Abstract
Bispecific antibodies (bsAbs) and antibody-drug conjugates (ADCs) have already demonstrated benefits for the treatment of cancer in several clinical studies, showing improved drug selectivity and efficacy. In particular, simultaneous targeting of prominent cancer antigens, such as EGF receptor (EGFR) and c-MET, by bsAbs has raised increasing interest for potentially circumventing receptor cross-talk and c-MET-mediated acquired resistance during anti-EGFR monotherapy. In this study, we combined the selectivity of EGFR × c-MET bsAbs with the potency of cytotoxic agents via bispecific antibody-toxin conjugation. Affinity-attenuated bispecific EGFR × c-MET antibody-drug conjugates demonstrated high in vitro selectivity toward tumor cells overexpressing both antigens and potent anti-tumor efficacy. Due to basal EGFR expression in the skin, ADCs targeting EGFR in general warrant early safety assessments. Reduction in EGFR affinity led to decreased toxicity in keratinocytes. Thus, the combination of bsAb affinity engineering with the concept of toxin conjugation may be a viable route to improve the safety profile of ADCs targeting ubiquitously expressed antigens.
Keywords: antibody engineering, anticancer drug, epidermal growth factor receptor (EGFR), sortase A, toxicity
Introduction
Next generation antibodies such as bispecific antibodies (bsAbs)3 and antibody-drug conjugates (ADCs) are among the most promising and fastest growing classes of therapeutic modalities with demonstrated benefit for patients (1–4). In comparison with combination therapies, bsAbs offer several potential advantages, such as (a) improved efficacy due to synergistic effects via simultaneous targeting (5), (b) increased tumor cell specificity (6), (c) increased target cross-linking and internalization (7), (d) reduced tumor resistance, (e) tumor-specific effector cell recruitment (e.g. bispecific T-cell engagers, or BiTEs) (8), and (f) lower cost during the development and approval process (1). The therapeutic success of monospecific ADCs is based on the selective intracellular delivery of highly potent cytotoxic agents (4). In combination with the advantages of bsAbs, bispecific ADCs might further increase potency and selectivity, opening a broader and novel target space. However, good internalization capabilities and high tumor selectivity are prerequisites for the utilized mAbs to mediate therapeutic activity and to avoid toxic side effects (9, 10).
The epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (HGFR, c-MET) are two clinically validated anti-cancer targets that demonstrated antibody-mediated internalization but poor tumor specificity. Notwithstanding, overexpression of these receptor tyrosine kinases is correlated with the development, progression, and metastasis of cancer as well as poor prognosis (11, 12). Therapies targeting both cancer antigens are currently under investigation in preclinical and clinical studies for monotherapy as well as combination therapy (13–15). Due to receptor cross-talk and redundancy, EGFR-targeted monotherapies are confronted with either the presence of intrinsic resistances or the development of acquired resistances, thus leading to patient relapse (16–19). In particular, up-regulation and amplification of c-MET and its ligand hepatocyte growth factor (HGF) are regarded as one major escape route during anti-EGFR therapy in non-small cell lung (16, 17), gastric (18, 20, 21), and breast cancer (22). Hence, combining the bsAb approach for targeting c-MET and EGFR with the conjugation of potent cytotoxic agents could be advantageous.
Recently, several bsAbs targeting c-MET and EGFR have been developed that demonstrated synergistic effects regarding inhibition of tumor proliferation and metastasis (5, 7, 23–27). Epitope combinations need to be carefully evaluated to assess antibody-mediated receptor activation. In particular, c-MET-directed antibodies struggle with full or partial agonism when employing a bivalent format (28–30). Also, in a bispecific format, suitable epitope combinations have to be identified aimed at avoiding bsAb-induced agonistic activity via c-MET/EGFR heterodimerization (19, 28).
The development of ADCs based on EGFR binding is hampered by the ubiquitous basal EGFR expression. Inhibition of EGFR signaling in keratinocytes by cetuximab, for example, leads to severe skin toxicities (31). Thus, engineering of tumor selectivity is of great importance for EGFR × c-MET bispecific ADC generation. Robinson et al. (32) reported that bispecificity per se can increase selectivity (e.g. by engineering of bsAbs against Her2 and Her3). Specific targeting of a distinct cell population can be further improved by use of affinity-attenuated binders displaying decreased binding to cell types with low target expression, whereas the binding of mutual overexpressing cells is increased via avidity effects (6, 33).
We generated affinity-optimized binders targeting different epitopes on c-MET and EGFR by applying display technologies and in silico screening. The bsAbs and bispecific ADCs were engineered with the strand exchange engineered domain (SEED) technology (34, 35) based on two asymmetric, non-identical CH3 chains, denoted as AG and GA. These chains were constructed from alternating segments of IgG and IgA so that heterodimerization is favored (35). Here, we describe for the first time the development of potent bispecific ADCs that display high selectivity in killing of EGFR- and/or c-MET-expressing tumor cell lines in comparison with normal tissue models.
Our study represents an illustrative data set for the in vitro assessment of balancing anti-tumor efficacy and high selectivity when targeting two tumor-associated rather than tumor-specific antigens.
Results
Generation and Characterization of EGFR Binders
With regard to the engineering of EGFR binders, cetuximab (C225) and matuzumab (mAb 425) were selected as starting material, because these two antibodies target two separate epitopes on the same domain of EGFR (36, 37). Due to possible toxicity related to EGFR-targeting issues, C225 mutants with varying affinities were generated by computational design using the Rosetta protein structure prediction and design suite.
Starting from the available crystal structure of C225 bound to the extracellular domain of EGFR (36), residues at or near the antigen binding interface were individually mutated to all amino acids except cysteine to create a virtual saturating scanning mutagenesis library. Each library member was optimized to find the lowest energy rotamer of the mutated residue and interacting neighbors according to the Rosetta energy function (38–40). In the case of the low affinity mutant, a variation of the energy function with a softened repulsive component (41) was used instead. The calculated binding energies of several computationally designed mutants used for this study are shown in Table 1 together with the in vitro measured affinities of the purified proteins. It was found that the calculated hydrogen bond energies together with the pair energy (representing electrostatic interactions) performed quite well in discriminating affinity-enhancing polar substitutions. Of the two affinity-lowering designed mutants (designated C225-L and C225-M for low and medium affinity), C225-M has a substantially higher calculated energy for the antibody alone, suggesting that the fixed backbone approximation was not appropriate for this substitution. Three affinity-enhancing substitutions were combined, and the additivity of the enhancements was confirmed in vitro and in silico, resulting in variant C225-H (Table 1).
TABLE 1.
Experimental and calculated binding affinity for computationally designed point mutants of C225
| Mutation | KDa | ΔΔGexpb | ΔΔGcalcc | ΔΔEpaird | ΔΔEHbnde | EHbnd_scf | ΔΔGAbg |
|---|---|---|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | |||||
| WT C225 | 1.52 ± 0.29 | 0 | 0 | 0 | 0 | 0 | 0 |
| C225-L | NQh | −0.7 | −0.4 | −0.4 | −1.1 | 1.6 | |
| C225-M | 15.26 | 1.36 | −0.7 | 0.1 | −0.8 | −1.1 | 6.3 |
| C225-H1 | 0.77 ± 0.09 | −0.41 | −2.0 | −1.1 | −1.2 | −2.2 | −0.7 |
| C225-H2 | 1.09 ± 0.13 | −0.20 | −0.2 | −0.1 | −0.7 | −1.1 | 3.1 |
| C225-H3 | 0.37 ± 0.04 | −0.84 | −0.9 | −0.6 | −0.2 | −0.2 | 0.4 |
| C225-H | 0.08 ± 0.03 | −1.75 | −3.2 | −6.3 | |||
| F06 | 5.5 | ||||||
| CS06 | 0.13 | ||||||
| B10 | 24.3 | ||||||
| B10v5 | 0.19 |
a The KD (nm) for wild type (WT) C225 and mutant mAbs was determined by SPR as described under “Experimental Procedures.” Where n > 1, the S.D. is given. KD values (nm) for c-MET interaction with bivalent anti-c-MET mAbs were determined by BLI. CS06 is the affinity-matured variant of F06, and B10v5 is the derivative of B10.
b Experimental binding affinity relative to wild type (kcal/mol).
c Predicted binding affinity relative to wild type using Rosetta.
d Predicted change in Rosetta pair energy across the interface.
e Predicted change in hydrogen bond energy across the interface.
f Calculated hydrogen bond energy of mutated residue side chain.
g Predicted change in folding energy of the isolated antibody.
h Not quantifiable; very weak binding.
c-MET Binder Generation and Characterization
Panning of naive human scFv phage display libraries (HAL 7/8) against the c-MET extracellular or SEMA domain was carried out as described previously (42, 43). After three selection rounds, subcloning into the bivalent IgG format, and expression in EXPI293FTM cells, two specific anti-c-MET antibodies, B10 and F06, with dissociation equilibrium constant (KD) values of 24.3 and 5.5 nm, respectively, for c-MET determined by biolayer interferometry (BLI) were identified and subsequently subjected to in vitro affinity maturation. To this end, Fab fragment sublibraries of F06 and B10 were generated by error-prone PCR of the variable regions, randomization of complementary-determining region 3 of the heavy chain (CDR-H3), and light chain shuffling. Due to fast dissociation rates of parental binders, an off-rate selection and screening strategy was performed by yeast surface and phage display (44, 45). The affinity maturation strategies resulted in slower off-rates and an up to 127-fold affinity improvement of the B10 derivative B10v5 and 42-fold affinity enhancement of CS06 in comparison with F06, respectively (Table 1), while retaining HGF competition as well as inhibition of receptor phosphorylation (data not shown). Epitope binning experiments revealed an overlapping epitope of B10 and B10v5 with the previously described anti-c-MET antibody LY2875358 (29) within the c-MET SEMA domain, whereas F06 and CS06 competed with antibody h224G11 (ABT-700) (46) in the c-MET stalk domain (data not shown) (14, 47). Thus, we generated antibodies covering two distinct c-MET domains enabling direct comparison of distinct epitopes and off-rates for evaluated bsAbs.
bsAbs Show Differentiated Affinities and Simultaneous Binding to c-MET and EGFR
The four anti-EGFR binders 225-L, -M, -H, and 425 and the four anti-c-MET binders B10, B10v5, F06, and CS06 were used to set up a broad panel of 16 bsAb constructs covering distinct epitopes on both receptor tyrosine kinases with varying affinities to analyze tumor selectivity. Generation of bsAbs was based on the SEED technology (35). To avoid light chain mispairing in the bispecific format (34, 35, 48), anti-EGFR binders 225-L, -M, -H, and 425 were converted to a single chain Fv format (scFv), where the variable domain of the light chain is linked through a linker sequence to the variable domain of the heavy chain. As control constructs, monovalent one-armed SEEDbodies were generated carrying either one c-MET Fab (at the AG chain; Fig. 1A) or one anti-EGFR scFv (at the GA chain; Fig. 1B). Production was carried out by transient transfection of EXPI293FTM cells followed by purification via protein A affinity chromatography, yielding bsAb homogeneity of >95%, as confirmed by SDS-gel electrophoresis (data not shown) and analytical size exclusion high performance liquid chromatography (SE-HPLC) (supplemental Fig. S1).
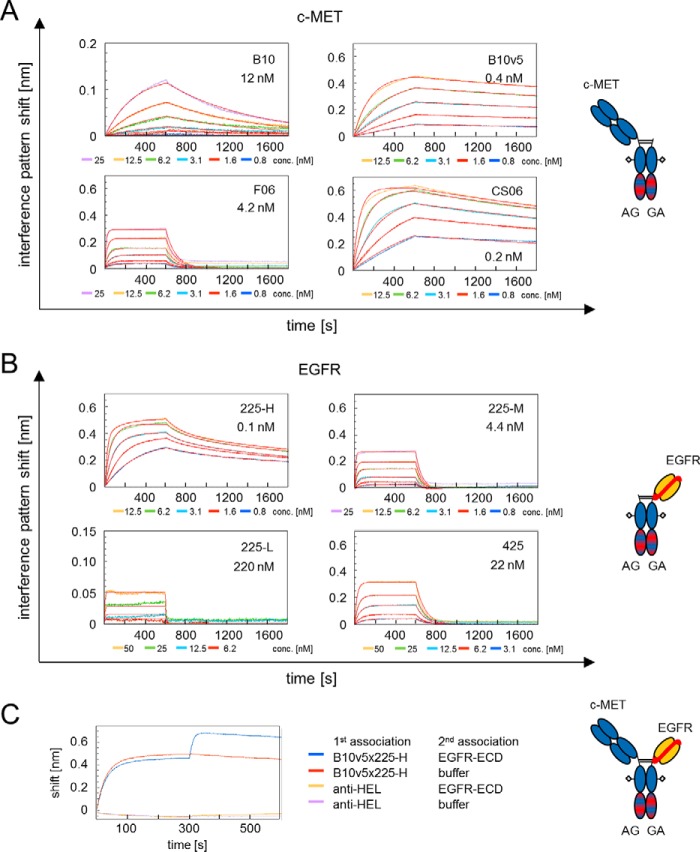
FIGURE 1.
A, BLI of monovalent oa Fab SEED (AG) antibodies to soluble c-MET ECD, including a schematic antibody representation. B10 and F06 are parental and B10v5 and CS06 are the affinity-matured variants thereof. Affinities for c-MET are given as the dissociation equilibrium constant (KD) (nm). B, BLI of monovalent oa scFv SEED (GA) antibodies to soluble EGFR ECD, including a schematic antibody representation. Variants of humanized cetuximab with low (225-L), medium (225-M), and high (225-H) affinity as well as humanized matuzumab (mAb 425) were analyzed. Affinities for EGFR are given in nm. C, simultaneous binding of soluble recombinant c-MET and EGFR by bsAb B10v5 × 225-H analyzed by BLI. Biotinylated c-MET ECD is captured to streptavidin octet biosensors. bsAb and EGFR-ECD are associated in two steps employing buffer and non-related isotype control (anti-HEL). Shown is a schematic representation of bispecific EGFR × c-MET antibody using the SEED technology.
Kinetic parameters, including on- and off-rates of monovalent mAbs, were determined by BLI and found to be retained when formatted into bsAbs (Fig. 1, A and B, and Table 2). The melting temperatures (Tm) of all generated EGFR × c-MET bsAbs were >62.2 °C (cf. Table 2), and bsAbs demonstrated long term stability in human serum up to 24 days (supplemental Fig. S1E).
TABLE 2.
Kinetic parameters of monovalent parental SEED antibodies and EGFR × c-MET bsAbs binding to soluble c-MET and EGFR extracellular domains
Additionally, kinetic constants were determined for cetuximab (C225) and matuzumab (mAb425) as references. Antibodies were captured by anti-human Fc Octet biosensors, and binding kinetics were analyzed at the indicated analyte concentrations (25 to 0.8 nm or alternatively 50 to 3.1 nm). Melting temperatures (Tm) were determined by thermal shift assays. ND, not determined.
| Antibody | Analyte | KD | KD error | ka | ka error | kd | kd error | Tm |
|---|---|---|---|---|---|---|---|---|
| m | m | m−1 s−1 | m−1 s−1 | s−1 | s−1 | ° C | ||
| oa B10 | c-MET | 1.2E − 08 | 1.2E − 10 | 1.0 E + 05 | 1.0E + 03 | 1.2E − 03 | 2.6E − 06 | 65.7 ± 0.001 |
| oa B10v5 | c-MET | 3.8E − 10 | 2.2E − 12 | 4.0E + 05 | 1.2E + 03 | 1.6E − 04 | 7.6E − 07 | 64.0 ± 0.172 |
| B10v5 × hu225-M | c-MET | 3.7E − 10 | 2.1E − 12 | 4.0E + 05 | 1.1E + 03 | 1.5E − 04 | 7.2E − 07 | 64.5 ± 0.001 |
| B10v5 × hu225-H | c-MET | 3.6E − 10 | 2.2E − 12 | 4.0E + 05 | 1.2E + 03 | 1.5E − 04 | 7.8E − 07 | 62.8 ± 0.174 |
| oa F06 | c-MET | 4.2E − 09 | 1.3E − 10 | 2.5E + 06 | 7.2E + 04 | 1.1E − 02 | 1.0E − 04 | 64.8 ± 0.172 |
| oa CS06 | c-MET | 1.9E − 10 | 1.0E − 12 | 1.1E + 06 | 3.0E + 03 | 2.1E − 04 | 6.5E − 07 | 64.9 ± 0.001 |
| CS06 × hu225-M | c-MET | 2.1E − 10 | 1.0E − 12 | 1.1E + 06 | 2.5E + 03 | 2.2E − 04 | 6.0E − 07 | 64.8 ± 0.126 |
| CS06 × hu225-H | c-MET | 1.2E − 10 | 1.0E − 12 | 3.3E + 06 | 9.7E + 03 | 3.9E − 04 | 8.4E − 07 | 62.2 ± 0.172 |
| oa hu225-L | EGFR | 2.2E − 07 | 1.8E − 08 | 6.1E + 05 | 4.2E + 04 | 1.4E − 01 | 5.2E − 03 | ND |
| oa hu225-M | EGFR | 4.4E − 09 | 1.6E − 10 | 3.7E + 06 | 1.3E + 05 | 1.6E − 02 | 1.9E − 04 | 65.2 ± 0.174 |
| B10v5 × hu225-M | EGFR | 4.7E − 09 | 1.3E − 10 | 3.6E + 06 | 9.7E + 04 | 1.7E − 02 | 1.5E − 04 | 64.5 ± 0.001 |
| CS06 × hu225-M | EGFR | 3.9E − 09 | 1.1E − 10 | 4.6E + 06 | 1.3E + 05 | 1.8E − 02 | 1.6E − 04 | 64.8 ± 0.126 |
| oa hu225-H | EGFR | 1.4E − 10 | 1.0E − 12 | 3.6E + 06 | 1.7E + 04 | 4.9E − 04 | 1.4E − 06 | 62.7 ± 0.001 |
| B10v5 × hu225-H | EGFR | 1.5E − 10 | 1.0E − 12 | 3.5E + 06 | 1.9E + 04 | 5.3E − 04 | 1.3E − 06 | 62.8 ± 0.174 |
| CS06 × hu225-H | EGFR | 1.2E − 10 | 1.0E − 12 | 3.3E + 06 | 9.7E + 03 | 3.9E − 04 | 8.4E − 07 | 62.2 ± 0.172 |
| Cetuximab (C225) | EGFR | 1.2E − 09 | 8.3E − 12 | 6.1E + 05 | 3.8E + 03 | 7.4E − 04 | 2.1E − 06 | 67.3 ± 0.172 |
| oa hu425 | EGFR | 2.2E − 08 | 3.8E − 10 | 5.3E + 05 | 8.7E + 03 | 1.2E − 02 | 6.3E − 05 | ND |
| Matuzumab (mAb425) | EGFR | 1.2E − 08 | 1.5E − 10 | 6.4E + 05 | 7.3E + 03 | 7.8E − 03 | 2.7E − 05 | ND |
Simultaneous binding of both antigens is critical for several modes of action of bsAbs and was analyzed via BLI. The bsAb and EGFR were stepwise associated to immobilized c-MET, yielding simultaneous binding, whereas the isotype control did not, as exemplarily shown for B10v5 × 225-H in Fig. 1C. This result demonstrated that simultaneous engagement of both recombinant target proteins by the generated bsAbs is sterically feasible.
To derive a mechanistic model for the biological activity of the bsAbs and to evaluate tumor selectivity of affinity-optimized antibodies, receptor cell surface levels on a variety of cancer cell lines from different indications were determined, including primary keratinocytes (NHEK) and the liver cell line HepG2, as models for normal tissue cells (Table 3). High EGFR levels on A431 and MDA-MB-468 cells, high surface c-MET levels in c-MET-amplified cell lines (EBC-1 and MKN45), and moderate EGFR levels on NHEK in the range of physiological EGFR expression were assessed and were similar to published data (5, 49). Employing flow cytometry, affinity-dependent and cell surface receptor level-dependent cellular binding of bsAbs to several tumor cell lines was demonstrated (supplemental Table S1). On NCI-H441 cells with equal c-MET and EGFR surface levels, bsAb displayed increased binding in comparison with corresponding mAbs, indicating an additive engagement of both antigens on the cell surface and, hence, simultaneous binding (supplemental Fig. S2).
TABLE 3.
Cell surface receptor levels of human c-MET and EGFR on several tumor cell lines from various indications
Keratinocytes (NHEK.f-c.) were used to evaluate EGFR-related skin toxicity, and the liver cell line HepG2 was used for c-MET-mediated liver toxicity. Receptor surface level values are presented as mean molecules per cell of triplicates with S.D. values given in percentages. ACA, adenocarcinoma; CA, carcinoma.
| Cell line | Origin | c-MET receptor surface level × 103 ± S.D.% | EGFR receptor surface level × 103 ± S.D.% |
|---|---|---|---|
| A431 | Epidermoid CA | 14.7 ± 0.2 | 661.0 ± 1.4 |
| A549 | Lung ACA | 18.0 ± 0.6 | 39.3 ± 0.6 |
| EBC-1 | Lung SCC | 261.6 ± 1.1 | 62.2 ± 1.1 |
| HepG2 | Hepatocellular CA | 11.1 ± 1.4 | 1.3 ± 4.7 |
| KP-4 | Pancreatic CA | 7.7 ± 0.5 | 50.8 ± 0.9 |
| MDA-MB-468 | Breast ACA | 14.2 ± 1.0 | 1825.5 ± 0.1 |
| MKN-45 | Gastric ACA | 171.7 ± 1.0 | 45.4 ± 0.3 |
| NCI-H1975 | Lung ACA | 35.5 ± 0.7 | 37.8 ± 0.7 |
| NCI-H441 | Lung ACA | 52.2 ± 0.8 | 46.6 ± 3.7 |
| NCI-H596 | Lung ACA | 6.7 ± 1.0 | 148.5 ± 1.4 |
| NHEK.f-c. | Keratinocytes | 7.1 ± 8.9 | 128.7 ± 8.7 |
| T47D | Breast ACA | 0.0 | 13.2 ± 0.9 |
bsAbs Synergistically Inhibit c-MET Signaling and EGFR Phosphorylation Dependent on Valence and Affinity
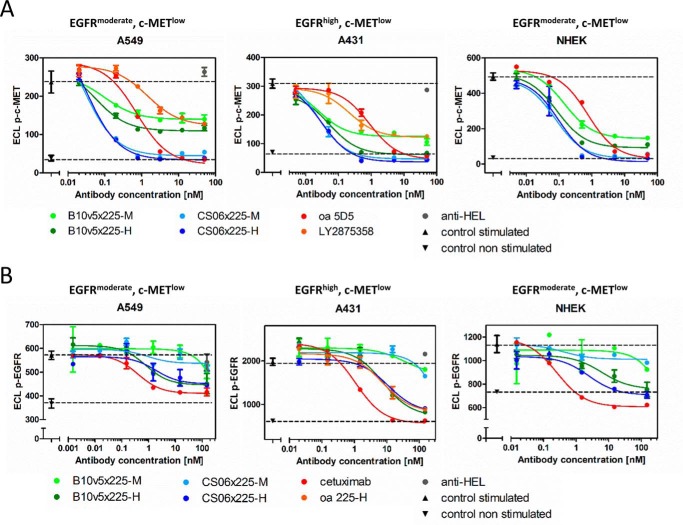
bsAbs retained capability to block ligand-induced receptor activation of c-MET and EGFR, as shown for several examples in Fig. 2 and supplemental Fig. S3A. All c-MET binders in the bsAb format efficiently inhibited c-MET phosphorylation in HGF-dependent (e.g. A549 and NCI-H596) and HGF autocrine cell lines (e.g. KP-4), whereas no inhibition of c-MET phosphorylation was observable in c-MET-amplified cell lines EBC-1 and MKN45 (data not shown). Comparable results were observed for the previously described monospecific reference antibody oa 5D5 (MetMAb) (28) and the anti-c-MET antibody LY2875358 (29). Similar c-MET surface expression levels in A549, A431, and keratinocytes correlated with comparable IC50 values for HGF-dependent c-MET inhibition independent of EGFR expression levels (Tables 3 and 4). Furthermore, simultaneous engagement of c-MET and EGFR by bsAbs significantly increased the potency of c-MET inhibition in comparison with one-armed CS06-Fab SEEDs (supplemental Fig. S3B). Interestingly, B10v5 bsAbs and mAb LY2875358 displayed a reduced efficacy compared with reference mAb oa 5D5 or bsAbs containing the c-MET binding moiety CS06 (Fig. 2). Furthermore, bsAbs containing B10v5 stimulated c-MET phosphorylation in the absence of the ligand (supplemental Fig. S3C) and therefore, unlike the CS06 construct, displayed some unwanted agonistic activity.
FIGURE 2.
Inhibition of c-MET and EGFR phosphorylation by EGFR × c-MET bsAbs during ligand stimulation. Phosphorylated c-MET (A) and phosphorylated EGFR (B) were quantified in A549, A431, and primary keratinocytes (NHEK) using ECL. Cells were treated with varying concentrations of bsAbs and a non-related isotype SEED control (anti-HEL) with subsequent stimulation with 100 ng/ml HGF (A) or 100 ng/ml EGF (B). Triangles and dotted lines indicate respective receptor phosphorylation levels for stimulated (upright triangle) and non-stimulated cells (inverted triangle). Dose-response curves were fitted using a 3PL model in GraphPad Prism version 5 (GraphPad Software). Error bars, S.D.
TABLE 4.
Inhibition of c-MET and EGFR phosphorylation by EGFR × c-MET bsAbs
IC50 values were calculated upon 3PL fitting of dose-response curves using GraphPad Prism. S.D. values were calculated for at least two independent experiments carried out in duplicates. NF, the fit returned by Prism is poor or ambiguous. n = number of independent experiments.
| Antibody | Receptor | A549 |
NHEK.f-c. |
||
|---|---|---|---|---|---|
| IC50 ± S.D. | n | IC50 ± S.D. | n | ||
| nm | nm | ||||
| CS06 × 225-H | Phospho-c-MET | 0.3 ± 0.2 | 4 | 0.1 | 1 |
| Phospho-EGFR | 0.8 | 1 | 0.8 | 1 | |
| B10v5 × 225-H | Phospho-c-MET | 0.3 ± 0.2 | 2 | 0.2 | 1 |
| Phospho-EGFR | 1.1 ± 0.4 | 2 | 1.7 ± 0.3 | 2 | |
| B10v5 × 225-M | Phospho-c-MET | 0.3 ± 0.2 | 2 | 0.14 | 1 |
| Phospho-EGFR | NF | 3 | NF | 2 | |
| oa 5D5 | Phospho-c-MET | 0.8 ± 0.5 | 4 | 0.7 | 1 |
| Cetuximab | Phospho-EGFR | 0.4 ± 0.1 | 3 | 0.3 ± 0.2 | 2 |
Regarding EGFR inhibition, monovalent 225-scFv-SEEDbodies and bsAbs with high (225-H) and medium affinity (225-M) to EGFR displayed similar efficacy compared with cetuximab (C225). The bsAbs harboring either the matuzumab (mAb425)-derived scFv or the low affinity EGFR binder (225-L) displayed low efficacy in blocking EGFR phosphorylation and were therefore not chosen for further evaluation (supplemental Figs. S3A and S4). The potency in inhibiting EGFR signaling of bsAb was dependent on affinity and valence of the EGFR binding moiety but was independent of receptor surface levels. bsAbs additionally blocked phosphorylation of downstream AKT, in contrast to cetuximab or anti-c-MET reference mAbs (supplemental Fig. S3D) (23). Furthermore, all bsAbs were able to induce antibody-dependent cellular cytotoxicity (ADCC) comparable with cetuximab (supplemental Fig. S5).
Bispecific Structure and Optimized Affinities Translate into High in Vitro Selectivity
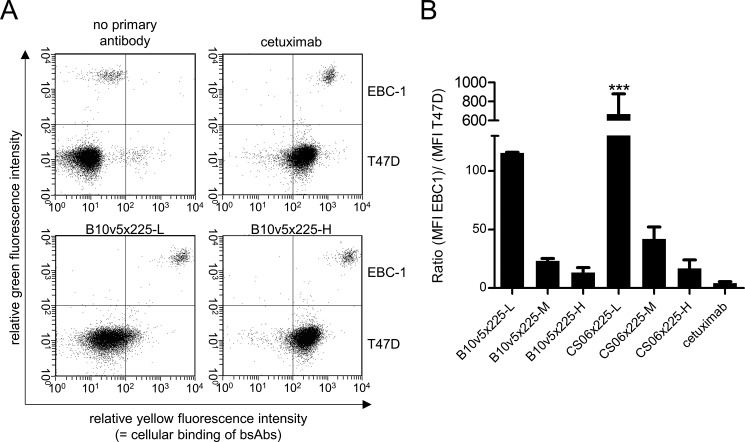
The impact of affinity on the in vitro tumor selectivity was evaluated with affinity-attenuated variants of anti-c-MET and anti-EGFR binders as mAbs or bsAbs. Treatment of patients with EGFR inhibitors mediates severe clinical adverse events in the skin; therefore, binding to EGFR was the focus of this study (31, 50). The in vitro binding selectivity of EGFR × c-MET bsAb to a cell mixture composed of a c-METhigh/EGFRhigh tumor model cell line (e.g. EBC-1) with an excess of an c-MET−/EGFRlow normal tissue model cell line (e.g. T47D) in a ratio of 1:30 was determined by flow cytometry. The breast cancer cell line T47D was therefore used as a model to represent expression levels on normal cells. This setup mimics the physiological situation based on experiments carried out by Robinson et al. (32). For cell line discrimination, EBC-1 cells were stained with a membrane dye, whereas T47D cells remained unstained. We observed that double-positive EBC-1 cells were more strongly bound by bsAbs than cetuximab, presumably due to simultaneous binding to both the c-MET and the EGF receptor, whereas binding to normal tissue model cells (T47D) was strongly dependent on EGFR affinity; B10v5 × 225-H bound with similar affinity as cetuximab, whereas reduced binding of B10v5 × 225-M and to a greater extent B10v5 × 225-L was observed (Fig. 3A). With in vitro tumor selectivity defined as the ratio of tumor to normal tissue model cellular binding (e.g. MFI EBC-1/MFI T47D), all bsAbs demonstrated higher selectivity compared with the reference EGFR binder cetuximab (C225; Fig. 3B).
FIGURE 3.
In vitro selectivity of EGFR × c-MET bsAbs in comparison with cetuximab. A, EBC-1 as a tumor model cell line with high to moderate c-MET and EGFR expression and T47D as an epithelial model cell line with low EGFR expression and no c-MET expression were mixed in a ratio of 1:30. To distinguish the two cell lines, EBC-1 cells were stained with the green membrane dye PKH2. The cell mixture was incubated with 30 nm bsAb or cetuximab and subjected to flow cytometric analysis. Antibody binding was detected by PE-labeled anti-human Fc secondary antibody. Representative dot plots for green versus yellow fluorescence are shown. B, in vitro selectivity was defined as the ratio of mean fluorescence intensity of the EBC-1 and the T47D cell population. Displayed are means with S.D. of two independent experiments. Asterisks, significant difference of groups in comparison with the selectivity of cetuximab (***, p < 0.001, with one-way analysis of variance with Dunnett's test). Error bars, S.D.
Bispecific ADCs for Enhanced Efficacy and Broadened in Vitro Therapeutic Index
Mutual overexpression of receptor tyrosine kinases, especially c-MET and EGFR, is associated with tumor growth, metastasis, and poor prognosis (11, 12) but could be exploited for the efficient delivery and internalization of toxins when applying bispecific ADCs. The EGFR × c-MET bsAbs fulfilled ADC requirements, because they (a) demonstrated internalization employing confocal microscopy and flow cytometry (supplemental Fig. S6) and (b) potentially increased tumor selectivity by affinity-attenuated variants while decreasing cytotoxic effects on normal cells (Fig. 3). The tubulin inhibitor monomethyl auristatin E (MMAE) was utilized for C-terminal sortase-mediated conjugation on both heavy chains via a protease-cleavable valine-citrulline (vc) linker with a drug-to-antibody ratio of 2, which was confirmed by MALDI-TOF and analytical size exclusion chromatography (data not shown) (51, 52). Furthermore, conjugation did not alter the antibody's binding characteristics to recombinant or cellular bound c-MET and EGFR (data not shown). Due to the presumable degradation mode of action and particularly murine cross-reactivity, an important feature for animal studies (data not shown), B10v5 was chosen for further analysis of bispecific ADCs despite partial agonism, which was not considered to be critical, because the anti-c-MET antibody emibetuzumab (LY2875358), which is currently being evaluated in clinical phase II, has not given rise to any safety concerns despite c-MET-related partial agonism (29).
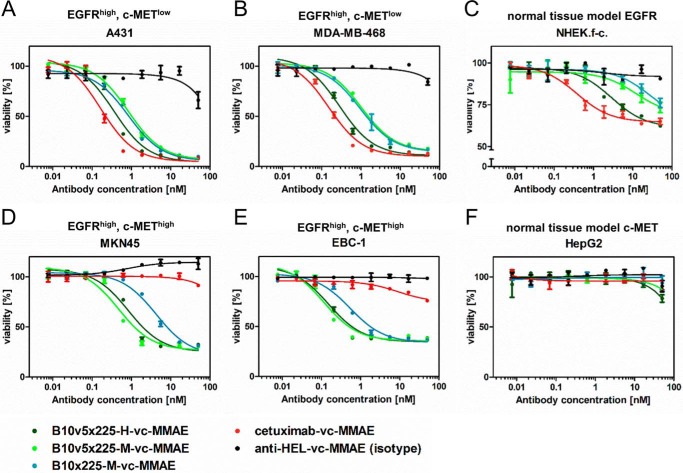
As a next step, the impact of bsAbs and bispecific ADCs on the cell viability was analyzed after 72-h treatment with an ATP-based luminescence assay (CellTiter-Glo®). Although bsAbs without the toxin showed little or no effect on cell viability after 72 h (data not shown), the same antibodies applied as ADCs induced significant cytotoxicity, as depicted in Fig. 4. For EGFR-overexpressing cells (A431 and MDA-MB-468), reduction of cell viability by >80% was achieved during treatment. The cytotoxic potency of B10v5 × 225-H-vc-MMAE was slightly reduced in comparison with reference ADC cetuximab-vc-MMAE (Fig. 4, A and B). Primary keratinocytes were affected with 30–40% reduced viability and lower potency when compared with tumor cells (Fig. 4C). Increasing incubation time on keratinocytes to 6 days enhanced efficacy but did not significantly alter potency (supplemental Fig. S7). Interestingly, similar cytotoxicity on keratinocytes was observed for antibodies devoid of toxin. This indicated that cell killing of keratinocytes could be caused by EGFR inhibition rather than by cytotoxic effects mediated by the ADC. To quantify the selectivity advantage of bispecific EGFR × c-MET ADCs in comparison with the reference cetuximab-vc-MMAE, an in vitro translational therapeutic index or therapeutic window was applied based on two formulas: (a) the difference of IC50 for NHEK (moderate EGFR surface level) and EC50 for A431 (high EGFR surface level) and (b) the ratio of the toxic dose on keratinocytes causing 20% cell killing (TD20) and the effective dose on A431 tumor cells leading to 80% cytotoxicity (ED80) (53). For B10v5 × 225-M-vc-MMAE and B10 × 225-M-vc-MMAE, dose-response curves did not reach saturated cell killing at high ADC concentrations. As a consequence, the resulting in vitro therapeutic window is an estimate only. Nevertheless, the data indicate that IC50 values will be >20 nm, supporting the notion that these bispecifics broaden the in vitro therapeutic window in comparison with cetuximab-vc-MMAE, which has a narrow in vitro therapeutic window demonstrated by the nearly identical TD20 and ED80 (Table 5). In contrast, the TD20/ED80 ratio of B10v5 × 225-M and B10 × 225-M conjugated to vc-MMAE was 5–6-fold broader compared with analogously generated cetuximab-ADC. Despite EGFR surface expression in c-MET-amplified cell lines MKN45 and EBC-1 (cf. Table 3), cetuximab-vc-MMAE did not influence cell viability in concentrations up to 50 nm in these cell lines (Fig. 4, D and E), whereas bispecific ADCs displayed potent cytotoxicity, depending on the affinity of the c-MET binding moiety (high affinity B10v5 versus medium affinity B10). In HepG2 cells, a hepatocellular carcinoma cell line that mimics receptor surface expression levels of liver cells and therefore was used as a model cell for liver toxicity, no significant cell killing was observed (Fig. 4F), probably due to low c-MET and EGFR cell surface levels. Similar results were detected for CS06 containing bispecific ADCs (data not shown).
FIGURE 4.
Cytotoxicity of EGFR × c-MET bispecific SEED antibody-drug conjugates generated by covalent, site-directed conjugation of the tubulin inhibitor MMAE C-terminally to both heavy chains in comparison with cetuximab as ADC and anti-HEL ADC as corresponding reference constructs. After incubation of bispecific ADCs on EGFR-overexpressing tumor cells A431 (A) and MDA-MB-468 (B), on primary keratinocytes (NHEK.f-c.; C) as normal epithelial cell line, and on c-MET-overexpressing cells MKN45 (D) and EBC-1 (E) as well as HepG2 (F) as liver cell line, cell viability was assessed using the ATP-based luminescence CellTiter-Glo® assay. The assay was run in duplicates in three independent experiments, and curves were fitted by sigmoidal curve fitting using GraphPad Prism version 5 (GraphPad Software). Error bars, S.D.
TABLE 5.
Cytotoxicity of bispecific EGFR × c-MET ADC on tumor cell line A431 and keratinocytes
EC50 values for A431 cells and IC50 values for keratinocytes (NHEK.f-c.) were calculated by sigmoidal curve fitting using GraphPad Prism version 5 (GraphPad Software). *, estimated values only due to poor fitting results. ED80 represents the ADC concentration at which 80% of cells are killed in A431 cells in comparison with untreated cells. TD20 stands for the dose where cell viability is reduced by 20% in keratinocytes. Two definitions for an in vitro translational therapeutic index or therapeutic window were calculated: the difference of IC50 and EC50 as well as the ratio of TD20 to ED80. For the calculation of the therapeutic window, error propagation was used. For statistical analysis, comparison with cetuximab-vc-MMAE is given.
| Antibody | A431 |
NHEK.f-c. |
Therapeutic window |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EC50 | ED80 | Cell killing at 50 nm | n | IC50 | TD20 | Cell killing at 50 nm | n | IC50 − EC50 | TD20/ED80 | |
| nm | nm | % | nm | nm | % | nm | ||||
| B10v5 × 225-H-vc-MMAE | 0.4 ± 0.1a | 2.1 ± 0.7a | 93 ± 1.3 | 5 | 5.9 ± 2.9b | 4 ± 1c | 37 ± 12 | 5 | 5.6 ± 2.8 | 2 ± 0.06 |
| B10v5 × 225-M-vc-MMAE | 1.0 ± 0.3d | 4.4 ± 1.0d | 91 ± 0.4 | 3 | >19* | 25 ± 8d | 30 ± 9 | 3 | >28* | 6 ± 0.5 |
| B10 × 225-M-vc-MMAE | 0.7 ± 0.2d | 3.6 ± 0.9e | 90 ± 0.5 | 3 | >20* | 19 ± 8d | 27 ± 11 | 3 | >18* | 5 ± 0.9 |
| Cetuximab-vc-MMAE | 0.1 ± 0.04 | 0.7 ± 0.3 | 92 ± 1.4 | 5 | 1.0 ± 0.5 | 0.8 ± 0.5 | 34 ± 10 | 5 | 0.9 ± 0.5 | 1 ± 0.2 |
a p < 0.05.
b p < 0.01, unpaired t test.
c Not significant.
d p < 0.001.
e p < 0.01 (all analysis of variance).
Taken together, we show that engineering of affinity-optimized variants and the generation of bispecific EGFR × c-MET ADCs enabled potent and selective cytotoxicity on EGFR- and c-MET-overexpressing cells while reducing toxicity on normal tissue model cells, such as HepG2 and keratinocytes.
Discussion
This study analyzed the balance between in vitro selectivity and efficacy of bsAb and bispecific ADCs directed against c-MET and EGFR by employing affinity-optimized binding moieties and different epitope combinations. Mutual overexpression of c-MET and EGFR is linked with tumor proliferation and poor prognosis, rendering both antigens appealing targets for therapeutic application in various indications (11, 12, 17, 54). Furthermore, a close proximity and interaction of c-MET and EGFR on the cell surface has already been confirmed by co-immunoprecipitation (55). Due to receptor cross-talk and signaling redundancies, combination therapy targeting both receptor tyrosine kinases seems to be advantageous because monotherapies can favor the growth of drug-resistant tumor cells by selection pressure. One major escape mechanism during EGFR monotherapy is related to enhanced c-MET signaling via receptor amplification or ligand up-regulation (16, 21, 56). Furthermore, EGFR inhibitors struggle with adverse events mediated by inhibition of basally expressed EGFR in normal tissues (50, 57). Bispecific molecules might therefore on the one hand improve the efficacy compared with a combination of monospecific mAbs via potential synergistic effects and on the other hand increase the selectivity via simultaneous targeting of both receptors favoring overexpressing cells by avidity effects.
The first EGFR × c-MET directed bsAb was described by Castoldi et al. (23) via C-terminal fusion of the anti-c-MET antibody fragment oa 5D5 to cetuximab IgG, demonstrating pharmacodynamic activities comparable with the combination of the respective parental antibodies. Our study demonstrated significantly increased potency for the inhibition of c-MET and AKT phosphorylation by CS06 × 225-H in comparison with the combination of monovalent control mAbs. Additionally, bsAbs also mediated ADCC similarly to cetuximab, which might be advantageous for applications without conjugated toxin because cetuximab is acting, inter alia, via ADCC (58). For ADC applications, however, an Fc devoid of effector functions might be beneficial.
Tumor selectivity can be achieved either by the choice of a tumor-specific target or by the application of affinity-attenuated variants for a ubiquitously expressed cancer antigen. For example, the antibody ABT-806 (or ABT-414 as ADC) targets a tumor-specific, misfolded EGFR epitope accessible during enhanced EGFR expression with minimal expression in normal tissue (59, 60). However, ABT-806 demonstrated decreased binding to the EGFR-overexpressing cell line A431 and reduced potency in inhibiting tumor growth in an A431 xenograft study in comparison with cetuximab (59). The present study, in contrast, aims to utilize affinity-attenuated cetuximab variants to drive tumor selectivity in the bispecific EGFR × c-MET mAb and ADC format. Work by Mazor et al. (6, 33) revealed that (a) bispecificity per se can increase tumor selectivity via simultaneous targeting of two cancer antigens and (b) bsAb employing affinity-attenuated variants can drive selectivity to a cell population with high expression in both targets, whereas bsAb binding to cell lines with single target expression is reduced. The present study indicates that bsAb containing affinity-attenuated variants of cetuximab display increased selectivity toward cell lines mutually overexpressing c-MET and EGFR in comparison with normal tissue models with basal EGFR expression, although the potency in inhibiting EGFR phosphorylation was concurrently reduced (Figs. 2 and 3).
Because the cell killing ability of mAbs per se is limited, ADC methodologies can be applied for combining high selectivity of antibodies with high potency of cytotoxic agents. Besides high tumor selectivity, antibodies require suitable internalization rates and appropriate affinities (9, 10). The bsAbs described herein exhibit appropriate internalization comparable with cetuximab for which internalization has already been investigated (61). In general, internalization of bsAbs might also correlate with the antibodies' ability to cross-link c-MET and EGFR. Modeling the binding of bsAbs to interacting receptors is highly complex; especially the consideration of relative target antigen epitope orientation within the cell membrane makes experimental evaluation of bsAb binding inevitable (62–64). Lee et al. revealed co-internalization of EGFR and c-MET induced by the bsAb ME22S, whereas the c-MET binding moiety, SAIT-301, was probably the driving force (25). In addition, internalization was also shown for the anti-c-MET antibody LY2875358, which occupied an overlapping epitope on c-MET with B10 and B10v5 (data not shown) (29). Due to the presumable degrading mode of action and murine cross-reactivity, B10 and B10v5 were favored for the ADC approach. Murine cross-reactivity is of importance for evaluation in preclinical mouse xenograft models. It is noteworthy that the in vitro cytotoxicity of CS06 containing bispecific ADCs was similar to that of B10v5 containing bispecific ADCs (data not shown).
Given the improved tumor selectivity and the suitable internalization by the presented bsAbs, conjugation of the tubulin inhibitor MMAE to both antibody heavy chains was conducted. Auristatins are currently the most widely validated class of ADC payloads in clinical development (65). Toxin and linker selection added thereby a second layer of selectivity because MMAE acts specifically on fast proliferating cells like tumor cells, and the protease-cleavable peptide linker, valine-citrulline, ensures intracellular release of the toxic compound by lysosomal proteases (e.g. cathepsin or plasmin) (9). The present study demonstrated highly potent and efficacious killing of EGFR- and c-MET-overexpressing cell lines by bispecific ADCs with reduced toxicity in keratinocytes. After 3 days of incubation with bispecific ADCs, the viability of NHEK cells was reduced by 50%. Extending the incubation time to 6 days did not significantly alter the IC50 values but increased the reduction in cell viability to 80%. Treatment of keratinocytes with naked bsAbs revealed similar cytotoxicity compared with the corresponding bispecific ADCs, suggesting that cytotoxic effects are caused by inherent EGFR inhibition rather than toxin delivery by the ADC. Taken together, B10v5 × 225-M and B10 × 225-M conjugated to vc-MMAE displayed a 5–6-fold broader in vitro therapeutic index compared with cetuximab-ADC (defined as the TD20/ED80 ratio). Further analysis is needed to determine the threshold of EGFR and c-MET expression required for potent cell killing, and xenograft studies are needed to evaluate maximum tolerated doses as well as dose-limiting toxicities of bispecific ADCs. It will be interesting to see whether the observed in vitro selectivity translates into an improved in vivo safety profile. Furthermore, stability and pharmacokinetic studies will be required before in vivo experiments.
The multifactorial nature of cancer based on receptor cross-talk and redundancies can drive the development of an acquired resistance mechanism during single EGFR blockade involving c-MET amplification (16) or ligand up-regulation (18, 56, 66) as well as activating mutations in EGFR (67, 68) (e.g. T790M) and downstream signaling molecules (e.g. KRAS) (67), giving rise to tumor heterogeneity. The application of bispecific EGFR × c-MET ADCs could potentially overcome these challenges. The generated bispecific ADCs efficiently induced cytotoxicity in c-MET-amplified cell lines (Fig. 4). Furthermore, cetuximab and potentially also the generated bispecific ADCs might inhibit growth of cells carrying the gatekeeper mutation T790M within the EGFR kinase domain. Although first generation EGFR kinase inhibitors (e.g. gefitinib and erlotinib) are not able to inhibit T790M-mutated EGFR, cetuximab recognizes a distinct extracellular epitope and therefore induces inhibition and internalization independent of T790M EGFR mutational status (69). Due to the ADC mechanism of action via intracellular delivery of the cytotoxic agent, bispecific ADCs might be independent of mutational status of KRAS. Nevertheless, resistance mechanisms are hard to model in vivo, and further evaluation of EGFR × c-MET bsAbs and ADCs is needed.
Taken together, the results presented in this study demonstrate that different antibody engineering approaches, both in silico and in vitro by phage and yeast surface display, can result in affinity-optimized binders with desired higher or attenuated affinities. The selection and combination of affinity-optimized variants in bispecific EGFR × c-MET ADCs targeting distinct epitopes can drive in vitro tumor selectivity while retaining highly potent anti-tumor efficacy. The appropriate balance between high selectivity and suitable potency in EGFR × c-MET bispecific ADCs could potentially broaden the therapeutic window. To fully exploit the therapeutic potential of these affinity-optimized bispecific ADCs, in vivo evaluation applying suitable models is necessary. For this, the strategy of employing affinity-attenuated variants may increase the space of available targets for ADC.
Experimental Procedures
Generation of Anti-c-MET Binders
Panning of naive phage display antibody gene libraries HAL7/8 against human c-MET was performed according to Hust et al. (42, 43). Briefly, after preselection with panning buffer (1% skim milk powder, 1% BSA, 0.05% Tween 20 in PBS) in MaxiSorp 96-well plates (Nunc), scFv-displaying phages were selected on 1 μg of immobilized c-MET-Fc (R&D Systems, 358-MT/CF) or c-MET SEMA domain (produced in house) and eluted with trypsin. After 2–3 rounds of panning, c-MET-specific binders were enriched and screened by capture c-MET ELISA of produced scFv.
For affinity maturation, (a) error-prone PCR for variable domains using the GeneMorph II random mutagenesis kits (Agilent Technologies) according to the manufacturer's instructions, (b) randomization of complementarity-determining region 3 of the heavy chain (CDR-H3) ordered by GeneArt applying a parsimonious mutagenesis strategy (70), and (c) light chain shuffling using the diversity of the HAL7/8 were conducted. Panning was carried out using phage and yeast surface display for F06 and B10, respectively. For clone F06, an off-rate screening strategy was applied by stringent washing (10 times) with 100 μl of panning buffer per well as well as adding soluble c-MET for competition (starting in the second round). CS06 was based on rational combination of abundant mutations from approaches a and b. B10v5 was derived from approach c using yeast surface display as described previously (44, 45).
Generation of Anti-EGFR Binders
The structure of C225 bound to the extracellular domain of EGFR (36) was optimized with the Rosetta Protein Structure and Design program (version 2.3.0) (38) using a fixed backbone protocol and side chain optimization to minimize the energy of the starting model for design according to the Rosetta energy function. Interfacial water molecules observed in the crystal structure were retained during the minimization but not during subsequent design calculations. Thirty-seven residues at or near the antibody-antigen interface were selected for a saturating, in silico point mutagenesis. At each of these residues, 19 variants were created (wild type and 18 mutations, no cysteine) optimizing the rotamer of the mutated residue while keeping the backbone fixed. Using these preliminary models, neighbor residues were identified as any residue with at least three heavy atoms within 5.5 Å of a heavy atom on the design residue. The rotamer of the mutated residue and its neighbors were optimized using the standard Rosetta score function (a linear combination of terms including a Lennard-Jones potential, an orientation-dependent hydrogen bonding potential, an implicit solvation model, and statistical terms that capture backbone-dependent amino acid and rotamer preferences) (38–40). The polar substitutions were filtered to only those variants with improvements of at least 0.5 Rosetta energy units in either the orientation-dependent hydrogen bonding score or the pair potential relative to the repacked native to select improved variants. The three affinity-enhancing point substitutions were combined into a triple mutant, and this was repacked and scored by Rosetta as described above for the point mutants. The affinity of the selected variants was measured in vitro by surface plasmon resonance (SPR). The variants were also transferred to the hu225 scFv (34), and the affinities in this context were verified by BLI.
Generation, Expression, and Purification of bsAbs
Several combinations of EGFR and c-MET antibody fragments were converted to bsAbs using the SEED technology (35). Briefly, sequences for variable regions were subcloned from phagemid or yeast display vectors into pTT5 mammalian expression vectors (National Research Council Canada) encoding for either constant SEED heavy chain or constant light chain domains. For this, standard cloning procedures were used either with SalI/NheI (New England Biolabs) or the Expresso CMV base system (Lucigen) followed by transformation into One Shot® TOP10 chemically competent Escherichia coli cells (Life Technologies, Inc.). All cloned constructs were verified by Sanger sequencing.
Antibodies were expressed by transient co-transfection of antibody chains in Expi293FTM cells using the corresponding transfection kit and media (all Life Technologies) according to the manufacturer's instructions. EXPI293FTM cells are a derivative of the human embryonic kidney cell line HEK293. Briefly, Expi293FTM cells were seeded with a final density of 2 × 106 viable cells/ml. Supernatants were harvested 5 days post-transfection and antibody constructs were harvested by centrifugation and filtration through 0.22-μm Stericup or Steriflip devices (Millipore).
Small scale productions were performed in a volume of 25 ml, and purification was carried out with PROSEP® A centrifugal Protein A columns (Millipore, catalog no. P36486) according to the manufacturer's instructions followed by dialysis to PBS, pH 7.4, using a Pur-A-LyzerTM dialysis kit (Sigma-Aldrich).
Midscale productions were performed in an expression volume of 200 ml. Supernatants were purified by affinity chromatography (5 ml of HiTrap MabSelect SuRe, GE Healthcare) on an ÄKTA Explorer 100 system (GE Healthcare) with subsequent preparative size exclusion chromatography (HiLoad 26/60 Superdex 200 pg, GE Healthcare). Protein concentrations were determined by UV A280 spectroscopy and purity was analyzed by gel electrophoresis with 4%/8% NuPAGE BisTris gels (Life Technologies) and Coomassie staining as well as analytical size exclusion high performance liquid chromatography (TSK Super SW3000, Tosoh). Endotoxin levels were assessed by Limulus amebocyte lysate Endosafe® PTS cartridges and an Endosafe® PTS reader (Charles River).
Anti-c-MET reference binders, including humanized oa 5D5 (MetMAb, onartuzumab) (28), LY2875358 (LA480_vC8H241, emibetuzumab) (29), and h224G11 (ABT-700) (46), were reproduced with antibody VH and VL sequences derived from publicly available information (71–73). Sequences were cloned in mammalian expression vectors containing constant IgG1 light and heavy chain fragments, except in the case of oa 5D5, knob-into-hole technology was applied (74). All anti-c-MET reference mAbs as well as cetuximab (C225, Erbitux) and matuzumab were produced in-house (Merck) in HEK293E cells using a standard transfection and purification procedure as described above. Note that anti-c-MET reference binders were reproduced and not obtained from the respective laboratories of the original publication or patent.
Surface Plasmon Resonance
The affinity and kinetic parameters of the in silico designed C225 variants were verified by surface plasmon resonance. Computationally guided substitutions were introduced into the wild-type C225 using the QuikChangeII kit (Stratagene) with mutagenic primers. The variant antibodies were expressed in HEK-293–6E cells. SPR was performed on a Biacore A-100 (GE Healthcare). CM5 chips were coupled with goat anti-human IgG antibody (Jackson ImmunoResearch, Inc., 109-005-098) and used to capture the wild-type C225 or designed variants. Human EGFR (extracellular domain, R&D Systems, 1095-ER) was used as analyte. The affinity was determined by titrating the analyte from 0 to 40 nm and determining kinetic rate constants using the BiaEvaluation software to fit the association and dissociation phases using a 1:1 Langmuir binding model. The KD was determined as the ratio of the kinetic constants.
Thermal Shift Assay
Thermal stability of antibodies was measured using a StepOnePlus real-time PCR system (Life Technologies) according to the manufacturer's instructions. Briefly, 1 μm protein was mixed with a 20-fold excess of SYPRO Orange (Life Technologies) in PBS, pH 7.4. Melting curves were recorded from 25 to 99 °C with an increment of 1 °C/60 s. Data were analyzed with the Protein Thermal ShiftTM Software (Life Technologies) by calculating the maximum of the second derivative curve.
Biolayer Interferometry
Kinetic parameters of antibodies were determined on the Octet Red96 system using Octet Data Acquisition software (version 8.2, Forté Bio, Pall). All data were collected at 30 °C in kinetics buffer (PBS, pH 7.4, 0.1% BSA, 0.02% Tween 20; Merck) with 1000-rpm orbital senor agitation in a volume of 200 μl using black 96-well microtiter plates (Greiner Bio One). Human c-MET ECD and EGFR ECD were produced and purified in house. Anti-human IgG Fc capture biosensor tips (Forté Bio, Pall) were equilibrated for 30 s in Dulbecco's PBS (Life Technologies). Then 5 μg/ml mAbs diluted in PBS were immobilized on biosensor tips for 120 s, and a baseline was recorded for 60 s in kinetics buffer followed by stepwise association and dissociation of the analyte for 600 and 1200 s, respectively. Buffer controls were subtracted as background, and binding parameters were calculated assuming a 1:1 Langmuir binding model performing global fitting algorithm provided by the Octet data analysis software (version 8.2, Forté Bio, Pall).
For evaluating simultaneous binding, 5 μg/ml biotinylated c-MET ECD was captured on streptavidin biosensor tips (Forté Bio, Pall) for 40 s. Biotinylation was performed with the EZ-LinkTM sulfo-NHS biotinylation kit (Thermo Scientific). Biosensors with captured c-MET were first blocked with 1% milk powder, 1% BSA, 0.1% Tween® 20, and 10 μg/ml biocytin for 60 s and then stepwise subjected to 50 nm bsAbs and 50 nm EGFR-ECD for 300 s each. As controls, the non-related isotype control anti-HEL mAb or buffer controls were implemented to exclude unspecific binding.
Cell Culture
Human cancer cell lines were obtained from the American Type Culture Collection (A431, A549, MDA-MB-468, NCI-H1975, NCI-H441, NCI-H596, and T47D), the Riken Biorescourse Center Cell Bank (EBC-1 and KP-4), Lipha (HepG2), and the German Collection of Microorganims and Cell Cultures (MKN45) and maintained according to standard culture conditions (37 °C, 5% CO2, 95% humidity) using recommended medium formulations. A549 and A431 were cultivated in minimal essential medium (Life Technologies) containing 10% FBS (Life Technologies). MDA-MB-468, NCI-H1975, HepG2, and MKN45 were maintained in RPMI 1640 (Life Technologies) supplemented with 10% FBS, 2 mm l-glutamine, and 1 mm sodium pyruvate (both from Life Technologies). NCI-H441 and NCI-H596 were cultivated in RPMI 1640 with 10% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate, 2.5 g/liter d-(+)-glucose (Sigma-Aldrich), and 10 mm HEPES (Life Technologies). T47D cells were cultivated in RPMI 1640 medium with 10% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate, and 10 μg/ml insulin (Sigma-Aldrich). KP-4 cells were cultivated in DMEM/F-12 with 10% FBS. EBC-1 cells were maintained in minimal essential medium with 10% FBS and 2 mm l-glutamine. NHEK.f-c. cells (PromoCell, catalog no. C-12007) were obtained from PromoCell and propagated in recommended keratinocyte growth medium with supplements (PromoCell, catalog no. C-20111) and with the DetachKit (PromoCell, catalog no. C-41210) for cell detachment. Expi293FTM cells were purchased from Life Technologies and cultivated in corresponding Expi293FTM expression medium. All cell lines were shown to be sterile, were certified mycoplasma-free, and never exceeded passage 20.
Quantification of Cell Surface Receptor Levels
Receptor surface expression levels on selected cell lines were determined using the QFIKIT (Dako K0078) employing flow cytometry. Briefly, five populations of calibration beads presenting different numbers of mouse mAb molecules on their surfaces were used as a calibration standard. 1.5 × 105 cells/well were labeled with primary mouse anti-EGFR (ab187287, Abcam) and mouse anti-c-MET antibodies (MAB3582, R&D Systems) at saturating doses (5 μg/ml). Then beads and cells were stained with secondary goat anti-mouse Fc F(ab′)2 FITC conjugate (10 μg/ml; Jackson ImmunoResearch) and were subjected to flow cytometry measurement using a Guava easyCyte HT cytometer (Millipore). Beads and cells were measured on the same day using the same settings. Based on a calibration line for fluorescence of beads versus bead surface level, antigen cell surface levels for c-MET and EGFR were calculated.
Receptor Phosphorylation Assay
Phosphorylation levels were determined by c-MET or EGFR capture electrochemiluminescence (ECL) ELISA (MSD assay). All reagents were obtained from Meso Scale Discovery and prepared according to the manufacturer's instructions unless stated otherwise. Briefly, cells were plated in 96-well tissue culture plates (Sigma-Aldrich) 1 day before treatment, serum-starved, and treated with serially diluted antibodies (0–167 nm in starvation medium) for 1 h at 37 °C, 5% CO2. Upon stimulation with 100 ng/ml HGF and/or EGF (both from R&D Systems) for 5 min at 37 °C, cells were lysed with ice-cold lysis buffer supplemented with protease and phosphatase inhibitors (Calbiochem). High bind 96-well plates including electrodes (Meso Scale Discovery) were coated with capture anti-total c-MET (Cell Signaling Technologies) or anti-total EGFR antibodies (Abcam) followed by blocking with 3% Block A in PBS supplemented with 0.05% Tween 20. After incubation with cell lysates, detection was carried out with anti-phospho-c-MET (Cell Signaling Technologies), anti-phospho-tyrosine antibodies (R&D Systems), and the supplier-recommended detection substances. Measurements were performed with the SECTOR® Imager 6000 (Meso Scale Discovery). For quantification of phospho-AKT levels, the Phospho(Ser473)/Total AKT Assay Whole Cell Lysate Kit (Meso Scale Discovery) was used. Dose-response curves were plotted as the logarithm of mAb concentration versus ECL signal. IC50 values were calculated by a 3PL fitting model using GraphPad Prism version 5 (GraphPad Software, Inc.). Data from at least two experiments were used to calculate mean IC50 ± S.D.
In Vitro Selectivity
The selectivity of bsAbs was assessed by analyzing binding of antibodies to a cell mixture composed of a tumor model cell line (EBC-1) with high expression of both EGFR and c-MET in the presence of an excess of an epithelial model cell line (T47D) with low EGFR expression and no c-MET expression. For this, cells were trypsinated and counted with a CEDEX cell counting device (Beckman Coulter). To discriminate the two cell lines during flow cytometry, EBC-1 cells were stained with the PKH2 green fluorescent cell linker kit (Sigma-Aldrich, PKH2GL) according to the manufacturer's instructions using 2 × 107 cells with 4 μl of dye. Unstained T47D and stained EBC-1 cells were mixed in a ratio of 1:30, 1.3 × 105 total cells/well seeded in 96-well round bottom plates (BD Biosciences) and incubated with 30 nm mAb for 1 h at 4 °C. After washing with 1% BSA-Dulbecco's PBS, binding of mAbs was detected with PE-conjugated goat anti-human Fc-specific Fab2 (Jackson ImmunoResearch). The samples were subjected to flow cytometric analysis using a Guava easyCyte HT (Merck Millipore) with 30,000 counts collected per sample. The membrane dye PKH2 was detected in the green channel, and antibody binding was detected in the PE channel (yellow). Due to cross-talk between the green and yellow channels, green emission was compensated in the yellow channel. Cytometry data were analyzed using guavaSoft ExpressPro and InCyte (version 2.2.3, Millipore).
ADC Generation
Sortase-mediated site-directed conjugation of vc-MMAE to antibody Fc was performed as described elsewhere (51). Briefly, antibodies carrying enzyme recognition sites C-terminally on both heavy chains were generated, transfected, and purified by affinity chromatography. Then 1 eq of antibody was incubated with 11 eq of substrate-vc-MMAE conjugate in the presence of 5 μm sortase and 5 mm CaCl2 in reaction buffer (50 mm Tris, 150 mm NaCl, pH 7.5) for 30 min at 22 °C. The reaction was stopped with 10 mm EDTA as calcium ion chelator. The resulting ADC was purified by size exclusion chromatography as described above.
Cytotoxicity Assay
Cell viability was quantified using the CellTiter-Glo® assay (Promega), which was performed according to the manufacturer's instructions. Briefly, cells were detached and seeded in the inner wells of opaque white tissue culture-treated 96-well plates (PerkinElmer Life Sciences). The seeding cell number ranged from 8000 to 15,000 viable cells/well, depending on the cell line, in 80 μl of cell line-specific medium. Cells were allowed to attach for at least 3 h in a humidified chamber at 37 °C, 5% CO2 before ADC treatment (ranging from 50 to 0.01 nm final concentration) in duplicates in cell line-specific medium. After 72 h, viability of cells was detected by adding 100 μl/well of CellTiter-Glo® reagent with subsequent mixing on a plate shaker for 2 min at 350 rpm and a 10-min incubation in the dark at room temperature. Luminescence was measured with a Synergy 5 reader (Biotek) with a read time of 0.5 s/well (sensitivity: 170). Background luminescence in wells with only medium plus the CellTiter-Glo® reagent was subtracted. Data were plotted as percentage of untreated cell viability versus the logarithm of antibody concentration and fitted with a 3PL model using GraphPad Prism version 5 (GraphPad Software). Data from at least three independent experiments were used to calculate mean IC50 ± S.D.
Statistical Analysis
Kinetic parameters and dose-response curves were displayed as means ± S.D. and plotted as a function of time. Statistical comparison of groups was performed using the analysis of variance test followed by Dunnett's test for the comparison of the mean of each group with the mean of a control group. Statistical calculations were performed with GraphPad Prism version 5 (GraphPad Software).
Author Contributions
C. S., A. D., L. T., H. K., C. K., and V. S. conceived and designed the study. C. S., N. R., S. K., L. R., A. M., and J. W. performed the experiments. C. S., A. D., C. K., L. T., B. H., M. S., S. B., and H. K. analyzed the data. C. S., A. D., M. S., V. S., H. K., and C. K. wrote the paper and prepared the figures. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful for support and advice from a number of scientists at EMD Serono and Merck KGaA, in particular Friedhelm Bladt and Christina Esdar for scientific support; Iris Mannberger for experimental design input; Deniz Demir, Birgit Piater, Angelika-Nicole Helfrich, Daniela Pur, Konstanze Waurisch, and Dominik Reitz for laboratory support and parental c-MET antibody generation; Suming Kong, Yue Li, Hien Nguyen, and Yaping Sun for assistance with cloning and purification for EGFR binder generation; Dirk Müller-Pompalla and Jens Hannewald for advice in mAb purification; Stephan Dickgiesser for help with confocal microscopy; and Doreen Könning and Christian Schröter for general advice.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1–S7.
- bsAb
- bispecific antibody
- ADC
- antibody-drug conjugate
- ADCC
- antibody-dependent cellular cytotoxicity
- BLI
- biolayer interferometry
- VH
- variable domain of the heavy chain
- VL
- variable domain of the light chain
- scFv
- single-chain variable fragment
- oa
- one-armed
- SEED
- strand exchange engineered domain
- MFI
- mean fluorescence intensity
- EGFR
- EGF receptor
- c-MET or HGFR
- hepatocyte growth factor receptor
- HGF
- hepatocyte growth factor
- PE
- phycoerythrin
- vc
- valine-citrulline
- MMAE
- monomethyl auristatin E
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HEL
- hen egg lysozyme
- NHEK
- normal human epidermal keratinocytes.
References
- 1. Holmes D. (2011) Buy buy bispecific antibodies. Nat. Rev. Drug Discov. 10, 798–800 [DOI] [PubMed] [Google Scholar]
- 2. Kontermann R. E., and Brinkmann U. (2015) Bispecific antibodies. Drug Discov. Today 20, 838–847 [DOI] [PubMed] [Google Scholar]
- 3. Garber K. (2014) Bispecific antibodies rise again. Nat. Rev. Drug Discov. 13, 799–801 [DOI] [PubMed] [Google Scholar]
- 4. Diamantis N., and Banerji U. (2016) Antibody-drug conjugates: an emerging class of cancer treatment. Br. J. Cancer 114, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jarantow S. W., Bushey B. S., Pardinas J. R., Boakye K., Lacy E. R., Sanders R., Sepulveda M. A., Moores S. L., and Chiu M. L. (2015) Impact of cell-surface antigen expression on target engagement and function of an epidermal growth factor receptor × c-MET bispecific antibody. J. Biol. Chem. 290, 24689–24704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mazor Y., Oganesyan V., Yang C., Hansen A., Wang J., Liu H., Sachsenmeier K., Carlson M., Gadre D. V., Borrok M. J., Yu X. Q., Dall'Acqua W., Wu H., and Chowdhury P. S. (2015) Improving target cell specificity using a novel monovalent bispecific IgG design. MAbs 7, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J. M., Lee S. H., Hwang J. W., Oh S. J., Kim B., Jung S., Shim S. H., Lin P. W., Lee S. B., Cho M. Y., Koh Y. J., Kim S. Y., Ahn S., Lee J., Kim K. M., et al. (2016) Novel strategy for a bispecific antibody: induction of dual target internalization and degradation. Oncogene 35, 4437–4446 [DOI] [PubMed] [Google Scholar]
- 8. Wolf E., Hofmeister R., Kufer P., Schlereth B., and Baeuerle P. A. (2005) BiTEs: bispecific antibody constructs with unique anti-tumor activity. Drug Discov. Today 10, 1237–1244 [DOI] [PubMed] [Google Scholar]
- 9. Ritchie M., Tchistiakova L., and Scott N. (2013) Implications of receptor-mediated endocytosis and intracellular trafficking dynamics in the development of antibody drug conjugates. MAbs 5, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perez H. L., Cardarelli P. M., Deshpande S., Gangwar S., Schroeder G. M., Vite G. D., and Borzilleri R. M. (2014) Antibody-drug conjugates: current status and future directions. Drug Discov. Today 19, 869–881 [DOI] [PubMed] [Google Scholar]
- 11. Sierra J. R., and Tsao M. S. (2011) c-MET as a potential therapeutic target and biomarker in cancer. Ther. Adv. Med. Oncol. 3, S21–S35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholson R. I., Gee J. M., and Harper M. E. (2001) EGFR and cancer prognosis. Eur. J. Cancer 37, S9–S15 [DOI] [PubMed] [Google Scholar]
- 13. Yewale C., Baradia D., Vhora I., Patil S., and Misra A. (2013) Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials 34, 8690–8707 [DOI] [PubMed] [Google Scholar]
- 14. Prat M., Oltolina F., and Basilico C. (2014) Monoclonal antibodies against the MET/HGF receptor and its ligand: multitask tools with applications form basic research to therapy. Biomedicines 2, 359–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma N., and Adjei A. A. (2011) In the clinic: ongoing clinical trials evaluating c-MET-inhibiting drugs. Ther. Adv. Med. Oncol. 3, S37–S50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., et al. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 17. Bean J., Brennan C., Shih J. Y., Riely G., Viale A., Wang L., Chitale D., Motoi N., Szoke J., Broderick S., Balak M., Chang W. C., Yu C. J., Gazdar A., Pass H., et al. (2007) MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. U.S.A. 104, 20932–20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Troiani T., Martinelli E., Napolitano S., Vitagliano D., Ciuffreda L. P., Costantino S., Morgillo F., Capasso A., Sforza V., Nappi A., De Palma R., D'Aiuto E., Berrino L., Bianco R., and Ciardiello F. (2013) Increased TGF-α as a mechanism of acquired resistance to the anti-EGFR inhibitor cetuximab through EGFR-MET interaction and activation of MET signaling in colon cancer cells. Clin. Cancer Res. 19, 6751–6765 [DOI] [PubMed] [Google Scholar]
- 19. Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., et al. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bardelli A., Corso S., Bertotti A., Hobor S., Valtorta E., Siravegna G., Sartore-Bianchi A., Scala E., Cassingena A., Zecchin D., Apicella M., Migliardi G., Galimi F., Lauricella C., Zanon C., et al. (2013) Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 3, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liska D., Chen C. T., Bachleitner-Hofmann T., Christensen J. G., and Weiser M. R. (2011) HGF rescues colorectal cancer cells from EGFR inhibition via MET activation. Clin. Cancer Res. 17, 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim Y. J., Choi J. S., Seo J., Song J. Y., Lee S. E., Kwon M. J., Kwon M. J., Kundu J., Jung K., Oh E., Shin Y. K., and Choi Y. L. (2014) MET is a potential target for use in combination therapy with EGFR inhibition in triple-negative/basal-like breast cancer. Int. J. Cancer 134, 2424–2436 [DOI] [PubMed] [Google Scholar]
- 23. Castoldi R., Ecker V., Wiehle L., Majety M., Busl-Schuller R., Asmussen M., Nopora A., Jucknischke U., Osl F., Kobold S., Scheuer W., Venturi M., Klein C., Niederfellner G., and Sustmann C. (2013) A novel bispecific EGFR/Met antibody blocks tumor-promoting phenotypic effects induced by resistance to EGFR inhibition and has potent antitumor activity. Oncogene 32, 5593–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spiess C., Merchant M., Huang A., Zheng Z., Yang N. Y., Peng J., Ellerman D., Shatz W., Reilly D., Yansura D. G., and Scheer J. M. (2013) Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat. Biotechnol. 31, 753–758 [DOI] [PubMed] [Google Scholar]
- 25. Lee B. S., Kim H. J., Hwang J. W., Cheong K. H., Kim K. A., Cha H. Y., Lee J. M., and Kim C. H. (2016) The dual inhibition of Met and EGFR by ME22S, a novel Met/EGFR bispecific monoclonal antibody, suppresses the proliferation and invasion of laryngeal cancer. Ann. Surg. Oncol. 23, 2046–2053 [DOI] [PubMed] [Google Scholar]
- 26. Castoldi R., Jucknischke U., Pradel L. P., Arnold E., Klein C., Scheiblich S., Niederfellner G., and Sustmann C. (2012) Molecular characterization of novel trispecific ErbB-cMet-IGF1R antibodies and their antigen-binding properties. Protein Eng. Des. Sel. 25, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis S. M., Wu X., Pustilnik A., Sereno A., Huang F., Rick H. L., Guntas G., Leaver-Fay A., Smith E. M., Ho C., Hansen-Estruch C., Chamberlain A. K., Truhlar S. M., Conner E. M., Atwell S., et al. (2014) Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat. Biotechnol. 32, 191–198 [DOI] [PubMed] [Google Scholar]
- 28. Merchant M., Ma X., Maun H. R., Zheng Z., Peng J., Romero M., Huang A., Yang N. Y., Nishimura M., Greve J., Santell L., Zhang Y. W., Su Y., Kaufman D. W., Billeci K. L., et al. (2013) Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent. Proc. Natl. Acad. Sci. U.S.A. 110, E2987–E2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L., Zeng W., Wortinger M. A., Yan S. B., Cornwell P., Peek V. L., Stephens J. R., Tetreault J. W., Xia J., Manro J. R., Credille K. M., Ballard D. W., Brown-Augsburger P., Wacheck V., Chow C. K., et al. (2014) LY2875358, a neutralizing and internalizing anti-MET bivalent antibody, inhibits HGF-dependent and HGF-independent MET activation and tumor growth. Clin. Cancer Res. 20, 6059–6070 [DOI] [PubMed] [Google Scholar]
- 30. Oh Y. M., Song Y. J., Lee S. B., Jeong Y., Kim B., Kim G. W., Kim K. E., Lee J. M., Cho M. Y., Choi J., Nam D. H., Song P. H., Cheong K. H., and Kim K. A. (2012) A new anti-c-Met antibody selected by a mechanism-based dual-screening method: therapeutic potential in cancer. Mol. Cells 34, 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cunningham D., Humblet Y., Siena S., Khayat D., Bleiberg H., Santoro A., Bets D., Mueser M., Harstrick A., Verslype C., Chau I., and Van Cutsem E. (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 351, 337–345 [DOI] [PubMed] [Google Scholar]
- 32. Robinson M. K., Hodge K. M., Horak E., Sundberg A. L., Russeva M., Shaller C. C., von Mehren M., Shchaveleva I., Simmons H. H., Marks J. D., and Adams G. P. (2008) Targeting ErbB2 and ErbB3 with a bispecific single-chain Fv enhances targeting selectivity and induces a therapeutic effect in vitro. Br. J. Cancer 99, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazor Y., Hansen A., Yang C., Chowdhury P. S., Wang J., Stephens G., Wu H., and Dall'Acqua W. F. (2015) Insights into the molecular basis of a bispecific antibody's target selectivity. MAbs 7, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muda M., Gross A. W., Dawson J. P., He C., Kurosawa E., Schweickhardt R., Dugas M., Soloviev M., Bernhardt A., Fischer D., Wesolowski J. S., Kelton C., Neuteboom B., and Hock B. (2011) Therapeutic assessment of SEED: a new engineered antibody platform designed to generate mono- and bispecific antibodies. Protein Eng. Des. Sel. 24, 447–454 [DOI] [PubMed] [Google Scholar]
- 35. Davis J. H., Aperlo C., Li Y., Kurosawa E., Lan Y., Lo K. M., and Huston J. S. (2010) SEEDbodies: fusion proteins based on strand-exchange engineered domain (SEED) CH3 heterodimers in an Fc analogue platform for asymmetric binders or immunofusions and bispecific antibodies. Protein Eng. Des. Sel. 23, 195–202 [DOI] [PubMed] [Google Scholar]
- 36. Li S., Schmitz K. R., Jeffrey P. D., Wiltzius J. J., Kussie P., and Ferguson K. M. (2005) Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 7, 301–311 [DOI] [PubMed] [Google Scholar]
- 37. Schmiedel J., Blaukat A., Li S., Knöchel T., and Ferguson K. M. (2008) Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer Cell 13, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., and Baker D. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 39. Kortemme T., Morozov A. V., and Baker D. (2003) An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326, 1239–1259 [DOI] [PubMed] [Google Scholar]
- 40. Lazaridis T., and Karplus M. (1999) Effective energy function for proteins in solution. Proteins 35, 133–152 [DOI] [PubMed] [Google Scholar]
- 41. Dantas G., Corrent C., Reichow S. L., Havranek J. J., Eletr Z. M., Isern N. G., Kuhlman B., Varani G., Merritt E. A., and Baker D. (2007) High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J. Mol. Biol. 366, 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hust M., Meyer T., Voedisch B., Rülker T., Thie H., El-Ghezal A., Kirsch M. I., Schütte M., Helmsing S., Meier D., Schirrmann T., and Dübel S. (2011) A human scFv antibody generation pipeline for proteome research. J. Biotechnol. 152, 159–170 [DOI] [PubMed] [Google Scholar]
- 43. Hust M., Dübel S., and Schirrmann T. (2007) Selection of recombinant antibodies from antibody gene libraries. Methods Mol. Biol. 408, 243–255 [DOI] [PubMed] [Google Scholar]
- 44. Rakestraw J. A., Sazinsky S. L., Piatesi A., Antipov E., and Wittrup K. D. (2009) Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol. Bioeng. 103, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benatuil L., Perez J. M., Belk J., and Hsieh C. M. (2010) An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 23, 155–159 [DOI] [PubMed] [Google Scholar]
- 46. Wang J., Goetsch L., Tucker L., Zhang Q., Gonzalez A., Vaidya K. S., Oleksijew A., Boghaert E., Song M., Sokolova I., Pestova E., Anderson M., Pappano W. N., Ansell P., Bhathena A., et al. (2016) Anti-c-Met monoclonal antibody ABT-700 breaks oncogene addiction in tumors with MET amplification. BMC Cancer 16, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Basilico C., Hultberg A., Blanchetot C., de Jonge N., Festjens E., Hanssens V., Osepa S. I., De Boeck G., Mira A., Cazzanti M., Morello V., Dreier T., Saunders M., de Haard H., and Michieli P. (2014) Four individually druggable MET hotspots mediate HGF-driven tumor progression. J. Clin. Invest. 124, 3172–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bird R. E., and Walker B. W. (1991) Single chain antibody variable regions. Trends Biotechnol. 9, 132–137 [DOI] [PubMed] [Google Scholar]
- 49. Hyatt D. C., and Ceresa B. P. (2008) Cellular localization of the activated EGFR determines its effect on cell growth in MDA-MB-468 cells. Exp. Cell Res. 314, 3415–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Melosky B., Burkes R., Rayson D., Alcindor T., Shear N., and Lacouture M. (2009) Management of skin rash during EGFR-targeted monoclonal antibody treatment for gastrointestinal malignancies: Canadian recommendations. Curr. Oncol. 16, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dickgiesser S., Rasche N., Nasu D., Middel S., Hörner S., Avrutina O., Diederichsen U., and Kolmar H. (2015) Self-assembled hybrid aptamer-Fc conjugates for targeted delivery: a modular chemoenzymatic approach. ACS Chem. Biol. 10, 2158–2165 [DOI] [PubMed] [Google Scholar]
- 52. Mullard A. (2013) Maturing antibody-drug conjugate pipeline hits 30. Nat. Rev. Drug Discov. 12, 329–332 [DOI] [PubMed] [Google Scholar]
- 53. Muller P. Y., and Milton M. N. (2012) The determination and interpretation of the therapeutic index in drug development. Nat. Rev. Drug Discov. 11, 751–761 [DOI] [PubMed] [Google Scholar]
- 54. Peng Z., Zhu Y., Wang Q., Gao J., Li Y., Li Y., Ge S., and Shen L. (2014) Prognostic significance of MET amplification and expression in gastric cancer: a systematic review with meta-analysis. PLoS One 9, e84502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jo M., Stolz D. B., Esplen J. E., Dorko K., Michalopoulos G. K., and Strom S. C. (2000) Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J. Biol. Chem. 275, 8806–8811 [DOI] [PubMed] [Google Scholar]
- 56. Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z. R., Du J., Davis A., Mongare M. M., Gould J., Frederick D. T., Cooper Z. A., Chapman P. B., Solit D. B., Ribas A., Lo R. S., Flaherty K. T., Ogino S., Wargo J. A., and Golub T. R. (2012) Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lacouture M. E. (2006) Mechanisms of cutaneous toxicities to EGFR inhibitors. Nat. Rev. Cancer 6, 803–812 [DOI] [PubMed] [Google Scholar]
- 58. Kimura H., Sakai K., Arao T., Shimoyama T., Tamura T., and Nishio K. (2007) Antibody-dependent cellular cytotoxicity of cetuximab against tumor cells with wild-type or mutant epidermal growth factor receptor. Cancer Sci. 98, 1275–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reilly E. B., Phillips A. C., Buchanan F. G., Kingsbury G., Zhang Y., Meulbroek J. A., Cole T. B., DeVries P. J., Falls H. D., Beam C., Gu J., Digiammarino E. L., Palma J. P., Donawho C. K., Goodwin N. C., and Scott A. M. (2015) Characterization of ABT-806, a humanized tumor-specific anti-EGFR monoclonal antibody. Mol. Cancer Ther. 14, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 60. Phillips A. C., Boghaert E. R., Vaidya K. S., Mitten M. J., Norvell S., Falls H. D., DeVries P. J., Cheng D., Meulbroek J. A., Buchanan F. G., McKay L. M., Goodwin N. C., and Reilly E. B. (2016) ABT-414, an antibody-drug conjugate targeting a tumor-selective EGFR epitope. Mol. Cancer Ther. 15, 661–669 [DOI] [PubMed] [Google Scholar]
- 61. Mendelsohn J., and Baselga J. (2003) Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J. Clin. Oncol. 21, 2787–2799 [DOI] [PubMed] [Google Scholar]
- 62. Sengers B. G., McGinty S., Nouri F. Z., Argungu M., Hawkins E., Hadji A., Weber A., Taylor A., and Sepp A. (2016) Modelling bispecific monoclonal antibody interaction with two cell membrane targets indicates the importance of surface diffusion. MAbs 8, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zheng S., Moores S., Jarantow S., Pardinas J., Chiu M., Zhou H., and Wang W. (2016) Cross-arm binding efficiency of an EGFR × c-Met bispecific antibody. MAbs 8, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rhoden J. J., Dyas G. L., and Wroblewski V. J. (2016) A modeling and experimental investigation of the effects of antigen density, binding affinity, and antigen expression ratio on bispecific antibody binding to cell surface targets. J. Biol. Chem. 291, 11337–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leal M., Sapra P., Hurvitz S. A., Senter P., Wahl A., Schutten M., Shah D. K., Haddish-Berhane N., and Kabbarah O. (2014) Antibody-drug conjugates: an emerging modality for the treatment of cancer. Ann. N.Y. Acad. Sci. 1321, 41–54 [DOI] [PubMed] [Google Scholar]
- 66. Wilson T. R., Fridlyand J., Yan Y., Penuel E., Burton L., Chan E., Peng J., Lin E., Wang Y., Sosman J., Ribas A., Li J., Moffat J., Sutherlin D. P., Koeppen H., et al. (2012) Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., and Varmus H. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tang Z., Du R., Jiang S., Wu C., Barkauskas D. S., Richey J., Molter J., Lam M., Flask C., Gerson S., Dowlati A., Liu L., Lee Z., Halmos B., Wang Y., et al. (2008) Dual MET-EGFR combinatorial inhibition against T790M-EGFR-mediated erlotinib-resistant lung cancer. Br. J. Cancer 99, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martin P., Stewart E., Pham N. A., Mascaux C., Panchal D., Li M., Kim L., Sakashita S., Wang D., Sykes J., Friess T., Shepherd F. A., Liu G., and Tsao M. S. (2016) Cetuximab inhibits T790M-mediated resistance to epidermal growth factor receptor tyrosine kinase inhibitor in a lung adenocarcinoma patient-derived xenograft mouse model. Clin. Lung Cancer 17, 375–383.e2 [DOI] [PubMed] [Google Scholar]
- 70. Balint R. F., and Larrick J. W. (1993) Antibody engineering by parsimonious mutagenesis. Gene 137, 109–118 [DOI] [PubMed] [Google Scholar]
- 71. Schwall R. H., and Tabor K. H. (April 10, 2001) Hepatocyte growth factor receptor antagonists and uses thereof. U. S. Patent 6,214,344 B1
- 72. Davies J., Liu L., Lu J., Vaillancourt P. E., Wortinger M. A., and Zeng W. (March 19, 2013) c-Met antibodies. U. S. Patent 8,398,974 B2
- 73. Goetsch L. (December 11, 2012) Antibodies inhibiting c-Met dimerization and uses thereof. U. S. Patent 8,329,173 B2
- 74. Ridgway J. B., Presta L. G., and Carter P. (1996) “Knobs-into-holes” engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 9, 617–621 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.