Abstract
Cereblon (CRBN) is a substrate receptor of the E3 ubiquitin ligase complex that is highly conserved in animals and plants. CRBN proteins have been implicated in various biological processes such as development, metabolism, learning, and memory formation, and their impairment has been linked to autosomal recessive non-syndromic intellectual disability and cancer. Furthermore, human CRBN was identified as the primary target of thalidomide teratogenicity. Data on functional analysis of CRBN family members in vivo, however, are still scarce. Here we identify Ohgata (OHGT), the Drosophila ortholog of CRBN, as a regulator of insulin signaling-mediated growth. Using ohgt mutants that we generated by targeted mutagenesis, we show that its loss results in increased body weight and organ size without changes of the body proportions. We demonstrate that ohgt knockdown in the fat body, an organ analogous to mammalian liver and adipose tissue, phenocopies the growth phenotypes. We further show that overgrowth is due to an elevation of insulin signaling in ohgt mutants and to the down-regulation of inhibitory cofactors of circulating Drosophila insulin-like peptides (DILPs), named acid-labile subunit and imaginal morphogenesis protein-late 2. The two inhibitory proteins were previously shown to be components of a heterotrimeric complex with growth-promoting DILP2 and DILP5. Our study reveals OHGT as a novel regulator of insulin-dependent organismic growth in Drosophila.
Keywords: CRISPR/Cas, development, Drosophila genetics, E3 ubiquitin ligase, insulin, Cereblon
Introduction
Body size and shape of animals are determined by genetic and environmental factors during development. The insulin/insulin-like growth factor (IGF)4 signaling (IIS) pathway is an evolutionarily conserved pathway that plays a key role in coupling body size and shape to extrinsic cues such as nutrient availability. Genetic and physiological studies using Drosophila melanogaster have contributed to provide information about how insulin signaling controls nutrients use, cell proliferation, and cell size to regulate final organismal size (1, 2).
In Drosophila, eight insulin-like peptides called DILPs function as ligands of a common insulin-like receptor (InR), initiating the signal cascade that resembles the vertebrate IIS pathway. The activated InR stimulates downstream effectors including phosphoinositide 3-kinase (PI3K), serine-threonine kinase Akt, and forkhead transcription factor FOXO (1, 2). Functional defects or altered expression of the pathway's component affects both cell size and cell number, leading to altered body and organ size (3–5).
At least four DILPs (DILPs 1, 2, 3, and 5) are produced in the two symmetric clusters of neurosecretory cells in the brain hemispheres called the insulin-producing cells (IPCs) (5, 6). The IPCs utilize endocrine mechanisms to couple DILP secretion with nutritional conditions. Previous studies indicate that the functional homolog of vertebrate liver and adipose tissue, called the fat body, senses nutrients and relays humoral signals to the IPCs (7, 8). Changes in nutrient influx into fat body cells result in altered expression of genes encoding humoral factors that are involved in systemic insulin signaling regulation (9–16). However, the details of how the fat body converts nutritional inputs into output signals still remain largely elusive.
Cereblon (CRBN) is a protein that is conserved across the animal and plant kingdoms. Human CRBN was first identified as a candidate gene for onset of an intellectual disability (17). Up to now, CRBN orthologs have been implicated in a variety of biological phenomena including development (18), metabolism (19), cytoprotective response against extracellular stresses (20, 21), and cancer cell biology (22–25) (for a review, see Ref. 26).
CRBN is a substrate receptor of the Cullin4-RING E3 ubiquitin ligase (CRL4) complex (18). CRL4 is a member of the largest E3 ligase family in eukaryotes, and it consists of a core catalytic complex (RBX1-Cullin4-DNA damage-binding protein 1 (DDB1) and a subset of substrate-specific receptors (27).
CRBN has gained prominence as the direct target of thalidomide and its derivatives (18, 22, 28). The presence of these molecules antagonizes endogenous substrate(s) in a mutually exclusive manner (29), and simultaneously CRBN promotes recruitment of neomorphic substrates (23, 24, 30, 31).
Previous studies indicate that CRBN is widely expressed throughout the body and is localized in the nucleus (18), cytoplasm (18) and mitochondria (21). In the nuclear compartment, CRBN regulates transcription by modulating protein levels of transcription factors (23, 32). In the cytoplasmic compartment, CRBN maintains intracellular glutamine concentration by targeting glutamine synthetase (33). We previously reported that exogenous CRBN partially localizes to mitochondria, and overexpressing CRBN with mitochondrial targeting signal shows a cytoprotective effect against oxidative stress (21).
CRBN was identified as a regulator of the intracellular energy sensor AMP-activated protein kinase (AMPK) (34). Loss of CRBN enhances phosphorylation and activation of AMPK both in mammalian culture cells and in murine liver (19, 34).
Mutations or inhibition of CRBN in vertebrates has been implicated in developmental or metabolic phenotypes. In human, a homozygous nonsense mutation that causes truncation at the C terminus of the product (R419X) has been identified from patients associated with a mild type of intellectual disability (17). In zebrafish, zcrbn knockdown by using morpholino antisense oligonucleotides leads to limb and otic vesicle malformation (18). In contrast, crbn-null mice do not show morphological anomalies; instead they are resistant against high fat diet challenge, characterized by less accumulation of fats in the epididymal tissues and liver and improved glucose homeostasis and insulin sensitivity (19). Despite recent progress, functions and regulation of CRBN in vivo are not fully elucidated.
To gain more insight into physiological functions of CRBN, we decided to utilize a genetically tractable model organism, Drosophila. Here we show for the first time that Ohgata (OHGT), the Drosophila ortholog of CRBN, is a novel regulator of insulin signaling-mediated growth. The ohgt mutant flies show increased body and organ size due to elevation of insulin signaling. Furthermore, we demonstrate that genetic inhibition of ohgt in the fat body phenocopies the mutant phenotype. Down-regulation of genes encoding inhibitory cofactors of DILPs in the circulation is associated with loss of ohgt. Our study provides a novel implication of a link between OHGT and insulin signaling-dependent growth in Drosophila.
Experimental Procedures
Antibodies
To generate anti-OHGT(1–187), a peptide corresponding to amino acids 1–187 of OHGT (OHGT(1–187)) with a glutathione S-transferase (GST) at the N terminus, was used as immunogen in rabbits (Pineda Antibody Service, Berlin, Germany). To generate anti-OHGT(15–33), a synthetic peptide corresponding to amino acid 15–33 of OHGT with a cysteine residue (OHGT(15–33)-Cys; RDEDVQLEDQQSQGLQDRQC), was covalently coupled to the carrier protein keyhole limpet hemocyanin, and it was used as immunogen in guinea pigs (Pineda Antibody Service). Antibodies were purified from antisera with OHGT(1–187)- or OHGT(15–33)-Cys-conjugated affinity columns prior to use. Specificity of the antibodies was tested by Western blotting. Anti-OHGT(1–187) and anti-OHGT(15–33) were used at dilutions of 1:1,000 and 1:100, respectively.
Anti-α-spectrin (3A9 developed by Drs. D. Branton and R. Dubreuil) (1:10) was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health, and maintained at the University of Iowa, Department of Biology. Anti-DILP2 (1:500) was a kind gift from Dr. M. Pankratz. The rest of the antibodies were purchased from the indicated manufacturers: anti-actin (1:500) from Sigma Aldrich, anti-phospho-Akt (1:1,000) from Cell Signaling Technology, anti-DDB1 (1:100) from Abcam, anti-ubiquitin (1:1,000) from Life Sensors, anti-GFP (1:600) and horseradish peroxidase-conjugated secondary antibodies (1:15,000) from Santa Cruz Biotechnology, and fluorophore-conjugated secondary antibodies (dilutions according to the manufacturer's instructions) from Life Technologies.
Fly Stocks
Flies were raised on standard fly food at 25 °C if not mentioned otherwise. The following fly lines were used in the experiments: w* (as a control), vasa-Cas9 (Bloomington stock number 51323); DrMio/TM3, Sb1, Ser1, eGFP (referred to as TM3-GFP balancer line; Bloomington stock number 6663); Df(3R)Exel6155/TM6B (referred to as ohgtDf; Bloomington stock number 7634); Cg-Gal4 (Bloomington stock number 7011); dilp2-Gal4 (Bloomington stock number 37516); FB-Gal4 (a kind gift from Dr. R. Kuehnlein); y,hs-FLP; UAS-Dicer2; act-FRT-CD2-FRT-Gal4,UAS-GFP/TM6b,Tb,Hu (a stock established in the group using a line described previously (35); UAS-Dcr2 (Bloomington stock number 24650); UAS-GFP.nls (Bloomington stock number 4775); UAS-ohgtRNAiv40486 (Vienna Drosophila Resource Center stock number 40486); UAS-ohgtRNAiNIG3 (National Institute of Genetics Fly stock number 3925R-3), UAS-picRNAi (Kyoto Drosophila Genomics Resource Center stock number 109605); and UAS-dCullin4RNAi (Vienna Drosophila Resource Center stock number 44829).
Generation of an ohgt Loss of Function Allele
To generate a fly line with the ohgt loss of function allele, clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9-mediated targeted mutagenesis using vasa-Cas9 was performed (36). In brief, we searched for potential target sequences within ohgt locus using the CRISPR Optimal Target Finder. Among candidates, a sense sequence within exon 2 (GCAGGACGACACAGCAA) was chosen as the target. An oligonucleotide pair containing the target sequence (F, 5′-CTTCGCAGGACGACACAGCAAGCG-3′; R, 5′-AAACCGCTTGCTGTGTCGTCCTGC-3′) was 5′-phosphorylated, annealed, and subsequently inserted into pU6-BbsI-chiRNA vector via BbsI sites. We named the generated plasmid pU6-ohgtEx2-gRNA.
The pU6-ohgtEx2-gRNA was injected into vasa-Cas9 preblastoderm embryos at 250 ng/μl according to standard protocols. Injected animals were raised on standard fly food at 25 °C. The eclosed flies were collected and crossed with TM3-GFP balancer flies. Once enough progenies from each cross were obtained, candidates were sacrificed to obtain their genomic DNAs by homogenizing in squishing buffer (10 mm Tris-HCl, pH 8.2, 1 mm EDTA, 25 mm NaCl, and proteinase K). Using the genomic DNAs, targeted modifications in each sample were screened by high resolution melt analysis (HRMA) (37) using Precision Melt Supermix reagent (Bio-Rad), and the following primer pair: F, 5′-ATGCCTTCCAAGATCCACTG-3′; R, 5′-TGTCACTGCTGGGATGACTG-3′. These processes were repeated for another generation to assess germ line transmission of the modification. Amplicons produced during HRMA were cloned using the TOPO TA cloning kit (Thermo Fisher) to characterize the modification.
Developmental Timing Assay
100 larvae from 2-h egg collections were transferred to apple agar plates with yeast paste at 24 h after egg deposition (AED) (25 larvae per plate). The number of pupae on each plate was counted every 12 h. Timing of pupariation was defined as the time point at which more than 50% of the larvae had pupated (38).
Average Weight, Length, Wing Area, and Cell Number Quantification
To determine average weights, staged animals were pooled in a 1.5-ml Eppendorf tube and weighed. Images of animals were obtained using an Olympus SZ12 binocular via SIS Analysis 2.1 software. Pupal lengths and wing areas were quantified using ImageJ (National Institutes of Health). To count cell numbers in the posterior compartment of the wing, numbers of hairs in 10,000 μm2 were counted because each hair is produced from a single epithelial cell. The cell density was multiplied by the area of the posterior compartment to calculate the number of cells in the compartment.
Food Intake Quantification
Larval food intake assay was performed as described previously (39).
Immunohistochemical Analysis of Drosophila Larval Tissues
Immunohistochemical samples of larval tissues were prepared, and their images were obtained using a confocal microscope (Zeiss LSM 710) as described previously (40).
Clonal analysis was performed as described previously (41). Virgin females of the genotype y,hs-FLP; UAS-Dicer2; act-FRT-CD2-FRT-Gal4,UAS-GFP/TM6b,Tb,Hu were crossed with males of the genotype UAS-ohgtRNAiv40486 to obtain larvae of the genotype y,hs-FLP/+; Dicer2/+; act-FRT-CD2-FRT-Gal4,UAS-GFP/UAS-ohgtRNAiv40486. Normalized staining intensities were measured via ImageJ. Nuclear and whole cell regions were marked, and the cytoplasmic regions were determined as whole cell region minus nuclear region. Pixel intensity values were normalized to the average wild type intensity per experiment. Nuclear to cytoplasm ratios were determined from raw data and normalized per experiment.
Sample Preparation and Western Blotting
Animals were washed with ice-cold PBS before sample preparation. To prepare the carcasses (larval bodies devoid of the fat body), larvae were dissected in ice-cold PBS, and their fat bodies were removed. Total animals or the carcasses were homogenized in ice-cold RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% IGEPAL CA-630 (Sigma), 0.5% sodium deoxycholate, and 0.1% SDS) containing Complete protease inhibitor mixture (Roche Applied Science). 5× Laemmli buffer with 10% β-mercaptoethanol was added to the lysates, and the mixtures were heated at 95 °C for 7 min.
To detect phosphorylated dAkt, at least 20 larvae or carcasses of each genotype were pooled and lysed in ice-cold RIPA buffer containing PhosStop phosphatase inhibitor (Roche Applied Science) and Complete protease inhibitor mixture. The homogenates were incubated at 75 °C for 10 min and centrifuged at 15,000 × g for 20 min. The supernatants after centrifugation were mixed with 5× Laemmli buffer with 10% β-mercaptoethanol. Western blotting was performed as described previously (42).
Quantitative Real Time PCR
Isolation of mRNA, synthesis of cDNA, and quantification by quantitative real time PCR (qRT-PCR) was performed as described previously (4). Approximately 10 animals were used to extract mRNA from total larvae, and 15–20 animals were used to extract mRNA from the carcasses or the fat body, respectively. The following primer pairs were used in the experiment: ohgt: F, 5′-AAGATGCCTTCCAAGATCCAC-3′; R, 5′-TCGCTTGCTGTGTCGTC-3′ (designed in this work); InR: F, 5′-AACAGTGGCGGATTCGGTT-3′; R, 5′-TACTCGGAGCATTGGAGGCAT-3′ (4); Thor (eIF-4E-binding protein (4EBP)): F, 5′-CATGCAGCAACTGCCAAATC-3′; R, 5′-CCGAGAGAACAAACAAGGTGG-3′ (4); Lipase 3 (Lip3): F, 5′-TGAGTACGGCAGCTACTTCCCT-3′; R, 5′-TCAACTTGCGGACATCGCT-3′ (4); acid-labile subunit (dALS): F, 5′-ATGCGGTGGCTGTCATGTC-3′; R, 5′-GGCAGCTTACCAAAGGCACTT-3′ (13); imaginal morphogenesis protein-late 2 (Imp-L2): F, 5′-AAGAGCCGTGGACCTGGTA-3′; R, 5′-TTGGTGAACTTGAGCCAGTCG-3′ (43); unpaired 2 (upd2): F, 5′-CGGAACATCACGATGAGCGAAT-3′; R, 5′-TCGGCAGGAACTTGTACTCG-3′ (9); ribosomal protein 49 (Rp49): F, 5′-GCTAAGCTGTCGCACAAATG-3′; R, 5′-GTTCGATCCGTAACCGATGT-3′ (4).
Cell Culture and Transfection
Drosophila Schneider 2 (S2) cells were grown in Schneider's Drosophila medium (Thermo Fisher) containing 10% fetal bovine serum and 1% penicillin-streptomycin at 25 °C. Transfection was performed using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. To generate a tagged OHGT expression vector, the cDNA coding for full-length OHGT protein with HA tag at the N terminus was amplified by PCR using the CG3925 cDNA clone (LD28592 from the Drosophila Genomics Resource Center) as a template with following primer pair: F, 5′-GGAATTCATGTACCCATACGACGTCCCAGACTACGCTGACGAAGAGGAGAAC-3′; R, 5′-CCGCTCGAGTCATTCCATATCGCTTGAGATC-3′. The cDNA was then inserted into pUASTattB backbone vector via EcoRI and XhoI sites. The generated expression vector was co-transfected with pAct-gal4 plasmid vector.
Co-immunoprecipitation
To immunoprecipitate HA-OHGT immunocomplex, S2 cells expressing HA-OHGT were lysed in ice-cold lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2.5 mm MgCl2, 5 mm EDTA, and 1% IGEPAL CA-630 with Complete protease inhibitor mixture). Co-immunoprecipitation was performed using Protein G-Sepharose (GE Healthcare) and 1 μg of anti-HA (3F10, Roche Applied Science) according to the manufacturers' instructions. For immunoprecipitating endogenous OHGT, the experiment was performed as described above with 2 μg of rabbit anti-OHGT(1–187).
In Vivo Ubiquitination Assay
S2 cells were treated with 25 μm MG132 overnight. The cells were lysed in ice-cold RIPA buffer with 5 mm N-ethylmaleimide (Sigma), 1 mm DTT, and Complete protease inhibitor mixture. After vigorous vortexing and sonication, the lysates were incubated for 30 min at 4 °C with constant rotation. Immunoprecipitation was performed using Protein G-Sepharose (GE healthcare) and rabbit anti-OHGT(1–187) according to the manufacturer's instructions.
3D Modeling of the OHGT-Piccolo Complex
3D models for the OHGT-Piccolo (PIC) complex were created using the SWISS-MODEL service (44). The models were subsequently repositioned to the Protein Data Bank code 4TZ4 model, and their images were obtained using UCSF Chimera (a molecular graphics program developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS, National Institutes of Health Grant P41-GM103311) (45).
Bioinformatics Analysis
The phylogenetic analysis was carried out by Bayesian inference (Markov chain Monte Carlo method, Jones amino acid model) via MrBayes (46–48). Analysis was carried out for eight chains until they converged (average standard deviation of split frequencies, 0.004868; potential scale reduction factor, 1.000; average effective sample size, 2432.32). The following bioinformatics tools were used: the ExPASy ScanProsite tool (for predicting phosphorylation sites and myristoylation sites (49, 50)), iUbiq-Lys (for predicting lysine ubiquitination sites (51, 52)), GPS-SUMO (for predicting lysine sumoylation sites and SUMO interaction motifs (53, 54)), PSORT II Prediction (for predicting subcellular localization), and NucPred (for predicting nuclear localization of the protein (55)).
Results
Cereblon Homologs Are Evolutionary Conserved
CRBN homologs are highly conserved in evolution as indicated by phylogenetic analysis (Fig. 1A), however most proteins are rather distant from each other. Especially the mammalian proteins seem to have split off rather early and there is a large set of modification between Caenorhabditis elegans and the insects. Also within the insects, the Drosophila proteins are rather remote from those of other insect species like Bombyx mori, Nasonia vitripennis, Apis mellifera, and Tribolium castaneum. They differ greatly in the N-terminal protein stretch, suggesting that they may also vary in function to some extent.
FIGURE 1.
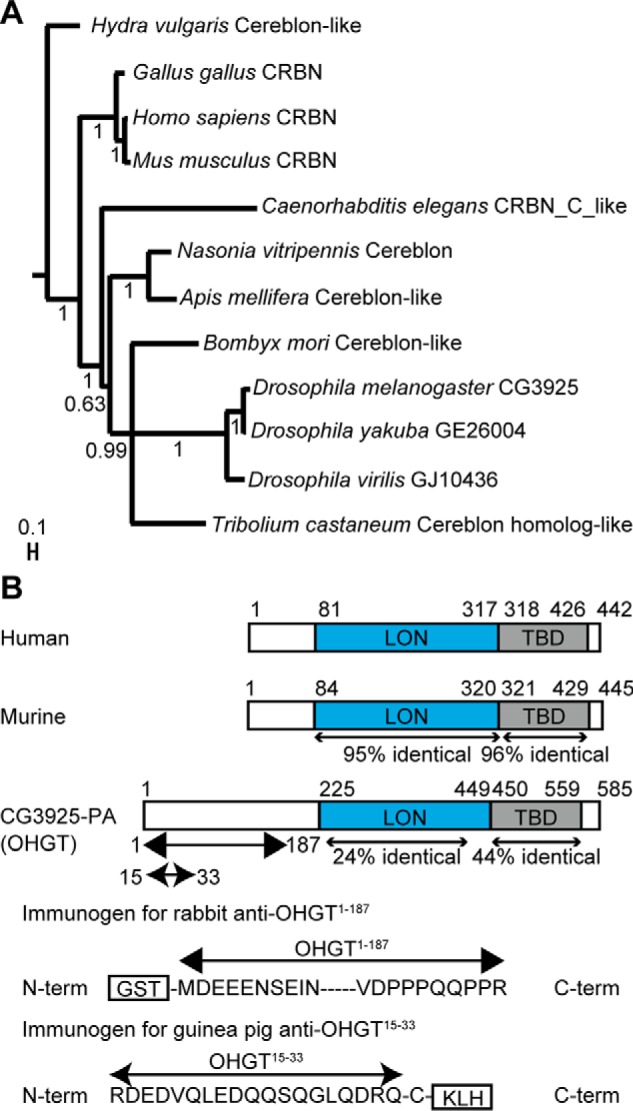
CRBN is conserved across the animal kingdom. A, phylogenetic tree of CRBN homologs. Depicted values indicate posterior probabilities. The scale bar references branch lengths. B, schemes representing the domain structure of human CRBN and its murine and Drosophila homologs (the structure of human CRBN is based on a previous study (64) and structures of murine and Drosophila homologs are based on those described in the Universal Protein Resource database (entries Q8C7D2 and Q9VH36, respectively). The two evolutionary conserved domains, LON domain and TBD, are indicated as filled boxes (blue and gray, respectively). Percentages of identity at the amino acid level are noted. Two immunogens used to generate OHGT-specific antibodies (rabbit anti-OHGT(1–187) and guinea pig anti-OHGT(15–33)) are represented. KLH, keyhole limpet hemocyanin.
A single CRBN family member in Drosophila is encoded by CG3925 (FlyBase ID FBgn0037780). Its putative protein product (CG3925-PA) contains two evolutionarily conserved domains characteristic for CRBN protein family members: a central LON protease-like (LON) domain that has been found to mediate interaction with the CRL4 adaptor protein DDB1 in mammals and a C-terminal thalidomide-binding domain (TBD) that mediates interaction with substrates and thalidomide-like molecules (each domain shows 24 and 44% identity with the counterparts of human CRBN, respectively) (Fig. 1B). A ubiquitination site (lysine residue at position 315), a SUMO interaction site in the N terminus (amino acid residues at positions 34–38), a sumoylation site (lysine residue at position 448), five myristoylation sites (amino acid residues at positions 87–92, 206–211, 259–264, and 549–554), and a cAMP- and cGMP-dependent protein kinase phosphorylation site (amino acid residues at positions 160–163) are predicted according to online bioinformatics tools (not presented in Fig. 1).
Targeted Mutagenesis of the Drosophila CRBN Ortholog
CG3925, the putative Drosophila ortholog of human CRBN gene, is located on chromosome 3R (cytological location, 85F8) and encompasses five exons (Fig. 2A). Only one annotated transcript (CG3925-RA) is reported in the FlyBase (56).
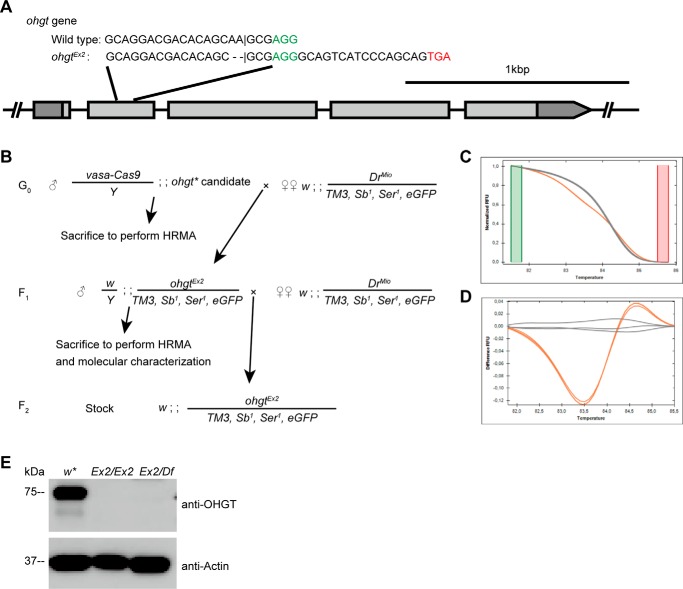
FIGURE 2.
Generating an ohgt mutant. A, a scheme representing the ohgt locus and the target site. A 20-bp-long sequence within exon 2 was chosen for targeting by the CRISPR-Cas9 system. The predicted cleavage site is indicated by a vertical bar. Two nucleotides deleted by Cas9 activity are indicated by horizontal bars (-). The protospacer adjacent motif (PAM) sequence is shown in green. The putative stop codon after the frameshift is shown in red. (B) A scheme to recover flies with ohgtEx2 allele. Microinjection survivors were crossed with TM3-GFP balancer flies and subsequently sacrificed to perform the HRMA-based mutagenesis screening. The F1 progenies from the cross were again crossed to the same strain, and further HRMA was performed to confirm germ line transmission of the mutation. The amplicon was inserted into pCRII-TOPO vector and subjected to molecular characterization. C, HRMA of an F1 mutant candidate. Melt curves of ohgtEx2/TM3-GFP (orange) and TM3-GFP balancer (gray) can be easily distinguished due to heteroduplex formation in candidate-derived sample. D, change in relative fluorescence units (RFU) relative to TM3-GFP balancer emphasizes the change of melt curve shown in C. E, total lysates were prepared from control (w*), homozygous ohgtEx2 (Ex2/Ex2), and transheterozygous ohgtEx2/ohgtDf (Ex2/Df) third instar larvae. Proteins from each lysate were subjected to Western blotting with antibodies to the indicated proteins.
To investigate the in vivo function of CG3925, we generated mutants by CRISPR-Cas9-mediated targeted mutagenesis (57). We utilized a method using vasa-Cas9, a transgenic fly line expressing Cas9 endonuclease in a germ line-specific manner (36). Among potential candidates, we chose a sense sequence within exon 2 of CG3925 as the target site (Fig. 2A). Because the evolutionarily conserved LON domain and TBD of CG3925-PA are encoded by exons 3–5, a 20-bp sequence within exon 2 was chosen for targeting. Using a mutagenesis screening strategy based on HRMA (Fig. 2, B–D), we successfully recovered flies with a Cas9-induced modification. Molecular characterization identified a mutation causing a frameshift in exon 2 of CG3925 in these animals. This would result in a putative truncated protein that contains only 15% of the full-length CG3925-PA protein (84 amino acid residues from the N terminus plus 7 amino acid residues (RGQSSQQ) induced by the frameshift). Because we later determined that this mutant allele leads to an overgrowth phenotype (see below), we propose to call the gene ohgata (meaning “large” in Japanese; abbreviated as ohgt) and the mutant allele ohgtEx2.
We raised two different sets of polyclonal OHGT-specific antibodies directed toward amino acid residues 1–187 and 15–33 (for details see “Experimental Procedures”; Fig. 1B) and could show that endogenous OHGT protein of ∼75-kDa size could not be detected from either ohgtEx2/Ex2 or transheterozygous (ohgtEx2 over a deficiency; referred to as ohgtEx2/Df) mutant larvae (Fig. 2E). This result provides evidence that full-length OHGT is absent in ohgtEx2/Ex2 mutant conditions and identifies ohgtEx2 as a strong allele.
ohgt Mutation Leads to Increased Organismal Size
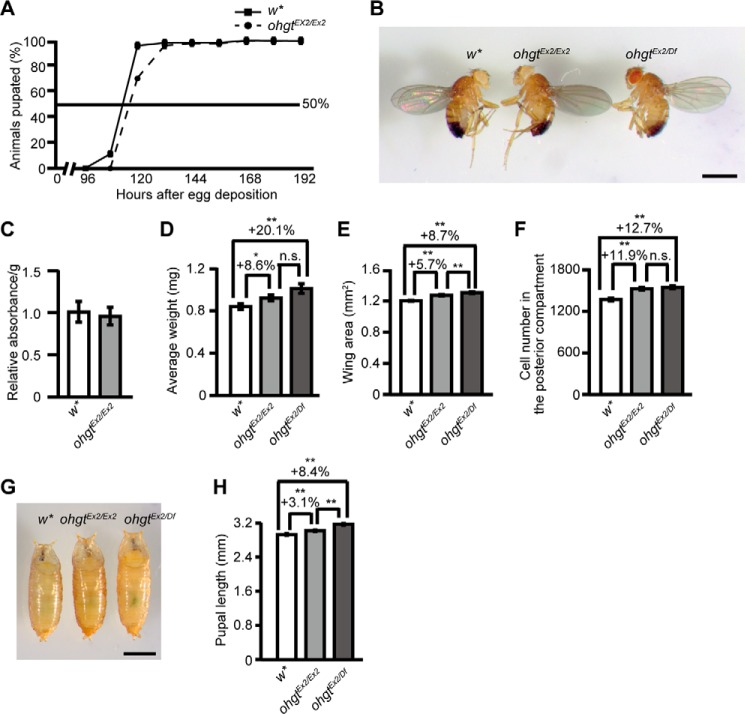
In an analysis of the phenotype of ohgtEx2/Ex2 mutants, we found that 30% of the homozygous animals showed a 12-h delay to pupate when raised on standard fly food (Fig. 3A). In most of the homozygous animals, however, developmental timing seems not to be majorly affected as compared with control animals. Furthermore, the homozygous ohgtEx2/Ex2 flies were viable and fertile. To our surprise, however, ohgtEx2/Ex2 flies were larger than the control animals (Fig. 3B). This is not due to increased food intake (Fig. 3C) as we showed by applying feeding assays (39). To further investigate the overgrowth phenotype, we quantified average wet weight and wing area of mutant male flies. Indeed, ohgtEx2/Ex2 flies showed a significant increase in average wet weight (Fig. 3D; p < 0.05) and wing area (Fig. 3E; p < 0.01). To test whether organ size also is affected, we analyzed the posterior compartment of the wing and found that the number of wing cells is increased (Fig. 3F; p < 0.01). The growth phenotype of ohgtEx2/Df flies is only slightly enhanced (Fig. 3, D–F; all p < 0.01), further confirming that ohgtEx2 is a strong allele (but most likely not a functional null) and that the overgrowth phenotype observed in ohgtEx2/Ex2 flies is not due to off-target effects of Cas9-mediated mutagenesis. Female mutant flies exhibited similar phenotypes (data not shown), suggesting that these overgrowth phenotypes are independent of gender. Consistent with the phenotypes at adult stage, ohgtEx2/Ex2 and ohgtEx2/Df pupae were significantly longer than control animals (Fig. 3, G and H; p < 0.01). Taken together, these findings identify the Drosophila CRBN family member ohgt as a new regulator of organismal growth and show that impairment of its function results in larger body and organ size associated with increased cell number.
FIGURE 3.
The ohgtEx2 mutants show overgrowth phenotype. A, percentage of w* and ohgtEx2 animals pupated at each time point after egg deposition. B, w*, ohgtEx2/Ex2, and ohgtEx2/ohgtDf (ohgtEx2/Df) adult male flies. The scale bar represents 1 mm. C, the ohgtEx2 larvae showed food intake similar to that of w* larvae (n = 4). D, average wet weight of w*, ohgtEx2/Ex2, and ohgtEx2/Df adult males. Animals were weighed in batches of 10, and average weight per animal was calculated (n = 10). E, average wing area of w*, ohgtEx2/Ex2, and ohgtEx2/Df adult males (n = 30). F, average number of cells in the posterior compartment of the wing of w*, ohgtEx2/Ex2, and ohgtEx2/Df adult males (n = 30). G, w*, ohgtEx2/Ex2, and ohgtEx2/Df pupae. An animal whose body length is closest to the average was chosen from each genotype. The scale bar represents 1 mm. H, average body length of w* (n = 66), ohgtEx2/Ex2 (n = 80), and ohgtEx2/Df (n = 73) pupae. Error bars indicate S.E. * indicates p < 0.05, and ** indicates p < 0.01. p values were calculated by Student's t test. n.s. indicates no significance.
ohgt Product Is Expressed in the Fat Body, and Its Knockdown Phenocopies the Overgrowth Phenotype of ohgt Mutants
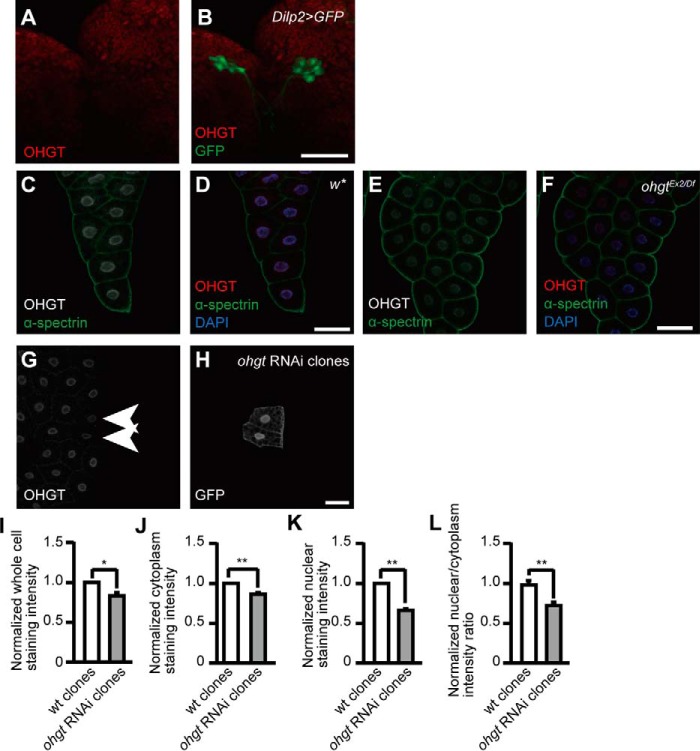
Organismal size of adult Drosophila flies is largely determined in the third instar and premetamorphic stages (2). The IPCs of the brain and the fat body, a peripheral endocrine tissue with mammalian adipose- and liver-like functions, play critical roles in linking growth regulation to nutrient conditions (7, 8). Immunohistochemical analysis using anti-OHGT antibodies indicates that, although OHGT seems not to be expressed in the IPCs (Fig. 4, A and B), we found a prominent expression in the fat body (Fig. 4, C and D) that is strongly reduced in the fat bodies of ohgtEx2/Df third instar larvae (Fig. 4, E and F) and in ohgt RNAi clones (Fig. 4, G–L). Subcellularly, we found OHGT to be localized almost exclusively to the nucleus, consistent with bioinformatics prediction algorithms for subcellular localization such as the Reinhardt NCNN algorithm and NucPred program, which predict nuclear and cytoplasmic localization based on the percentage of positively charged amino acids and similarity to other nuclear proteins.
FIGURE 4.
OHGT is expressed in the nucleus of fat body cells. A and B, co-immunostaining of OHGT and GFP in the IPCs of dilp2>GFP third instar larva. C and D, co-immunostaining of OHGT and plasma membrane marker α-spectrin in the fat body of w* early third instar larva (72 h AED). E and F, co-immunostaining of OHGT and α-spectrin in the fat body of ohgtEx2/Df early third instar larva (72h AED). G and H, co-immunostaining of OHGT and GFP after clonal ohgt RNAi induction in fat body cells. Genotype, y,hs-flp/+; Dcr-2/+; act-FRT-CD2-FRT-Gal4, UAS-GFP/UAS-ohgtRNAiv40486. The scale bars in images represent 50 μm. I–L, normalized staining intensities in whole cells, the cytoplasm, and the nucleus as well as the nuclear to cytoplasm ratio of wild type clones (n = 34) and ohgt RNAi induced clones (n = 17). Error bars indicate S.E. * indicates p < 0.05, and ** indicates p < 0.01. p values were calculated by Mann-Whitney U test.
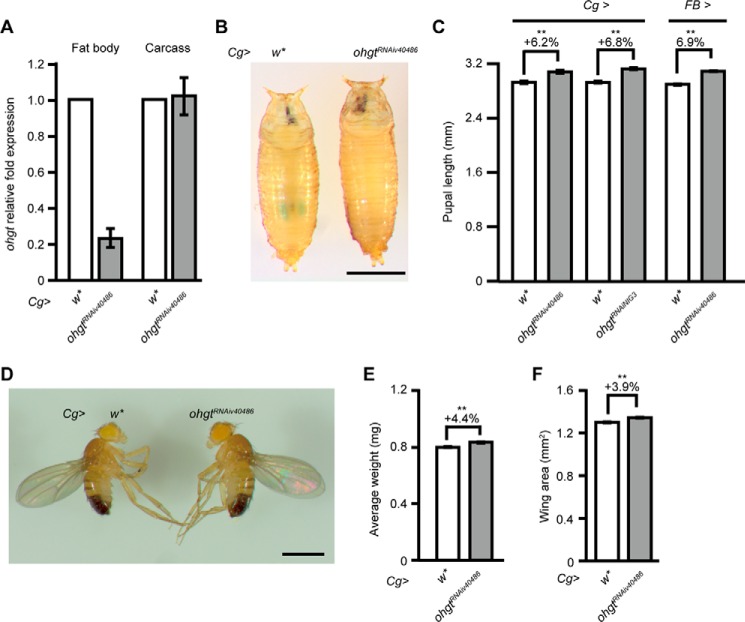
To address whether impairment of ohgt function in the fat body affects organismal size, we used an ohgt RNAi line (UAS-ohgtRNAiv40486) and a fat body driver line (Cg-Gal4) to specifically knock down ohgt in the fat body (Cg>ohgtRNAiv40486). Molecular analysis indicates that ohgt RNAi induction reduced ohgt transcripts in the fat body by about 80%, whereas its transcript level in the carcasses (larval bodies devoid of the fat body) was almost unaffected (Fig. 5A). Cg>ohgtRNAiv40486 animals showed increased pupal length (Fig. 5, B and C; p < 0.01) as well as increased adult body weight (Fig. 5, D and E; p < 0.01) and posterior wing area (Fig. 5F; p < 0.01), very similar to the overgrowth phenotypes of ohgtEx2/Ex2 animals (Fig. 3).
FIGURE 5.
Fat body-specific ohgt knockdown phenocopies the ohgt mutant phenotype. A, transcript levels of ohgt in the fat body and carcass of Cg>w* and Cg>ohgtRNAiv40486 third instar larvae were quantified using qRT-PCR (n = 4). B, Cg>w* and Cg>ohgtRNAiv40486 pupae. An animal whose length is closest to the average was chosen from each genotype. The scale bar represents 1 mm. C, average body length of Cg>w* (n = 48) versus Cg>ohgtRNAiv40486 (n = 66) pupae, Cg>w* (n = 40) versus Cg>ohgtRNAiNIG3 (n = 48) pupae, and FB>w* (n = 48) versus FB>ohgtRNAiv40486 (n = 52) pupae. D, Cg>w* and Cg>ohgtRNAiv40486 adult male flies. The scale bar represents 1 mm. E, average wet weight of Cg>w* and Cg>ohgtRNAiv40486 adult male flies. Animals were weighed in batches of 10, and average weight per animal was calculated (n = 8). F, average wing area of Cg>w* and Cg>ohgtRNAiv40486 adult males (n = 20). Error bars indicate S.E. * indicates p < 0.05, and ** indicates p < 0.01. Each p value was calculated by Student's t test. n.s. indicates no significance.
Using a different ohgt RNAi line (UAS-ohgtRNAiNIG3) or a different fat body driver line (FB-Gal4) also led to increased pupal length (Fig. 5C; both p < 0.01) and adult weight (not shown), suggesting that these phenotypes are due to the down-regulation of ohgt in the fat body. These results point to a specific function of ohgt in the fat body to control organismal size.
Loss of ohgt Causes Elevated Insulin Signaling
A major pathway involved in controlling body and organ size is the insulin-like signaling cascade (ILS), and we therefore examined whether ILS is altered in the ohgt mutants. ILS is activated by DILPs, which display distinct temporal and spatial expression patterns in the body. The major site of DILP production is the IPCs where dilp1, dilp2, dilp3, and dilp5 are expressed (6). ILS is mediated through the InR, and its activation eventually leads to induction of a kinase cascade involving PI3K and protein kinase B/Akt (58). This kinase cascade eventually phosphorylates the forkhead transcription factor FOXO, resulting in FOXO retention in the cytoplasm via binding to 14-3-3 proteins (59). Down-regulation of ILS results in increased FOXO dephosphorylation and its accumulation in the nucleus (60).
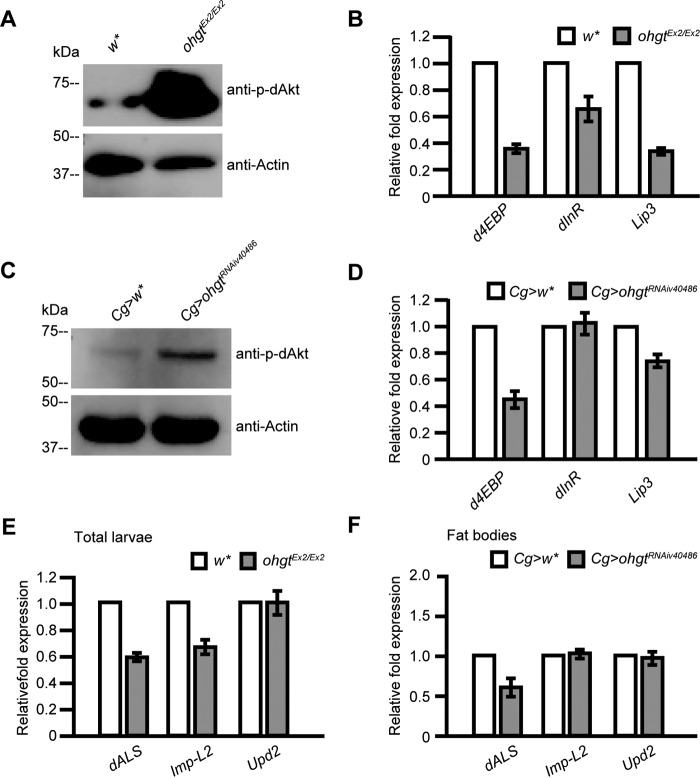
We first analyzed the phosphorylation level of dAkt, the central regulator of the pathway (61). Because dAkt is phosphorylated under active insulin signaling (62), the level of phospho-dAkt reflects activity of the signaling pathway. We subjected total protein from ohgtEx2/Ex2 third instar larvae to Western blotting with a phospho-Akt-specific antibody and found that the phospho-dAkt level was indeed increased in ohgtEx2/Ex2 larvae compared with age-matched controls (Fig. 6A). A major role of dAkt is to regulate the nuclear localization of dFOXO (60). We therefore quantified expression levels of two dFOXO target genes, d4EBP and dInR, in ohgt mutants by using qRT-PCR. The transcription of both genes is up-regulated when dFOXO is nuclear (low ILS), whereas their expression is reduced when dFOXO is cytoplasmic (high ILS activity) (60). We found that both d4EBP and dInR were strongly down-regulated in ohgtEx2/Ex2 third instar larvae compared with control animals (Fig. 6B), consistent with the increased phospho-dAkt level in the mutants. Similar results were also obtained by tissue-specific inhibition of ohgt in the fat body using Cg>ohgtRNAiv40486 third instar larvae (Fig. 6, C and D). From these results, we conclude that OHGT activity in the fat body regulates organismal growth through negatively regulating insulin signaling.
FIGURE 6.
The ohgt mutation and ohgt knockdown in the fat body are associated with insulin signaling up-regulation and altered expression of fat body-derived cofactors of DILPs. A, the expression level of phospho-dAkt (p-dAkt) was increased in ohgtEx2/Ex2 compared with w*. The experiment was performed in triplicate. B, transcription of the genes d4EBP, dInR, and Lip3 (included as a starvation marker) in ohgtEx2/Ex2 and w* (n = 4). C, the expression level of phospho-dAkt was increased in Cg>ohgtRNAiv40486 compared with Cg>w* early third instar larval carcasses. The experiment was performed in triplicate. D, transcription of d4EBP, dInR, and Lip3 in Cg>ohgtRNAiv40486 and Cg>w* in early third instar larval carcasses (n = 4). E, transcription of the genes dALS, Imp-L2, and upd2 in the total ohgtEx2/Ex2 and w* early third instar larvae was quantified (n = 4). F, transcription of dALS, Imp-L2, and upd2 in the fat bodies of Cg>ohgtRNAiv40486 and Cg>w* early third instar larvae was quantified (n = 4). Error bars indicate S.E.
Genes Encoding DILPs Cofactors Are Down-regulated in ohgt Mutants
Several proteins have been identified in mammals and insects that interact with IGFs and ILPs after their release into circulation. Among these are two inhibitory cofactors of circulating DILPs named dALS and IMP-L2. The two inhibitory proteins are components of a heterotrimeric complex with DILP2 and DILP5 (13, 14). In larvae, dALS is expressed in the IPCs and in the fat body, and it was shown to functionally antagonize the role of DILPs in controlling body growth as well as the metabolism of carbohydrates and lipids (13). IMP-L2 is already expressed in the cellular blastoderm stage and can be observed in several cell types throughout development (63). Its expression becomes abundant at the onset of metamorphosis, reaching maximal levels in pupae ∼14 h after puparium formation (14). IMP-L2 was shown to counteract ILS and to be essential for tolerance to starvation stress (14). IMP-L2 was also shown to be capable of interacting with some of the DILPs and is proposed to act as a secreted antagonist of ILS in Drosophila.
To address whether reduction of ohgt affects the expression levels of dALS and Imp-L2, we performed qRT-PCR experiments using cDNAs synthesized from total ohgtEx2/Ex2 third instar larvae and cDNAs synthesized from the fat body of Cg>ohgtRNAiv40486 third instar larvae. We found that Imp-L2 was reduced only in the total fraction, and dALS was reduced in both total and fat body-specific fractions (Fig. 6, E and F). These results are consistent with previous studies showing that dALS is predominantly expressed in the fat body and that its fat body-specific knockdown under physiological conditions results in increased body mass (13). Thus, our data suggest that the overgrowth phenotype in ohgt mutants is likely caused by a feedback loop: ILS activity both systemically and in the fat body is elevated in ohgt mutants, which in turn causes a reduction of the secreted inhibitory cofactor dALS (and IMP-L2 to a minor extent) from the fat body, thus allowing enhanced activity of circulating DILPs.
OHGT May Be Part of a Putative Drosophila Cullin4 Complex
What is the biochemical function of OHGT? It has been demonstrated for mammalian CRBN that it acts as a substrate receptor of the CRL4 complex (18) to regulate the stability and turnover of proteins via ubiquitination.
CRBN is known to interact with the core catalytic complex (RBX1-Cullin4-DDB1) by directly binding to DDB1 (64), and the CRBN-DDB1 interaction has been recently crystallized, identifying conserved helices and amino acid residues mediating the contact of both proteins. A Drosophila homolog of DDB1 has been identified by sequence comparison (named PIC), and a previous study revealed its putative role in development and maintenance of genome stability based on the phenotypes of knockdown animals (65, 66).
To analyze whether OHGT may bind to PIC in a manner analogous to the CRBN-DDB1 interaction, we first modeled the structure of the putative interaction site according to the crystallization data and found a similar interface structure with a number of conserved amino acid residues (Fig. 7, A–D). Next, we performed series of biochemical experiments to confirm the interaction. A commercial polyclonal anti-human DDB1 antibody could detect a PIC protein of predicted molecular mass (126 kDa) (Fig. 7E). Using this antibody, we performed co-immunoprecipitation experiments in S2 tissue culture cells. HA-tagged OHGT was co-precipitated using HA-specific antibodies, and a protein that is expected to be PIC was included in the HA-OHGT-containing immunocomplex (Fig. 7E). The protein was also included in the endogenous OHGT-containing immunocomplex (Fig. 7F). In addition, we found that OHGT was polyubiquitinated in the presence of the proteasome inhibitor MG132 (Fig. 7G), similar to human CRBN (18, 20). Together, these results suggest that the CRL4CRBN complex may be conserved in Drosophila. Unfortunately, both pic and dCullin4 RNAi induction in the fat body using the Cg-Gal4 driver caused lethality during early larval stage (data not shown). Therefore, we could not extend our in vitro studies toward functional experiments with the putative interaction partners.
FIGURE 7.
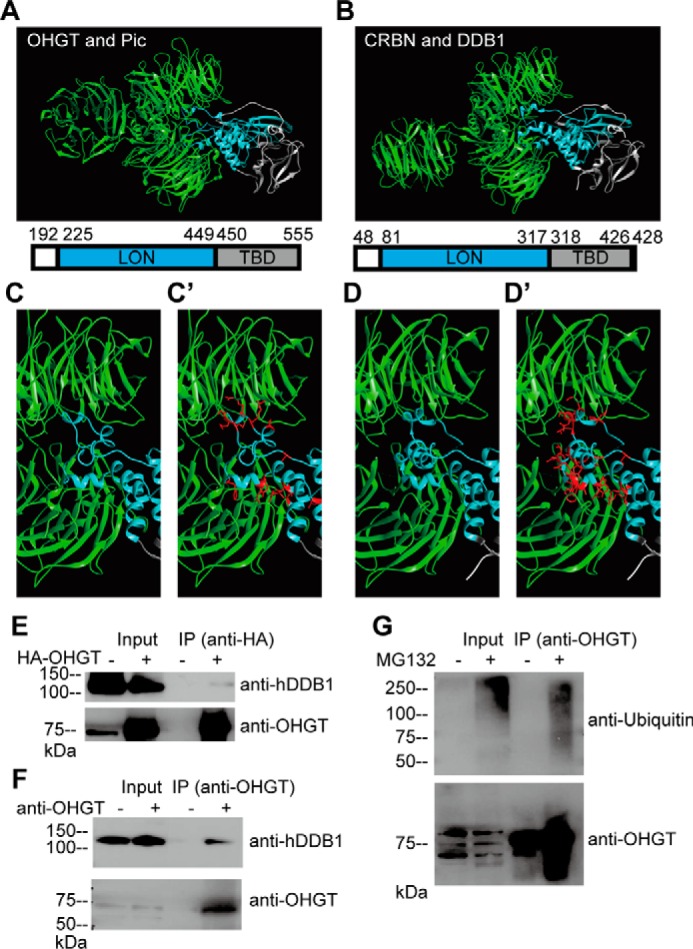
OHGT interacts with PIC, the Drosophila homolog of human DDB1. A, 3D model for OHGT-PIC complex was created using the SWISS-MODEL service and then aligned to respective positions in Protein Data Bank code 4TZ4. Amino acid residues and domains depicted in the structures are shown. B, the crystal structure of the human CRBN-DDB1 complex (Protein Data Bank code 4TZ4). C and D, a close-up view of the OHGT-PIC/CRBN-DDB1 interaction interfaces. C′ and D′, amino acid side chains of OHGT/CRBN that point toward the PIC/DDB1 interface are shown in red. E, S2 cells were transfected with HA-OHGT expression vector, and protein complexes containing HA-OHGT were immunoprecipitated using HA tag-specific antibody. Total lysates (Input) and immunoprecipitates (IP) were subjected to Western blotting with antibodies to the indicated proteins (for detecting OHGT, guinea pig OHGT-specific antibody was used). F, protein complexes containing endogenous OHGT were immunoprecipitated using rabbit OHGT-specific antibody. Total lysates (Input) and immunoprecipitates (IP) were subjected to Western blotting with antibodies to the indicated proteins (for detecting OHGT, guinea pig OHGT-specific antibody was used). G, S2 cells were treated with MG132, and endogenous OHGT was immunoprecipitated using rabbit OHGT-specific antibody. Total lysates (Input) and immunoprecipitates (IP) were subjected to Western blotting with antibodies to the indicated proteins (for detecting OHGT, guinea pig OHGT-specific antibody was used).
Discussion
Growth regulation in animals is controlled by paracrine and endocrine systems, which regulate tissue and organ size to determine the proper proportions of the body. The genetic programs underlying growth are dependent on nutrient access of the organisms, which must be able to adapt their metabolic and growth programs to changes in environmental conditions. A key growth regulatory pathway is the conserved IIS cascade. When nutrients are abundant, IIS activity is high to activate growth, whereas upon nutrient shortage, IIS restricts growth via activation of FOXO target genes.
Our work unveiled OHGT as a novel regulator of body size and tissue growth. We show that OHGT acts in the fat body and negatively regulates insulin signaling. In ohgt mutants, which we generated by targeted mutagenesis using the CRISPR-Cas9 system, phospho-dAkt levels are elevated, leading to the down-regulation of dFOXO target genes such as d4EBP and dInR, thereby promoting overgrowth. In vertebrates, the majority of IGF-1 accumulates in body fluids as a ternary complex with ALS and either IGF-binding protein 3 or 5 (67). Formation of the complex enhances the stability of IGF-1, prolonging its half-life in circulation (67). A similar ternary complex is also present in Drosophila as DILP2 interacts with dALS and IMP-L2 in hemolymph (13). Our studies revealed that the expression of both dALS and Imp-L2 is reduced in ohgt mutants, thus allowing enhanced activity of circulating DILPs. Thus, our results suggest that the overgrowth phenotype of the ohgt mutants may likely be caused by a feedback loop: ILS activity both systemically and in the fat body is elevated in ohgt mutants, which in turn causes a reduction of the secreted inhibitory cofactor dALS (and IMP-L2 to a minor extent) from the fat body, thus allowing enhanced activity of circulating DILPs. Of note, we also observed an up-regulation of dilp6 upon fat body-specific knockdown of ohgt (data not shown). Because loss of dilp6 is known to cause a growth defect resulting in smaller body size (15, 16), an elevation of dilp6 observed in ohgt mutants may further enhance the overgrowth phenotype of ohgt mutants.
Human CRBN is a substrate receptor of the E3 ubiquitin ligase complex and known to interact with CRL4, the core catalytic complex consisting of the RBX1-Cullin4-DDB1 proteins to control ubiquitin-mediated degradation. The mammalian CRL4CRBN is localized in the nucleus (18), and previous studies demonstrated that mammalian CRBN regulates transcription by two distinct mechanisms: by directly targeting transcriptional factors and by regulating histone modification. CRL4CRBN was shown to target transcription factors including lymphoid tissue-specific Ikaros and Aiolos as well as homeobox protein MEIS2 for polyubiquitination and degradation (23, 24, 29). Also, a recent publication identified that in T cells CRBN directly binds to a regulatory region of a gene and modulates histone modification by recruiting the CRL4 and a histone methyltransferase, EZH1 (68). Only five in vivo substrates of the mammalian CRL4CRBN complex have been identified so far: the large conductance Ca2+ and voltage-activated K+ channels (69, 70), voltage-gated CLC-1 chloride channels (71), AMPK (19, 34), the homeobox protein MEIS2 (29), and glutamine synthetase (33).
All the components of the mammalian CRL4 complex are conserved in Drosophila including the Cullin 4 homolog and the DDB1 homolog PIC (72). Down-regulation of both components results in severe larval growth defects and early larval lethality (72). Consistent with the conservation of the Cullin4-PIC complex, the modeling of the structure of the putative binding interface between OHGT and PIC according to the CRBN-DDB1 crystallization data revealed very similar structural properties of the binding region and conserved amino acids. In addition, our immunoprecipitation data further suggest the possibility that the interaction between CRBN/OHGT and DDB1/PIC may also be conserved in Drosophila. Many more experiments that go beyond the scope of this study will have to be performed to further support this proposal. If a CRL4OHGT complex exists in Drosophila, what are the targets in respect to insulin signaling? Of note, another CRL complex, the Skp1-Cullin1-Skp2 complex, has been shown to regulate insulin signaling via targeting the major downstream effector of the pathway, FOXO, for degradation. Under high insulin signaling activity, FOXO1 is phosphorylated at Ser-256, and phosphorylated FOXO1 in the cytoplasm is degraded and lost, leading to increased cell proliferation of tumor cells (73). Unfortunately, we could not investigate the status of dFOXO in ohgt mutants due to a lack of anti-FOXO antibodies.
One intriguing aspect of CRBN is that a certain class of small molecules can modulate interaction with its substrates. Previous studies have shown that thalidomide and its derivatives modulate interaction between CRBN and a number of substrates (23, 24, 30, 31, 33). CRBN orthologs across the animal and plant kingdoms exhibit 100% sequence conservation in three tryptophan residues that form a hydrophobic pocket occupied by the small molecules (64), indication that endogenous modulators of these proteins may exist. Further studies will be required to elucidate whether functions of OHGT can be regulated by such still unidentified modulators.
Author Contributions
M. H., T. A., and N. S. conceived and supervised the project. M. H., M. B., and S. W. designed experiments. S. W. performed all experiments. A. V. created 3D models of the OHGT-PIC complex, performed bioinformatics analysis, and analyzed data. M. B. provided reagents for biochemical experiments and interpreted the results. All authors discussed the results. M. H. and S. W. wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We thank Dr. Pilar Carrera, Dominic Gosejacob, Mirco Brondolin, and Melanie Thielisch for technical supports and suggestions; Sabine Büttner for embryo microinjection; members of the Hoch laboratory and the Asahi/Sawamura laboratory for suggestions; Dr. Michael Pankratz for DILP2 antibody; Dr. Ronald Kuehnlein for the FB-Gal4 line; the Drosophila Genomics Resource Center for the CG3925 cDNA clone; the Bloomington Drosophila Stock Center, Kyoto Stock Center, National Institute of Genetics, and Vienna Drosophila RNAi Center for fly stocks; the Developmental Studies Hybridoma Bank for α-spectrin antibody; and the Resource for Biocomputing, Visualization and Informatics at the University of California San Francisco for the UCSF Chimera package.
This work was supported by the Leading Graduate Program in Science and Engineering, Waseda University from MEXT, Japan; the European Union Institute in Japan at Waseda University (EUIJ Waseda); Deutsche Forschungsgemeinschaft (DFG) Grant VO 2071/1-1 (to A. V.); and DFG Grants SFB 645 and TRR83, the Helmholtz cross program topic “Metabolic Dysfunction,” and the DFG Excellence Cluster ImmunoSensation (to M. H.). The authors declare that they have no conflicts of interest with the contents of this article.
- IGF
- insulin-like growth factor
- ALS
- acid-labile subunit
- AMPK
- AMP-activated protein kinase
- CRBN
- Cereblon
- CRISPR
- clustered regularly interspaced short palindromic repeat
- CRL4
- Cullin4-RING E3 ubiquitin ligase
- DDB1
- DNA damage-binding protein 1
- DILP
- Drosophila insulin-like peptide
- HRMA
- high resolution melt analysis
- IIS
- insulin/IGF signaling
- ILS
- insulin-like signaling
- IMP-L2
- imaginal morphogenesis protein-late 2
- InR
- insulin-like receptor
- IPCs
- insulin-producing cells
- LIP3
- Lipase 3
- OHGT
- Ohgata
- PIC
- Piccolo
- S2
- Schneider 2
- TBD
- thalidomide-binding domain
- UPD2
- Unpaired 2
- AED
- after egg deposition
- RIPA
- radioimmune precipitation assay
- qRT-PCR
- quantitative real time PCR
- F
- forward
- R
- reverse
- 4EBP
- eIF-4E-binding protein
- SUMO
- small ubiquitin-like modifier
- d
- Drosophila.
References
- 1. Garofalo R. S. (2002) Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 13, 156–162 [DOI] [PubMed] [Google Scholar]
- 2. Edgar B. A. (2006) How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907–916 [DOI] [PubMed] [Google Scholar]
- 3. Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., and Hafen E. (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 [DOI] [PubMed] [Google Scholar]
- 4. Fuss B., Becker T., Zinke I., and Hoch M. (2006) The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature. 444, 945–948 [DOI] [PubMed] [Google Scholar]
- 5. Ikeya T., Galic M., Belawat P., Nairz K., and Hafen E. (2002) Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293–1300 [DOI] [PubMed] [Google Scholar]
- 6. Rulifson E. J., Kim S. K., and Nusse R. (2002) Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118–1120 [DOI] [PubMed] [Google Scholar]
- 7. Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J., and Léopold P. (2003) A nutrient sensor mechanism controls Drosophila growth. Cell. 114, 739–749 [DOI] [PubMed] [Google Scholar]
- 8. Géminard C., Rulifson E. J., and Léopold P. (2009) Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10, 199–207 [DOI] [PubMed] [Google Scholar]
- 9. Rajan A., and Perrimon N. (2012) Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sano H., Nakamura A., Texada M. J., Truman J. W., Ishimoto H., Kamikouchi A., Nibu Y., Kume K., Ida T., and Kojima M. (2015) The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 11, e1005481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koyama T., and Mirth C. K. (2016) Growth-blocking peptides as nutrition-sensitive signals for insulin secretion and body size regulation. PLoS Biol. 14, e1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agrawal N., Delanoue R., Mauri A., Basco D., Pasco M., Thorens B., and Léopold P. (2016) The Drosophila TNF Eiger is an adipokine that acts on insulin-producing cells to mediate nutrient response. Cell Metab. 23, 675–684 [DOI] [PubMed] [Google Scholar]
- 13. Arquier N., Géminard C., Bourouis M., Jarretou G., Honegger B., Paix A., and Léopold P. (2008) Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 7, 333–338 [DOI] [PubMed] [Google Scholar]
- 14. Honegger B., Galic M., Köhler K., Wittwer F., Brogiolo W., Hafen E., and Stocker H. (2008) Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H., O'Connor M. B., and Mizoguchi A. (2009) A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Slaidina M., Delanoue R., Gronke S., Partridge L., and Léopold P. (2009) A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell 17, 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higgins J. J., Pucilowska J., Lombardi R. Q., and Rooney J. P. (2004) A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology 63, 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., and Handa H. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 19. Lee K. M., Yang S. J., Kim Y. D., Choi Y. D., Nam J. H., Choi C. S., Choi H. S., and Park C. S. (2013) Disruption of the cereblon gene enhances hepatic AMPK activity and prevents high-fat diet-induced obesity and insulin resistance in mice. Diabetes 62, 1855–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sawamura N., Wakabayashi S., Matsumoto K., Yamada H., and Asahi T. (2015) Cereblon is recruited to aggresome and shows cytoprotective effect against ubiquitin-proteasome system dysfunction. Biochem. Biophys. Res. Commun. 464, 1054–1059 [DOI] [PubMed] [Google Scholar]
- 21. Kataoka K., Nakamura C., Asahi T., and Sawamura N. (2016) Mitochondrial cereblon functions as a Lon-type protease. Sci. Rep. 6, 29986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez-Girona A., Mendy D., Ito T., Miller K., Gandhi A. K., Kang J., Karasawa S., Carmel G., Jackson P., Abbasian M., Mahmoudi A., Cathers B., Rychak E., Gaidarova S., Chen R., et al. (2012) Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26, 2326–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krönke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., Schreiber S. L., et al. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu G., Middleton R. E., Sun H., Naniong M., Ott C. J., Mitsiades C. S., Wong K.-K., Bradner J. E., and Kaelin W. G. Jr. (2014) The myeloma drug lenalidomide promotes the Cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eichner R., Heider M., Fernández-Sáiz V., van Bebber F., Garz A. K., Lemeer S., Rudelius M., Targosz B. S., Jacobs L., Knorn A. M., Slawska J., Platzbecker U., Germing U., Langer C., Knop S., et al. (2016) Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat. Med. 22, 735–743 [DOI] [PubMed] [Google Scholar]
- 26. Kim H. K., Ko T. H., Nyamaa B., Lee S. R., Kim N., Ko K. S., Rhee B. D., Park C.-S., Nilius B., and Han J. (2016) Cereblon in health and disease. Pflugers Arch. 468, 1299–1309 [DOI] [PubMed] [Google Scholar]
- 27. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., and Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 28. Zhu Y. X., Braggio E., Shi C. X., Bruins L. A., Schmidt J. E., Van Wier S., Chang X. B., Bjorklund C. C., Fonseca R., Bergsagel P. L., Orlowski R. Z., and Stewart A. K. (2011) Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood 118, 4771–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fischer E. S., Böhm K., Lydeard J. R., Yang H., Stadler M. B., Cavadini S., Nagel J., Serluca F., Acker V., Lingaraju G. M., Tichkule R. B., Schebesta M., Forrester W. C., Schirle M., Hassiepen U., et al. (2014) Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krönke J., Fink E. C., Hollenbach P. W., MacBeth K. J., Hurst S. N., Udeshi N. D., Chamberlain P. P., Mani D. R., Man H. W., Gandhi A. K., Svinkina T., Schneider R. K., McConkey M., Järås M., Griffiths E., et al. (2015) Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matyskiela M. E., Lu G., Ito T., Pagarigan B., Lu C. C., Miller K., Fang W., Wang N. Y., Nguyen D., Houston J., Carmel G., Tran T., Riley M., Nosaka L., Lander G. C., et al. (2016) A novel cereblon modulator recruits GSPT1 to the CRL4 CRBN ubiquitin ligase. Nature 535, 252–257 [DOI] [PubMed] [Google Scholar]
- 32. Wada T., Asahi T., and Sawamura N. (2016) Nuclear cereblon modulates transcriptional activity of Ikaros and regulates its downstream target, enkephalin, in human neuroblastoma cells. Biochem. Biophys. Res. Commun. 477, 388–394 [DOI] [PubMed] [Google Scholar]
- 33. Nguyen T. V., Lee J. E., Sweredoski M. J., Yang S. J., Jeon S. J., Harrison J. S., Yim J. H., Lee S. G., Handa H., Kuhlman B., Jeong J. S., Reitsma J. M., Park C. S., Hess S., and Deshaies R. J. (2016) Glutamine triggers acetylation-dependent degradation of glutamine synthetase via the thalidomide receptor Cereblon. Mol. Cell 61, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee K. M., Jo S., Kim H., Lee J., and Park C.-S. (2011) Functional modulation of AMP-activated protein kinase by cereblon. Biochim. Biophys. Acta 1813, 448–455 [DOI] [PubMed] [Google Scholar]
- 35. Pignoni F., and Zipursky S. L. (1997) Induction of Drosophila eye development by decapentaplegic. Development 124, 271–278 [DOI] [PubMed] [Google Scholar]
- 36. Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., and O'Connor-Giles K. M. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bassett A. R., Tibbit C., Ponting C. P., and Liu J. L. (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodenfels J., Lavrynenko O., Ayciriex S., Sampaio J. L., Carvalho M., Shevchenko A., and Eaton S. (2014) Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev. 28, 2636–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gündner A. L., Hahn I., Sendscheid O., Aberle H., and Hoch M. (2014) The PIKE homolog Centaurin γ regulates developmental timing in Drosophila. PLoS One 9, e97332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hahn I., Fuss B., Peters A., Werner T., Sieberg A., Gosejacob D., and Hoch M. (2013) The Drosophila Arf GEF Steppke controls MAPK activation in EGFR signaling. J. Cell Sci. 126, 2470–2479 [DOI] [PubMed] [Google Scholar]
- 41. Becker T., Loch G., Beyer M., Zinke I., Aschenbrenner A. C., Carrera P., Inhester T., Schultze J. L., and Hoch M. (2010) FOXO-dependent regulation of innate immune homeostasis. Nature 463, 369–373 [DOI] [PubMed] [Google Scholar]
- 42. Bauer R., Weimbs A., Lechner H., and Hoch M. (2006) DE-cadherin, a core component of the adherens junction complex modifies subcellular localization of the Drosophila gap junction protein innexin2. Cell Commun. Adhes. 13, 103–114 [DOI] [PubMed] [Google Scholar]
- 43. Kwon Y., Song W., Droujinine I. A., Hu Y., Asara J. M., and Perrimon N. (2015) Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell. 33, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 46. Ronquist F., and Huelsenbeck J. P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 47. Huelsenbeck J. P., and Ronquist F. (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 48. Altekar G., Dwarkadas S., Huelsenbeck J. P., and Ronquist F. (2004) Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20, 407–415 [DOI] [PubMed] [Google Scholar]
- 49. Sigrist C. J., de Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., Bougueleret L., and Xenarios I. (2013) New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., and Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu W.-R., Xiao X., Lin W.-Z., and Chou K.-C. (2015) iUbiq-Lys: prediction of lysine ubiquitination sites in proteins by extracting sequence evolution information via a gray system model. J. Biomol. Struct. Dyn. 33, 1731–1742 [DOI] [PubMed] [Google Scholar]
- 52. Wang P., Xiao X., and Chou K. C. (2011) NR-2l: a two-level predictor for identifying nuclear receptor subfamilies based on sequence-derived features. PLoS One 6, e23505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., and Ren J. (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., and Xue Y. (2009) Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 55. Brameier M., Krings A., and MacCallum R. M. (2007) NucPred—predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160 [DOI] [PubMed] [Google Scholar]
- 56. Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., Rey A. J., Marygold S. J., and FlyBase Consortium (2016) FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44, D786–D792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hsu P. D., Lander E. S., and Zhang F. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Teleman A. A. (2010) Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13–26 [DOI] [PubMed] [Google Scholar]
- 59. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., and Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 60. Puig O., Marr M. T., Ruhf M. L., and Tjian R. (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verdu J., Buratovich M. A., Wilder E. L., and Birnbaum M. J. (1999) Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1, 500–506 [DOI] [PubMed] [Google Scholar]
- 62. Scanga S. E., Ruel L., Binari R. C., Snow B., Stambolic V., Bouchard D., Peters M., Calvieri B., Mak T. W., Woodgett J. R., and Manoukian A. S. (2000) The conserved PI3′K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19, 3971–3977 [DOI] [PubMed] [Google Scholar]
- 63. Garbe J. C., Yang E., and Fristrom J. W. (1993) IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development 119, 1237–1250 [DOI] [PubMed] [Google Scholar]
- 64. Chamberlain P. P., Lopez-Girona A., Miller K., Carmel G., Pagarigan B., Chie-Leon B., Rychak E., Corral L. G., Ren Y. J., Wang M., Riley M., Delker S. L., Ito T., Ando H., Mori T., et al. (2014) Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 21, 803–809 [DOI] [PubMed] [Google Scholar]
- 65. Shimanouchi K., Takata K., Yamaguchi M., Murakami S., Ishikawa G., Takeuchi R., Kanai Y., Ruike T., Nakamura R., Abe Y., and Sakaguchi K. (2006) Drosophila damaged DNA binding protein 1 contributes to genome stability in somatic cells. J. Biochem. 139, 51–58 [DOI] [PubMed] [Google Scholar]
- 66. Takata K., Yoshida H., Yamaguchi M., and Sakaguchi K. (2004) Drosophila damaged DNA-binding protein 1 is an essential factor for development. Genetics 168, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Boisclair Y. R., Rhoads R. P., Ueki I., Wang J., and Ooi G. T. (2001) The acid-labile subunit (ALS) of the 150 kDa IGF-binding protein complex: an important but forgotten component of the circulating IGF system. J. Endocrinol. 170, 63–70 [DOI] [PubMed] [Google Scholar]
- 68. Kang J. A., Park S. H., Jeong S. P., Han M. H., Lee C. R., Lee K. M., Kim N., Song M. R., Choi M., Ye M., Jung G., Lee W. W., Eom S. H., Park C. S., and Park S. G. (2016) Epigenetic regulation of Kcna3-encoding Kv1.3 potassium channel by cereblon contributes to regulation of CD4+ T-cell activation. Proc. Natl. Acad. Sci. U.S.A. 113, 8771–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jo S., Lee K. H., Song S., Jung Y. K., and Park C. S. (2005) Identification and functional characterization of cereblon as a binding protein for large-conductance calcium-activated potassium channel in rat brain. J. Neurochem. 94, 1212–1224 [DOI] [PubMed] [Google Scholar]
- 70. Liu J., Ye J., Zou X., Xu Z., Feng Y., Zou X., Chen Z., Li Y., and Cang Y. (2014) CRL4A(CRBN) E3 ubiquitin ligase restricts BK channel activity and prevents epileptogenesis. Nat. Commun. 5, 3924. [DOI] [PubMed] [Google Scholar]
- 71. Chen Y. A., Peng Y. J., Hu M. C., Huang J. J., Chien Y. C., Wu J. T., Chen T. Y., and Tang C. Y. (2015) The Cullin 4A/B-DDB1-Cereblon E3 ubiquitin ligase complex mediates the degradation of CLC-1 chloride channels. Sci. Rep. 5, 10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu J., Zacharek S., He Y. J., Lee H., Shumway S., Duronio R. J., and Xiong Y. (2008) WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 22, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang H., Regan K. M., Wang F., Wang D., Smith D. I., van Deursen J. M., and Tindall D. J. (2005) Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 102, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]