Abstract
The genus Microbacterium contains bacteria that are ubiquitously distributed in various environments and includes plant-associated bacteria that are able to colonize tissue of agricultural crop plants. Here, we report the 3,508,491 bp complete genome sequence of Microbacterium sp. strain BH-3-3-3, isolated from conventionally grown lettuce (Lactuca sativa) from a field in Vestfold, Norway. The nucleotide sequence of this genome was deposited into NCBI GenBank under the accession CP017674.
Keywords: Epiphytic bacteria, Lettuce, Microbacterium sp., Whole genome sequencing
| Specifications | |
|---|---|
| Organism/cell line/tissue | Microbacterium sp. |
| Strain | BH-3-3-3 |
| Sequencer or array type | PacBio RS II |
| Data format | Analyzed |
| Experimental factors | Bacterial strain |
| Experimental features | Whole genome analysis and gene annotation of BH-3-3-3 |
| Sample source location | Lettuce (Lactuca sativa) from a conventional field in Vestfold, Norway |
1. Direct link to deposited data
2. Experimental design, materials and methods
The Gram-positive genus Microbacterium belongs to the family Microbacteriaceae, within the phylum Actinobacteria. Microbacterium spp. have been isolated from diverse environments including agricultural crop plants [1], [2], [3], [4]. An orange-pigmented Microbacterium sp. strain (BH-3-3-3) was isolated from the leaf surface of lettuce (Lactuca sativa) originating from a conventional field in Vestfold, Norway [5]. Genomic DNA was extracted using Genomic-tip 500/G kit (Qiagen GmbH, Hilden, Germany), a library was created using PacBio (Pacific Biosciences, California, USA) 20 kb library preparation protocol and whole genome sequencing was performed using PacBio RS II. The library was sequenced using P6-C4 chemistry with 360 min movie time on one single-molecule real-time (SMRT) cell. The reads were assembled using HGAP v3 (Pacific Biosciences, SMRT Analysis Software v2.3.0). The minimus2 software of the Amos package was used to circularize the contig, which was confirmed by a dot plot to contain the same sequence at the beginning and end of the contig. RS_Resequencing.1 software (SMRT Analysis version v2.3.0) was used to map reads back to the assembled and circularized sequence in order to correct the sequence after circularization. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
3. Data description
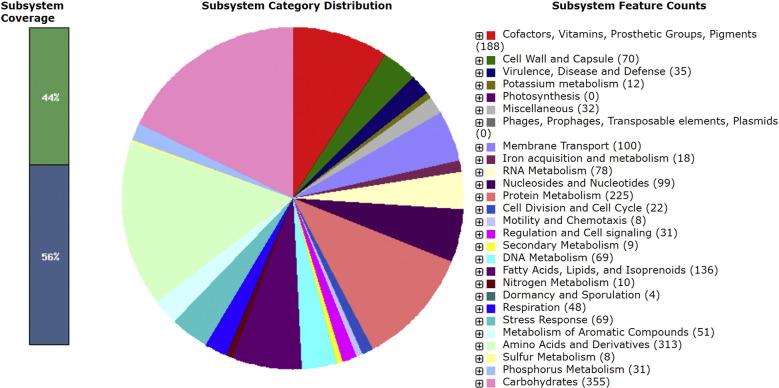
The genome of Microbacterium sp. BH 3-3-3 was annotated using NCBI Prokaryotic Genome Annotation Pipeline [6], GeneMarkS + v 3.3 and the Rapid Annotation System Technology (RAST) server [7]. Fig. 1 presents an overview of the count of each subsystem feature and the subsystem coverage. The circular chromosome has a GC content of 70.5%, consisted of 3,508,491 bp and contained 3113 coding sequences (CDSs), 9 rRNA genes, 45 tRNAs, and 3 noncoding RNA (ncRNA) genes.
Fig. 1.
Subsystem category distribution of major protein coding genes of Microbacterium sp. strain BH 3-3-3 as annotated by the RAST annotation server. The bar chart shows the subsystem coverage in percentage (blue bar corresponds to percentage of proteins included). The pie chart shows percentage distribution of the 25 most abundant subsystem categories.
4. Nucleotide accession number
This whole genome project has been deposited at NCBI GenBank under the accession number CP017674.
Acknowledgements
We are grateful to Hege Særvold Steen for preparing the DNA. This study was supported by a grant from the Norwegian Research Council (221663/F40).
References
- 1.Ferreras E.R., De Maayer P., Makhalanyane T.P., Guerrero L.D., Aislabie J.M., Cowan D.A. Draft genome sequence of Microbacterium sp. strain CH12i, isolated from shallow groundwater in Cape Hallett, Antarctica. Genome Announc. 2014;2 doi: 10.1128/genomeA.00789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morohoshi T., Wang W.Z., Someya N., Ikeda T. Genome sequence of Microbacterium testaceum StLB037, an N-acylhomoserine lactone-degrading bacterium isolated from potato leaves. J. Bacteriol. 2011;193:2072–2073. doi: 10.1128/JB.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinniel D.K., Lambrecht P., Harris N.B., Feng Z., Kuczmarski D., Higley P., Ishimaru C.A., Arunakumari A., Barletta R.G., Vidaver A.K. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 2002;68:2198–2208. doi: 10.1128/AEM.68.5.2198-2208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P., Diene S.M., Gimenez G., Robert C., Rolain J.M. Genome sequence of Microbacterium yannicii, a bacterium isolated from a cystic fibrosis patient. J. Bacteriol. 2012;194:4785. doi: 10.1128/JB.01088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dees M.W., Lysoe E., Nordskog B., Brurberg M.B. Bacterial communities associated with surfaces of leafy greens: shift in composition and decrease in richness over time. Appl. Environ. Microbiol. 2015;81:1530–1539. doi: 10.1128/AEM.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Ciufo S., Li W. The NCBI Handbook. second ed. National Center for Biotechnology Information; Bethesda, MD: 2013. Prokaryotic genome annotation pipeline. [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A.…Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]