Abstract
Triple-negative breast cancer (TNBC) was regarded as the most aggressive and mortal subtype of breast cancer (BC) since the molecular subtype system has been established. Abundant studies have revealed that epithelial-mesenchymal transition (EMT) played a pivotal role during breast cancer metastasis and progression, especially in TNBC. Herein, we showed that inhibition the expression of replication factor C subunit 3 (RFC3) significantly attenuated TNBC metastasis and progression, which was associated with EMT signal pathway. In TNBC cells, knockdown of RFC3 can down-regulate mesenchymal markers and up-regulate epithelial markers, significantly attenuated cell proliferation, migration and invasion. Additionally, silencing RFC3 expression can decrease nude mice tumor volume, weight and relieve lung metastasis in vivo. Furthermore, we also demonstrated that overexpression of RFC3 in TNBC showed increased metastasis, progression and poor prognosis. We confirmed all of these results by immunohistochemistry analysis in 127 human TNBC tissues and found that RFC3 expression was significantly associated with poor prognosis in TNBC. Taken all these findings into consideration, we can conclude that up-regulation of RFC3 promotes TNBC progression through EMT signal pathway. Therefore, RFC3 could be an independent prognostic factor and therapeutic target for TNBC.
Introduction
Breast cancer is one of the most common types of malignant cancer and accounted for the second leading cause of mortal tumors in women worldwide [1], [2]. Based on the distinct gene expression profiles, several molecular subtypes of breast cancer have been divided [3], [4], [5]. One of these subtypes is triple-negative breast cancer (TNBC), which is characterized by the absence or negative expression in estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 (HER2). TNBC accounted for approximately 10% to 20% of all breast cancer cases and considered to be the most aggressive clinical outcome subtypes of breast cancer [5], [6], [7]. Since metastatic TNBC responded weakly to chemotherapy and no effective targeted therapeutic drugs, TNBC has been regarded as the poorest prognosis breast cancer type [6], [8], [9]. Therefore, it is essential and necessary to understanding the underlying molecular mechanisms which involved in TNBC progression.
Epithelial mesenchymal-transition has been illustrated as a significant procession which characterized as cells loosing polarity and cells contact [10], [11], [12]. During EMT, cells obtain a spindle liked phenotype and lose epithelial cell adhesion. Meanwhile, cells undergoing EMT gain motile features with mesenchymal markers up-regulation and epithelial markers down-regulation. Increasing evidences reveal that EMT plays a significant role in the tumor metastasis and progression [13], [14]. Thus, EMT can be an important event, even the trigger, involving in the tumor metastasis and progression.
Replication factor C (RFC) was a big family, comprising of five subunits (RFC1-5). At previously, abundant researches have revealed that RFC played a pivotal role in DNA replication, DNA damage repair and checkpoint control [15], [16], [17]. In addition, the abnormal amplification of RFC gene has also been demonstrated to be involved in cancer cell proliferation. Recently, several studies have reported that the abnormal activation of RFC3 subunit was associated with progression of esophageal adenocarcinoma and ovarian tumor cells, suggesting that RFC3 was a potential oncogenic gene implicated in tumorigenesis [18], [19], [20]. It has been found that RFC3 can preferentially bind to proliferating cell nuclear antigen (PCNA) forming a complex and attenuating of expression of RFC3 can inhibit tumor cells proliferation [21]. Although the up-regulation of RFC3 has been reported in several malignant tumors, the correlation between RFC3 expression profiles and TNBC progression has not been reported yet. Therefore, the underlying mechanism and the roles of RFC3 in TNBC should be illustrated.
In current study, we provided the evidences that the overexpression of RFC3 promotes TNBC proliferation and invasion in vitro. Silencing of RFC3 can decrease nude mice tumor volume, weight and relieve lung metastasis in vivo. We also demonstrated that the overexpression of RFC3 was associated with metastasis and poor prognosis in TNBC patients.
Materials and Methods
Patient Information and Tissue Specimens
In this study, a total of 127 breast cancer and 30 adjacent normal tissue samples were examined with informed consent under the institutional board-approved protocols from the Sun. Yat-sen University Cancer Center, Sun. Yat-sen University (Guangzhou, China). This study was approved by the institutional research ethics committee of the Sun. Yat-sen University Cancer Center, Sun. Yat-sen University. The patients were histopathologically and clinically diagnosed at Sun. Yat-sen University Cancer Center between 2003 and 2005; the pathological diagnosis was verified for each case. The clinicopathological features of the patients are summarized in Table 1.
Table 1.
Clinicopathological parameters and RFC3 expression in 127 primary breast cancer
| Total | Low expression (RFC3) | High expression (RFC3) | P value | |
|---|---|---|---|---|
| Number | 127 | 51 | 76 | |
| Age(year) | .4028 | |||
| ≥49 | 59 | 26(51.0%) | 33(43.4%) | |
| <49 | 68 | 25(49%) | 43(56.6%) | |
| Tumor size (cm) | .0208 | |||
| <2 | 93 | 43(84.3%) | 50(65.8%) | |
| ≥2 | 34 | 8(15.7%) | 26(34.2%) | |
| Lymph node metastasis | .6033 | |||
| negative | 94 | 39(76.5%) | 55(72.4%) | |
| positive | 33 | 12(23.5%) | 21(27.6%) | |
| Histological grade | .0204 | |||
| I | 19 | 11(21.6%) | 8(10.5%) | |
| II | 89 | 36(70.6%) | 53(69.8%) | |
| III | 19 | 4(7.8%) | 15(19.7%) | |
| Ki67 | .0003 | |||
| negative | 46 | 28(54.9%) | 18(23.7%) | |
| positive | 81 | 23(45.1%) | 58(76.3%) |
Immunohistochemistry
Immunohistochemical analysis was performed to investigate the expression of RFC3 in TNBC. Briefly, immunohistochemistry was performed on the paraffin-embedded human TNBC tissue sections using mouse monoclonal anti-RFC3 (1:50, ab154899; Abcam, Cambridge, MA). For the negative controls, isotype-matched antibodies were applied. The tissue sections were observed under a Zeiss AX10-Imager A1 microscope e (Carl Zeiss, Thornwood, NY) and all images were captured using AxioVision 4.7 microscopy software (Carl Zeiss, Thornwood, NY). Immunoreactivity for RFC3 protein was scored by semi-quantitative method by evaluating the number of positive cells over the total number of cells, which was reported by Cai et al. [22]. Scores were assigned by using 5% increments (0%, 5%, 10%...100%). RFC3 expression was assessed by three independent pathologists, if two or all of them agreed with the results they scored, the value was selected. If the results were completely different, then all of them would work collaboratively to confirm the score.
Cell Line and Cell Culture
Two human TNBC cell lines (MDA-MB-231 and MDA-MB-468), five non-TNBC cell lines (MCF-10A, MCF-7, ZR-75-1, T47D and MDA-MB-436) were used in this study and all the cell lines were obtained from American Type Culture Collection. MDA-MB-231, MDA-MB-436, MCF7, and MDA-MB-468 were cultured in DMEM, T47D and ZR-75-1 was cultured in Roswell Park Memorial Institute (RPMI)-1640, and MCF10A in DMEM/F12 (1:1) medium, with all recommended supplements, respectively. All cells were maintained at 37°C in a humidified incubator with 5% CO2.
RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR
Total RNA was extracted from tissues or cells using the Trizol reagent (Invitrogen, Carlsbad, California, USA), and the reverse transcription was performed using the Prime Script™ RT reagent kit according to the manufacturer's instructions (Takara Biotechnology, China). For amplification of RFC3, reverse transcription PCR was programmed as follows: 95°C for 2 minutes, 30 cycles of 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 45 seconds, 72°C for 10 minutes, hold at 4°C. RT-PCR products were analyzed via 2.0% agarose gel electrophoresis and stained with ethidium bromide for visualization using ultraviolet light. Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Takara, China). The primers used as follows: RFC3-F: 5′-GCCTGCAGAGTGCAACAATA-3′; RFC3-R: 5′-TCAAGGAGCCTTTGTGGAGT-3′; GAPDH-F: 5′-GAGTCAACGGATTTGGTCGT-3′, GAPDH-R: 5′-GACAAGCTTCCCGTTCTCAG-3′;
Western Blotting
Cells were lysed in sample buffer and the protein concentrations were measured using the BCA kit (Thermo, USA). Equal amounts of proteins were separated by 10% polyacrylamide SDS gels (SDS-PAGE), transferred on polyvinylidene fluoride (PVDF) membranes (Amersham Pharmacia Biotech) and the membranes were probed with anti-RFC3 antibody (1:500, ab154899; Abcam, Cambridge, MA), ZO-1 (1:1000, Invitrogen), E-cadherin, N-Cadherin, Vimentin, (1:500, Santa Cruz, Santa Cruz, CA, USA), or GAPDH (1:3000, Proteintech, Chicago, IL, USA), and then with peroxidase-conjugated secondary antibody (1:3000, Proteintech). Then, the signals were visualized by enhanced chemiluminescence kit (GE, Fairfield, CT, USA) according to the manufacturer's instructions. Anti-GAPDH antibody (Proteintech) was used as a loading control.
Lentivirus Construction and Gene Silencing
Lentiviral short hairpin RNA (shRNAs) in LV-008 vector containing a GFP reporter (Forevergen Biosciences, Guangzhou, China) was used to express short hairpin RNA (shRNA). The RNAi sequence targeting to RFC3 gene was (5′-AAGTAACTACCACCTTGAAGTTA-3′) and negative control (5′-TGGTTTACATGTCGACTAA-3′). Lentivirus was produced in 293 T cells. Briefly, LV-008-shRFC3 plasmids were transfected into HEK 293 T cells together with the lentiviral packaging vectors. Infection lentiviruses were collected 72 h after transfection and concentrated by ultracentrifugation using Beckman Instruments (Fullerton, CA, USA). MDA-MB-231 and MDA-MB-468 cells were seeded in six-well plate and infected with RFC3-lentiviruse or NC-shRNA lentivirus in the presence of 5 μg/ml of polybrene, respectively. After 5 days, the knockdown efficiency was validated by real-time PCR and western blot.
Colony Formation Assay
Cells were digested and seeded into 6-well plates with 200 cells per well. After two weeks of culture, the cells were washed and fixed with 4% paraformaldehyde for 30 minutes at room temperature, and then stained with crystal violet. The number of colonies was counted under a fluorescence microscope. Each experiment was performed in triplicate and repeated three times.
Boyden Chamber Assay
Cell migration and invasion ability was examined using insert transwell with 8 μm pores (BD Biosciences, San Jose, CA, USA). For the invasion assays, 1 × 105 cells were added into the upper boyden chamber pre-coated with matrigel (Corning, New York, NY, USA). After 24 h, the non-invading cells were removed from the upper chamber with a soft cotton swab, and the cells that had invaded through the membrane to the lower compartment of the transwell were fixed, stained, photographed and counted. For migration assay, the procedures were almost similar, except that 1 × 105 cells were placed into the top chamber without matrigel pre-coated. Finally, the cells in lower compartment of the chamber that had invaded to the bottom of the transwell were fixed, stained, photographed and counted.
In Vivo Mice Xenograft Assay
Four- to six-week old BALB/c nu/nu mice were used for tumor implantation and all the mice were purchased from Shanghai Slac Laboratory Animal Co., Ltd. (Shanghai, China). All the animal experiments were strictly complied with the Regulations for the Administration of Affairs Concerning Experimental Animals, the Chinese national guideline for animal experiment. The animal experiments were approved by The Institute Research Medical Ethics Committee of Sun. Yat-Sen University. To generate the tumor implantation mouse model, LV-shNC/LV-shRFC3 and MDA-MB-231 cell were subcutaneous injected into the mice fat pat. The tumor size and weight were measured every week. All the harvested tumors were imaged after sacrifice, and then the tumor tissues were analyzed with H&E staining.
Statistical Analysis
All statistical analyses were carried out using SPSS 19.0 statistical software (SPSS, Chicago, IL, USA). The chi-square and Fisher's exact tests were used to analyze the relationship between RFC3 expression and clinicopathologic features. Survival curves were plotted by the Kaplan-Meier method and compared using the log-rank test. Survival data were evaluated using univariate and multivariate Cox regression analyses. P < .05 was considered statistically significant in all cases.
Results
RFC3 is Overexpressed in Primary Breast Cancer Tissues
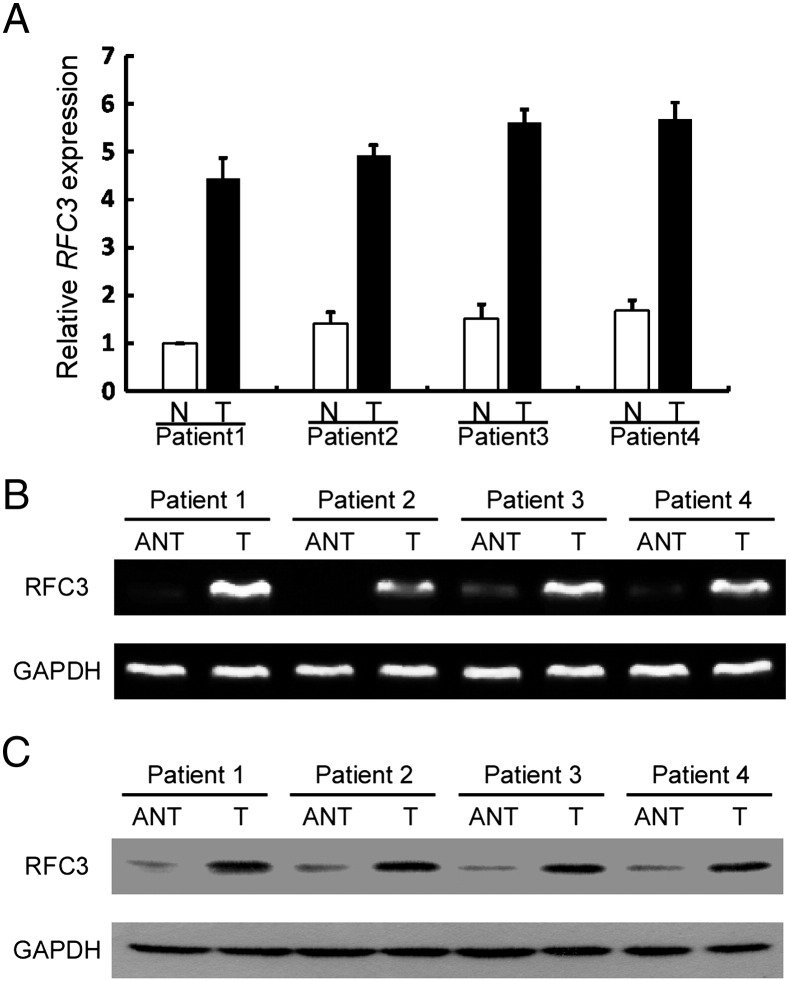
Overexpression of RFC3 has been previously reported in several human cancers. However, its expression status in breast cancer was still unknown. To measure the RFC3 mRNA expression profile, quantitative real time PCR analysis was carried out to quantify the expression level of RFC3 in paired primary breast cancer and matched adjacent normal breast tissues. As shown in Figure 1A, RFC3 was overexpressed in all four human primary breast cancer samples compared with matched adjacent non-tumor tissues from the same patients. This result is also can be seen in RT-PCR (Figure 1B) and western blotting (Figure 1C).
Figure 1.
RFC3 is up-regulated in primary breast cancer.
(A) Quantitative real time RT-PCR analysis of RFC3 mRNA from the same four pairs of breast cancer and adjacent non-tumor breast tissues. Error bars represent SDs calculated from triplicate experiments. (B) RT-PCR analysis of RFC3 protein in four pairs of human primary breast tumor tissues (T) and paired adjacent non-tumor breast tissue (N). (C) Western blotting analysis of RFC3 protein in four human primary breast cancer (T) and paired adjacent non-tumor breast tissues (N), with each pair taken from a same patient.
Up-Regulation of RFC3 is Associated With Poor Prognostic Phenotype in BC
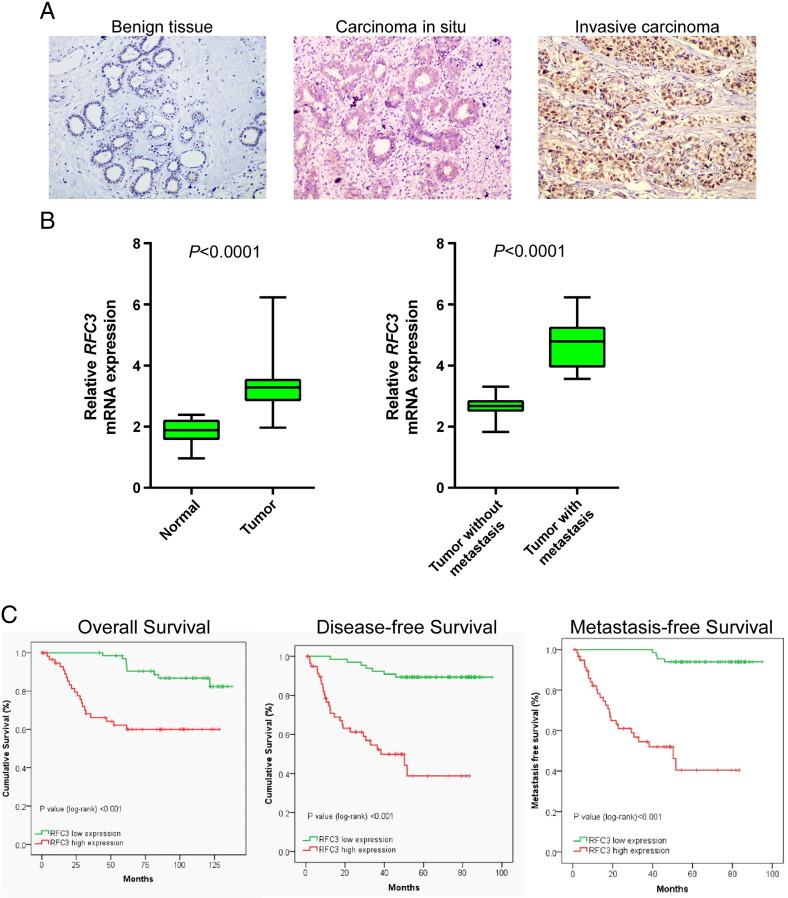
To further investigate the clinicopathological and prognostic significance of RFC3 expression level in patients with breast cancer, the levels of RFC3 in a large cohort of 127 BC tissues were examined by quantitative real-time PCR and then verified by immunohistochemistry. As shown in Figure 2A, the RFC3 exhibited various expression profiles in different differentiation stage and the poorer differentiated stage tissues showed the higher expression of RFC3. Additionally, using quantitative real-time PCR, 30 cases of paired primary BC samples and matched adjacent non-tumor breast tissues were analyzed, most of cases had much higher RFC3 expression levels, compared with adjacent non-neoplastic breast tissues (Figure 2B). Then, the correlation between RFC3 expression and metastatic status were analyzed in 127 BC samples by quantitative real-time PCR, the result showed that, most BC had much higher expression of RFC3, which was significantly associated with a more aggressive tumor phenotype (P < .0001, Table 1, Figure 2C).
Figure 2.
Up-regulation of RFC3 is associated with poor prognostic phenotype in breast cancer.
(A) RFC3 expression level in different differentiation stage (benign tissue, carcinoma in situ, invasive carcinoma) analyzed by immunohistochemistry. (B) Expression levels of RFC3 in 30 paired breast cancer and adjacent non-tumor breast tissues. Alteration of expression is shown as box plot presentations and the mean level of RFC3 expression in breast cancer were significantly higher than that in non-tumor tissues. (P < .0001, independent t test). Expression levels of RFC3 between 127 BCs with and without metastasis. The mean level of RFC3 expression in BCs with metastasis was significantly higher than that in BCs without metastasis (P < .0001, independent t test). (C) Overall survival, disease-free survival and metastasis-free survival rate for cases with high RFC3 expression versus that of cases with low RFC3 expression.
To further verify the results above, immunohistochemistry was carried out in all the 127 BC samples. We found that 76/127 (59.8%) BCs had high expression of RFC3, while 51/127 (40.2%) BCs had low expression of RFC3 (Table 1). Furthermore, as shown in Table 1, Table 2, RFC3 expression strongly correlated with tumor size (P = .0208), lymph histological grade (P = .0204), Ki-67 expression (P = .0003) and metastasis (P < .001) in patients with breast cancer; however, the analysis data indicated that RFC3 expression was not correlated with age and lymph node status. Taken together, our analyses revealed that the expression of RFC3 was up-regulated during the clinical progression of breast cancer, indicating that the expression of RFC3 may promote the progression of breast cancer.
Table 2.
Cox regression analyses for contribution of various potential prognostic factors to survival in BC patients.
| Variable factors | Disease-free survival HR(95%CI) | P value | Overall survival HR(95%CI) | P value | Metastasis-free survival HR(95%CI) | P value |
|---|---|---|---|---|---|---|
| Univariate analysis | ||||||
| RFC3 expression (high vs low) | 4.484 (1.695-13.864) | .003 | 4.154 (1.889-9.136) | <.001 | 14.515 (4.985-42.263) | <.001 |
| Age (≥49 vs <49) | 1.016 (0.975-1.059) | .455 | 1.006 (0.974-1.040) | .708 | 0.997 (0.965-1.029) | .843 |
| Tumor size (≥2 cm vs <2 cm) | 2.770 (1.294-5.927) | .009 | 2.535 (1.081-5.943) | .032 | 3.168 (1.455-6.899) | .004 |
| Nodel status (positive vs negative) | 3.732 (1.921-7.250) | <.001 | 3.313 (1.617-6.788) | <.001 | 4.138 (2.038-8.042) | <.001 |
| Ki67 expression (positive vs negative) | 0.996 (0.460-2.154) | .991 | 0.945 (0.408-2.191) | .896 | 1.105 (0.485-2.522) | .812 |
| Histologic grade (I/II vs III) | 0.821 (0.408-1.651) | .58 | 1.186 (0.572-2.460) | .646 | 0.758 (0.358-1.604) | .468 |
| Multivariate analysis | ||||||
| RFC3 expression (high vs. low) | 11.646 (4.772-28.420) | <.001 | 5.037 (2.184-11.622) | <.001 | 20.429 (6.743-61.892) | <.001 |
| Tumor size (≥2 cm vs <2 cm) | 1.346 (0.414-4.373) | .622 | 0.839 (0.319-2.207) | .722 | 1.051 (0.409-2.699) | .918 |
| Nodel status (positive vs negative) | 7.265 (3.154-16.738) | .000 | 3.444 (1.365-8.687) | .009 | 8.773 (3.552-21.669) | <.001 |
Analysis was conducted on 127 cases. HRs (95% CI) and P values were calculated using univariate or multivariate Cox proportional hazard regression.
Association Between RFC3 Expression and Patient Survival
Patient survival analysis presented in Table 2 indicated a clear positive correlation between RFC3 protein expression level and the disease-free, overall and metastasis-free survival time in breast cancer patients. The effects of classic clinicopathologic features, including age, tumor size, lymph node status, Ki-67 status and histological status, in conjunction with RFC3 protein expression, on patient survival, were examined with Kaplan-Meier analysis and the log-rank test. As shown in Figure 2C, the length of overall survival time varied significantly different between patients with low and high RFC3 expression (P < .001), with the low RFC3 expression group having a longer overall survival time, compared with those with high level expression of RFC3. In addition, the prognostic value of RFC3 expression in patient was also evaluated according to the disease free and metastasis free survival. The analysis results revealed that the patients with tumors exhibiting low expression of RFC3 have more possibility of disease free and metastasis free.
When univariate and multivariate analyses were done to determine whether RFC3 expression is an independent prognostic factor of patient outcomes, as Table 2 shows, high level expression of RFC3 was recognized as an independent prognostic factors for poor survival of patients with BC (P < .001). Taken together, our data suggest that RFC3 might represent a novel and potentially useful independent biomarker for the prognosis of patients with breast cancer.
RFC3 Expression is Up-Regulated in Breast Cancer Cells Lines
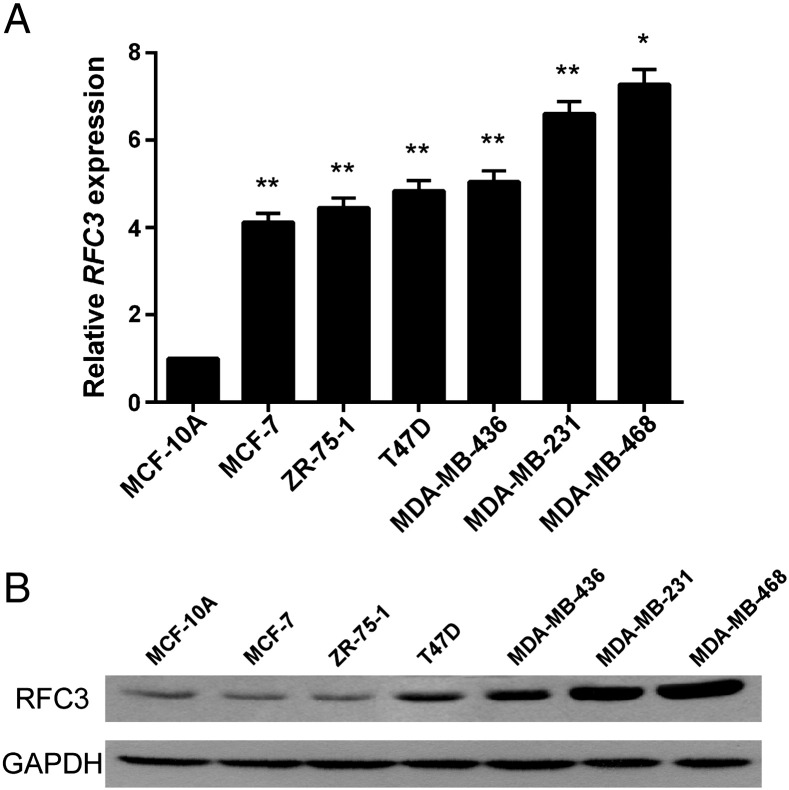
All of these results indicate that RFC3 was up-regulated in primary breast cancer tissues. To verify the RFC3 expression status above in breast cancer cell lines, quantitative real time PCR analysis was performed in 6 breast cancer cell lines and a non-tumor immortalized human breast epithelial cell line MCF-10A. The results showed that RFC3 was highly expressed in breast cancer cell lines, especially in the TNBC cell lines MDA-MB-231 and MDA-MB-468, compared to the non-tumor immortalized human breast epithelial cell line MCF-10A (Figure 3A). To confirm whether the RFC3 up-regulation was also at the protein level, western blotting was performed. As shown in Figure 3B, in parallel with up-regulation of the RFC3 mRNA gene, all the breast cancer cell lines exhibited higher levels of RFC3 expression, comparing with non-tumor immortalized human breast epithelial cell line MCF-10A. In addition, two TNBC cell lines MDA-MB-231 and MDA-MB-468 shared the highest expression status.
Figure 3.
RFC3 expression is up-regulated in breast cancer cells lines
(A) Quantitative real time PCR analysis was performed in 6 breast cancer cell lines and a non-tumor immortalized human breast epithelial cell line MCF-10A. (B) RFC3 protein level in 6 breast cancer cell lines and a non-tumor immortalized human breast epithelial cell line MCF-10A were analyzed by western blotting.
Down-Regulation RFC3 Expression Attenuated Breast Cancer Cell Proliferation, Migration and Invasion In Vitro
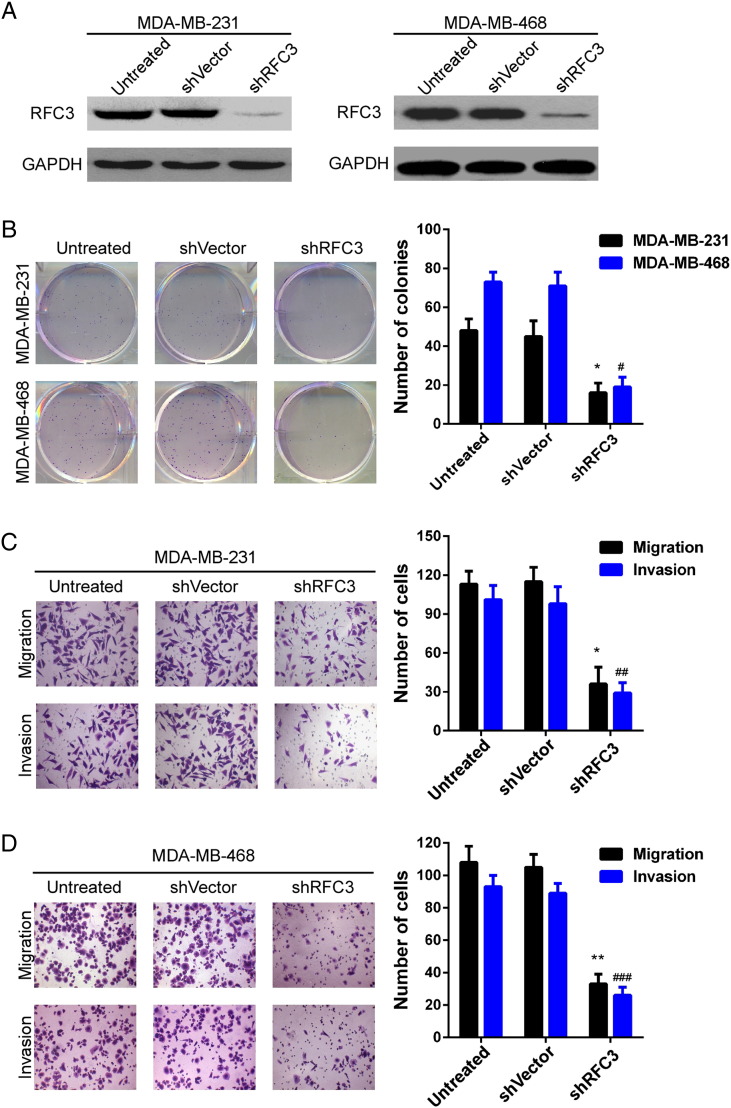
As RFC3 expression was positively related with metastasis and metastasis to distant organs contributes to poorer survival, next we investigate the effect of RFC3 on the proliferation, migration and invasion of TNBC cancer cell. Firstly, MDA-MB-231 and MDA-MB-468 stable cell lines with knockdown of RFC3 were constructed using lentivirus-mediated transduction and the silence efficiency was examined by western blotting in these two cell lines. As shown in Figure 4A, the shRNA efficiently knocked down the endogenous expression of RFC3 protein in MDA-MB-231 and MDA-MB-468 cells. Through colony formation assay, we found that the proliferation ability was severely impaired in both the two cell lines when knockdown the RFC3 expression (Figure 4B). Meanwhile, using the transwell chamber model, we observed that compared to untreated or vector, silencing of RFC3 expression severely inhibited the migration and invasion ability of MDA-MB-231 and MDA-MB-468 cells (Figure 4, C and D). These results indicated that the expression of RFC3 promotes the metastatic ability of breast cancer cells.
Figure 4.
Inhibition of RFC3 attenuated BC cell proliferation, motility and invasion.
(A) The knockdown efficiency of shRNA against RFC3 was examined by Western blotting in MDA-MB-231 and MDA-MB-468 cells. (B) The proliferation ability was measured in MDA-MB-231 and MDA-MB-468 cells using colony formation assay. (C) The migration and (D) invasiveness abilities were analyzed in MDA-MB-231 and MDA-MB-468 cells by Boyden chamber assay (*P < .05, **P < .01).
RFC3 Knockdown Attenuated Tumor Growth and Metastasis in Breast Cancer In Vivo
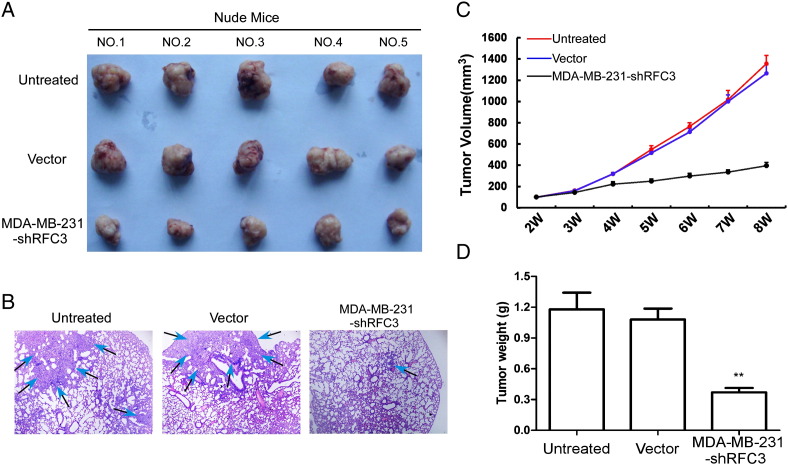
To further investigated the effects of RFC3 knockdown on the growth of breast cancer xenograft tumors in vivo. The three groups of MDA-MB-231 cell lines (LV-ctrl, LV-NC and LV-shRFC3) were subcutaneously implanted in the fat pat of nude mice. Tumors became palpable from 3 weeks to 8 weeks and continued to grow. A significant reduction in tumor size, tumor weight and lung metastasis were observed in the RFC3 knockdown group, respectively, compared with the control groups (Figure 5, A–D).
Figure 5.
Knockdown of RFC3 can relieve xenograph tumor progression in null mice.
Xenograft model in nude mice. MDA-MB-231, shVector and shRFC3 cells were inoculated in the fat pat of nude mice (n = 5/group). (A) Images of the tumors from all mice in each group. (B) The lung metastasis staining in H&E, (C) tumor volume and (D) mean tumor weight was analyzed.
The EMT Pathway is Involved in the Progression of BC Regulated by RFC3
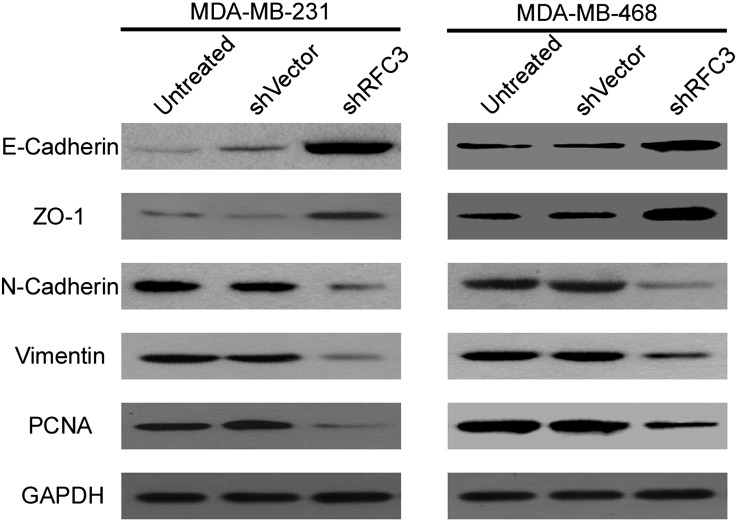
All the results above demonstrate that overexpression of RFC3 is positively associated with poor prognosis in patients with breast cancer; however, the underlying molecular mechanism which mediate this effect are little known. It has been well-known that EMT plays a pivotal role during tumor metastasis and progression. Moreover, abnormal activation of the EMT is a universal phenomenon and considered to contribute to cancer metastasis in multiple types of cancer. Therefore, Western blotting was performed to investigate the expression of EMT markers in MDA-MB-231 and MDA-MB-468 cells. We found that silencing of RFC3 inhibited the expression of PCNA. Meanwhile, the expression of two classical mesenchymal cell markers, N-cadherin and Vimentin, whereas the epithelial cell marker, E-cadherin and ZO-1 expression were elevated, indicating that there is a potential correlation between RFC3 and EMT (Figure 6).
Figure 6.
Knockdown of RFC3 can reversed epithelial-mesenchymal transition (EMT)
The expression of EMT markers of E-Cadherin, ZO-1, Vimentin and N-Cadherin were analyzed by western blotting both in MDA-MB-231 and MDA-MB-468 breast cancer cells. GAPDH was probed as the loading control.
Considering all the results above, our findings indicate that RFC3 may activate EMT and correlate significantly with the metastasis and progression of breast cancer.
Discussion
Triple negative breast cancer is regarded as the most aggressive and considerable threat to women health in the worldwide [23], [24]. Many researchers have attempted to reveal the underlying mechanisms which may trigger TNBC progression. It is believed that metastasis is a distinguishing feature of cancer and contributes to the most of tumor related deaths in humans and several signal pathways are involved during this procession, especially the EMT [25], [26], [27]. Epithelial-mesenchymal transition (EMT) is a process whereby tumor cells lose the epithelial features to acquire a mesenchymal phenotype and become invasive, which is closely associated with tumor metastasis [27], [28]. It has been illustrated that cancer cells can dedifferentiate to obtain the capability to invade, endowing cancer cells to disseminate to distant organs, via activating oncogene expression which associated with EMT. Meanwhile, EMT is closely regulated by other signal pathway and transcription factor network, such as Snail, Twist and ZEB, a regulator of E-cadherin expression, which is a major suppressor of tumor invasiveness and transcriptionally repressed during the EMT [29], [30], [31].
In current study, we demonstrated the importance of RFC3 expression in the survival of TNBC patients and the effect of RFC3-mediated tumor progression. So far, TNBC remains a challenging disease since the poorest prognosis of all of the breast cancer subtypes. It is extremely important that there are no efficient and therapeutic targets and drugs to conquer this subgroup of cancer. Identification of novel targets can potentially have a major impact on the disease and dramatically benefit breast cancer patients. Although RFC3 expression has been documented in some types of cancer, including esophageal adenocarcinoma and ovarian tumor, the correlative analysis to clinical outcome has been limited. Herein we show that RFC3 expression in breast cancer is higher in the TNBC subtype and that TNBC patients with high RFC3 expression have a higher risk of distant metastases, the main cause of death from breast cancer. We also found that the high expression of RFC3 was associated with an aggressive profile both in vitro and in vivo.
In summary, our study demonstrated that RFC3 was up-regulated and positively associated with the overall, disease- and metastasis-free survival. In addition, knockdown of RFC3 expression can severely impair breast cancer cells proliferation, invasion, EMT process and xenograft progression. This study suggests that RFC3 may play a pivotal role in tumor metastasis and can be a novel diagnostic marker and potential therapeutic target in breast cancer.
Ethical Approval
This study was approved by the institutional research ethics committee of the Sun. Yat-sen University Cancer Center, Sun. Yat-sen University. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All persons gave their informed consent prior to their inclusion in the study.
Author Contributions
Yong Bao and Ming Chen conceived and designed the experiments; Zhen-Yu He, San-Gang Wu and Fang Peng performed the experiments; Yong Bao, Ming Chen, Zhen-Yu He, San-Gang Wu and Fang Peng analyzed the data; Qun Zhang and Yin Luo contributed reagents/materials/analysis tools; Zhen-Yu He wrote the paper.
Conflicts of Interest
None.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81402527), the Sci-Tech Office of Guangdong Province (No. 2013B021800157, 2013B021800458, 2014A020212528), the Youth Foundation of Fujian Provincial Health and Family Planning Commission (No. 2014-2-63), and the Natural Science Foundation of Fujian Province (No. 2015 J01550, 2016J01635).
Contributor Information
Ming Chen, Email: chenming@zjcc.org.cn.
Yong Bao, Email: baoyong0205@sina.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2015;65:87–108. [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2014;64:9–29. [Google Scholar]
- 3.Haibekains B, Desmedt C, Loi S, Culhane AC, Bontempi G, Quackenbush J, Sotiriou C. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104:311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desmedt C, HaibeKains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological Processes Associated with Breast Cancer Clinical Outcome Depend on the Molecular Subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 5.Network CGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 7.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA, Jr, Ellis P. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 10.Acloque H, Adams MS, Fishwick K, Bronnerfraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. J Oral Maxillofac Pathol. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Kalluri R. EMT: When epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhim A, Mirek E, Aiello N, Maitra A, Bailey J, Mcallister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venclovas C, Colvin ME, Thelen MP. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 2002;11:2403–2416. doi: 10.1110/ps.0214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YR, Song SY, Kim SS, An CH, Lee SH, Yoo NJ. Mutational and expressional analysis of RFC3, a clamp loader in DNA replication, in gastric and colorectal cancers. Hum Pathol. 2010;41:1431–1437. doi: 10.1016/j.humpath.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Cai M, Zhao S, Wang H, Li M, Yao S, Jiang N. Overexpression of RFC3 is correlated with ovarian tumor development and poor prognosis. Tumour Biol. 2014;35:10259–10266. doi: 10.1007/s13277-014-2216-2. [DOI] [PubMed] [Google Scholar]
- 18.Lockwood WW, Thu KL, Lin L, Pikor LA, Chari R, Lam WL, Beer DG. Integrative genomics identified RFC3 as an amplified candidate oncogene in esophageal adenocarcinoma. Clin Cancer Res. 2012;18:1936–1946. doi: 10.1158/1078-0432.CCR-11-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hee-Don C, Bryan M, Lacayo NJ, Sakamoto KM. Replication factor C3 is a CREB Target gene that regulates cell cycle progression through modulation of chromatin loading of PCNA. Leukemia. 2014;29:1379–1389. doi: 10.1038/leu.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Xu J, Zhao S, Shi H, Yao S, Jiang N. ShRNA-mediated silencing of the RFC3 gene suppress ovarian tumor cells proliferation. Int J Clin Exp Pathol. 2014;8:8968–8975. [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Z, Hu K, Huang H, Xu S, Wang Q, Zhang P, Yang P, Liu B. shRNA-mediated silencing of the RFC3 gene suppresses hepatocellular carcinoma cell proliferation. Int J Mol Med. 2015;36:1393–1399. doi: 10.3892/ijmm.2015.2350. [DOI] [PubMed] [Google Scholar]
- 22.Cai MY, Zhang B, He WP, Yang GF, Rao HL, Rao ZY, Wu QL, Guan XY, Kung HF, Zeng YX. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci. 2010;101:1543–1549. doi: 10.1111/j.1349-7006.2010.01560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry DA, Cirrincione C, Henderson IC, Citron ML, Budman DR, Goldstein LJ, Martino S, Perez EA, Muss HB, Norton L. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA. 2006;295:1658–1667. doi: 10.1001/jama.295.14.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 25.Chambers AF, Groom AC, Macdonald IC. Metastasis: Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 26.Gupta GP, Massagué J. Cancer Metastasis: Building a Framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer Metastasis Is Accelerated through Immunosuppression during Snail-Induced EMT of Cancer Cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 28.Mantovani A. Cancer: Inflaming metastasis. Nature. 2009;457:597–605. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 29.Martin A, Cano A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat Cell Biol. 2010;12:924–925. doi: 10.1038/ncb1010-924. [DOI] [PubMed] [Google Scholar]
- 30.Wellner U, Schubert JU. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 31.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12:3433–3439. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]