Abstract
AMP-activated protein kinase (AMPK) is an energy sensor and master regulator of metabolism. AMPK functions as a fuel gauge monitoring systemic and cellular energy status. Activation of AMPK occurs when the intracellular AMP/ATP ratio increases and leads to a metabolic switch from anabolism to catabolism. AMPK phosphorylates and inhibits acetyl-CoA carboxylase (ACC), which catalyzes carboxylation of acetyl-CoA to malonyl-CoA, the first and rate-limiting reaction in de novo synthesis of fatty acids. AMPK thus regulates homeostasis of acetyl-CoA, a key metabolite at the crossroads of metabolism, signaling, chromatin structure, and transcription. Nucleocytosolic concentration of acetyl-CoA affects histone acetylation and links metabolism and chromatin structure. Here we show that activation of AMPK with the widely used antidiabetic drug metformin or with the AMP mimetic 5-aminoimidazole-4-carboxamide ribonucleotide increases the inhibitory phosphorylation of ACC and decreases the conversion of acetyl-CoA to malonyl-CoA, leading to increased protein acetylation and altered gene expression in prostate and ovarian cancer cells. Direct inhibition of ACC with allosteric inhibitor 5-(tetradecyloxy)-2-furoic acid also increases acetylation of histones and non-histone proteins. Because AMPK activation requires liver kinase B1, metformin does not induce protein acetylation in liver kinase B1-deficient cells. Together, our data indicate that AMPK regulates the availability of nucleocytosolic acetyl-CoA for protein acetylation and that AMPK activators, such as metformin, have the capacity to increase protein acetylation and alter patterns of gene expression, further expanding the plethora of metformin's physiological effects.
Keywords: acetyl coenzyme A (acetyl-CoA), AMP-activated kinase (AMPK), cancer, histone acetylation, metformin, transcription
Introduction
Acetylation is one of the epigenetic post-translational modifications of histones; it affects chromatin structure and regulates diverse cellular functions, such as gene expression, DNA replication and repair, and cellular proliferation (1, 2). Acetylation and deacetylation of chromatin histones, mediated by histone acetyltransferases (HATs)3 and histone deacetylases (HDACs), respectively, represent the major mechanisms for epigenetic gene regulation. The dynamic balance between histone acetylation and deacetylation, mediated by the activities of HATs and HDACs, is stringently regulated in healthy cells but is often dysregulated in cancer (3, 4).
Histone acetylation depends on intermediary metabolism for supplying acetyl-CoA in the nucleocytosolic compartment (5). In mammalian cells, the nucleocytosolic enzyme ATP-citrate lyase is the major source of acetyl-CoA for histone acetylation (6). Another mechanism for generation of acetyl-CoA in the nucleus involves translocation of pyruvate dehydrogenase from mitochondria to the nucleus (7). In yeast, global histone acetylation depends on nucleocytosolic acetyl-CoA produced by acetyl-CoA synthetase (5). In both yeast and mammalian cells, the nucleocytosolic acetyl-CoA is the link among cellular energy, carbon metabolism, histone acetylation, and chromatin regulation (8–11).
The nucleocytosolic acetyl-CoA is a critical precursor of several anabolic processes, including de novo synthesis of fatty acids. Acetyl-CoA carboxylase (ACC) catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, the first and rate-limiting reaction in the de novo synthesis of fatty acids (12). The ACC activity affects the concentration of nucleocytosolic acetyl-CoA. We have previously shown that attenuated expression of yeast ACC increases global acetylation of chromatin histones and alters transcriptional regulation (13). Moreover, chronic inhibition of ACC in mouse hepatocytes increases protein acetylation (14). The human genome encodes two tissue-specific ACC isoforms, ACCα (ACCA) and ACCβ (ACCB) (15). ACCA activity is controlled by AMP-activated protein kinase (AMPK), a conserved cellular energy sensor and master regulator of metabolism. A hallmark of AMPK activation is phosphorylation of ACCA at Ser79, which results in reduced activity of ACCA and inhibition of fatty acid synthesis (16, 17). In yeast, inactivation of SNF1, the budding yeast ortholog of mammalian AMPK, results in increased ACC activity, a reduced pool of cellular acetyl-CoA, and globally decreased histone acetylation (18).
The main objective of this study was to test the hypothesis that inhibition of ACC activity in human cells increases the nucleocytosolic pool of acetyl-CoA and histone acetylation. We show that suppression of ACC activity either by direct inhibition or by metformin-mediated AMPK activation increases acetylation of histones and non-histones proteins and induces transcriptional changes in prostate and ovarian cancer cells. Metformin, widely used for diabetes type 2 treatment, decreases ATP production by inhibiting mitochondrial respiratory chain complex I, leading to AMPK activation (19–23). The metformin therapy is associated with a reduced risk of cancer in diabetes type 2 patients; however, the mechanisms are not completely understood (24). Our results indicate that some of the physiological effects of metformin may involve increased acetylation of histone and non-histone proteins and altered patterns of transcriptional regulation.
Results
Inhibition of Acetyl-CoA Carboxylase Increases Protein Acetylation
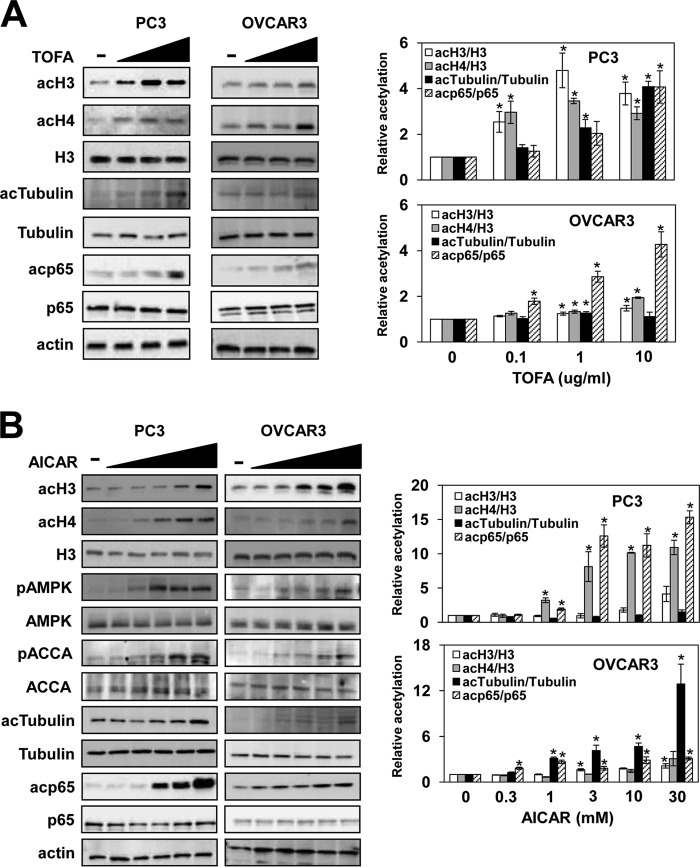
Histone acetylation depends on intermediary metabolism for supplying acetyl-CoA as a substrate for HATs in the nucleocytosolic compartment (5, 6). Cytosolic acetyl-CoA is also used by acetyl-CoA carboxylase to yield malonyl-CoA, a precursor for de novo synthesis of fatty acids (25, 26). We have previously shown that acetyl-CoA carboxylase Acc1p regulates homeostasis of nucleocytosolic acetyl-CoA and acetylation of histones and nonhistone proteins in yeast (13). To investigate whether ACCA regulates histone acetylation also in mammalian cells, we analyzed histone acetylation in prostate cancer PC3 and ovarian cancer OVCAR3 cells treated with 5-(tetradecyloxy)-2-furoic acid (TOFA), an allosteric ACCA inhibitor that decreases conversion of acetyl-CoA to malonyl-CoA and induces apoptosis in lung and colon cancer cells (27). Our results show that inhibition of ACCA significantly increases acetylation levels of histones H3 and H4 in PC3 cells and to a lesser extent in OVCAR3 cells (Fig. 1A).
FIGURE 1.
Inhibition of ACCA activity by TOFA or AMPK activation by AICAR increases protein acetylation in PC3 and OVCAR3 cells. PC3 and OVCAR3 cells were treated with 0, 0.1, 1.0, and 10 μg/ml TOFA for 48 h (A) or 0, 0.3, 1, 3, 10, and 30 mm AICAR for 24 h (B). Samples were analyzed by Western blotting with antibodies against acH3, acH4, total histone H3, AMPK, pAMPK, ACCA, pACCA, tubulin, acTubulin, p65, acp65, and actin. The figure represents typical results from three independent experiments. Quantitative evaluation of the Western blots was performed by densitometric analysis of the band intensities. The ratios of acH3/H3, acH4/H3, acTubulin/tubulin, and acp65/p65 were plotted; they represent means ± S.D. (error bars). Values that are statistically significantly different (p < 0.05) from the untreated samples are indicated by an asterisk.
In addition to histones, many other proteins are acetylated (3, 8). To determine whether ACCA inhibition selectively affects only histone acetylation or has a similar effect on acetylation of other proteins, we assayed acetylation of α-tubulin and p65 NFκB. α-Tubulin is acetylated at Lys40 by a conserved α-tubulin acetyltransferase, increasing stability of microtubules (28). The transcription factor NFκB regulates expression of genes involved in inflammation, growth, development, and apoptosis (29, 30). Acetylation of p65 at Lys310 is required for the full transcriptional activity of NFκB (31). Our results show that the acetylation levels of α-tubulin and p65 are increased after TOFA treatment in PC3 and OVCAR3 cells without affecting the total cellular levels of these proteins (Fig. 1A).
AMPK Activation Increases Acetylation of Histones and Non-histone Proteins
ACCA activity is inhibited by AMPK phosphorylation (32–34). A hallmark of AMPK activation is ACCA phosphorylation at Ser79, resulting in ACCA inactivation and inhibition of fatty acid biosynthesis (35). We have shown that inactivation of the yeast AMPK homolog SNF1 results in a decreased level of nucleocytosolic acetyl-CoA, leading to hypoacetylation of chromatin histones and non-histone proteins (18). Because SNF1 modulates acetyl-CoA homeostasis in yeast cells, we speculated that activation of AMPK in mammalian cells might decrease ACCA activity, leading to increased acetylation of histones.
As expected, stimulation of AMPK in PC3 and OVCAR3 cells with the AMP homolog 5-amino-1-β-d-ribofuranosyl-1H-imidazole-4-carboxamide (AICAR) increased AMPK phosphorylation at Thr172, a hallmark of AMPK activation by liver kinase B1 (LKB1), the primary upstream kinase that activates the AMPK pathway (34). Activation of AMPK resulted also in phosphorylation of ACCA at Ser79 (Fig. 1B), known to decrease ACCA enzymatic activity (35). Importantly, AMPK activation by AICAR increased acetylation of histones H3 and H4, α-tubulin, and p65 in both PC3 and OVCAR3 cells (Fig. 1B).
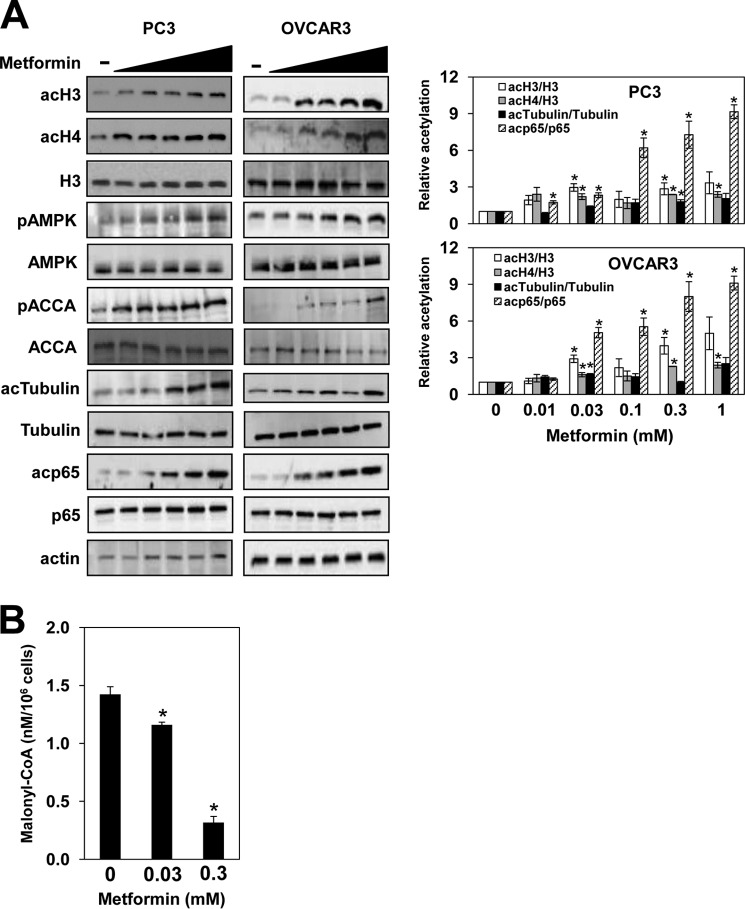
AMPK can also be activated by drugs that inhibit the mitochondrial electron transport pathway and oxidative phosphorylation and reduce the cellular ATP level. One of these drugs is metformin, a widely used antidiabetic drug that inhibits mitochondrial complex I, reducing ATP production and increasing AMP levels (20, 23). Upon AMP binding, AMPK becomes a better substrate for its activator kinase LKB1 (34). In PC3 and OVCAR3 cells, metformin activated AMPK as shown by increased AMPK phosphorylation at Thr172 and ACCA phosphorylation at Ser79 (Fig. 2A). In accordance with ACCA inactivation, 0.03 and 0.3 mm metformin decreased cellular malonyl-CoA levels to 80 and 20% compared with untreated cells, respectively (Fig. 2B). Similarly to TOFA and AICAR, treatment of PC3 and OVCAR3 cells with metformin increased acetylation of histones H3 and H4, α-tubulin, and p65 (Fig. 2A). The lowest concentration of metformin effective in increasing protein acetylation was about 30 μm (Fig. 2), which corresponds to the metformin concentration in human plasma following a therapeutic dose of around 30 mg/kg (36). Together, our results indicate that, by regulating ACCA activity, AMPK controls acetyl-CoA homeostasis and protein acetylation.
FIGURE 2.
AMPK activation by metformin increases protein acetylation in PC3 and OVCAR3 cells. A, PC3 and OVCAR3 cells were treated with 0, 0.01, 0.03, 0.1, 0.3, and 1 mm metformin for 72 h. Whole cell extracts were analyzed by Western blotting using antibodies against acH3, acH4, total histone H3, AMPK, pAMPK, ACCA, pACCA, tubulin, acTubulin, p65, acp65, and actin. The figure represents typical results from three independent experiments. Quantitative evaluation of the Western blots was performed by densitometric analysis of the band intensities. The ratios of acH3/H3, acH4/H3, acTubulin/tubulin, and acp65/p65 were plotted; they represent means ± S.D. (error bars). Values that are statistically significantly different (p < 0.05) from the untreated samples are indicated by an asterisk. B, metformin decreases the cellular malonyl-CoA level. PC3 cells were treated with 0, 0.03, and 0.3 mm metformin for 72 h. Malonyl-CoA was assayed in cell lysates by ELISA. The experiment was repeated three times, and the results are shown as means ± S.D. (error bars). Values that are statistically different (p < 0.05) from the control (0 mm metformin) are indicated by an asterisk.
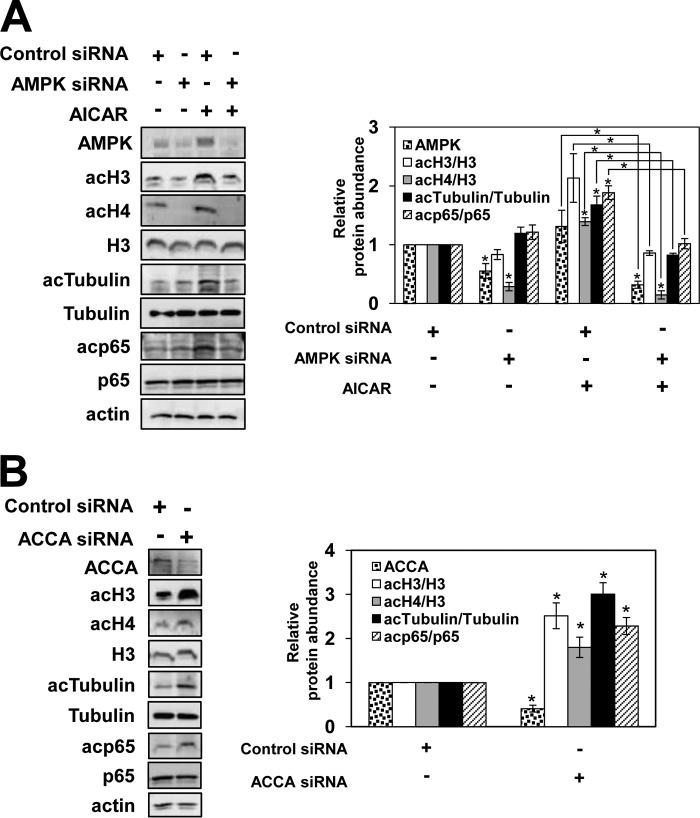
AMPK Silencing Impairs AICAR-induced Protein Acetylation
To investigate whether activation of AMPK, rather than a modulation of other cellular activities, accounts for the AICAR-induced increase in protein acetylation in PC3 cells, we analyzed global acetylation of histones and non-histone proteins after small interfering RNA (siRNA)-mediated silencing of both AMPKα1 and AMPKα2. As shown in Fig. 3A, AMPK siRNA silencing suppressed the cellular AMPK level by about 50% in untreated cells and by about 70% in AICAR-treated PC3 cells. Cells with suppressed AMPK expression exhibited significantly reduced acetylation of histones H3 and H4, tubulin, and p65 after AICAR treatment (Fig. 3A). These results indicate that activation of AMPK is responsible for the increased protein acetylation.
FIGURE 3.
AMPK and ACCA regulate protein acetylation. Shown is Western analysis of whole cell extracts using antibodies against AMPK, ACCA, acH3, acH4, total histone H3, tubulin, acTubulin, p65, acp65, and actin in PC3 cells transfected with AMPKα1/2-specific siRNA or control siRNA and treated for 48 h with 3 mm AICAR (A) or with ACCA-specific siRNA or control siRNA and incubated for 24 h (B). The figures represent typical results from three independent experiments. Quantitative evaluation of the Western blots was performed by densitometric analysis of the band intensities. The band intensities of AMPK and ACCA in cells transfected with control non-silencing siRNA were arbitrarily set at 1. The ratios of acH3/H3, acH4/H3, acTubulin/tubulin, and acp65/p65 were plotted; they represent means ± S.D. (error bars). Values that are statistically significantly different (p < 0.05) from the untreated samples are indicated by an asterisk. Values that are significantly different (p < 0.05) from each other are indicated by a bracket and an asterisk.
Treatment of PC3 and OVCAR3 cells with TOFA results in increased protein acetylation (Fig. 1A). To confirm that the mechanism responsible involves inhibition of the ACCA activity, we analyzed protein acetylation in PC3 cells transfected with ACCA siRNA as well as with control non-silencing siRNA. Cell transfection with ACCA siRNA suppressed the ACCA protein levels by about 60%. The ACCA suppression significantly increased acetylation of histones H3 and H4 as well as increased acetylation of tubulin and p65 (Fig. 3B). These results are consistent with the effect of TOFA on protein acetylation (Fig. 1A); we interpret these results to mean that decreased activity of ACCA results in increased protein acetylation. These results are also consistent with increased protein acetylation upon repression of yeast ACC (13).
LKB1 Is Required for Metformin-induced Protein Acetylation
The tumor suppressor serine/threonine LKB1 activates AMPK by phosphorylation at Thr172. LKB1 is a low energy sensor that regulates tumorigenesis and apoptosis by regulating AMPK and mTOR pathways (37). LKB1-deficient cells have increased mTOR signaling due to the lack of tuberous sclerosis 2 protein phosphorylation by AMPK, which results in increased growth and tumorigenic potential. To investigate whether metformin-induced protein acetylation requires LKB1-dependent activation of AMPK, we analyzed global protein acetylation of histones and non-histone proteins in PC3 cells transfected with LKB1 siRNA as well as with control non-silencing siRNA. As shown in Fig. 4A, LKB1 siRNA silencing suppressed the cellular LKB1 level by about 80% in untreated cells and by about 95% in metformin-treated PC3 cells. The LKB1 suppression abolished the increase in protein acetylation after metformin treatment (Fig. 4A), suggesting that LKB1 activity is required for metformin-induced protein acetylation.
FIGURE 4.
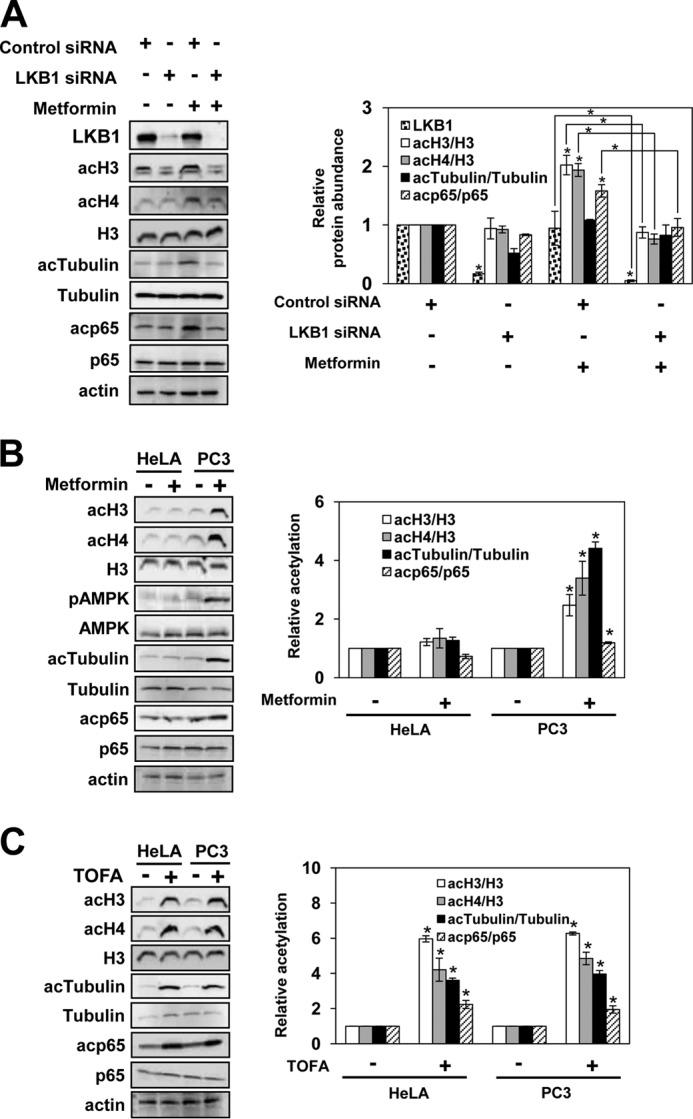
LKB1 is required for metformin-induced protein acetylation. A, PC3 cells were transfected with LKB1-specific siRNA or control siRNA and treated with 1 mm metformin for 48 h. HeLa S3 cells and PC3 cells were treated with 0 and 1 mm metformin for 72 h (B) or 0 and 1 μg/ml TOFA for 48 h (C). Samples were analyzed by Western blotting with the indicated antibodies. The figures represent typical results from three independent experiments. Quantitative evaluation of the Western blots was performed by densitometric analysis of the band intensities. The intensity of the LKB1 band in cells transfected with control non-silencing siRNA was arbitrarily set at 1. The ratios of acH3/H3, acH4/H3, acTubulin/tubulin, and acp65/p65 were plotted; they represent means ± S.D. (error bars). Values that are statistically significantly different (p < 0.05) from the untreated samples are indicated by an asterisk. Values that are significantly different (p < 0.05) from each other are indicated by a bracket and an asterisk.
To further investigate the role of LKB1 in metformin-induced protein acetylation, we used HeLa S3 cells that lack LKB1 expression (38). Metformin did not induce AMPK phosphorylation at Thr172 or ACCA phosphorylation at Ser79 and did not increase acetylation of histones H3 and H4, α-tubulin, and p65 in HeLa S3 cells (Fig. 4B). However, inhibition of ACCA with TOFA in HeLa S3 cells increased acetylation of histones H3 and H4, α-tubulin, and p65 (Fig. 4C). Taken together, our results suggest that the metformin-induced protein acetylation in PC3 and OVCAR3 cells is due to the LKB1-dependent activation of AMPK and AMPK-dependent inactivation of ACCA.
AMPK Activation Globally Increases Acetylation of Chromatin Histones and Alters Transcriptional Patterns
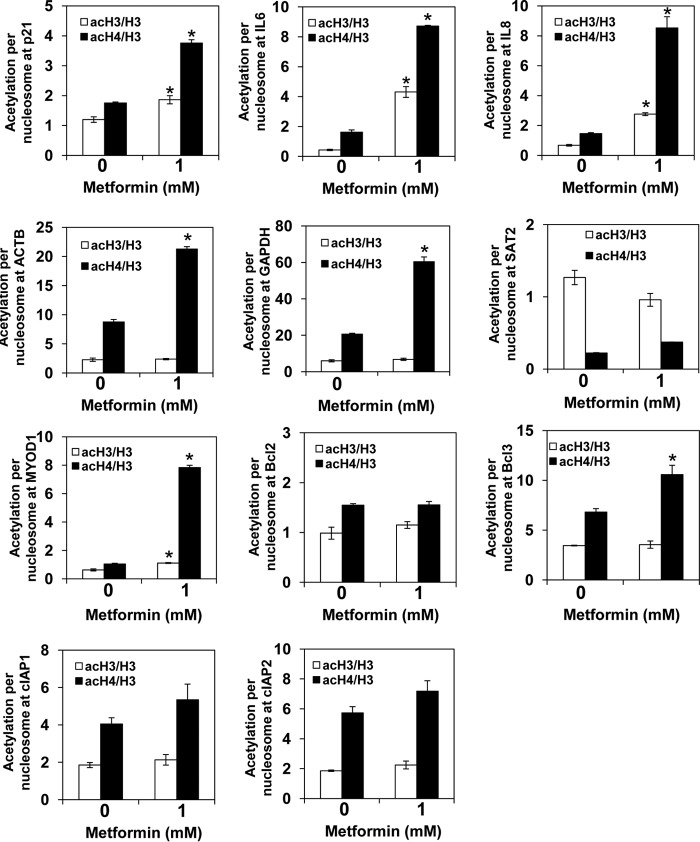
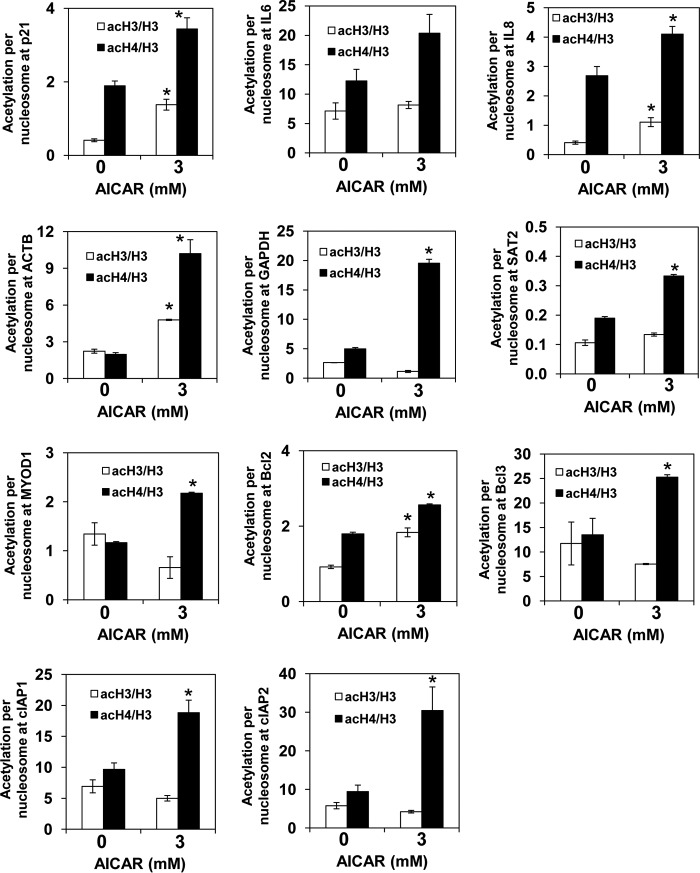
To test whether metformin regulates histone acetylation globally or only at specific loci, we used chromatin immunoprecipitation (ChIP) to evaluate the occupancy of histone H3 acetylated at Lys14 (acH3) as well as hyperacetylated histone H4 (acH4; acetylated at Lys5,8,12,16) in the promoter regions of β-actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cyclin-dependent kinase inhibitor p21, apoptosis regulator Bcl-2, proinflammatory genes IL6 and IL8, the transcription factor Bcl-3, the transcriptionally inactive euchromatin gene MYOD1 encoding myogenic differentiation 1 protein, and the transcriptionally inactive heterochromatin gene SAT2 encoding spermidine/spermine N1-acetyltransferase. We used anti-H3 antibody that recognizes the C-terminal region of histone H3, which is not post-translationally modified. The ChIP signal obtained with this antibody thus represents the total H3 occupancy and can be used to calculate the histone acetylation levels per nucleosome content (18). To account for differences in nucleosome density at different genomic loci, we corrected the acH3 and acH4 occupancies for histone H3 content and generated values that represent acetylation per nucleosome. In metformin-treated PC3 cells, acetylation of histone H3 was increased 4- and 10-fold in the promoters of IL8 and IL6, respectively. The acetylation status of histone H3 in the other promoters was not altered. Acetylation of histone H4 was increased 1.3–7.5 times in all examined promoters (Fig. 5). Upon treatment of PC3 cells with AICAR, acetylation of histone H3 was increased 2–3.4 times in the promoters of p21, IL8, ACTB, and Bcl2, whereas acetylation of histone H4 was increased 1.2–5.2 times in all examined promoters (Fig. 6). The fact that the increased acetylation of histone H4 was not always accompanied by increased acetylation of histone H3 is in an agreement with the notion that different acetylation levels of histones H3 and H4 are due to different affinity of individual HATs for acetyl-CoA (8, 39–42). Individual genes differed in the acetylation levels, and as expected the transcriptionally inactive heterochromatin gene SAT2 displayed the lowest acetylation. This result is consistent with the general correlation between acetylation of promoter histones and transcriptional activity (43).
FIGURE 5.
Metformin-treated cells display increased untargeted acetylation of chromatin histones. PC3 cells were treated with 0 and 1 mm metformin for 72 h. ChIP experiments were performed with antibodies against total histone H3, acH3, and acH4. Occupancies of H3, acH3, and acH4 were determined in the promoter regions of p21, IL6, IL8, ACTB, GAPDH, SAT2, MYOD1, Bcl-2, Bcl-3, cIAP1, and cIAP2 genes. Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. The experiments were repeated three times, and the results are shown as means ± S.D. (error bars). Values that are statistically different (p < 0.05) from the control (0 mm metformin) are indicated by an asterisk.
FIGURE 6.
AICAR-treated cells display increased untargeted acetylation of chromatin histones. PC3 cells were treated with 0 and 3 mm AICAR for 24 h. ChIP experiments were performed with antibodies against total histone H3, acH3, and acH4. Occupancies of H3, acH3, and acH4 were determined in the promoter regions of p21, IL6, IL8, ACTB, GAPDH, SAT2, MYOD1, Bcl-2, Bcl-3, cIAP1, and cIAP2 genes. Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. The experiments were repeated three times, and the results are shown as means ± S.D. (error bars). Values that are statistically different (p < 0.05) from the control (0 mm AICAR) are indicated by an asterisk.
Our observation that metformin or AICAR treatment increases histone acetylation raised the possibility that gene expression might be also altered upon AMPK activation. We found that treatment of PC3 cells with metformin increased expression of p21, IL8, Bcl-2, Bcl-3, and cIAP2 genes 1.3–2.0-fold. Similarly to metformin, AICAR treatment increased expression of p21, IL6, IL8, Bcl-2, Bcl-3, cIAP1, and cIAP2 genes 1.2–2.8-fold. However, expression of the highly expressed housekeeping genes GAPDH and ACTB was not affected after metformin or AICAR treatment (Fig. 7).
FIGURE 7.
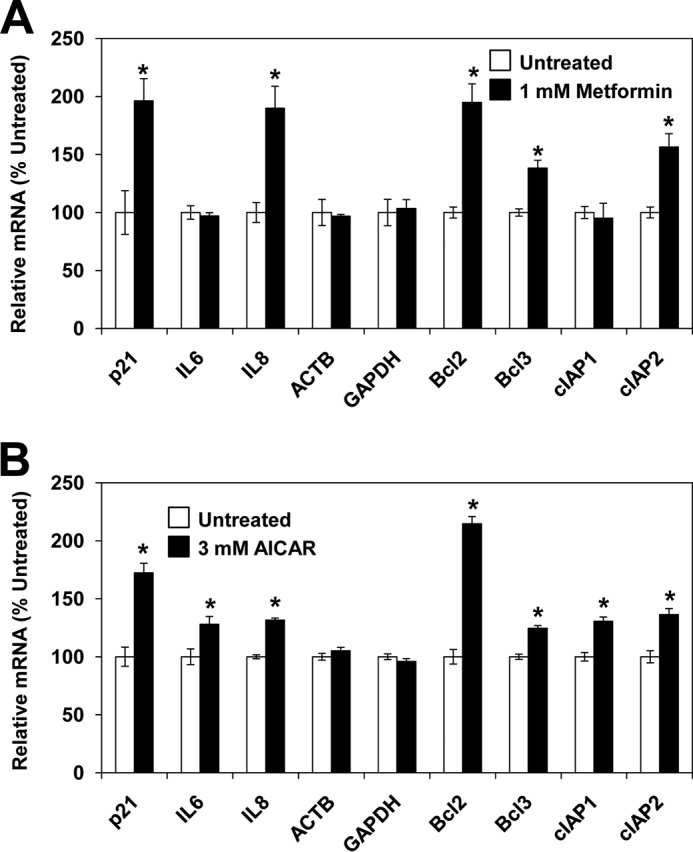
AMPK activation with metformin or AICAR alters gene expression patterns. PC3 cells were treated with 0 and 1 mm metformin for 72 h (A) or 0 and 3 mm AICAR for 24 h (B). Total RNA was isolated and assayed for 18S ribosomal subunit, p21, IL6, IL8, ACTB, GAPDH, Bcl-2, Bcl-3, cIAP1, and cIAP2 transcripts by real time RT-PCR. The results were normalized to 18S ribosomal RNA subunit and expressed relative to the value for untreated cells. The experiments were repeated three times, and the results are shown as means ± S.D. (error bars). Values that are statistically different (p < 0.05) from the control (0 mm metformin or AICAR) are indicated by an asterisk.
Metformin Increases Recruitment of Acetyl-p65 NFκB to Gene Promoters
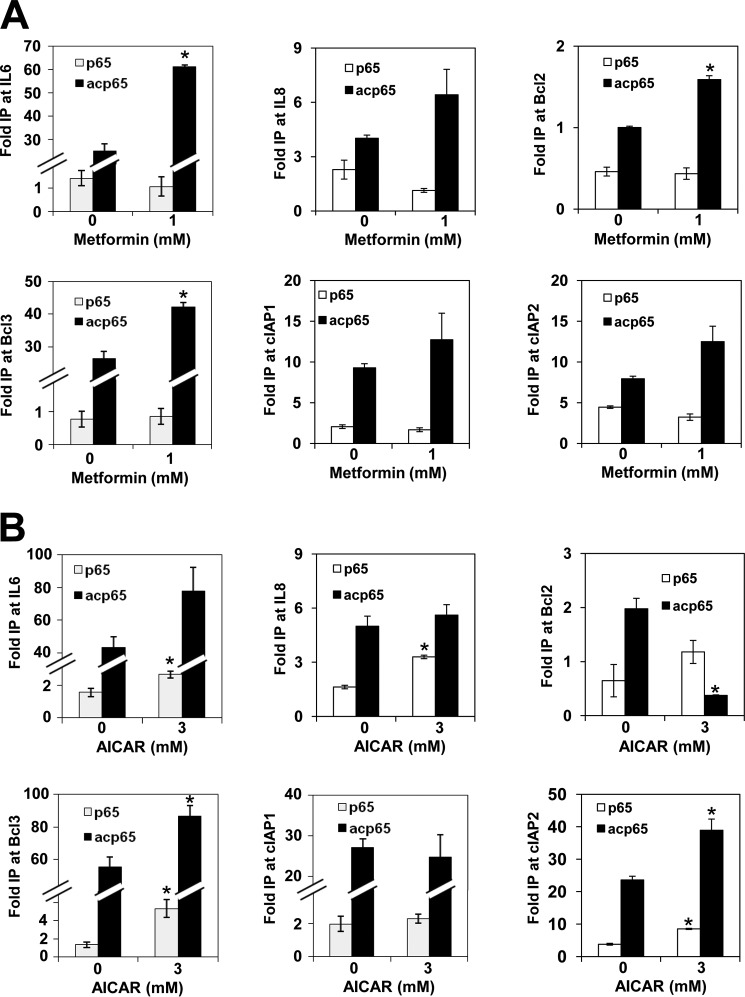
Many non-histone acetylated proteins are transcription factors, including p65 NFκB, p53, STAT1, STAT3, and MYC (3, 8). Acetylation of these proteins also regulates transcription, presumably independently of histone acetylation. For example, p65 can be acetylated at lysine residues 218, 221, and 310 by p300 and CBP acetyltransferases. Acetylation of Lys221 enhances DNA binding and impairs p65 binding to IκBα (44). Conversely, acetylation of Lys310 does not affect DNA or IκBα binding but is required for the full transcriptional activity of NFκB. We found that AMPK activation by metformin and AICAR increased acetylation of p65 NFκB at Lys310 in PC3 and OVCAR3 cells (Figs. 1B and 2). These results indicate that the NFκB activity might be increased as a result of AMPK activation. To determine whether p65 NFκB acetylated at Lys310 (acp65) recruitment to target promoters is also increased in metformin- or AICAR-treated cells, we measured occupancy of acp65 and p65 in the promoters of Bcl-2, Bcl-3, IL6, IL8, cIAP1, and cIAP2 genes. Our results show that even though metformin does not increase the total amount of p65 bound to the promoters, the occupancy of acp65 is 1.4–2.5 times higher in metformin-treated PC3 cells (Fig. 8A). Interestingly, AICAR treatment increased recruitment of acp65 only to promoters of Bcl-3, IL6, and cIAP2 but did not alter acp65 occupancy at IL8 and cIAP1 promoters and reduced acp65 occupancy at Bcl-2 promoter (Fig. 8B). acp65 NFκB is a substrate for the NAD+-dependent HDAC sirtuin SIRT1 (45), and AMPK activation with AICAR increases the NAD+/NADH ratio, resulting in SIRT1 activation (46, 47). However, by inhibiting complex I of the mitochondrial electron transport pathway and glycerophosphate dehydrogenase, metformin inhibits conversion of NADH to NAD+ and thus reduces the mitochondrial NAD+/NADH ratio (20, 48, 49). Indeed, treatment of lung carcinoma and osteosarcoma cells with metformin decreases the NAD+/NADH ratio almost 10-fold (50). It is possible that although AMPK activation by AICAR increases the NAD+/NADH ratio, resulting in SIRT1 activation and deacetylation of acp65, treatment with metformin does not increase the NAD+/NADH ratio, therefore maintaining high levels of acp65 NFκB at the Bcl-2 promoter.
FIGURE 8.
AMPK activation with metformin or AICAR increases recruitment of acetylated p65 to NFκB-regulated promoters. PC3 cells were treated with 0 and 1 mm metformin for 72 h (A) and 0 and 3 mm AICAR for 24 h (B). ChIP experiments were performed with antibodies against p65 and acp65. The experiments were repeated three times, and the results are shown as means ± S.D. (error bars). Values that are statistically different (p < 0.05) from the control (0 mm metformin or AICAR) are indicated by an asterisk. IP, immunoprecipitation.
Discussion
The key finding of this study is that the inexpensive and widely used antidiabetic drug metformin has a previously unrecognized effect of increasing acetylation of histones and non-histone proteins. The mechanism involves metformin-mediated AMPK activation, resulting in phosphorylation and inhibition of ACCA, reduced conversion of acetyl-CoA into malonyl-CoA, and increased acetylation of histone and non-histone proteins (Fig. 9). AMPK is an energy sensor and master regulator of metabolism and functions as a fuel gauge monitoring systemic and cellular energy status (17, 21). Activation of AMPK occurs when the intracellular AMP/ATP ratio increases and leads to a metabolic switch from anabolism to catabolism. AMPK activity is induced through phosphorylation of Thr172 by LKB1 (37). The metformin-mediated increase in protein acetylation is LKB1-dependent because suppression of LKB1 by LKB1 siRNA abolishes the metformin-induced protein acetylation. In addition, HeLa S3 cells that lack LKB1 expression and are unable to activate AMPK (38) fail to increase protein acetylation upon metformin treatment (Fig. 4B). When activated, AMPK phosphorylates key metabolic enzymes, such as ACCA, and transcription factors, thus inhibiting growth and synthesis of glucose, lipids, and proteins. At the same time, activated AMPK stimulates catabolism of fatty acids and glucose uptake. Additionally, AMPK inhibits cell proliferation by stabilizing the tumor suppressors tuberous sclerosis 2 protein and p53 and by regulating the cyclin-dependent kinase inhibitors p21 and p27; this implicates AMPK as a potential target for cancer treatment (51–54).
FIGURE 9.
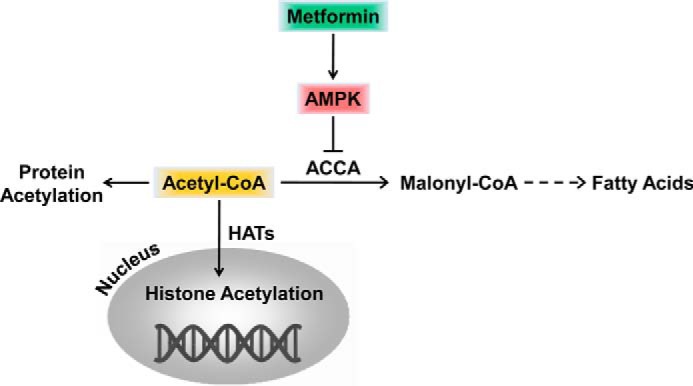
Model for the role of AMPK and metformin in the regulation of acetyl-CoA homeostasis and histone acetylation. Dashed arrows indicate more than one enzymatic conversion step. Solid arrows indicate direct reaction. Cytosolic and nuclear acetyl-CoA forms a single pool.
Although our results suggest that AMPK activation stimulates protein acetylation through ACCA inhibition, AMPK also regulates protein acetylation through the activity of HDACs. Activation of AMPK increases fatty acid oxidation, leading to production of ketone bodies, including β-hydroxybutyrate (βOHB), in the liver. The ketone bodies are the main energy source during starvation or prolonged exercise (55). Similarly to the HDAC inhibitor butyrate, βOHB specifically inhibits class I and II HDACs, increases acetylation of histones, and changes global transcription in kidney (56). Furthermore, class IIa HDACs (HDACs 4, 5, and 7) are hyperphosphorylated and excluded from the nucleus in the liver after metformin treatment in an AMPK-dependent manner (57). Another mechanism by which AMPK regulates protein acetylation involves sirtuin SIRT1 (58). AMPK enhances SIRT1 activity by increasing cellular NAD+ levels in skeletal muscle (46, 47). In this case, however, AMPK activation would be expected to produce a decrease in histone acetylation.
AMPK activation affects protein acetylation by four distinct mechanisms: (i) phosphorylation and inhibition of ACCA; (ii) inhibition of class I and II HDACs by increasing hepatic βOHB levels; (iii) inducing translocation of HDACs 4, 5, and 7 from the nucleus to the cytoplasm; and (iv) regulation of SIRT1 activity by modulating the NAD+/NADH ratio (59). Which mechanism AMPK utilizes probably depends on the particular cell type and the physiological conditions. Our results suggest that, in prostate and ovarian cancer cells with increased acetyl-CoA flux into fatty acids biosynthesis, activation of AMPK increases protein acetylation through phosphorylation and inhibition of ACCA, inhibition of fatty acid synthesis, and nucleocytosolic accumulation of acetyl-CoA. This conclusion is supported by the increased protein acetylation upon ACCA inhibition with TOFA or upon ACCA suppression by ACCA siRNA (Figs. 1 and 3). The same mechanism probably also operates in non-cancer cells with high acetyl-CoA flux into the fatty acid biosynthetic pathway. Because inhibition of ACCA in yeast also promotes histone acetylation due to increased availability of acetyl-CoA (13), it appears that regulation of protein acetylation by AMPK/ACCA is conserved in yeast and mammalian cells.
The recent interest in the use of AMPK agonists to support cancer prevention and treatment is based on clinical studies that show that the use of metformin is associated with significantly lower cancer incidence in diabetic patients (24, 60). The mechanism of metformin function in diabetes treatment consists of decreasing glucose production by gluconeogenesis in the liver through inhibition of mitochondrial respiratory chain complex I (19, 20, 23). The decrease in mitochondrial ATP production results in AMPK activation; however, the AMPK activation does not seem to be required for the antidiabetic effect of metformin (22, 23). Untreated diabetes type 2 is associated with a significantly increased risk of cancer, attributed mostly to the growth-promoting effect of chronically elevated plasma glucose and insulin levels (61, 62). The mechanism of metformin's antitumor effect is not completely understood. It appears that metformin inhibits tumor growth through both AMPK-independent and AMPK-dependent mechanisms. The AMPK-independent mechanism has been attributed to the improved glucose and insulin blood levels. The AMPK-dependent mechanism of metformin is mediated through the inhibition of mTORC1 signaling (63, 64) and the NFκB pathway (65). In addition, AMPK activation inhibits tumor growth through inhibition of fatty acid synthesis (66, 67). An increased rate of fatty acid synthesis is essential for tumor progression. Blocking lipid biosynthesis by inhibiting lipogenic enzymes, such as ACCA, fatty-acid synthase, ATP-citrate lyase, or stearoyl-CoA desaturase, decreases proliferation and increases apoptosis of cancer cells (68–71).
Our results show that, in addition to the above effects, AMPK activation results in increased histone acetylation. Active transcription generally correlates with increased acetylation of promoter histones; HDAC inhibitors have been developed for cancer treatment with the aim of increasing histone acetylation and restimulating expression of genes, such as tumor suppressor genes, that are silenced in cancer cells (43, 72, 73). In general, HDAC inhibitors increase histone acetylation and expression of p21 and proapoptotic genes and induce apoptosis (74, 75). Our results show that, similarly to HDAC inhibitors, activation of AMPK in PC3 cells increases histone acetylation within the p21 promoter and increases expression of p21 (Figs. 1, 2, 5, and 7).
The effect of AMPK on protein acetylation is not limited to histones. Our results show increased acetylation of α-tubulin upon AMPK activation with AICAR or metformin in both PC3 and OVCAR3 cells (Figs. 1 and 2). α-Tubulin is acetylated by tubulin acetyltransferase at Lys40 in the microtubule lumen and deacetylated by HDAC6 and SIRT2 (76–78). Tubulin acetylation marks stable microtubules and is required for polarity establishment and directional migration. Activation of HDAC6 results in a loss of α-tubulin acetylation and induces epithelial-mesenchymal transition, a hallmark of cancer progression (79). The increased acetylation of α-tubulin and stabilization of microtubules thus may represent additional mechanism of metformin's anticancer effect.
Our data indicate that AMPK regulates acetylation of histone and non-histone proteins. Activation of AMPK by the safe and inexpensive antidiabetic drug metformin results in increased acetylation of histones and altered transcriptional regulation, previously unrecognized effects of metformin. Metformin displays antiproliferative and proapoptotic properties toward cancer cells; however, the underlying mechanisms are not yet fully understood. The effect of metformin on protein acetylation and transcriptional regulation may represent one of these mechanisms and may provide a rationale for the development of novel combination cancer therapies involving metformin.
Experimental Procedures
Reagents
Metformin and TOFA were obtained from Cayman Chemical (Ann Arbor, MI). AICAR was obtained from LC Laboratories (Woburn, MA). All other reagents were molecular biology grade and were from Sigma.
Cell Culture
All cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA). Prostate cancer PC3 cells were cultured in Ham's F-12K (Kaighn's) medium (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) as described (80). HeLa S3 and ovarian cancer OVCAR3 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 20% FBS and antibiotics (81). Before treatment, cells were seeded (5 × 105 cells/ml) for 24 h in 6-well plates and grown at 37 °C with 5% CO2. Metformin was dissolved in PBS, pH 7.2; AICAR and TOFA were dissolved in DMSO and stored at −80 °C. An equivalent volume of either PBS or DMSO was used in all experiments as a solvent control. Cell viability was measured using trypan blue exclusion.
Western Blotting
Whole cell extracts were prepared as described previously (81). Denatured proteins were separated on 10 or 12% denaturing polyacrylamide gels. Western blotting was performed as described previously (82). Western blots were quantified using NIH ImageJ software (W. S. Rasband, National Institutes of Health, imagej.nih.gov/ij/, 1997–2011). The following primary antibodies were used: anti-histone H3 polyclonal antibody (ab1791, Abcam) at a dilution of 1:3000, anti-acH3 polyclonal antibody (07-353, Millipore) at a dilution of 1:2000, anti-acH4 (penta) polyclonal antibody (06-946, Millipore) at a dilution of 1:500, anti-AMPK monoclonal antibody (2603, Cell Signaling Technology) at a dilution of 1:1000, anti-AMPK phosphorylated at Thr172 (pAMPK) monoclonal antibody (2535, Cell Signaling Technology) at a dilution of 1:1000, anti-ACCA monoclonal antibody (3676, Cell Signaling Technology) at a dilution of 1:1000, anti-ACCA phosphorylated at Ser79 (pACCA) polyclonal antibody (3661, Cell Signaling Technology) at a dilution of 1:1000, anti-tubulin polyclonal antibody (2144, Cell Signaling Technology) at a dilution of 1:1000, anti-tubulin acetylated at Lys40 (acTubulin) polyclonal antibody (3971, Cell Signaling Technology) at a dilution of 1:500, anti-p65 NFκB polyclonal antibody (sc-372, Santa Cruz Biotechnology) at a dilution of 1:500, anti-acp65 polyclonal antibody (ab52175, Abcam) at a dilution of 1:500, and anti-actin polyclonal antibody (A5060, Sigma) at a dilution of 1:1000.
siRNA Transfections
Human AMPKα1/2 (sc-45312), ACCA (sc-40312), LKB1 (sc-35816), and non-silencing (sc-37007) siRNAs were obtained from Santa Cruz Biotechnology. Prior to transfection, PC3 cells were seeded into a 6-well plate and incubated in a humidified 5% CO2 atmosphere at 37 °C in antibiotic-free RPMI 1640 medium supplement with 10% FBS for 24 h to 80% confluence. For each transfection, an 80 nm final concentration of either non-silencing siRNA-A control or AMPK, ACCA, or LKB1 siRNA was used. Cells were transfected for 6 h in siRNA transfection medium (sc-36868) with siRNA transfection reagent (sc-29528) according to the manufacturer's instructions (Santa Cruz Biotechnology). After transfection, fresh medium with antibiotics was added, and the cells were grown for 24 h before treatment.
ChIP Assay
In vivo chromatin cross-linking and immunoprecipitation were performed essentially as described previously (83). Immunoprecipitation was performed with the following antibodies: anti-histone H3 antibody (ab1791, Abcam), anti-acH3 antibody (07-353, Millipore), anti-acH4 (penta) antibody (06-946, Millipore), anti-p65 NFκB antibody (sc-372, Santa Cruz Biotechnology), and anti-acp65 NFκB antibody (ab52175, Abcam). The primers used for real time PCR were as follows: p21, 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′; GAPDH-2, 71006 (Active Motif); MYOD1, GPH110002C(+)01A (SABiosciences); SAT2, GPH110003C(+)01A (SABiosciences); IGX1A (ChIP negative control), GPH100001C(−)01A (SABiosciences); ACTB, 71005 (Active Motif); Bcl-3, 5′-TTGCGGAGAGAAACACCTACT-3′ and 5′-CGCTCTCTCTGCCTCTGTT-3′; cIAP1, 5′-TGACTGGCAGGCAGAAATGA-3′ and 5′-TTTGCCCGTTGAATCCGAT-3′; cIAP2, 5′-TTCAGTAAATGCCGCGAAGAT-3′ and 5′-TGGTTTGCATGTGCACTGGT-3′; Bcl-2, 5′-TGCATCTCATGCCAAGGG-3′ and 5′-CCCCAGAGAAAGAAGAGGAGTT-3′; IL6, 5′-CCTCACCCTCCAACAAAGATTT-3′ and 5′-TTCATAGCTGGGCTCCTGGA-3′; and IL8, 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-GCTTGTGTGCTCTGCTGTCTC-3′.
Real Time RT-PCR
Total RNA was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA). The iScript one-step RT-PCR kit with SYBR Green (Bio-Rad) was used as a supermix, and 20 ng/μl RNA was used as template on a Bio-Rad MyIQ Single Color Real-time PCR Detection System (Bio-Rad). The primers used for mRNA quantification were as follows: Bcl-3, PPH02009D (Qiagen); Bcl-2, PPH00079B (Qiagen); IL6, PPH00560C (Qiagen); IL8, PPH0568A (Qiagen); cIAP1, PPH00340B (Qiagen); cIAP2, PPH00326B (Qiagen); and p21 and ribosomal subunit 18S (84). Primers for GAPDH (5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCTGTTGTCATACTTCTCATGG-3′) and ACTB (5′-CATGTACGTTGCTATCCAGGC-3′ and 5′-CTCCTTAATGTCACGCACGAT-3′) were obtained from PrimerBank (85).
Malonyl-CoA Assay
PC3 cells were grown to 70% confluence and treated with metformin for 72 h. Cells were harvested and lysed in 1 ml of radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.5 mm EGTA, 1% Triton, 0.1% sodium deoxycholate, 0.1% SDS, 140 mm NaCl, 1 mm PMSF). Malonyl-CoA was assayed in cell lysates by ELISA (ABIN366452, Antibodies-online GmbH) according to the manufacturer's instructions.
Statistical Analysis
The results represent at least three independent experiments. Numerical results are presented as means ± S.D. Data were analyzed using an InStat software package (GraphPad Software, San Diego, CA). Statistical significance was evaluated by one-way analysis of variance, and p < 0.05 was considered significant.
Author Contributions
L. G. conceived the project, conducted most of the experiments, analyzed the results, and wrote the paper. H. G. and I. V. developed the methods and analyzed the data. A. V. conceived the project, analyzed the data, wrote the paper, and coordinated the study.
Acknowledgments
We thank members of the Vancura and Vancurova laboratories for helpful comments.
This work was supported by National Institutes of Health Grants GM106324 and GM120710 (to A. V). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- AMPK
- AMP-activated protein kinase
- ACC
- acetyl-CoA carboxylase
- ACCA
- acetyl-CoA carboxylase-α
- LKB1
- liver kinase B1
- AICAR
- 5-amino-1-β-d-ribofuranosyl-1H-imidazole-4-carboxamide
- TOFA
- 5-(tetradecyloxy)-2-furoic acid
- mTOR
- mechanistic target of rapamycin
- CBP
- cAMP-response element-binding protein (CREB)-binding protein
- βOHB
- β-hydroxybutyrate
- acH3
- histone H3 acetylated at Lys14
- acH4
- hyperacetylated histone H4
- acp65
- p65 NFκB acetylated at Lys310
- acTubulin
- tubulin acetylated at Lys40
- pAMPK
- AMPK phosphorylated at Thr172
- pACCA
- ACCA phosphorylated at Ser79.
References
- 1. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 2. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 3. Farria A., Li W., and Dent S. Y. (2015) KATs in cancer: functions and therapies. Oncogene 34, 4901–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsu Y. C., Hsieh Y. H., Liao C. C., Chong L. W., Lee C. Y., Yu Y. L., and Chou R. H. (2015) Targeting post-translational modifications of histones in cancer therapy. Cell. Mol. Biol. 61, 69–84 [PubMed] [Google Scholar]
- 5. Takahashi H., McCaffery J. M., Irizarry R. A., and Boeke J. D. (2006) Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 [DOI] [PubMed] [Google Scholar]
- 6. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., and Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T. H., Haromy A., Hashimoto K., Zhang N., Flaim E., and Michelakis E. D. (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97 [DOI] [PubMed] [Google Scholar]
- 8. Pietrocola F., Galluzzi L., Bravo-San Pedro J. M., Madeo F., and Kroemer G. (2015) Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 [DOI] [PubMed] [Google Scholar]
- 9. Janke R., Dodson A. E., and Rine J. (2015) Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 31, 473–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai L., Sutter B. M., Li B., and Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wellen K. E., and Thompson C. B. (2012) A two-way street. Reciprocal regulation of metabolism and signaling. Nat. Rev. Mol. Cell Biol. 13, 270–276 [DOI] [PubMed] [Google Scholar]
- 12. Kim K. H. (1997) Regulation of mammalian acetyl-coenzyme A carboxylase. Annu. Rev. Nutr. 17, 77–99 [DOI] [PubMed] [Google Scholar]
- 13. Galdieri L., and Vancura A. (2012) Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 287, 23865–23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chow J. D., Lawrence R. T., Healy M. E., Dominy J. E., Liao J. A., Breen D. S., Byrne F. L., Kenwood B. M., Lackner C., Okutsu S., Mas V. R., Caldwell S. H., Tomsig J. L., Cooney G. J., Puigserver P. B., et al. (2014) Genetic inhibition of hepatic acetyl-CoA carboxylase activity increases liver fat and alters global protein acetylation. Mol. Metab. 3, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Travers M. T., and Barber M. C. (1997) Tissue-specific control of the acetyl-CoA carboxylase gene. Biochem. Soc. Trans. 25, 1215–1219 [DOI] [PubMed] [Google Scholar]
- 16. Carling D., Zammit V. A., and Hardie D. G. (1987) A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett. 223, 217–222 [DOI] [PubMed] [Google Scholar]
- 17. Hardie D. G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang M., Galdieri L., and Vancura A. (2013) The yeast AMPK homolog SNF1 regulates acetyl coenzyme A homeostasis and histone acetylation. Mol. Cell. Biol. 33, 4701–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El-Mir M. Y., Nogueira V., Fontaine E., Avéret N., Rigoulet M., and Leverve X. (2000) Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275, 223–228 [DOI] [PubMed] [Google Scholar]
- 20. Owen M. R., Doran E., and Halestrap A. P. (2000) Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 348, 607–614 [PMC free article] [PubMed] [Google Scholar]
- 21. Hardie D. G. (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 22. Foretz M., Hébrard S., Leclerc J., Zarrinpashneh E., Soty M., Mithieux G., Sakamoto K., Andreelli F., and Viollet B. (2010) Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Investig. 120, 2355–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Viollet B., Guigas B., Sanz Garcia N., Leclerc J., Foretz M., and Andreelli F. (2012) Cellular and molecular mechanisms of metformin: an overview. Clin. Sci. 122, 253–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans J. M., Donnelly L. A., Emslie-Smith A. M., Alessi D. R., and Morris A. D. (2005) Metformin and reduced risk of cancer in diabetic patients. BMJ 330, 1304–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tehlivets O., Scheuringer K., and Kohlwein S. D. (2007) Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771, 255–270 [DOI] [PubMed] [Google Scholar]
- 26. Beld J., Lee D. J., and Burkart M. D. (2015) Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Mol. Biosyst. 11, 38–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang C., Xu C., Sun M., Luo D., Liao D. F., and Cao D. (2009) Acetyl-CoA carboxylase-α inhibitor TOFA induces human cancer cell apoptosis. Biochem. Biophys. Res. Commun. 385, 302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al-Bassam J., and Corbett K. D. (2012) α-Tubulin acetylation from the inside out. Proc. Natl. Acad. Sci. U.S.A. 109, 19515–19516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verma I. M., Stevenson J. K., Schwarz E. M., Van Antwerp D., and Miyamoto S. (1995) Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 9, 2723–2735 [DOI] [PubMed] [Google Scholar]
- 30. Ghosh S., May M. J., and Kopp E. B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 31. Chen L. F., Mu Y., and Greene W. C. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies S. P., Carling D., Munday M. R., and Hardie D. G. (1992) Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP activated protein kinase, demonstrated using free-clamping. Effects of high fat diets. Eur. J. Biochem. 203, 615–623 [DOI] [PubMed] [Google Scholar]
- 33. Woods A., Munday M. R., Scott J., Yang X., Carlson M., and Carling D. (1994) Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269, 19509–19515 [PubMed] [Google Scholar]
- 34. Hardie D. G., Scott J. W., Pan D. A., and Hudson E. R. (2003) Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 546, 113–120 [DOI] [PubMed] [Google Scholar]
- 35. Ha J., Daniel S., Broyles S. S., and Kim K. H. (1994) Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 269, 22162–22168 [PubMed] [Google Scholar]
- 36. Fogarty S., and Hardie D. G. (2010) Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim. Biophys. Acta 1804, 581–591 [DOI] [PubMed] [Google Scholar]
- 37. Shaw R. J., Kosmatka M., Bardeesy N., Hurley R. L., Witters L. A., DePinho R. A., and Cantley L. C. (2004) The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. U.S.A. 101, 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tiainen M., Ylikorkala A., and Mäkelä T. P. (1999) Growth suppression by LKB1 is mediated by a G1 cell cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 96, 9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanner K. G., Langer M. R., Kim Y., and Denu J. M. (2000) Kinetic mechanisms of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 275, 22048–22055 [DOI] [PubMed] [Google Scholar]
- 40. Henry R. A., Kuo Y. M., and Andrews A. J. (2013) Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 52, 5746–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J. V., Carrer A., Shah S., Snyder N. W., Wei S., Venneti S., Worth A. J., Yuan Z.-F., Lim H.-W., Liu S., Jackson E., Aiello N. M., Haas N. B., Rebbeck T. R., Judkins A., et al. (2014) Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galdieri L., Zhang T., Rogerson D., Lleshi R., and Vancura A. (2014) Protein acetylation and acetyl-CoA metabolism in budding yeast. Eukaryot. Cell 13, 1472–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606 [DOI] [PubMed] [Google Scholar]
- 44. Chen L. f., Fischle W., Verdin E., and Greene W. C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 45. Salminen A., Kauppinen A., Suuronen T., and Kaarniranta K. (2008) SIRT1 longevity factor suppresses NF-κB-driven immune responses: regulation of aging via NF-κB acetylation? BioEssays 30, 939–942 [DOI] [PubMed] [Google Scholar]
- 46. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., and Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cantó C., Jiang L. Q., Deshmukh A. S., Mataki C., Coste A., Lagouge M., Zierath J. R., and Auwerx J. (2010) Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 11, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Madiraju A. K., Erion D. M., Rahimi Y., Zhang X.-M., Braddock D. T., Albright R. A., Prigaro B. J., Wood J. L., Bhanot S., MacDonald M. J., Jurzak M. J., Camporez J.-P., Lee H.-Y., Cline G. W., Samuel V. T., et al. (2014) Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Janzer A., German N. J., Gonzalez-Herrera K. N., Asara J. M., Haigis M. C., and Struhl K. (2014) Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 10574–10579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fendt S. M., Bell E. L., Keibler M. A., Olenchock B. A., Mayers J. R., Wasylenko T. M., Vokes N. I., Guarente L., Vander Heiden M. G., and Stephanopoulos G. (2013) Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jones R. G., Plas D. R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M. J., and Thompson C. B. (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 [DOI] [PubMed] [Google Scholar]
- 52. Mihaylova M. M., and Shaw R. J. (2011) The AMPK signaling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li W., Saud S. M., Young M. R., Chen G., and Hua B. (2015) Targeting AMPK for cancer prevention and treatment. Oncotarget 6, 7365–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Faubert B., Vincent E. E., Poffenberger M. C., and Jones R. G. (2015) The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 356, 165–170 [DOI] [PubMed] [Google Scholar]
- 55. Cahill G. F., Jr. (2006) Fuel metabolism in starvation. Annu. Rev. Nutr. 26, 1–22 [DOI] [PubMed] [Google Scholar]
- 56. Shimazu T., Hirschey M. D., Newman J., He W., Shirakawa K., Le Moan N., Grueter C. A., Lim H., Saunders L. R., Stevens R. D., Newgard C. B., Farese R. V. Jr., de Cabo R., Ulrich S., Akassoglou K., and Verdin E. (2013) Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., and Shaw R. J. (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lan F., Cacicedo J. M., Ruderman N., and Ido Y. (2008) SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 283, 27628–27635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Salminen A., Kauppinen A., and Kaarniranta K. (2016) AMPK/Snf1signaling regulates histone acetylation: impact on gene expression and epigenetic functions. Cell. Signal. 28, 887–895 [DOI] [PubMed] [Google Scholar]
- 60. Decensi A., Puntoni M., Goodwin P., Cazzaniga M., Gennari A., Bonanni B., and Gandini S. (2010) Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prev. Res. 3, 1451–1461 [DOI] [PubMed] [Google Scholar]
- 61. Currie C. J., Poole C. D., and Gale E. A. (2009) The influence of glucose-lowering therapies on cancer risk in type 2 diabetes. Diabetologia 52, 1766–1777 [DOI] [PubMed] [Google Scholar]
- 62. Jalving M., Gietema J. A., Lefrandt J. D., de Jong S., Reyners A. K., Gans R. O., and de Vries E. G. (2010) Metformin: taking away the candy for cancer? Eur. J. Cancer 46, 2369–2380 [DOI] [PubMed] [Google Scholar]
- 63. Dowling R. J., Zakikhani M., Fantus I. G., Pollak M., and Sonenberg N. (2007) Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 67, 10804–10812 [DOI] [PubMed] [Google Scholar]
- 64. Green A. S., Chapuis N., Maciel T. T., Willems L., Lambert M., Arnoult C., Boyer O., Bardet V., Park S., Foretz M., Viollet B., Ifrah N., Dreyfus F., Hermine O., Moura I. C., et al. (2010) The LKB1/AMPK signaling pathway has tumor suppressor activity in acute myeloid leukemia through the repression of mTOR-dependent oncogenic mRNA translation. Blood 116, 4262–4273 [DOI] [PubMed] [Google Scholar]
- 65. Salminen A., Hyttinen J. M., and Kaarniranta K. (2011) AMP-activated protein kinase inhibits NFκB signaling and inflammation: impact on healthspan and lifespan. J. Mol. Med. 89, 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zadra G., Priolo C., Patnaik A., and Loda M. (2010) New strategies in prostate cancer: targeting lipogenic pathways and the energy sensor AMPK. Clin. Cancer Res. 16, 3322–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flavin R., Zadra G., and Loda M. (2011) Metabolic alterations and targeted therapies in prostate cancer. J. Pathol. 223, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuhajda F. P. (2000) Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16, 202–208 [DOI] [PubMed] [Google Scholar]
- 69. Brusselmans K., Vrolix R., Verhoeven G., and Swinnen J. V. (2005) Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J. Biol. Chem. 280, 5636–5645 [DOI] [PubMed] [Google Scholar]
- 70. Brusselmans K., De Schrijver E., Verhoeven G., and Swinnen J. V. (2005) RNA interference-mediated silencing of the acetyl-CoA-carboxylase-α gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 65, 6719–6725 [DOI] [PubMed] [Google Scholar]
- 71. Fritz V., Benfodda Z., Rodier G., Henriquet C., Iborra F., Avancès C., Allory Y., de la Taille A., Culine S., Blancou H., Cristol J. P., Michel F., Sardet C., and Fajas L. (2010) Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 9, 1740–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Minucci S., and Pelicci P. G. (2006) Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 6, 38–51 [DOI] [PubMed] [Google Scholar]
- 73. Bolden J. E., Peart M. J., and Johnstone R. W. (2006) Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5, 769–784 [DOI] [PubMed] [Google Scholar]
- 74. Huang L., Sowa Y., Sakai T., and Pardee A. B. (2000) Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene 19, 5712–5719 [DOI] [PubMed] [Google Scholar]
- 75. Piekarz R. L., Robey R. W., Zhan Z., Kayastha G., Sayah A., and Abdeldaim A. H., Torrico S., and Bates S. E. (2004) T-cell lymphoma as a model for the use of histone deacetylase inhibitors in cancer therapy: impact of depsipeptide on molecular markers, therapeutic targets, and mechanisms of resistance. Blood 103, 4636–4643 [DOI] [PubMed] [Google Scholar]
- 76. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., and Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 77. Szyk A., Deaconescu A. M., Spector J., Goodman B., Valenstein M. L., Ziolkowska N. E., Kormendi V., Grigorieff N., and Roll-Mecak A. (2014) Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157, 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. North B. J., Marshall B. L., Borra M. T., Denu J. M., and Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 79. Gu S., Liu Y., Zhu B., Ding K., Yao T.-P., Chen F., Zhan L., Xu P., Ehrlich M., Liang T., Lin X., and Feng X.-H. (2016) Loss of α-tubulin acetylation is associated with TGF-β-induced epithelial-mesenchymal transition. J. Biol. Chem. 291, 5396–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Manna S., Singha B., Phyo S. A., Gatla H. R., Chang T. P., Sanacora S., Ramaswami S., and Vancurova I. (2013) Proteasome inhibition by bortezomib increases IL-8 expression in androgen-independent prostate cancer cells: the role of IKKα. J. Immunol. 191, 2837–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Singha B., Gatla H. R., Manna S., Chang T. P., Sanacora S., Poltoratsky V., Vancura A., and Vancurova I. (2014) Proteasome inhibition increases recruitment of IκB kinase β (IKKβ), S536P-p65, and transcription factor EGR1 to interleukin-8 (IL-8) promoter, resulting in increased IL-8 production in ovarian cancer cells. J. Biol. Chem. 289, 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Galdieri L., Zhang T., Rogerson D., and Vancura A. (2016) Reduced histone expression or a defect in chromatin assembly induces respiration. Mol. Cell. Biol. 36, 1064–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ramaswami S., Manna S., Juvekar A., Kennedy S., Vancura A., and Vancurova I. (2012) Chromatin immunoprecipitation analysis of NFκB transcriptional regulation by nuclear IκBα in human macrophages. Methods Mol. Biol. 809, 121–134 [DOI] [PubMed] [Google Scholar]
- 84. Laurenzana A., Balliu M., Cellai C., Romanelli M. N., and Paoletti F. (2013) Effectiveness of the histone deacetylase inhibitor (S)-2 against LNCaP and PC3 human prostate cancer cells. PLoS One 8, e58267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Spandidos A., Wang X., Wang H., and Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]