Abstract
Thermotolerant Bacillus coagulans is considered to be a more promising producer for bio-chemicals, due to its capacity to withstand harsh conditions. Two L-lactate dehydrogenase (LDH) encoding genes (ldhL1 and ldhL2) and one D-LDH encoding gene (ldhD) were annotated from the B. coagulans DSM1 genome. Transcriptional analysis revealed that the expression of ldhL2 was undetectable while the ldhL1 transcription level was much higher than that of ldhD at all growth phases. Deletion of the ldhL2 gene revealed no difference in fermentation profile compared to the wild-type strain, while ldhL1 single deletion or ldhL1ldhL2 double deletion completely blocked L-lactic acid production. Complementation of ldhL1 in the above knockout strains restored fermentation profiles to those observed in the wild-type strain. This study demonstrates ldhL1 is crucial for L-lactic acid production and NADH balance in B. coagulans DSM1 and lays the fundamental for engineering the thermotolerant B. coagulans strain as a platform chemicals producer.
Lactic acid has been produced by microbial fermentation for many years, and is used primarily in the food, pharmaceutical, cosmetic, and chemical industries1. It is also becoming a bulk building block for chemical production of the green polymer poly-lactic acid (PLA) that could be applied in bio-plastics2. A major advantage of microbial production of lactic acid over its chemical synthesis is that the enzyme-catalyzed microbial production results in a significantly higher optical purity1. Since only optically pure L- and D-lactic acid monomers could be used as PLA precursors, microbial fermentation is the preferred process for lactate production3,4.
Bacteria, fungi of the Rhizopus genus, yeast, cyanobacteria and various genetically modified strains have the ability to produce lactic acid5,6. Recently, several studies have shown that lactic-acid-producing thermophiles might have substantial advantages over the traditionally used mesophilic strains, such as Lactococcus lactis and Lactobacillus rhamnosus. Not only do these organisms have general advantages in terms of fermentation at high temperature (>50 °C), but can also utilize inexpensive carbon resources, require less complex nitrogen sources, and in most cases require no aeration7,8,9. Thermophilic Bacillus coagulans is one such microorganism displaying a number of these characteristics. Additionally, the ability of thermophilic B. coagulans to produce high levels of optically pure L-lactic acid at 50–55 °C is expected to minimize contamination in industrial-scale fermentations under non-sterile fermentation conditions, making it beneficial as an industrial strain and improving the commercial competitiveness of lactic acid production10,11.
L-Lactate dehydrogenase (L-LDH; EC 1.1.1.27) and D-lactate dehydrogenase (D-LDH; EC 1.1.1.28) are responsible for the conversion of pyruvic acid (produced by glycolysis) to L- and D-lactic acid, respectively12,13,14. Studies have shown that the relative catalytic efficiencies of ldhL- and ldhD-encoded products are crucial for the optical purity of lactic acid produced by Lactobacillus strains3. We previously showed that only L-LDH activity was detected in B. coagulans 2–6 (a L-lactic acid producer) under native conditions, and that ldhL transcription was much higher than that of other lactic-acid-metabolism-related genes at all growth phases. The high catalytic efficiency and high transcription levels of L-LDH may provide key explanations for the high optical purity of L-lactic acid produced by B. coagulans12. There are several ldhL genes annotated from the genome sequence in one B. coagulans strain while the individual functional role still remains unknown.
In this study, two genes encoding L-LDH (ldhL1 and ldhL2) were annotated from the whole-genome sequence of B. coagulans DSM1. Production of lactic acid and other metabolites was analyzed in both the wild-type and mutant strains. These results provide a first understanding of the roles played by different L-LDHs in thermophilic B. coagulans strains and lay the fundament for further engineering this strain to producing other useful bio-chemicals.
Results
Characterization of annotated LDH genes in B. coagulans DSM1
According to the whole-genome sequence of B. coagulans DSM1 (GenBank accession number: CP009709), three possible L-LDH-encoding genes (ldhL1, ldhL2, and ldhL3) and one D-LDH-encoding gene (ldhD) were discerned. Using the annotated genome sequence of two related B. coagulans strains, 36D1 (GenBank accession number: CP003056) and 2–6 (GenBank accession number: CP002472), as a refs 15, 16, 17, we determined that the enzyme encoded by ldhL3 was a leucine dehydrogenase (for strain 36D1) or a glutamate/leucine/phenylalanine/valine dehydrogenase (for strain 2–6). Based on these results, only the first two ldhL genes and one ldhD gene were chosen for subsequent experiments. After purification of the heterologously expressed His-tagged L-LDHs and D-LDH, the enzyme activities of the purified enzymes were determined. Both L-LDH1 (5.27 ± 0.14 U/mg) and L-LDH2 (4.53 ± 0.26 U/mg) were found to have catalytic activities. Furthermore, the activities of both enzymes were higher than that of D-LDH (1.87 ± 0.08 U/mg), indicating that the activity of L-LDHs in the conversion of pyruvate to L-lactic acid was higher than that of D-LDH in the conversion of pyruvate to D-lactic acid.
The transcription levels of ldhL1, ldhL2 and ldhD genes were also examined by quantitative real-time (RT)-PCR assay. Although enzymatic activity of L-LDH2 was detected in vitro, no transcription level was detected in ldhL2 in vivo. The transcription ratio of ldhL1 to ldhD was 80-fold at logarithmic phase, and the transcription levels of ldhL1 were higher than those of ldhD at different phase (data not shown), which is consistent with the results obtained in B. coagulans 2–612. The transcription level of ldhL1 per OD cells was also calculated. Results showed that the transcription level of ldhL1 was highest at the logarithmic phase and decreased when cell came to stationary phase and decline phase (Fig. 1).
Figure 1. The transcription level of ldhL1 per OD cells by RT-PCR analyses.
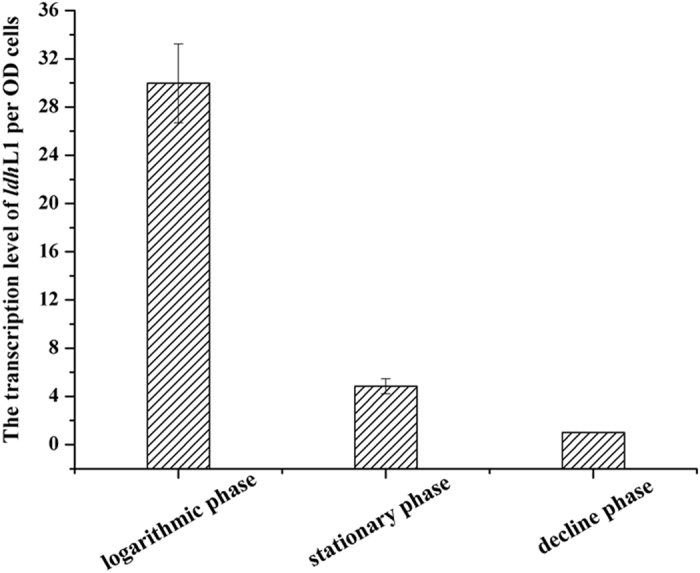
Error bars represent the standard deviations of the means for three independent experiments.
Although two L-LDH-encoding genes (ldhL1 and ldhL2) and one D-LDH-encoding gene (ldhD) were annotated in the complete genome of B. coagulans DSM1, active staining studies showed that D-LDH activity was not detectable (Fig. 2). Similar results were observed in B. coagulans 2–6 of our previous study, in which a D-LDH that catalyzes the production of D-lactic acid has been identified and the activity verified in vitro, while contribution of this enzyme to total lactic acid production appears to be minimal12. The in vitro enzymatic activities and active staining results observed in B. coagulans DSM1 were consistent with those of B. coagulans 2–6, while the individual role of ldhL genes remained unknown in both strains. The genes encoding L-LDH1 (ldhL1) was inferred to contribute to L-lactic acid synthesis in B. coagulans DSM1. To further determine the roles of two L-LDHs, ldhL1and ldhL2 were chosen as targets for gene deletion in the subsequent experiments.
Figure 2. Active staining of B. coagulans DSM1 wild-type and mutant strains following native-PAGE.
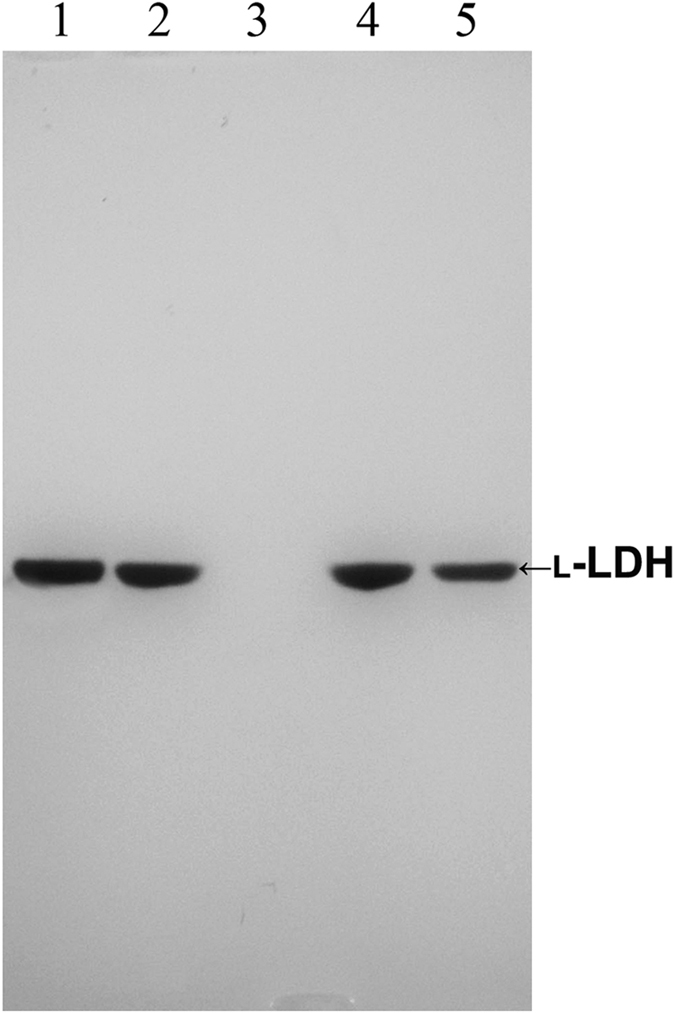
Cell extracts of B. coagulans DSM1 (lane 1), DSM1ΔldhL2 (lane 2), DSM1ΔldhL1ΔldhL2 (lane 3), and DSM1ΔldhL1ΔldhL2-ldhL1 (lane 4) were used for native-PAGE analysis. DL-Lactate was used as substrates for active staining. Lane 5, the commercial L-LDH enzyme (Sigma-Aldrich) used as a positive control.
Construction of the ldhL2-knockout strain
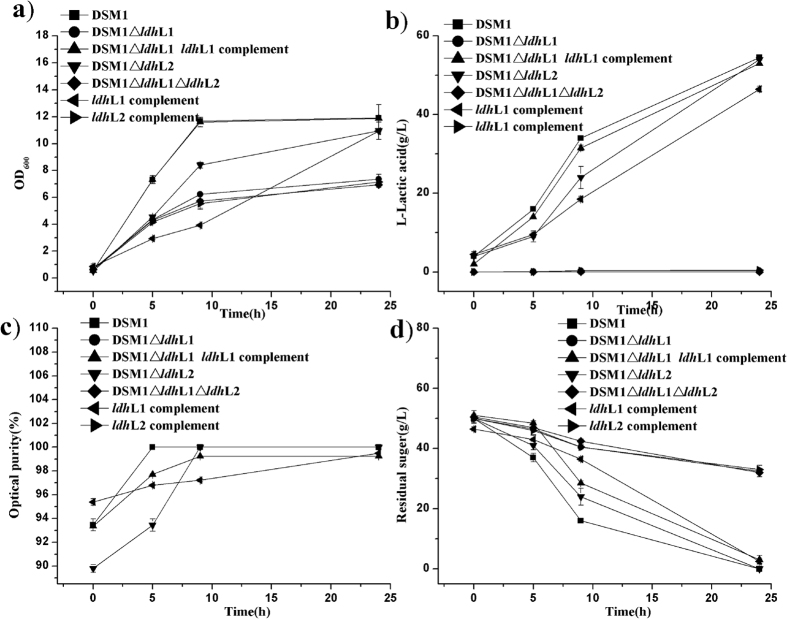
As no transcription level of ldhL2 was detected in vivo, the gene was first knocked out to confirm its role in lactate production. B. coagulans DSM1 (wild type) produces L-lactic acid as the primary fermentation product, with an optical purity of 99.8% and a specific productivity of 2.06 g/L/h after subtracting the initial concentration of L-lactate from seed culture (Fig. 3c). Acetic acid, formic acid, and ethanol were not detected in the fermentation broth (Table 1). Although a D-LDH that catalyzes the production of D-lactic acid has been identified and expressed in E. coli, the contribution of this enzyme to the total lactic acid produced appeared to be minimal in B. coagulans DSM1, since only trace amount (<0.1 g/L) of D-lactate was detected in the broth. Fermentation profile of DSM1ΔldhL2 was the same as that observed for the wild-type strain, except that the growth rate of DSM1ΔldhL2 was slightly slower than the parent strain (Fig. 3a), and trace amounts of acetic acid (0.39 g/L) were detected (Table 1). The same L-lactic acid optical purity (99.8%), with a consistent specific productivity of 2.04 g/L/h, was obtained in the fermentation broth of DSM1ΔldhL2, indicating that ldhL2 plays a minimal role in L-lactic acid produced by B. coagulans DSM1.
Figure 3. Fermentation profiles of B. coagulans DSM1 wild-type and mutant strains.
(a) A600 value, (b) L-lactic acid production, (c) optical purity of L-lactic acid, and (d) glucose consumption. Each data point represents the average of three replicates, with the error bars representing the standard deviation.
Table 1. Metabolite characteristics of B. coagulans DSM1wild-type and mutant strains.
| Products | DSM1 | DSM1△ldhL1 | DSM1△ldhL1 -ldhL1 | DSM1△ldhL2 | DSM1△ldhL1△ldhL2 | DSM1△ldhL1△ldhL2-ldhL1 | DSM1△ldhL1△ldhL2-ldhL2 |
|---|---|---|---|---|---|---|---|
| L-Lactic acid (g/L) | 49.40 ± 0.56 | 0.17 ± 0.01 | 50.39 ± 0.37 | 48.9 ± 0.38 | ND | 48.22 ± 0.32 | 0.49 ± 0.01 |
| Productivity of L-lactic acid (g/L/h) | 2.06 ± 0.02 | 0.01 ± 0 | 2.10 ± 0.01 | 2.04 ± 0.02 | ND | 2.01 ± 0.01 | 0.02 ± 0 |
| D-Lactic acid (g/L) | 0.10 ± 0 | 0.32 ± 0.01 | 0.39 ± 0.01 | 0.10 ± 0 | 0.26 ± 0.01 | 0.17 ± 0 | 0.65 ± 0.03 |
| Ethanol (g/L) | NDa | 1.40 ± 0.03 | 0.80 ± 0.04 | ND | 0.19 ± 0.01 | ND | 0.99 ± 0.05 |
| Oxaloacetic acid (g/L) | ND | ND | ND | ND | 0.02 ± 0 | ND | ND |
| Formic acid (g/L) | ND | 1.22 ± 0.09 | ND | ND | 1.14±0.02 | ND | 1.26 ± 0.08 |
| Acetic acid (g/L) | ND | 1.13 ± 0.12 | 0.90 ± 0.06 | 0.39 ± 0.03 | 2.54 ± 0.23 | 1.89 ± 0.09 | 1.08 ± 0.02 |
| Pyruvic acid (g/L) | 1.25 ± 0.12 | 1.01 ± 0.04 | 0.22 ± 0.03 | ND | 1.98 ± 0.11 | 0.21 ± 0.03 | 1.23 ± 0.03 |
aND, not detected.
Construction of the ldhL1-knockout and ldhL1 ldhL2 double deletion strains
Deletion of ldhL1 resulted in significantly reduced cell-growth rates to value ~50% of those observed in the wild-type strain (Fig. 3a), and only 34% glucose was consumed by the DSM1ΔldhL1 strain (Fig. 3d). Small amounts of L-lactic acid (0.17 g/L), with a productivity of 0.01 g/L/h, were detected. Compared with the wild type strain, ethanol (1.40 g/L), formic acid (1.22 g/L) and acetic acid (1.13 g/L) were produced by DSM1ΔldhL1 (Table 1). Double deletion of gene ldhL1ldhL2 (DSM1ΔldhL1ldhL2) significantly reduced cell-growth rates and glucose consumption, which was the same situation with strain DSM1ΔldhL1 (Fig. 3a and d). No L-lactic acid was detected in the fermentation broth, while acetic acid (2.54 g/L) become the major product, with only a little amount of D-lactic acid (0.26 g/L) present. The main product shifted from lactic acid to acetic acid, formic acid, and ethanol, as well as increased concentration of pyruvate as compared with DSM1 and DSM1ΔldhL2 results (Table 1). These results indicatied that ldhL1 plays a central role in producing optically pure L-lactic acid in B. coagulans DSM1.
LdhL1 is essential for optically pure L-lactic acid synthesis in strain DSM1
To determine whether ldhL1 is the key factor affecting L-lactic acid production in B. coagulans DSM1, ldhL-complemented strains (DSM1ΔldhL1-ldhL1, DSM1ΔldhL1ΔldhL2-ldhL1 and DSM1ΔldhL1ΔldhL2-ldhL2) were constructed. Complementation of gene ldhL1 (DSM1ΔldhL1-ldhL1 and DSM1ΔldhL1ΔldhL2-ldhL1) with its native promoter expressed in plasmid pNW33n restored cell growth and L-lactic acid production profiles to wild-type level (Fig. 3). The main products of two ldhL1 complemented strains were shifted back to L-lactic acid with an optical purity of 99.4% and specific productivities of 2.01 g/L/h and 2.10 g/L/h, which is consistent with that of the wild-type. Although trace amounts of acetic acid and ethanol were detected by HPLC, the amounts of formic acid were reduced to values below the detection limit. Notably, the concentration of D-lactic acid also decreased to the consistent level with the wild-type (Table 1). Meanwhile, ldhL2 complementation resulted in no significant change in glucose consumption and cell growth compared with double knockout strain (DSM1ΔldhL1ΔldhL2), except that small amounts of L-lactic acid (0.49 g/L) was produced. Complementing the double mutant with ldhL2 did not lead to high L-lactic acid production as the wild type strain.
Enzyme expression in cell extracts were also confirmed by active staining of LDHs after native-PAGE. As expected, L-LDH activity was detected in B. coagulans DSM1 and native-PAGE showed that L-LDH activity was still detected in B. coagulans DSM1ΔldhL2 (Fig. 2). Further gene deletion of ΔldhL1 resulted in no detectable L-LDH activity according to native-PAGE results, while L-LDH activity was detected in the ldhL1-complementary strain (DSM1ΔldhL1ΔldhL2-ldhL1). These results showed that although two L-LDH-encoding genes were annotated in the B. coagulans DSM1 genome, the catalytic efficiency of the enzyme encoded by ldhL1 involving L-lactic acid production was much higher than that observed in the enzyme encoded by ldhL2. For all strains, only one fragment corresponding to L-LDH activity was observed when gels were soaked with DL-lactate, indicating that only L-LDH activities were detected in the B. coagulans DSM1 strains, and that D-LDH activity might be too low to be detected in all the strains12.
Discussion
B. coagulans DSM1 is a homofermentative L-lactic acid producer, with a high optical purity of 99.8%. L-nLDH is responsible for the synthesis of L-lactic acid. However, two L-LDH-encoding genes (ldhL1 and ldhL2) and one D-LDH-encoding gene (ldhD) were annotated in the complete genome of B. coagulans DSM1. Similar results were observed in other B. coagulans strain12. The individual role of these genes in L-lactic acid production remained unknown. Thus, the genes responsible for L-lactic acid production and their mechanisms were the focus of this study.
Construction of a targeted gene deletion system is one of the most effective approaches for analyzing gene function in vivo. As the genetic manipulation of parent strain, such as B. coagulans 2–6, is currently not available, its phylogenetically close type strain B. coagulans DSM1 was used in this study since the genome sequence of B. coagulans 2–6 shares a high similarity (>99%) with B. coagulans DSM115,16. Deletion of ldhL1 led to alteration of the main product of B. coagulans DSM1 from L-lactic acid to acetic acid and other byproducts. Compared with ldhL2, ldhL1 plays a central role in producing optically pure L-lactic acid in B. coagulans DSM1.
LDHs play a complex role in B. coagulans by not only catalyzing pyruvate transformation to lactic acid, but also by catalyzing NADH oxidation. As a predominant redox product of catabolism, NADH plays an important role in over 700 biochemical reactions, a number of which constitute synthetically practical enzymatic reactions18. Under micro-aerobic conditions, NADH generated from glycolysis is mainly consumed by two metabolic pathways: the lactate-production pathway and the alcohol-production pathway19. The primary product of glucose fermentation in B. coagulans DSM1 is L-lactic acid (~98.5% of the fermentation products), indicating that the lactic acid-production pathway constitutes the main NADH-metabolism pathway in B. coagulans DSM1. The physiological role of LDHs in bacteria is to balance regeneration of NAD+ during fermentation, which is an important step in the metabolism and energy conversion of living cells, since it allows re-oxidation of NADH, which is necessary for glycolysis. In this study, ldhL1 knockout resulted in the complete loss of LDH function affecting intracellular NADH distribution and eliminating the primary route for NADH oxidation. The redox imbalance that resulted from the elimination of the main NAD+-regeneration pathway affected cellular metabolism and thus impaired growth rates20,21.
To maintain cellular redox balance, pyruvate produced from the glycolytic breakdown of carbohydrates would be metabolized by several alternative pathways in the absence of active LDH22. The common NADH-oxidation pathway in Gluconobacter oxydans involves pyruvate oxidation to acetyl-coenzyme A (CoA), followed by: 1) CoA entry into the TCA cycle, which provides carbon and energy resources for bacterial growth; or 2) accumulated pyruvate entry into a non-energy-generating pathway, with acetate as the final product23. It was also suggested that accumulation of intracellular NADH might enhance hydrogen production by a putative membrane-bound hydrogenase in Enterobacter aerogenes19. In this study, ldhL mutants exhibited increased yields of acetic acid as compared to the wild-type strain, which showed no acetic acid production (Table 1). The acetic acid yield of the DSM1ΔldhL1ΔldhL2 strain (2.54 g/L) was significantly higher than that of the DSM1ΔldhL2 strain (0.39 g/L). Compared with G. oxydans, B. coagulans DSM1 preferred to use an energy-generating pathway, with acetic acid production, to metabolize the accumulated NADH following loss of LDH function, because acetate kinase encoding gene exists in B. coagulans DSM1 genome. Formic acid and ethanol were the other products accumulated in the DSM1ΔldhL1ΔldhL2. Formic acid is produced by pyruvate formate lyase (PFL). In order to maintain redox balance, the PFL-produced acetyl-CoA needs to be converted to acetate and ethanol, which could well explain the product alteration in the LDH null strain (Figure S1 in Supporting Information)24. Furthermore, some glucose was consumed for CoA production, which entry into the TCA cycle for bacterial growth. The DSM1ΔldhL1ΔldhL2 strain also produced increased amount of D-lactic acid (0.26 g/L) that were twice higher than the values in wild-type strain or DSM1ΔldhL2. One possible explanation is that the elimination of the main NAD+-regeneration pathway and the apparent lack of an alternative pathway in the DSM1ΔldhL1ΔldhL2 strain provoked the increased expression of other NADH consumption enzymes, such as D-LDH25.
In conclusion, two L-LDH-encoding genes (ldhL1 and ldhL2) were identified in the B. coagulans DSM1 genome. Although the functional role of ldhL2 needs to be further investigated, the results obtained in this study confirmed that only ldhL1 is the key encoded gene enabling L-LDH activity and L-lactic acid production in B. coagulans DSM1. This study provides useful information for the further engineering of B. coagulans DSM1 into a platform for production of other value-added chemicals.
Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1 in Supporting Information. As the genetic manipulation of strain B. coagulans 2–6 is currently not available, its phylogenetically close type strain B. coagulans DSM1 was used in this study15,16. B. coagulans DSM1 is a homofermentative producer of L-lactic acid to an optical purity of 99.8%, which was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). For genetic engineering, B. coagulans DSM1 was grown in BC medium at 45 °C, 120 rpm26. For lactic acid fermentation, B. coagulans DSM1 was grown in glucose yeast-extract carbonate (GYC) medium (50 g glucose, 10 g yeast extract, and 30 g CaCO3 per liter) at 50 °C, 120 rpm9. The pH value was maintained at 6.2~6.5 with the addition of CaCO3 and the inoculum volume was 10% (v/v). Escherichia coli was grown aerobically in Luria-Bertani (LB) medium at 37 °C, 200 rpm. Kanamycin (Kan) and chloramphenicol (Cm) were added to the medium when required at concentrations of 40 μg/mL Kan for E. coli, and 7 μg/mL and 25 μg/mL Cm for B. coagulans DSM1 and E. coli, respectively.
Cloning, expression, and assay of enzymes responsible for lactic acid production
Homologous genes encoding both L-LDH (ldhL1 and ldhL2) and D-LDH (ldhD) were amplified by polymerase chain reaction (PCR) from B. coagulans DSM1 genomic DNA using Pfu DNA polymerase (Takara Bio Co., Dalian, China). The primers are listed in Table S2 in Supporting Information. Three DNA fragments were cloned into the pET-28a expression vector, and the recombinant plasmids were transformed into competent E. coli BL21 (DE3) cells, respectively. For protein expression, the cells were grown to an optical density of A600 = 0.6–0.8 at 37 °C, then induced with 0.5 mM isopropyl-β-d-1- thiogalactopyranoside and grown for an additional 12 h at 18 °C.
The purification procedures and activity assays of the enzymes were employed as previously described12. Enzymatic assays were performed in transparent 96-well plates, and the reaction mixture containing 100 mM sodium phosphate (pH 6.5), 200 μM NADH, and 0.1 mg/ml enzyme. The mixture was pre-incubated at 50 °C for 10 min. To start the reaction, sodium pyruvate was added to a final concentration of 20 mM, and NADH oxidation was monitored at 340 nm in a 96-well plate reader. One unit (1 U) of LDH activity was defined as the amount of enzyme required to reduce 1 μmol nicotinamide adenine dinucleotide (NAD) per minute.
Transcriptional analysis of lactate dehydrogenase gene expression
Determination of the transcription levels of ldhL1, ldhL2 and ldhD genes was analyzed by quantitative RT-PCR according to the literature27. Initially, B. coagulans DSM1 were inoculated in GYC broth at 50 °C to measure the growth curve and time courses of optical purity during fermentation for the determination of sample points. Cells of the three representative strains were harvested by centrifugation (5,000 × g for 10 min, 4 °C) for RNA isolation by using an E.Z.N.A.TM Bacterial RNA Kit (Omega). Total RNA concentration was determined from the absorbance at 260 nm (NanoVue, GE). cDNA copies were synthesized with a FastQuant RT Kit (with gDNase) (Tiangen, China) and amplified with SYBR Premix Ex Taq (TaKaRa, China). The threshold cycle for each PCR with different concentrations of cDNA was determined and compared against a standard DNA (16 S rRNA gene) that was also analyzed at the same time. The 2−△△Ct relative quantification method was used to determine the mRNA levels.
Construction of B. coagulans DSM1 ldhL gene knockout mutants
The ldhL gene knockouts were performed by using plasmid pMH77 with the thermosensitive lactococcal pSH71/pWV01 replicon and the procedures were conducted according to the reference 26. The primers used for gene deletion are listed in Table S2 in Supporting Information. For knocking out ldhL2 gene from DSM1, the flanking regions of the ldhL2 gene were first PCR amplified from B. coagulans DSM1 genomic DNA using the primers L2 up-For and L2 up-Rev (upstream, 1000 bp) and L2 down-For and L2 down-Rev (downstream, 1000 bp), respectively. After gel purification, overlap extension PCR was performed, wherein the upstream and downstream regions were fused using the primers L2 up-For and L2 down-Rev. The resulting PCR product was gel purified, digested with EcoRI and XhoI, and cloned into pMH77, resulting in plasmid pMH77-ΔldhL2. Plasmid pMH77-ΔldhL2 was first transformed into Lactococcus lactis MG1363. And then the extracted plasmid pMH77-ΔldhL2 was transformed into B. coagulans DSM1 by electroporation.
For integration, a colony harboring the integration plasmid was cultured overnight at 45 °C, followed by a temperature shift to 60 °C and further incubation for 12 h. A dilution series was plated on BC plates containing Cm and incubated at 60 °C until colonies appeared. Colonies were picked and tested for first single crossover recombination by PCR analysis. Then the right colonies were incubated in BC liquid broth without Cm overnight at 45 °C, 120 rpm, and then a dilution series was streaked onto BC plates without Cm and incubated overnight at 45 °C. Colonies were sequentially streaked onto BC plates with and without Cm and incubated overnight at 45 °C. The colonies that grew on BC plates without Cm, but did not grow on those with Cm, were picked out for PCR analysis using the primers L2 For and L2 Rev.
For knocking out ldhL1 gene, the involved steps are similar to those described for DSM1ΔldhL2. The ldhL1-flanking regions were first PCR amplified from B. coagulans DSM1 genomic DNA using the primers L1 up-For and L1 up-Rev (upstream, 1000 bp) and L1 down-For and L1 down-Rev (downstream, 1000 bp), respectively. After gel purification, overlap extension PCR was performed, wherein the upstream and downstream regions were fused using the primers L1 up-For and L1 down-Rev. Then, the ldhL1-flanking regions were cloned into the integration vector to construct the plasmid of pMH77-ΔldhL1, and introduced into B. coagulans DSM1 by electroporation to construct the double gene-deletion strain (DSM1ΔldhL1). The gene knockout mutant was confirmed by PCR using the primer pairs L1 For/L1 Rev.
The involved steps of ldhL1ldhL2 gene double deletion are similar to those described for DSM1ΔldhL1. Plasmid pMH77-ΔldhL1 was introduced into B. coagulans DSM1ΔldhL2 by electroporation to construct the double gene-deletion strain (DSM1ΔldhL1ΔldhL2). The gene knockout mutant was confirmed by PCR using the primer pairs L1 For/L1 Rev.
Construction of ldhL complementation strain
For construction of the ldhL1 complementation plasmid pNW33n-ldhL1, the ldhL1 gene with its native promoter (1182-bp upstream of the gene) was amplified from B. coagulans DSM1 genomic DNA using the primers ldhL1-For and ldhL1-Rev (Table S2 in Supporting Information). The fragment was cloned into pNW33n by using the restriction sites of HindIII and SacI. The resulting plasmid (pNW33n-ldhL1) was introduced into B. coagulans DSM1ΔldhL1 and B. coagulans DSM1ΔldhL1ΔldhL2 by electroporation to construct the ldhL1-complemented strains.
For construction of the ldhL2 complementation plasmid pNW33n-ldhL2, the ldhL2 gene with its native promoter sequence was amplified from B. coagulans DSM1 genomic DNA using the primers ldhL2-For and ldhL2-Rev (Table S2 in Supporting Information). The fragment was cloned into pNW33n, and the resulted plasmid (pNW33n-ldhL2) was introduced into B. coagulans DSM1ΔldhL1ΔldhL2 by electroporation to construct the ldhL2-complemented strain.
Active staining of L-LDHs
B. coagulans DSM1 wild-type, DSM1ΔldhL2, DSM1ΔldhL1ΔldhL2, and DSM1 ldhL1-complementation strains were grown in 50 mL GYC medium to logarithmic phase at 50 °C, 120 rpm. Exponentially growing cells were harvested by centrifugation (10,540 g, 10 min, 4 °C) and washed with 0.85% (w/v) physiological saline. Cell pellets were subsequently suspended in 100 mM potassium phosphate buffer (pH 7.0) and disrupted by sonication in an ice bath. After centrifugation at 12,000 g for 10 min, the supernatants were used as the crude enzymes.
Active staining of L-LDHs was performed according to a previous report with some modifications12. Briefly, crude enzymes of the four representative strains were concentrated by ultrafiltration. and separated by native-polyacrylamide gel electrophoresis (PAGE) on gradient 4–20% native polyacrylamide gels. Protein concentrations were determined with a Bradford protein assay kit (Bio-Rad). Loading quantity of each sample was 5 μg. After electrophoresis, gels were soaked with 100 mM Tris-HCl buffer (pH 8.0) containing 0.1 mM phenazinemethosulfate, 0.1 mM nitrotetrazolium blue chloride, 2 mM NAD, and 100 mM DL-lactate.
Analytical methods
The growth curves were measured at A600 by a 7200 Visible Spectrophotometer (UNICO, Shanghai, China). L-Lactic acid concentrations were measured using a SBA-40C biosensor analyzer (Institute of Biology, Shandong Academy of Sciences, China). The optical purity of L-lactic acid was analyzed using high-performance liquid chromatography (HPLC, Agilent 1260 Series, Hewlett-Packard, Palo Alto, CA, USA) equipped with a chiral column (MCI GEL CRS10W, Tokyo, Japan)28. The mobile phase consisted of 2 mM CuSO4 at a flow rate of 0.5 mL/min (25 °C), with UV detection at 254 nm. The optical purity of L-lactic acid was defined as [L-lactic acid/(L-lactic acid + D-lactic acid) × 100%]. Glucose, ethanol and organic acids (oxaloacetic acid, formic acid, acetic acid, pyruvic acid, maletate acid and lactic acid) were measured at 210 nm by using an HPLC system equipped with an organic-acid column (MCI GEL CRS10W) with a UV and differential detector. The mobile phase consisted of 6 mM H2SO4 at a flow rate of 0.5 mL/min (55 °C).
Additional Information
How to cite this article: Sun, L. et al. Contributory roles of two L-lactate dehydrogenases for L-lactic acid production in thermotolerant Bacillus coagulans. Sci. Rep. 6, 37916; doi: 10.1038/srep37916 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (31270108) and Key International Cooperation Project from Chinese Academy of Sciences (155112KYSB20160010). B.Y. is supported by the Youth Innovation Promotion Association, Chinese Academy of Sciences.
Footnotes
Author Contributions B.Y. and L.W. designed the project; L.S., and C.Z. performed experiments; L.W., L.S., Y.W. and P. L. analyzed the data; L.W. and B.Y. wrote the manuscript. All authors read and approved the final manuscript.
References
- Bosma E. F., van der Oost J., de Vos W. M. & van Kranenburg R. Sustainable production of bio-based chemicals by extremophiles. Curr. Biotech. 2, 360–379 (2013). [Google Scholar]
- Ma K., Maeda T., You H. & Shirai Y. Open fermentative production of L-lactic acid with high optical purity by thermophilic Bacillus coagulans using excess sludge as nutrient. Bioresour. Technol. 151, 28–35 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng Z. et al. Relative catalytic efficiencies of ldhL- and ldhD-encoded products is crucial for optical purity of lactic acid produced by Lactobacillus strains. Appl. Environ. Microbiol. 78, 3480–3483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang L. M., Ju J. S., Yu B. & Ma Y. H. Efficient production of polymer-grade D-lactate by Sporolactobacillus laevolacticus DSM442 with agricultural waste cottonseed as the sole nitrogen source. Bioresour. Technol. 142, 186–191 (2013). [DOI] [PubMed] [Google Scholar]
- Okano K., Tanaka T., Ogino C., Fukuda H. & Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 85, 413–423 (2010). [DOI] [PubMed] [Google Scholar]
- Mohamed A. A., Yukihiro T. & Kenji S. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 31, 877–902 (2013). [DOI] [PubMed] [Google Scholar]
- Qin J. Y. et al. Non-sterilized fermentative production of polymer-grade L-lactic acid by a newly isolated thermophilic strain Bacillus sp. 2-6. PLoS One 4, e4359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. M. et al. Efficient production of L-lactic acid from corncob molasses, a waste by-product in xylitol production, by a newly isolated xylose utilizing Bacillus sp. strain. Bioresour. Technol. 101, 7908–7915 (2010). [DOI] [PubMed] [Google Scholar]
- Peng L. L. et al. Bacillus sp. strain P38: an efficient producer of L-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour. Technol. 49, 169–176 (2013). [DOI] [PubMed] [Google Scholar]
- Wang L. M. et al. Jerusalem artichoke powder: a useful material in producing high-optical-purity L-lactate using an efficient sugar-utilizing thermophilic Bacillus coagulans strain. Bioresour. Technol. 130, 174–180 (2013). [DOI] [PubMed] [Google Scholar]
- Peng L. L. et al. Efficient open fermentative production of polymer-grade L-lactate from sugarcane bagasse hydrolysate by thermotolerant Bacillus sp. strain P38. PLoS One 9, e107143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. M., Cai Y. M., Zhu L. F., Guo H. L. & Yu B. Major role of NAD-dependent lactate dehydrogenases in high optically pure L-lactic acid production by thermophilic Bacillus coagulans. Appl. Environ. Microbiol. 80, 7134–7141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. F. et al. NADP+-preferring D-lactate dehydrogenase from Sporolactobacillus inulinus. Appl. Environ. Microbiol. 81, 6294–6301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. Efficient production of enantiomerically pure D-phenyllactate from phenylpyruvate by structure-guided design of an engineered D-lactate dehydrogenase. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-016-7456-1 (2016). [DOI] [PubMed] [Google Scholar]
- Su F. et al. Genome sequence of thermophilic strain Bacillus coagulans 2-6, an efficient producer of high optical purity L-lactic acid. J. Bacteriol. 193, 4563–4564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L. et al. Complete genome sequences for 35 biothreat assay-relevant Bacillus species. Genome Announc. 3, e00151–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee M. S. et al. Complete genome sequence of a thermotolerant sporogenic lactic acid bacterium, Bacillus coagulans strain 36D1. Stand. Genomic Sci. 5, 331–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckbecker A., Gröger H. & Hummel W. Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Adv. Biochem. Engin./Biotechnol. 120, 195–242 (2010). [DOI] [PubMed] [Google Scholar]
- Zhao H. et al. Disruption of lactate dehydrogenase and alcohol dehydrogenase for increased hydrogen production and its effect on metabolic flux in Enterobacter aerogenes. Bioresour. Technol. 194, 99–107 (2015). [DOI] [PubMed] [Google Scholar]
- Romero S., Merino E., Bolívar F., Gosset G. & Martinez A. Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl. Environ. Microbiol. 73, 5190–5198 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma E. F. et al. Establishment of markerless gene deletion tools in thermophilic Bacillus smithii and construction of multiple mutant strains. Microb. Cell Fact. 14, 99–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Yebra M. J., Galán J. L., Monedero V. & Pérez-Martínez G. Pleiotropic effects of lactate dehydrogenase in activation in Lactobacillus casei. Res. Microbiol. 156, 641–649 (2005). [DOI] [PubMed] [Google Scholar]
- Sheng B. et al. Utilization of D-lactate as an energy source supports the growth of Gluconobacter oxydans. Appl. Environ. Microbiol. 81, 4098–4110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Rhee M. S., Ingram L. O. & Shanmugam K. T. Physiological and fermentation properties of Bacillus coagulans and a mutant lacking fermentative lactate dehydrogenase activity. J. Ind. Microbiol. Biotechnol. 38, 441–450 (2010). [DOI] [PubMed] [Google Scholar]
- Okano K. et al. Efficient production of optically pure D-lactic acid from raw corn starch by using a genetically modified L-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum strain. Appl. Environ. Microbiol. 75, 462–467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács Á. T., van Hartskamp M., Kuipers O. P. & van Kranenburg R. Genetic tool development for a new host for biotechnology, the thermotolerant bacterium Bacillus coagulans. Appl. Environ. Microbiol. 76, 4085–4088 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. F., Li Y. F., Wang L. M., Wang Y. P. & Yu B. Diammonium phosphate stimulates transcription of L-lactate dehydrogenase leading to increased L-lactate production in the thermotolerant Bacillus coagulans strain. Appl. Microbiol. Biotechnol. 100, 6653–6660 (2016). [DOI] [PubMed] [Google Scholar]
- Zhu L. F., Xu X., Wang L. M., Dong H. & Yu B. The D-lactate dehydrogenase from Sporolactobacillus inulinus also possessing reversible deamination activity. PLoS One 10, e0139066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.