activity at 50 μg/liter of HOCl (Table 6). In contrast, C. intestini A911T, E. coli K12, and P. alcalifaciens JCM1673T were highly sensitive to HOCl under the same conditions (Table 6).
TABLE 6.
HOCl resistance of the isolated gut and established bacteria
HOCl resistance activity is expressed as relative cfu on plates with the number of cfu in the untreated bacteria arbitrarily set to 100%. Data represent means ± S.E. (n = 3).
| Bacterial strains | Resistance activity |
|
|---|---|---|
| 15 μg/liter HOCl | 50 μg/liter HOCl | |
| % | ||
| A. persici SK1 | 110 ± 12 | 84 ± 12 |
| A. indonesiensis SK2 | 110 ± 10 | 36 ± 1.1 |
| L. pentosus SK3 | 87 ± 9.5 | 9.3 ± 1.3 |
| P. rettgeri SK4 | 130 ± 7.0 | 56 ± 2.9 |
| G. morbifer G707T | 105 ± 3.0 | 87 ± 10 |
| C. intestini A911T | 77 ± 3.6 | 0.0 ± 0.0 |
| A. pomorum | 124 ± 11 | 112 ± 0.8 |
| L. brevis EW | 104 ± 2.7 | 101 ± 0.6 |
| L. plantarum WJL | 97 ± 2.1 | 83 ± 6.0 |
| Ecc15 2141 | 100 ± 8.5 | 100 ± 11 |
| P. azotoformans IAM1603 | 100 ± 1.8 | 19 ± 1.4 |
| E. coli K12 | 69 ± 11 | 0.3 ± 0.1 |
| P. alcalifaciens JCM1673T | 6.6 ± 1.1 | 0.0 ± 0.0 |
Effects of Long Term Ingestion of Strains SK1–4 on Survival Rates and Midgut Apoptosis
Gut bacteria are important for maintaining host homeostasis by generating nutrients and inhibiting the growth of invading pathogens (2, 33, 34). In Drosophila, a strain of A. pomorum plays an important role in host homeostatic programs, such as body size and energy metabolism, by activating insulin/insulin-like growth factor signaling (35). To examine the effects of the ingestion of strains SK1–4 on the fly survival rate, gnotobiotic flies were prepared by long term feeding in germ-free flies. Survival rate experiments using gnotobiotic flies revealed that mono-ingestion of strains SK1–4 for 25 days had little effect on the survival rates (Fig. 6A). Interestingly, in gnotobiotic flies, co-ingestion of SK4 and SK1 significantly decreased the survival rate, whereas co-ingestion of SK4 and SK2 had little effect. To examine the colonization ability of strains SK1–4 in the long term ingestion experiments, bacterial loads of gnotobiotic flies that mono-ingested or co-ingested strains SK1–4 were counted at 15 and 25 days after ingestion (Fig. 6B). Twenty-five days after continuous ingestion, each strain exhibited sufficient colonization with 104 to 105 cfu/gut, whether mono- or co-ingested. Bacterial loads of mono-ingested strains SK1–3 after 25 days of ingestion were 10 times greater than those after 15 days of ingestion. The phenomenon of an age-dependent bacterial load increase in the midgut was reported previously (29).
FIGURE 6.
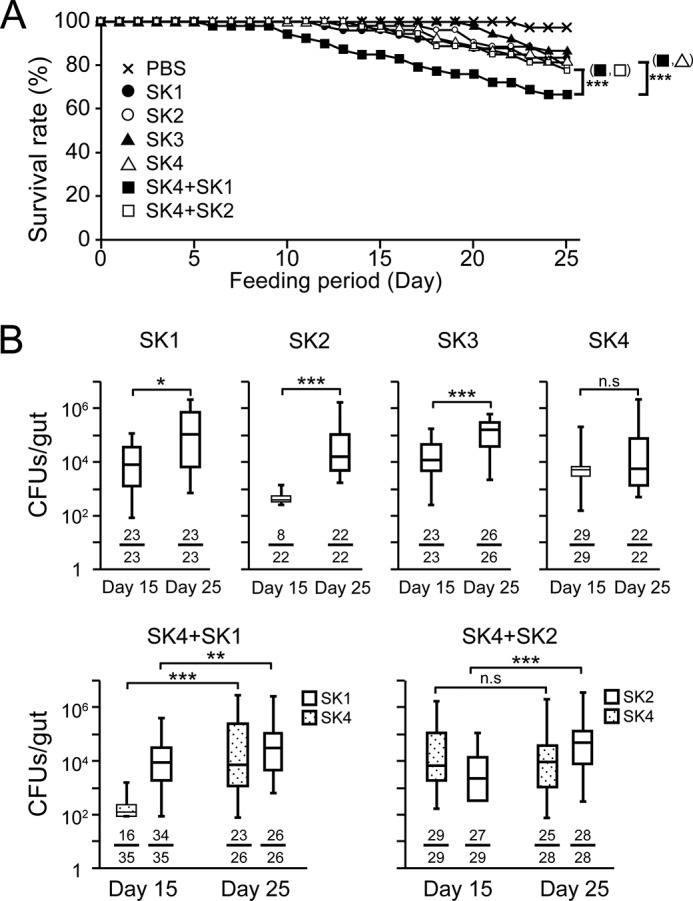
Survival rates and bacterial colonization in the gut after long term mono- or co-ingestions of strains SK1–4. A, survival rates of w1118 flies after long term mono- or co-ingestion. Ingestion groups were as follows: PBS (n = 40) (crosses); SK1 (n = 48) (closed circles); SK2 (n = 54) (open circles); SK3 (n = 54) (closed triangles); SK4 (n = 54) (open triangles); mixture of SK1 and SK4 (n = 54) (closed squares); mixture of SK2 and SK4 (n = 54) (open squares). p values were calculated with the log-rank test. ***, p < 0.005. B, bacterial colonization ability in w1118 germ-free flies after 15 and 25 days of continuous mono-ingestion or co-ingestion. Box plots indicate the interquartile range (boxes) and the median (a horizontal line in each box); whiskers extend toward the minimum and maximum values. Each denominator and numerator of the fractions shows the number of all guts used for the experiments and the number of guts with bacteria growing on agarose plates, respectively. p values were calculated by the non-parametric Mann-Whitney U test. *, p < 0.05; **, p < 0.01; ***, p < 0.005; n.s, not significant.
We previously performed TUNEL assays and detected apoptotic cells in the middle midgut of conventionally reared TG-RNAi flies, but not in conventionally reared non-TG-RNAi flies or germ-free TG-RNAi flies (23). To evaluate the effects of long term ingestion of strains SK1–4 on apoptosis of midgut cells, we performed TUNEL assays after 18 days of ingestion. The number of TUNEL-positive guts was significantly increased in the middle midgut region of gnotobiotic flies that co-ingested SK4 and SK1 compared with gnotobiotic flies that mono-ingested SK4 or co-ingested SK4 and SK2 (Fig. 7). Therefore, the cause of apoptosis in the midguts is not simply due to a larger number of bacteria in the gut.
FIGURE 7.
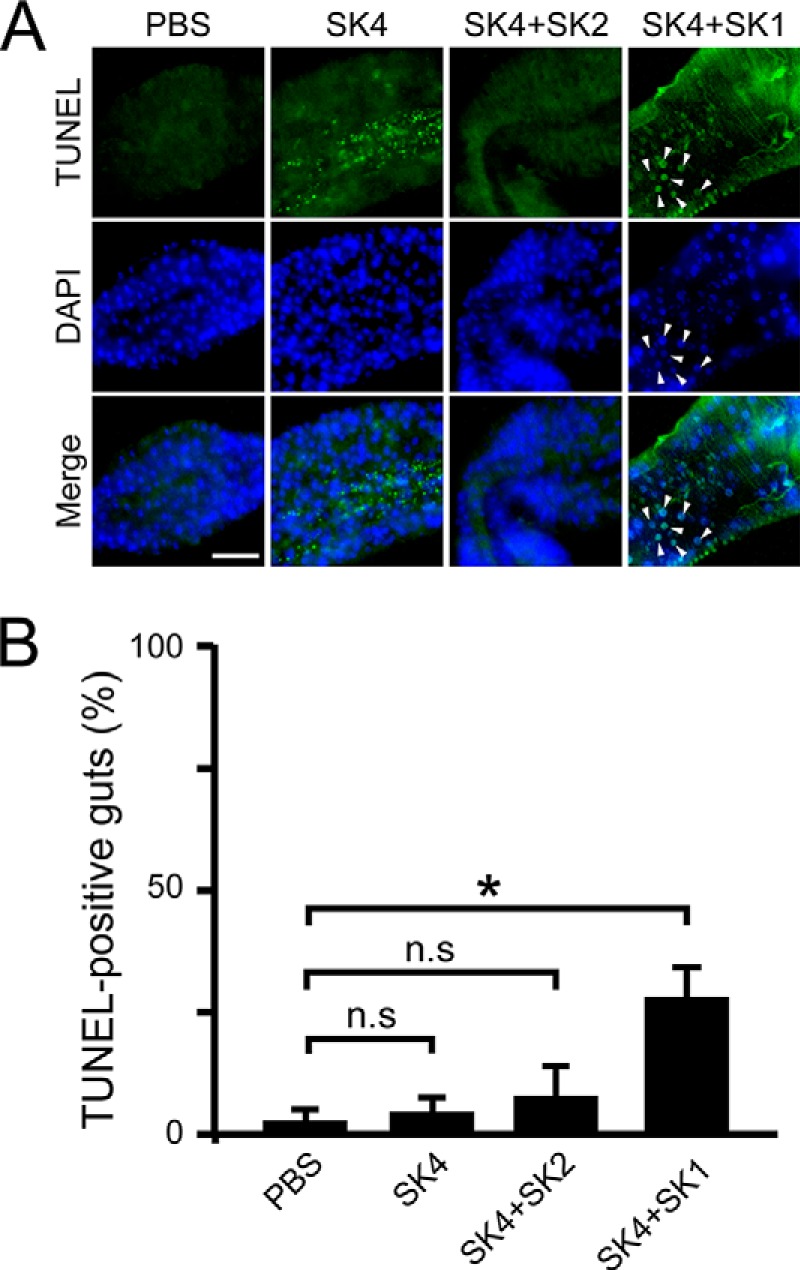
TUNEL assays in the midguts of gnotobiotic flies that mono- or co-ingested strains SK1–4. A, TUNEL staining of apoptotic cells in the fly midgut after 18 days of continuous ingestion. Images show the copper cell region of the midgut. Arrowheads, representative TUNEL-positive cells. Green, TUNEL-positive cells; blue, DAPI nuclear stain; scale bar, 50 μm. B, the percentage of guts showing apoptosis-positive signals was calculated. The values shown are means ± S.E. (error bars) (n = 3). p values were calculated by one-way ANOVA followed by the Bonferroni post hoc test. *, p < 0.05; n.s, not significant.
ROS Production in the Midgut following Long Term Ingestion of Strains SK1–4
Bacteria-derived uracil, which is secreted by pathogenic or opportunistic bacteria but not by commensal bacteria, acts as a signal to trigger the ROS production of DUOX through a G-protein-coupled receptor-mediated signaling pathway in the fly gut (16). ROS production in the midgut of gnotobiotic flies was examined after 18 days ingestion, using a fluorescent probe, CellROX green. The signal intensity of CellROX induced by ROS production was measured quantitatively by a fluorescent cell imager. ROS production was statistically greater in the midgut of gnotobiotic flies that co-ingested SK4 and SK1 compared with that of gnotobiotic flies that mono-ingested SK4 (Fig. 8A). Interestingly, gnotobiotic flies that co-ingested SK4 and SK2 also exhibited high ROS production in the midgut (Fig. 8A). To confirm that ROS production was induced by co-ingestion of SK4 and SK1 or SK2, uracil-sensitive Drosophila S2 cells were treated with microbial extracts derived from mono- or co-incubated bacterial strains, as described under “Experimental Procedures.” ROS production in S2 cells was examined using another fluorescent probe, 2′,7′-dichlorofluorescin-diacetate (DCF-DA). The number of fluorescence-positive cells was greater in S2 cells treated with the extract derived from co-incubated SK4 and SK1 or SK2, but not with the extracts derived from mono-incubated SK4, SK1, or SK2 (Fig. 8B).
FIGURE 8.
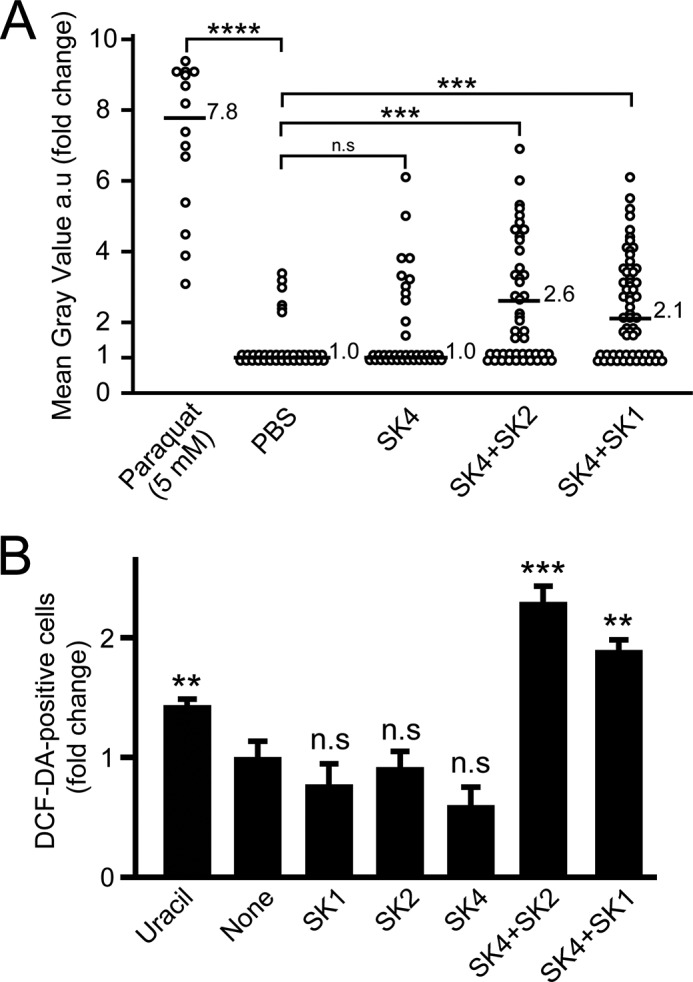
ROS production in the midguts of gnotobiotic flies that mono- and co-ingested strains SK1–4. A, as a positive control for ROS production, flies ingested 5 mm paraquat. -Fold change in mean gray value of CellROX signal intensity in the anterior midgut was evaluated and normalized to the negative control (average signal intensity of negative control was set to 1). Horizontal bars, median -fold change for each ingestion group with the -fold change value. Ingestion groups were as follows: paraquat (n = 14); PBS (n = 36); SK4 (n = 38); SK4 + SK2 (n = 46); SK4 + SK1 (n = 61). Statistical analysis of the differences between the five ingestion groups was performed using the non-parametric Kruskal-Wallis test followed by the Bonferroni post hoc test. ***, p < 0.005; ****, p < 0.001; n.s, not significant. B, ROS production in S2 cells treated with microbial extracts. The -fold change of DCF-DA-positive cell number was analyzed (1,000 S2 cells in each group). The values shown are the means ± S.E. (error bars) (n = 3). p values were calculated by one-way ANOVA followed by the Bonferroni post hoc test. **, p < 0.01; ***, p < 0.005; n.s, not significant.
Expression of IMD-AMP Genes in Gnotobiotic Flies That Mono- or Co-ingested Strains SK1–4
TG-RNAi enhances the expression of IMD-AMP genes, such as Cecropin A1 and Diptericin (23) (Fig. 2D). Here we examined the expression of these genes in gnotobiotic flies that mono- or co-ingested strains SK1–4 (Fig. 9). Unexpectedly, expression of both genes was markedly induced in gnotobiotic files that co-ingested SK4 and SK2, but not gnotobiotic flies that co-ingested SK4 and SK1 or mono-ingested SK4. These findings suggest that the release of microbe-derived elicitors, peptidoglycans, to induce the expression of IMD-AMP genes was suppressed in the midgut in gnotobiotic flies that co-ingested SK4 and SK1 or mono-ingested SK4.
FIGURE 9.
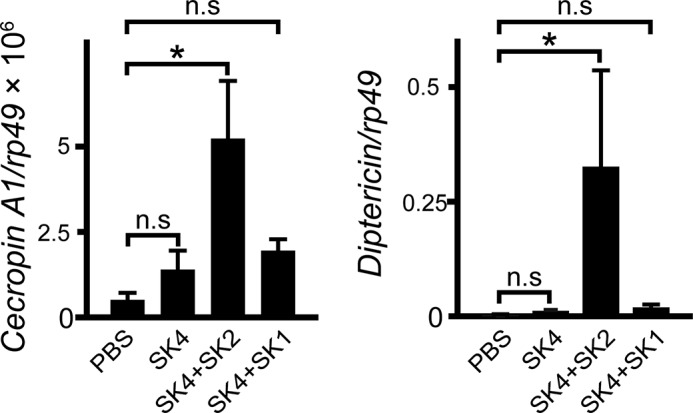
Gene expression of IMD-AMPs in gnotobiotic flies that mono- and co-ingested strains SK1–4. Gene expression of Cecropin A1 or Diptericin was analyzed by quantitative PCR in the fly midgut after 18 days of continuous ingestion. Results are shown as the Cecropin A1/rp49 and Diptericin/rp49 ratio. Values shown are means ± S.E. (error bars) (n = 3). p values were calculated by one-way ANOVA followed by the Bonferroni post hoc test. *, p < 0.05; n.s, not significant.
Discussion
In the present study, 16S rDNA sequencing analysis and characterization of bacteria isolated from the gut revealed that microbiota comprising the single dominant genus of Acetobacter in non-TG-RNAi flies was shifted to that comprising the two dominant genera of Acetobacter and Providencia in TG-RNAi flies, leading to gut dysbiosis (Fig. 2, A and B). Four bacterial strains, including Acetobacter persici SK1, Acetobacter indonesiensis SK2, Lactobacillus pentosus SK3, and Providencia rettgeri SK4, were isolated from the midgut of TG-RNAi flies. SK1 and SK3 were also isolated from the midgut of non-TG-RNAi flies. Acetobacter-dominated gut microbiota of Drosophila has been reported, and multiple factors could contribute to the establishment of the composition of microbiota, such as environmental circumstances, compatibility of bacteria, and transmission of bacteria from mother flies to their offspring (28, 30, 36). SK1 exhibited the strongest resistance against IMD-AMPs, including Cecropin A1 and Diptericin, and ROS, compared with the other three isolated bacteria (Figs. 4 and 5 and Tables 4 and 6), which could explain why the genus Acetobacter is dominant in non-TG-RNAi flies. Why the genus Providencia becomes another dominant genus in TG-RNAi flies, however, remains unclear, because SK4 exhibited very low resistance against Cecropin A1 with an IC50 value of 0.07 μg/ml, similar to the established bacteria P. alcalifaciens JCM 1673T with an IC50 of 0.05 μg/ml (Table 4). In humans, P. rettgeri and P. alcalifaciens are opportunistic pathogens known to cause diarrhea (37). The resistance of strains SK1–4 against IMD-AMPs in in vitro assays could not explain the shift of microbiota in the gut in TG-RNAi flies. Interestingly, gene expression of IMD-AMPs was significantly suppressed in gnotobiotic flies that co-ingested SK4 and SK1 or mono-ingested SK4 compared with that of gnotobiotic flies that co-ingested SK4 and SK2 (Fig. 9). It is possible that SK4 belonging to the opportunistic species P. rettgeri displays a cryptic defense mechanism against IMD-AMP gene expression, depending on the environmental conditions in the midgut, but not in the in vitro culture conditions. SK4 exhibited much higher resistance to ROS than P. alcalifaciens JCM 1673T (Table 6). According to a septic injury model in Drosophila, P. rettgeri Dmel and P. alcalifaciens Dmel are moderately and highly virulent pathogens, respectively (38, 39), suggesting that SK4 becomes a virulent opportunistic bacterial strain in the fly gut.
In addition to the shift in the gut microbiota, 10-day-old TG-RNAi flies retained a higher bacterial load in the gut than 10-day-old non-TG-RNAi flies (Fig. 2C). We previously reported that systemic TG-RNAi causes a peritrophic matrix defect that allows for the penetration of dextran beads from the gut lumen into the ectoperitrophic space (23) and that TG-catalyzed cross-linking of peritrophic matrix protein is important for protection against pathogenic bacteria (40). The peritrophic matrix in the fly gut is a non-cellular sieve-like structure that lines the midgut epithelium and provides protection from food particles and enteric pathogens (10, 41), and therefore, the defect in the peritrophic matrix by TG-RNAi is a possible cause of the high bacterial load in TG-RNAi flies.
One of the important findings in this study is that gnotobiotic flies that co-ingested SK4 and SK1 had a short lifespan, but not gnotobiotic flies that mono-ingested each of the strains SK1–4 (Fig. 6A). In Drosophila, ingestion of pathogenic bacteria promotes excess generation of ROS to cause apoptosis, leading to a short lifespan (16). In gnotobiotic flies that co-ingested SK4 and SK1, the percentage of TUNEL-positive guts and ROS production in the midgut were both statistically significant, but not in flies that mono-ingested SK4 (Figs. 7B and 8A). ROS production was also observed in S2 cells by adding the microbial extract derived from co-incubated SK4 and SK1, but not in the flies that mono-ingested SK4 or SK1 (Fig. 8B). These findings suggest that microbiota comprising the two dominant bacteria of the genus Acetobacter and Providencia in TG-RNAi flies may produce ROS, resulting in a short life span. Interestingly, gnotobiotic flies that co-ingested SK4 and SK2 exhibited significant ROS production in the midgut (Fig. 8A), and ROS production was also induced in S2 cells by treatment with microbial extract derived from co-incubated SK4 and SK2 (Fig. 8B). Gnotobiotic flies that co-ingested SK4 and SK2, however, exhibited no significant apoptosis of the midgut cells (Fig. 7) and did not have a significantly shorter lifespan (Fig. 6A). To our knowledge, uracil is the only metabolite to trigger DUOX-dependent ROS production, and it is released from pathobionts, not commensal bacteria (16, 42), suggesting that uracil is secreted from the opportunistic strain P. rettgeri SK4 in the midgut of gnotobiotic flies that co-ingested SK4 and SK1 or SK2. In gnotobiotic flies that co-ingested SK4 and SK1, an additional unknown factor(s) may be required to cause midgut apoptosis, leading to a short lifespan.
Experimental Procedures
Fly Stocks
Flies were maintained at 25 °C on standard Drosophila medium: 3.2% (w/v) dry yeast (Oriental Yeast Co., Tokyo, Japan), 3.2% (w/v) corn flour (GABAN, Tokyo, Japan), 8.0% (w/v) glucose (Nacalai Tesque, Kyoto, Japan), 0.64% (w/v) agar (Nacalai Tesque), 0.24% (w/v) propionic acid (Nacalai Tesque), and 0.04% (w/v) methyl p-hydroxybenzoate (Wako, Osaka, Japan). Da-GAL4 and w1118 were obtained from the Bloomington Stock Center (Bloomington, IN). UAS-TG IR flies were obtained from the National Institute of Genetics (Mishima, Japan). To obtain the TG-RNAi flies, Da-GAL4 flies were crossed with UAS-TG IR flies, and to obtain the non-TG-RNAi flies, Da-GAL4 flies were crossed with w1118 flies, as described previously (22). The fly strain RelE20 was described previously (43).
Bacterial Strains
P. alcalifaciens JCM 1673T and P. azotoformans IAM 1603 (JCM 2777) were obtained from the Japan Collection of Microorganisms, RIKEN BRC, participating in the National BioResource Project of MEXT, Japan. Ecc15 2141 (44) was provided by S. Kurata (Tohoku University, Sendai, Japan). G. morbifer G707T, C. intestini A911T, A. pomorum, L. brevis EW, and L. plantarum WJL (24) were provided by W.-J. Lee (Seoul National University, Seoul, Korea).
Bacterial 16S rDNA Analysis
Genomic DNAs from midguts of 0.5- and 10-day-old TG-RNAi (Da>TG IR) or non-TG-RNAi (Da>+) flies maintained under conventional rearing conditions were extracted using NucleoSpin® Tissue XS (Macherey-Nagel GmbH & Co. KG, Düren, Germany). PCR amplification was performed for each sample with Tks GflexTM DNA Polymerase (Takara Bio, Shiga, Japan) using the following primers (45): forward primer, 515F 5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT ATG GTA ATT GTG TGC CAG CMG CCG CGG TAA-3′; reverse primer, 806R 5′-CAA GCA GAA GAC GGC ATA CGA GAT NNN NNN NNN NNN AGT CAG TCA GCC GGA CTA CHV GGG TWT CTA AT-3′. The N repeated sequence shows the Golay barcode (46). Cycling conditions were 1 min at 94 °C, followed by 28 cycles of 10 s at 98 °C, 15 s at 60 °C, and 15 s at 68 °C, and 5 min at 68 °C at the final elongation step. PCR products were quantified by an Agilent 2100 Bioanalyzer (Agilent Technologies) and then sequenced using an Illumina MiSeq sequencer (150-bp paired end reads of the V4 region of the 16S rDNA) at Takara Bio. A total of 24,437,440 reads (2,500,000 ± 350,000 reads/sample) were obtained, and quality filtering, removal of chimeras, and denoising were performed by cluster data base of high identity with tolerance operation taxonomic units 0.01 (47) with default parameters. Sequences from Read 1 and Read 2 were clustered into operational taxonomic units at 97% identity using the cluster data base of high identity with tolerance operation taxonomic units pipeline, and then the sequences were taxonomically classified down to the genus level with the EzTaxon server (48).
Bacteria contain multiple heterogeneous 16S rRNA genes in their genomes, and the gene copy number information was obtained from the rRNA Operon Copy Number Database (rrnDB). To estimate the effective number of reads, read count data of the next-generation sequencing were corrected using representative copy numbers of the corresponding genera, five copies for Acetobacter and seven copies for Providencia. The copy number of genus Lactobacillus varies from one to nine, depending on the species, and the median number of gene copies (5 copies) was used.
Quantitative PCR
To quantify the number of bacteria, 16S rRNA bacteria-specific oligonucleotide primers, HDA1 (5′-ACT CCT ACG GGA GGC AGC AGT-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′), were used (23). The theoretical coverage rate of the HDA1-HDA2 primer set at the genus level was evaluated to be 99.4% for Acetobacter, 98.3% for Providencia, and 96.1% for Lactobacillus, using the TestPrime 1.0 program. Standard calibration curves were generated using 10-fold serial dilutions of a plasmid containing a segment of the rp49 or 16S rRNA genes. Genomic DNA was extracted from the midguts of 0.5- and 10-day-old non-TG-RNAi and TG-RNAi flies (4 guts/sample) by phenol/chloroform extraction and used as templates. The number of gut bacteria was estimated after correction using the representative copy numbers, as described under “Bacterial 16S rDNA Analysis,” and then normalized to the amount of genomic rp49 and estimated per gut. To quantify the copy number of Cecropin A1 and Diptericin, total RNA from conventionally reared adult midguts or germ-free adult midguts following 18 days of ingestion were extracted with RNAiso (Takara Bio) and treated with deoxyribonuclease I, and then total RNA (400–450 ng) was used as a template for reverse transcription using SuperScript III (Life Technologies, Inc.). Absolute copy numbers analyzed for Cecropin A1 and Diptericin were normalized to absolute control rp49 mRNA copy numbers. Standard calibration curves were generated using 10-fold serial dilutions of a plasmid containing a segment of the rp49, Cecropin A1, and Diptericin. The set of PCR primers was described previously (23). All qPCR was performed with FastStart Essential DNA Green Master (Roche Applied Science), and the reactions were performed on a LightCycler Nano (Roche Applied Science).
Isolation of Gut Microbes
To obtain strains belonging to the genera Acetobacter and Lactobacillus, homogenates of midguts (1–2 guts dissected from adult 10–12-day-old TG-RNAi or non-TG-RNAi flies) were plated on Hestrin-Schramm medium plates containing 2.0% glucose, 0.5% Bacto Peptone (Difco), 0.5% yeast extract (Difco), 0.68% Na2HPO4·12H2O, 0.15% citric acid monohydrate, and 1.5% agar adjusted to pH 5.0 with 1 m CH3COOH (49) and incubated for 2 days at 30 °C. To obtain strains belonging the genera Providencia species, homogenates of midguts (1–2 guts dissected from adult 10–12-day-old TG-RNAi or non-TG-RNAi flies) were plated on nutrient agar medium plates containing 5.0% Bacto Peptone, 3.0% beef extract (MP Biomedicals), 1.5% agar, 0.05% phenol red, 0.04% 2,3,5-triphenyl tetrazolium chloride (50, 51) and incubated for 16 h at 37 °C. Bacterial colonies (24 and 57 colonies for non-TG-RNAi and TG-RNAi flies, respectively) were randomly isolated and sequenced. Bacterial 16S rDNA was sequenced using 16S universal primers, 8FE (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGM TAG CTT GTT ACG ACT T-3′). The similarity of the sequenced data was determined using an EzTaxon server (48).
Antimicrobial Activity
Peptide synthesis of Cecropin A1 and Diptericin was outsourced to Genenet Co., Ltd. (Fukuoka, Japan). The peptide sequence of Cecropin A1 is GWLKK IGKKI ERVGQ HTRDA TIQGL GIAQQ AANVA ATAR-NH2 (39 residues), and the peptide sequence of Diptericin is DDMTM KPTPP PQYPL NLQGG GGGQS GDGFG FAVQG HQKVW TSDNG RHEIG LNGGY GQHLG GPYGN SEPSW KVGST YTYRF PNF-NH2 (83 residues). Antimicrobial activities were assayed as described previously (52). In brief, bacteria were grown under the following conditions: Hestrin-Schramm medium for Acetobacter and Lactobacillus isolated from the gut and incubated for 2 days at 30 °C. G. morbifer G707T, C. intestini A911T, and A. pomorum were cultured for 2 days at 30 °C in mannitol broth containing 2.5% mannitol (Nacalai Tesque), 0.3% Bacto Peptone, and 0.5% yeast extract (53). L. brevis EW and L. plantarum WJL were cultured for 2 days at 30 °C in MRS broth containing 2.0% glucose, 1.0% Bacto Tryptone (Difco), 1.0% beef extract, 0.5% yeast extract, 0.5% CH3COONa, 0.2% K2HPO4, 0.2% diammonium hydrogen citrate, 0.1% polysorbate 80, 0.02% MgSO4·7H2O, and 0.005% MnSO4·5H2O (54). Providencia, E. coli K12, Ecc15, and P. azotoformans IAM 1603 were cultured in Luria-Bertani medium and incubated for 16 h at 37 °C. Bacteria collected by centrifugation were washed three times with PBS containing 8.0% NaCl, 0.2% KCl, 2.9% Na2HPO4·12H2O, and 0.2% KH2PO4 and suspended in 10 mm HEPES-NaOH, pH 7.5. Then 50 μl of antimicrobial peptide solution in 10 mm HEPES-NaOH, pH 7.5, was added to 450 μl of the bacterial suspensions, and the mixtures were incubated at 37 °C for 1 h and plated onto three agar plates (100 μl each). Each plate was incubated at either 30 °C for 2 days or 37 °C for 16 h.
HOCl Resistance Assay
Hypochlorous acid (HOCl) resistance assays were performed as described previously (55). The concentration of available chlorine was measured using N,N-diethylphenylenediamine (56). In brief, bacteria collected by centrifugation were washed three times with PBS and suspended in 10 mm HEPES-NaOH, pH 7.5. Then 50 μl of the freshly prepared NaOCl solution in 10 mm HEPES-NaOH, pH 7.5, containing 150 or 500 μg/liter available chlorine was added to 450 μl of the bacteria suspensions, and the mixture was incubated at 37 °C for 40 min. To stop the reaction, aliquots (5 μl each) of the incubated samples were transferred to 495 μl of PBS and plated onto three agar plates (100 μl each).
Bacterial Ingestion and Survival Experiments
Survival rate experiments were performed using conventionally reared, germ-free, and long term ingested germ-free flies. Germ-free flies were maintained as described previously (23). Bacterial contamination in germ-free flies was checked by PCR analysis using the 16S rDNA universal primers (8FE and 1492R). For long term ingestion experiments, 1–3-day-old germ-free flies were transferred to a fresh vial containing in total 4.0 × 109 cells of isolated bacteria or a mixture of the isolated bacteria (Providencia/Acetobacter ratio was 1:1) and then transferred to a fresh vial containing 4.0 × 109 cells of bacteria every 3 days.
Bacterial Colonization Ability in the Midgut of Drosophila
To examine transient colonization of flies with bacteria, 3–5-day-old adult germ-free flies were transferred to a fresh vial containing 4.0 × 109 bacterial cells in 5% sucrose solution for 24 h and then transferred into sterile vials containing sterile food. After 5 h of incubation, midguts were dissected into three parts and homogenized in 100 μl of sterile PBS. To examine the colonization ability in flies after long term ingestion of bacteria, the midguts were dissected after 15 and 25 days of ingestion and homogenized in 100 μl of sterile PBS. The homogenates were 2-, 20-, 200-, or 2000-fold diluted with sterile PBS, and then 5-μl aliquots of the homogenates were spotted onto agar plates and incubated under the following conditions: Hestrin-Schramm medium plates for flies that mono-ingested SK1, SK2, and SK3 and then incubation for 2 days at 30 °C; Luria-Bertani medium plates for flies that mono-ingested SK4 and then incubation for 16 h at 37 °C. For flies that co-ingested the bacteria, 5-μl aliquots of the serial dilutions were spotted onto Hestrin-Schramm medium plates and incubated for 2 days at 30 °C, and a second 5-μl aliquot of each dilution was spotted onto Luria-Bertani medium plates and incubated for 16 h at 37 °C. The number of cfu in two or three selected spots was counted, and the average cfu were calculated.
TUNEL Imaging Analysis
For detection of apoptosis in midgut cells, the TUNEL method was performed using an In Situ Cell Death Detection Kit, Fluorescein (Roche Applied Science), as described previously (23). The guts were stained with DAPI (1:2,000; Dojindo Molecular Technologies, Kumamoto, Japan). Midguts were dissected from adult flies following 18 days of ingestion. The samples were analyzed with a ZOETM fluorescent cell imager (Bio-Rad). Each experiment was repeated at least three times with samples dissected on different dates.
ROS Measurement
To measure ROS production in vivo, midguts were dissected from adult flies following 18 days of ingestion in the presence of 20 mm N-acetyl cysteine (Sigma). Midguts were incubated with 5 μm CellROX (Molecular Probes, Inc.) for 30 min at 37 °C and then fixed by 4% paraformaldehyde. As a positive control for oxidative stress, the ROS inducer paraquat (Nacalai Tesque) was fed to adult germ-free flies for 3 h before dissection. As a negative control, midguts were dissected from 1–2-day-old germ-free flies. After mounting with 70% glycerol, samples were analyzed with a ZOETM fluorescent cell imager. The resulting signal intensity was measured by ImageJ (National Institutes of Health, Bethesda, MD) and normalized to the average of negative control gut signal intensity. Each experiment was repeated at least three times with samples dissected on different dates. To measure ROS production in vitro, S2 cells were maintained in Insect-XPRESS protein-free insect cell medium (Lonza, Switzerland) at 27 °C. Soluble microbial extracts was prepared as described previously (57). In brief, bacteria were grown under the following conditions. Hestrin-Schramm medium was used for the genus Acetobacter, and the plates were incubated for 2 days at 30 °C. Luria-Bertani medium was used for the genus Providencia, and the plates were incubated for 16 h at 37 °C. After washing with PBS, bacteria were adjusted to 7.0 × 109 cells. For a mixture of two bacterial strains, one strain (3.5 × 109 cells) was mixed with another strain (3.5 × 109 cells) in 5 ml of PBS buffer. The bacterial solutions were incubated for 1 h at 30 °C at 200 rpm and then collected by centrifugation. S2 cells were treated with 10 μg/ml soluble microbial extract in the medium for 1 h. Uracil at 20 nm was added as a positive control to produce ROS. The cells were stained with Hoechst 33342 (1:1,000; Dojindo Molecular Technologies) and 2 μm DCF-DA (Sigma). After washing with PBS, the fluorescent signal was observed using a ZOE fluorescent cell imager. From these pictures, Hoechst 33342-stained nuclei, representing all cells, were randomly defined, and the number of DCF-DA-positive cells, representing ROS-positive cells, was determined.
Author Contributions
S. S. performed all of the experiments except for those in Figs. 2D, 8B, and 9. M. H. performed mRNA expression experiments in Fig. 2D. T. S. performed experiments in Figs. 8B and 9. S. S., T. S., and S. K. analyzed the data and interpreted the experimental results. S. S., T. S., and S. K. designated the experiments and wrote the paper. All authors approved the final version of the manuscript.
Acknowledgments
We are grateful to R. Ueda (National Institute of Genetics, Mishima, Japan) for providing the fly strains; S. Kurata (Tohoku University, Sendai, Japan), T. Kuraishi (Kanazawa University, Kanazawa, Japan), and W.-J. Lee (Seoul National University, Seoul, Korea) for providing the bacterial strains; and T. Koshiba (Kyushu University, Fukuoka, Japan) for helpful discussions regarding this study.
This work was supported by Grant-in-Aid for Scientific Research (C) 24570164 (to S. K.); Grant-in-Aid for Scientific Research (B) 15H04353 (to S. K.); Grant-in-Aid for Scientific Research on Innovative Areas 24117712 (to S. K.); the Uehara Memorial Foundation (to S. K.); Grant-in-Aid for Young Scientists (B) 26860333 (to T. S.); and the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development Fund (TT type) (Grant 26701) (to T. S.). The authors declare that they have no conflicts of interest regarding the content of this article.
- AMP
- antimicrobial peptide
- TG
- transglutaminase
- IMD
- immune deficiency pathway
- ROS
- reactive oxygen species
- DUOX
- dual oxidase
- IMD-AMP
- immune deficiency pathway-controlled antimicrobial peptide
- qPCR
- quantitative PCR
- DCF-DA
- 2′,7′-dichlorofluorescin-diacetate.
References
- 1. Guinane C. M., and Cotter P. D. (2013) Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 6, 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee W.-J., and Hase K. (2014) Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424 [DOI] [PubMed] [Google Scholar]
- 3. Lee K.-A., and Lee W.-J. (2014) Drosophila as a model for intestinal dysbiosis and chronic inflammatory diseases. Dev. Comp. Immunol. 42, 102–110 [DOI] [PubMed] [Google Scholar]
- 4. Ma D., Storelli G., Mitchell M., and Leulier F. (2015) Studying host-microbiota mutualism in Drosophila: harnessing the power of gnotobiotic flies. Biomed. J. 38, 285–293 [DOI] [PubMed] [Google Scholar]
- 5. Ren C., Webster P., Finkel S. E., and Tower J. (2007) Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6, 144–152 [DOI] [PubMed] [Google Scholar]
- 6. Cox C. R., and Gilmore M. S. (2007) Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong C. N. A., Ng P., and Douglas A. E. (2011) Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchon N., Broderick N. A., and Lemaitre B. (2013) Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol. 11, 615–626 [DOI] [PubMed] [Google Scholar]
- 9. Buchon N., Osman D., David F. P. A., Fang H. Y., Boquete J.-P., Deplancke B., and Lemaitre B. (2013) Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 [DOI] [PubMed] [Google Scholar]
- 10. Kuraishi T., Hori A., and Kurata S. (2013) Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S.-H., and Lee W.-J. (2014) Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front. Cell. Infect. Microbiol. 3, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryu J.-H., Ha E.-M., and Lee W.-J. (2010) Innate immunity and gut-microbe mutualism in Drosophila. Dev. Comp. Immunol. 34, 369–376 [DOI] [PubMed] [Google Scholar]
- 13. Ryu J.-H., Ha E.-M., Oh C.-T., Seol J.-H., Brey P. T., Jin I., Lee D. G., Kim J., Lee D., and Lee W.-J. (2006) An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 25, 3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha E.-M., Oh C.-T., Ryu J.-H., Bae Y.-S., Kang S.-W., Jang I., Brey P. T., and Lee W.-J. (2005) An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell. 8, 125–132 [DOI] [PubMed] [Google Scholar]
- 15. Capo F., Charroux B., and Royet J. (2016) Bacteria sensing mechanisms in Drosophila gut: Local and systemic consequences. Dev. Comp. Immunol. 64, 11–21 [DOI] [PubMed] [Google Scholar]
- 16. Lee K.-A., Kim S.-H., Kim E.-K., Ha E.-M., You H., Kim B., Kim M.-J., Kwon Y., Ryu J.-H., and Lee W.-J. (2013) Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811 [DOI] [PubMed] [Google Scholar]
- 17. Lorand L., and Graham R. M. (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 18. Beninati S., and Piacentini M. (2004) The transglutaminase family: an overview: minireview article. Amino Acids 26, 367–372 [DOI] [PubMed] [Google Scholar]
- 19. Kopácek P., Hall M., and Söderhäll K. (1993) Characterization of a clotting protein, isolated from plasma of the freshwater crayfish Pacifastacus leniusculus. Eur. J. Biochem. 213, 591–597 [DOI] [PubMed] [Google Scholar]
- 20. Theopold U., Schmidt O., Söderhäll K., and Dushay M. S. (2004) Coagulation in arthropods: defence, wound closure and healing. Trends Immunol. 25, 289–294 [DOI] [PubMed] [Google Scholar]
- 21. Kawabata S. (2010) Immunocompetent molecules and their response network in horseshoe crabs. in Invertebrate Immunity (Söderhäll K., ed) pp. 122–136, Springer Science + Business Media, New York: [DOI] [PubMed] [Google Scholar]
- 22. Shibata T., Ariki S., Shinzawa N., Miyaji R., Suyama H., Sako M., Inomata N., Koshiba T., Kanuka H., and Kawabata S. (2010) Protein crosslinking by transglutaminase controls cuticle morphogenesis in Drosophila. PLoS One 5, e13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shibata T., Sekihara S., Fujikawa T., Miyaji R., Maki K., Ishihara T., Koshiba T., and Kawabata S. (2013) Transglutaminase-catalyzed protein-protein cross-linking suppresses the activity of the NF-κB-like transcription factor Relish. Sci. Signal. 6, ra61. [DOI] [PubMed] [Google Scholar]
- 24. Ryu J.-H., Kim S.-H., Lee H.-Y., Bai J. Y., Nam Y.-D., Bae J.-W., Lee D. G., Shin S. C., Ha E.-M., and Lee W.-J. (2008) Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782 [DOI] [PubMed] [Google Scholar]
- 25. Chandler J. A., Lang J. M., Bhatnagar S., Eisen J. A., and Kopp A. (2011) Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7, e1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blum J. E., Fischer C. N., Miles J., and Handelsman J. (2013) Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio. 4, e00860–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amir A., Zeisel A., Zuk O., Elgart M., Stern S., Shamir O., Turnbaugh P. J., Soen Y., and Shental N. (2013) High-resolution microbial community reconstruction by integrating short reads from multiple 16S rRNA regions. Nucleic Acids Res. 41, e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Staubach F., Baines J. F., Künzel S., Bik E. M., and Petrov D. A. (2013) Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS One 8, e70749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Broderick N. A., Buchon N., and Lemaitre B. (2014) Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5, e01117–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong A. C.-N., Luo Y., Jing X., Franzenburg S., Bost A., and Douglas A. E. (2015) The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl. Environ. Microbiol. 81, 6232–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hertig M., and Wolbach S. B. (1924) Studies on Rickettsia-like micro-organisms in insects. J. Med. Res. 44, 329–374.7 [PMC free article] [PubMed] [Google Scholar]
- 32. Ha E.-M., Oh C.-T., Bae Y. S., and Lee W.-J. (2005) A direct role for Dual oxidase in Drosophila gut immunity. Science 310, 847–850 [DOI] [PubMed] [Google Scholar]
- 33. Macpherson A. J., and Harris N. L. (2004) Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485 [DOI] [PubMed] [Google Scholar]
- 34. Spasova D. S., and Surh C. D. (2014) Blowing on embers: Commensal microbiota and our immune system. Front. Immunol. 5, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shin S. C., Kim S.-H., You H., Kim B., Kim A. C., Lee K.-A., Yoon J.-H., Ryu J.-H., and Lee W.-J. (2011) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674 [DOI] [PubMed] [Google Scholar]
- 36. Erkosar B., and Leulier F. (2014) Transient adult microbiota, gut homeostasis and longevity: novel insights from the Drosophila model. FEBS Lett. 588, 4250–4257 [DOI] [PubMed] [Google Scholar]
- 37. Yoh M., Matsuyama J., Ohnishi M., Takagi K., Miyagi H., Mori K., Park K.-S., Ono T., and Honda T. (2005) Importance of Providencia species as a major cause of travellers' diarrhoea. J. Med. Microbiol. 54, 1077–1082 [DOI] [PubMed] [Google Scholar]
- 38. Galac M. R., and Lazzaro B. P. (2011) Comparative pathology of bacteria in the genus Providencia to a natural host, Drosophila melanogaster. Microbes Infect. 13, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chambers M. C., Jacobson E., Khalil S., and Lazzaro B. P. (2014) Thorax injury lowers resistance to infection in Drosophila melanogaster. Infect. Immun. 82, 4380–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shibata T., Maki K., Hadano J., Fujikawa T., Kitazaki K., Koshiba T., and Kawabata S. (2015) Crosslinking of a peritrophic matrix protein protects gut epithelia from bacterial exotoxins. PLoS Pathog. 11, e1005244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elvin C. M., Vuocolo T., Pearson R. D., East I. J., Riding G. A., Eisemann C. H., and Tellam R. L. (1996) Characterization of a major peritrophic membrane protein, Peritrophin-44, from the larvae of Lucilia cuprina: cDNA and deduced amino acid sequences. J. Biol. Chem. 271, 8925–8935 [DOI] [PubMed] [Google Scholar]
- 42. Lee K.-A., Kim B., Bhin J., Kim D. H., You H., Kim E.-K., Kim S.-H., Ryu J.-H., Hwang D., and Lee W.-J. (2015) Bacterial uracil modulates Drosophila DUOX-dependent gut immunity via Hedgehog-induced signaling endosomes. Cell Host Microbe 17, 191–204 [DOI] [PubMed] [Google Scholar]
- 43. Hedengren M., Asling B., Dushay M. S., Ando I., Ekengren S., Wihlborg M., and Hultmark D. (1999) Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell. 4, 827–837 [DOI] [PubMed] [Google Scholar]
- 44. Basset A., Khush R. S., Braun A., Gardan L., Boccard F., Hoffmann J. A., and Lemaitre B. (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. U.S.A. 97, 3376–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chassaing B., Koren O., Goodrich J. K., Poole A. C., Srinivasan S., Ley R. E., and Gewirtz A. T. (2015) Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., and Knight R. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W., Fu L., Niu B., Wu S., and Wooley J. (2012) Ultrafast clustering algorithms for metagenomic sequence analysis. Brief. Bioinform. 13, 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chun J., Lee J.-H., Jung Y., Kim M., Kim S., Kim B. K., and Lim Y.-W. (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 [DOI] [PubMed] [Google Scholar]
- 49. Hestrin S., and Schramm M. (1954) Synthesis of cellulose by Acetobacter xylinum. 2. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem. J. 58, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Somvanshi V. S., Lang E., Sträubler B., Spröer C., Schumann P., Ganguly S., Saxena A. K., and Stackebrandt E. (2006) Providencia vermicola sp. nov., isolated from infective juveniles of the entomopathogenic nematode Steinernema thermophilum. Int. J. Syst. Evol. Microbiol. 56, 629–633 [DOI] [PubMed] [Google Scholar]
- 51. Senior B. W. (1997) Media for the detection and recognition of the enteropathogen Providencia alcalifaciens in faeces. J. Med. Microbiol. 46, 524–527 [DOI] [PubMed] [Google Scholar]
- 52. Kawabata S., Nagayama R., Hirata M., Shigenaga T., Agarwala K. L., Saito T., Cho J., Nakajima H., Takagi T., and Iwanaga S. (1996) Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J. Biochem. 120, 1253–1260 [DOI] [PubMed] [Google Scholar]
- 53. American Society for Microbiology, Committee on Bacteriological Technic. (1957) Manual of Microbiological Methods (Conn H. J., ed.) pp. 99–119, McGraw-Hill, New York [Google Scholar]
- 54. De Man J. C., Rogosa M., and Sharpe M. E. (1960) A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 23, 130–135 [Google Scholar]
- 55. Fukuzaki S., Urano H., and Yamada S. (2007) Effect of pH on the efficacy of sodium hypochlorite solution as cleaning and bactericidal agents. J. Surf. Finish. Soc. Jpn. 58, 465–469 [Google Scholar]
- 56. Urano H., Ishikawa H., and Fukuzaki S. (2006) Involvement of radical species in inactivation of Vibrio parahaemolyticus in saline solutions by direct-current electric treatment. J. Biosci. Bioeng. 102, 457–463 [DOI] [PubMed] [Google Scholar]
- 57. Ha E.-M., Lee K.-A., Park S. H., Kim S.-H., Nam H.-J., Lee H.-Y., Kang D., and Lee W.-J. (2009) Regulation of DUOX by the Gαq-phospholipase Cβ-Ca2+ pathway in Drosophila gut immunity. Dev. Cell 16, 386–397 [DOI] [PubMed] [Google Scholar]


