Abstract
Background
MicroRNAs (miRNAs or miRs) are post-transcriptional regulators of eukaryotic cells and knowledge of differences in miR levels may provide new approaches to diagnosis and therapy.
Methods
The present study measured the levels of nine miRs in head and neck squamous cell carcinomas (HNSCC) and determined whether clinical pathological features are associated with differences in miR levels. SET (I2PP2A) and PTEN protein levels were also measured, since their levels can be regulated by miR-199b and miR-21, respectively. Nine miRs (miR-15a, miR-21, miR-29b, miR-34c, miR-100, miR-125b, miR-137, miR-133b and miR-199b) were measured by real time qRT-PCR in HNSCC samples from 32 patients and eight resection margins. SET (I2PP2A) and PTEN protein levels were estimated by immunohistochemistry in paired HNSCC tissues and their matched resection margins.
Results
In HNSCC, the presence of lymph node invasion was associated with low miR-15a, miR-34c and miR-199b levels, whereas the presence of perineural invasion was associated with low miR-199b levels. In addition, miR-21 levels were high whereas miR-100 and miR-125b levels were low in HNSCC compared to the resection margins. When HNSCC line HN12, with or without knockdown of SET, were transfected with miR-34c inhibitor or miR-34c mimic, the miR-34c inhibitor increased cell invasion capacity while miR-34c mimic decreased the cell invasion.
Conclusions
We showed that the levels of specific miRs in tumor tissue can provide insight into the maintenance and progression of HNSCC.
General significance
MiRNAs are up- or down-regulated during cancer development and progression; they can be prognosis markers and therapeutic targets in HNSCC.
Keywords: miR-15a, miR-34c, miR-199b, Lymph node invasion, Perineural invasion
Graphical abstract
Highlights
-
•
Association between miR-15a, miR-34c and miR-199b levels and lymph node metastasis in HNSCC.
-
•
miR-199b is reduced while SET protein is accumulated in HNSCC.
-
•
Levels of miR-34c control invasion in HNSCC.
1. Introduction
Since the discovery of small RNAs as regulators of mRNA levels in C. elegans [1], the miRNAs have emerged as one of the most important class of post-transcriptional regulators of eukaryotic organisms [2]. The estimated number of coding genes ranges from 2000 [3] to > 3000 in humans [4] and the mature form of these molecules is approximately 22 nucleotides in length. They recognize their targets by base-pairing interactions between the miRNA 5′-end (the “seed region”) and complementary sequences in the 3′ untranslated regions (3′-UTRs) of mRNA targets [5]. Among other activities, miRs regulate immune cell differentiation [6], cardiac ageing and function [7], and proliferation/invasion in cancer cells [8].
The analysis of miRNA levels in tumors can provide important information about deregulated metabolic pathways in cancer. This methodological approach can contribute to the discovery of therapeutic targets, or even biomarkers [9]. In the present study we quantified nine miRNAs (miR-15a, miR-21, miR-29b, miR-34c, miR-100, miR-125b, miR-133b, miR-137 and miR-199b) in 32 samples from HNSCC, and eight of their resection margins by qRT-PCR, in order to determine whether their levels are associated with clinical pathological features. These specific miRs were chosen because they participate in the regulation of pathways considered to be “hallmarks of cancers”, such as cell death, proliferation and migration/invasion [10], [11], [12], [13], [14]. We also assessed by immunohistochemistry in six selected HNSCC samples and their matched resection margins the SET (I2PP2A) protein, involved in the promotion of cancer cell proliferation [15], and PTEN protein, a classic tumor suppressor [16]. In HNSCC samples, lower levels of miR-15a, miR-34c and miR-199b were associated with lymph node invasion, while lower levels of miR-199b were associated with perineural invasion. By comparing HNSCC samples regarding resection margins, miR-21 levels were increased while miR-100 and miR-125b levels were decreased in tumors. The anti-tumorigenic miR-34c was chosen for functional tests in the HNSCC cell line HN12. MiR-34c inhibitor and miR-34c mimic were transfected in HN12 cells, promoting either increase or decrease in HN12 cell invasion capacity, respectively. We concluded that the levels of miR-15a, miR-34c, miR-199b, miR-21, miR-100 and miR-125b could provide insights regarding HNSCC behavior, highlighting these molecules as potential new targets in cancer.
2. Methods
2.1. Patients and sample information
Thirty-two HNSCC tissue fragments and eight their resection margins were obtained from the “Sample Bank for Clinical Laboratory Studies and Molecular Genetics and Bioinformatics Laboratory of the Genetics Department, Ribeirão Preto Medical School.” The Sample Bank was created with the approval of the Research Ethics Committee of the Ribeirão Preto Medical School Hospital (HCRP) (Case No. 4911/2008 HCRP). Some clinical pathological features of patients and tumors are listed in Table 1.
Table 1.
Epidemiologic and pathological features of 32 patients with head and neck squamous carcinoma cell included in the present study.
| Feature | N (%) | |
|---|---|---|
| Sex | Female | 1 (3.1) |
| Male | 31 (96.9) | |
| Tumor site | Larynx | 9 (28.1) |
| Larynx (glottic and supraglottic) | 1 (3.1) | |
| Larynx (glottic) | 10 (31.2) | |
| Larynx (glottic and subglottic) | 1 (3.1) | |
| Larynx (supraglottic) | 7 (21.9) | |
| Oropharynx | 2 (6.2) | |
| Epiglottis | 1 (3.1) | |
| Oral cavity | 1 (3.1) | |
| Age (years) | Average (SD) | 61.1 (10.7) |
| Median (min–max) | 58.5 (40–83) | |
| Smokea | No | – |
| Yes | 27 (100) | |
| Alcoholb | No | – |
| Yes | 26 (100) | |
| Tumor sizec | T1 | 6 (18.8) |
| T2 | 7 (21.8) | |
| T3 | 5 (15.6) | |
| T4 | 13 (40.6) | |
| Lynph node invasionc | N0 | 23 (71.9) |
| N1 | 1 (3.1) | |
| N2 | 1 (3.1) | |
| N2a | 1 (3.1) | |
| N2b | 3 (9.4) | |
| N2c | 2 (6.3) | |
| Inflammatory infiltrate | Peritumoral | 32 (100) |
| Intratumoral | 2 (6.3) | |
| Perineural invasion | No | 27 (84.4) |
| Yes | 5 (15.6) | |
| Differentiation grade | I | 4 (12.5) |
| II | 14 (43.75) | |
| III | 14 (43.75) |
Four cases without information.
Five cases without information.
One case without information.
2.2. Real-time quantitative reverse transcription PCR (real time qRT-PCR)
Total RNA was extracted from 32 tumors and 8 margins with TRIzol® reagent (15596-26, Life technologies, USA). RNA concentration was determined using absorbance (A) at 230 nm; values ranging from 2.0 to 2.2 for A260/A280 and A260/A230 ratios were considered for RNA quality. Two measurements were carried out and differences of < 5% were observed. Samples were stored at − 80 °C.
The TaqMan® MicroRNA reverse transcription kit (4366596, Applied Biosystems, USA) was used to convert 5.0 ng of total RNA to complementary DNA (cDNA) for each mature miRNA studied. The TaqMan® Small RNA Assays kit (4364031E, Applied Biosystems) was used to identify and measure by qRT-PCR miR-15a (assay: 000389, Applied Biosystems), miR-21 (assay: 000397), miR-29b (assay: 000413), miR-34c (assay: 000428), miR-100 (assay: 000437), miR-125b (assay: 000449), miR-133b (assay: 591), miR-137 (assay: 000593) and miR-199b (assay: 000500) in tumor and resection margin tissues. The relative level of each miRNA was normalized to the small nucleolar RNA RNU6b (assay: 4427975, Applied Biosystems), used as endogenous control to normalize real time qRT-PCR data and to compare the 2− ΔCt values of tumor and resection margin samples, using the equation 2− ΔCt, where ΔCt = (Ct miRNA − Ct RNU6b). The assays were performed using an Eppendorf Mastercycler Realplex 4S apparatus (Eppendorf, Germany). Data are reported as median (min–max) 2− ΔCt of two independent experiments, each one carried out in triplicate.
2.3. Immunohistochemistry for SET and PTEN in HNSCC samples and their respective resection margins
Formalin-fixed and paraffin-embedded (FFPE) sections (10 μm) obtained by microdissection of 6 tumors and their respective resection margins were subjected to immunostaining, as previously described [17]. The antibody against SET protein (sc-5655; Santa Cruz Biotechnology, USA) was used at 1:200 dilution, and the antibody against PTEN (9559 - Cell Signaling, USA) at 1:100 dilution. The Dako LSAB + System-HRP (K0690, Dako North America, USA) kit was used as secondary antibody. Immunoreactive cells were visualized after incubation with the chromogenic substrate diaminobenzidine (DAB) (K3468, Dako) for 10 min at room temperature. Photomicrographs of the sections at 20 × magnifications were obtained using a digital camera (AxioCam MRC, Carl Zeiss, Germany) coupled to an Axiovert 40 bright field light microscope (Carl Zeiss). Five fields were scored. The immunostaining intensity was scored as follows: 0 = negative, 1 = weak and 2 = moderate or strong. The score was multiplied by 1.0 or 2.0 to correct for the percentage of cells stained: 1 corresponds to < 50% cells stained and 2 correspond to ≥ 50% cells stained [18]. Data are reported as median and min–max score.
2.4. Cell culture
The tumorigenic and metastatic HNSCC cell line HN12 was cultured in Dulbecco's Modified Eagle Medium (D5648 DMEM Sigma-Aldrich, USA) supplemented with 10% fetal bovine serum (FBS - GIBCO), 1% antimycotic and antibacterial agents (10,000 U/mL penicillin stock solution, 10 mg/mL streptomycin, and 25 mg/mL amphotericin B - Sigma A5955) at 37 °C, in the presence of 5% CO2, in a CO2 incubator.
2.5. MiRNA transfection in HNSCC cells
HN12shCTRL and HN12shSET cells, established by Sobral et al. [15], were transfected with 50 nM HSA-miRNA-34c miRNA inhibitor (4464084 Ambion, USA) or 50 nM HSA-miR-34c miRNA-mimic (4464066 Ambion) for functional studies. MiR-34c inhibitor and miR-34c mimic were transfected by using the HiPerFect® reagent (Qiagen - 301705), according to manufacturer's instructions.
2.6. HN12shControl and HN12shSET cell invasion assay
For the invasion assay, Matrigel (BD Biosciences, catalogue number: 354230) was diluted 1:1 with serum-free medium, and used to coat the filter membranes. HN12shControl and HN12shSET cells (1 × 105) transfected or not with the miR-34c inhibitor and miR-34c mimic were suspended in 250 μL serum-free DMEM, and seeded into the upper compartment of the transwell chamber (8.0 μm transparent PET membrane, Falcon, catalogue number: 353097); DMEM containing 10% FBS was used in the lower chamber for stimulation. After 72 h incubation, the medium in the upper chamber was removed, and filters were fixed in 10% formalin for 15 min. The cells on the lower surface were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma). Five fields were photographed at × 200 original magnification using a Zeiss Axiovert 40 inverted microscope, and processed using the AxioVision Rel. 4.8.2 software. Cells were counted using the ImageJ software. The invasion data were reported as mean (± SD) number of cells per microscopic field; five fields were analyzed.
2.7. Statistical analysis
The SPSS Statistics© version 19.0 and GraphPad Prism 5.03 softwares were used for statistical analysis. The level of significance was set at p < 0.05.
3. Results
3.1. Low miR-15a, miR-34c and miR-199b levels were observed in HNSCC samples from patients with lymph node invasion, and low miR-199b levels in HNSCC samples from patients with perineural invasion
Significant clinical pathological associations were found in the present study concerning miR-15a, miR-34c and miR-199b levels (Table 2). Their levels were significantly lower in tumors from patients with lymph node invasion (LNI +) compared to patients without LNI (LNI −). The median 2− ΔCt miR-15a value was 3.63 (0.40–13.18) for LNI − patients and 2.28 (0.87–6.32) for LNI + patients (p = 0.032). The median 2− ΔCt miR-34c value was 0.57 (0.01–4.26) for LNI − patients and 0.37 (0.04–1.21) for LNI + patients (p = 0.022). The median 2− ΔCt miR-199b value was 0.24 (0.02–1.39) for LNI − patients and 0.15 (0.01–0.58) for LNI + patients (p = 0.011). Specifically regarding miR-199b, the levels were higher in tumors from patients without perineural invasion (2− ΔCt median = 0.24 (0.02–1.39) compared to tumors from patients with perineural invasion (2− ΔCt median = 0.11 (0.01–0.37) (p = 0.040).
Table 2.
MiRNA content of HNSCC tissue with or without lymph node invasion (LNI + or LNI −) or perineural invasion (PNI + or PNI −), obtained by qRT-PCR. Data are reported as 2− ΔCt median (min–max). The Mann-Whitney test was used to compare data for tissue with or without invasion (two categories) using the SPSS Statistics© version 19.0 software. The level of significance was set at p < 0.05 (*). Statistically different values for each miR are given in bold type.
| Lymph node invasion |
Perineural invasion |
|||
|---|---|---|---|---|
| −(N = 23) |
+(N = 8) |
−(N = 27) |
+(N = 5) |
|
| 2− ΔCt median (min–max) | 2− ΔCt median (min–max) | |||
| miR-15a | 3.63 (0.40–13.18) | 2.28 (0.87–6.32) | 2.99 (0.4–13.18) | 1.43 (0.95–12.47) |
| p = 0.032 (*) | p = 0.944 | |||
| miR-21 | 382.68 (43.11–1820.35) | 242.19 (71.01–1038.29) | 382.68 (43.11–1820.35) | 196.72 (71.01–625.99) |
| p = 0.327 | p = 0.170 | |||
| miR-29b | 2.53 (0.50–13.18) | 0.74 (0.33–5.46) | 1.88 (0.33–13.18) | 1.12 (0.33–8.57) |
| p = 0.070 | p = 0.754 | |||
| miR-34c | 0.57 (0.01–4.26) | 0.37 (0.04–1.21) | 0.51 (0.01–4.26) | 0.17 (0.04–0.83) |
| p = 0.022 (*) | p = 0.139 | |||
| miR-100 | 32.90 (3.05–122.79) | 14.83 (5.06–91.77) | 26.72 (3.05–122.79) | 21.71 (5.06–73.52) |
| p = 0.319 | p = 0.631 | |||
| miR-125b | 43.11 (10.13–198.09) | 30.91 (9.58–172.45) | 43.11 (10.13–198.09) | 13.74 (9.58–163.14) |
| p = 0.463 | p = 0.082 | |||
| miR-133b | 0.624 (0.136–1370.04) | 0.790 (0.067–269.57) | 0.624 (0.136–1370.04) | 1.11 (0.067–259.57) |
| p = 0.557 | p = 0.815 | |||
| miR-137 | 0.103 (0.021–2.321) | 0.072 (0.007–0.362) | 0.088 (0.007–2.321) | 0.135 (0.009–0.242) |
| p = 0.528 | p = 0.603 | |||
| miR-199b | 0.24 (0.02–1.39) | 0.15 (0.01–0.58) | 0.24 (0.02–1.39) | 0.11 (0.01–0.37) |
| p = 0.011 (*) | p = 0.040 (*) | |||
3.2. MiR-21 levels were higher whereas miR-100 and miR-125b levels were lower in HNSCC tissue samples compared to resection margins
The comparison of miRNAs levels between HNSCC samples and resection margins revealed the following: in tumors, miR-21 levels were significantly higher, whereas miR-100 and miR-125b levels were significantly lower compared to resection margins (Table 3). Median (min–max) miR-21 2− ΔCt was 371.0 (43.1–1820.3) in tumors and 136.7 (16.1–744.4) in margins (p = 0.036). The miR-100 value was 24.7 (3.0–122.8) and 93.0 (27.3–171.2) in tumors and margins (p = 0.017), respectively. Another miRNA, miR-125b, showed a median 2− ΔCt of 37.5 (9.6–198.1) and 92.8 (26.0–352.1) in tumors and margins (p = 0.018), respectively.
Table 3.
Comparison of miRNA content of tumors (N = 32) and non-tumor resection margins (N = 8). Data were obtained by qRT-PCR and are reported as 2− ΔCt median (min–max). The Wilcoxon test was used to compare the data for each miR using the SPSS Statistics© version 19.0 software. The level of significance was set at p < 0.05 (*). Statistically different values for each miR are given in bold type.
| Tumor (N = 32) |
Margin (N = 8) |
p value | |
|---|---|---|---|
| 2− ΔCt median (min–max) | 2− ΔCt median (min–max) | ||
| miR-15a | 2.86 (0.40–13.18) | 2.73 (1.45–5.28) | 0.123 |
| miR-21 | 371.04 (43.11–1820.35) | 136.72 (16.11–744.43) | 0.036 (*) |
| miR-29b | 1.38 (0.33–13.18) | 2.40 (1.27–5.17) | 0.208 |
| miR-34c | 0.46 (0.012–4.26) | 0.17 (0.066–15.89) | 0.866 |
| miR-100 | 24.75 (3.05–122.79) | 93.05 (27.28–171.25) | 0.017 (*) |
| miR-125b | 37.53 (9.58–198.09) | 92.85 (25.99–352.14) | 0.018 (*) |
| miR-133b | 0.64 (0.067–1370.04) | 2.44 (0.39–23.59) | 0.161 |
| miR-137 | 0.10 (0.007–2.32) | 0.071 (0.028–0.166) | 0.285 |
| miR-199b | 0.23 (0.013–1.39) | 0.35 (0.093–1.92) | 0.069 |
3.3. PTEN protein score was lower and miR-21 levels were higher in HNSCC samples compared to their respective resection margins
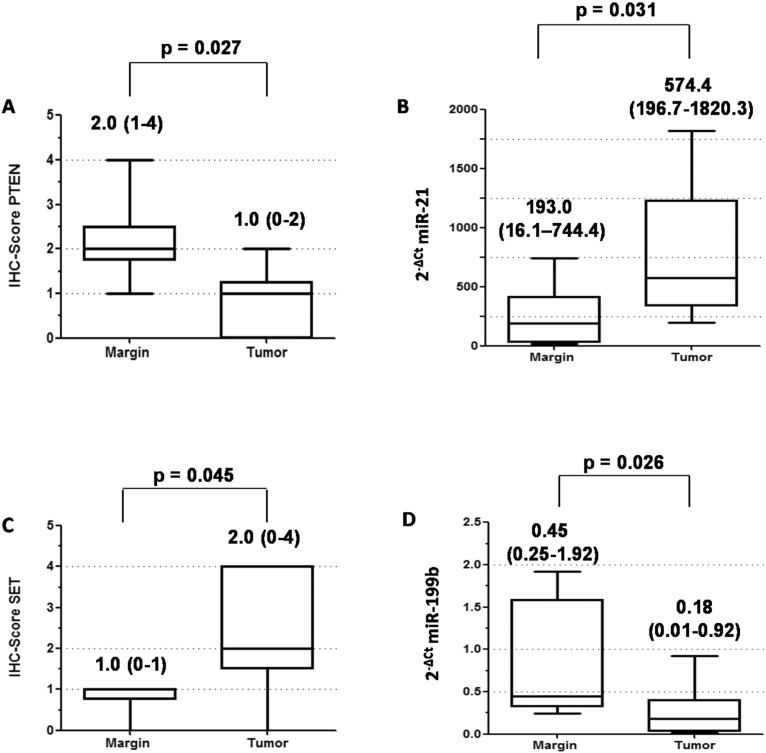
Meng et al. [19] reported increased miR-21 levels in human hepatocellular carcinoma compared to margin cells. These higher levels were capable of inducing growth and spread of cancer cells through the reduction of mRNA coding for Tumor Supressor Protein “PTEN”, an established miR-21 target. The PTEN score was significantly lower in tumors compared to resection margins. The median PTEN score value was 1 (0–2) in tumors and 2 [1], [2], [3], [4] in margins (p = 0.027) (Fig. 1-A and Supplementary Fig. 1-A). Conversely, the values for miR-21 were 574.4 (196.7–1820.3) and 193.0 (16.1–744.4) in tumors and margins (p = 0.031), respectively (Fig. 1-B).
Fig. 1.
(A) Comparison of median PTEN score values obtained by immunohistochemistry (IHC) and of (B) median miR-21 levels by real time qRT-PCR in HNSCC tumors and their respective non-tumor resection margins from the same patients (N = 6). Values above the box plots are (A) median (min–max) PTEN scores and (B) median (min–max) miR-21 2− ΔCt values. (C) Comparison of median (min–max) SET score obtained by immunohistochemistry and of (D) median (min–max) miR-199b 2− ΔCt values in HNSCC tumors and their respective non-tumor resection margins from the same patients (N = 6). Values above the box plots are (C) median (min–max) SET scores and (D) median (min–max) miR-199b 2− ΔCt values. The significance of the differences between tumors and their respective resection margins was determined by the Mann-Whitney test using the GraphPad Prism 5.03 software.
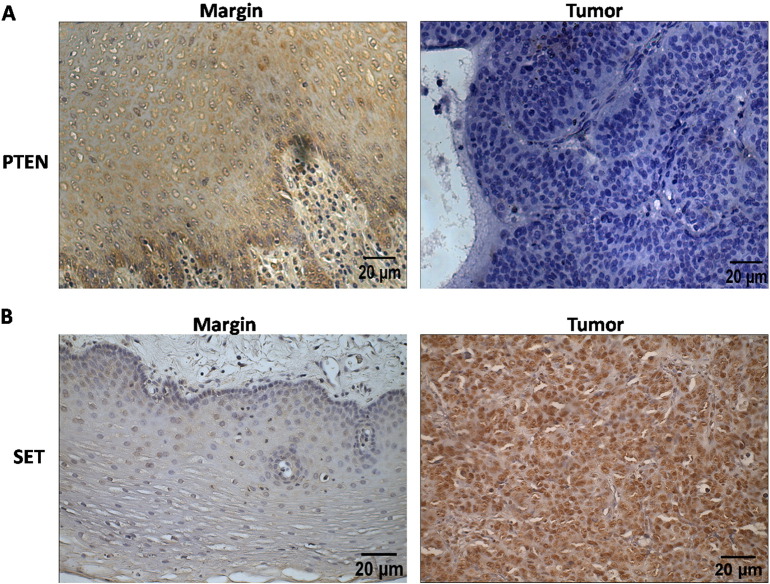
Supplementary Fig. 1.
HNSCC tissues compared to non-tumor resection margin present lower PTEN and higher SET scores. (A) Representative micrographs of PTEN immunohistochemical labeling in tumor tissues (N = 6) and their respective resection margin tissues (N = 6). Images were obtained at 20 × magnification using a digital camera (AxioCam MRC, Carl Zeiss, Germany) coupled to a bright field Axiovert 40 light microscope (Carl Zeiss). (B) Micrographs are representative of SET staining in tumor tissues (N = 6) and their resection margin (N = 6). Images were obtained at 20 × magnification using the same equipment reported above in (A).
3.4. HNSCC samples showed a higher SET protein level and lower miR-199b levels compared to their respective resection margins
Chao et al. [20] reported that mRNA coding for SET protein is a target of miR-199b, which is present at lower levels in choreocarcinoma than in normal cells. In order to determine whether HNSCC tumors also present such an inverse relationship, six paired tumors and their respective resection margins, chosen based on miR-199b levels, were assessed As shown in Fig. 1-C and illustrated in Supplementary Fig. 1-B, while the median SET score was 2 (0–4) in tumors and 1 (0–1) in margins (p = 0.045), the median miR-199b 2− ΔCt value was 0.18 (0.01–0.92) in tumors and 0.45 (0.25–1.92) in margins (p = 0.026) (Fig. 1-D).
3.5. MiR-34c controls cell invasion in HNSCC
We have reported that SET knockdown in the HNSCC cell line HN12 increased its invasion capability [15]. Here, we have used HN12 cell line either with (HN12shSET) or without (HN12shControl) SET knockdown in order to determine the miR-34c impact in their invasiveness. The transfection of miR-34c inhibitor caused an increase from 13.5 (± 9.7) to 289.3 (± 80.7) in HN12shControl cell invasion, against an increase from 238.0 (± 86.3) to 408 (± 130.0) in HN12shSET cell invasion (Fig. 2). On the other hand, while the transfection of miR-34c mimic did not significantly alter HN12shControl cell invasion capacity (13.5 ± 9.7 to 25.3 ± 42.2 values), it strongly reduced the HN12shSET cell invasion capacity (238.0 ± 86.3 to 34.3 ± 30.2 values) (Fig. 2).
Fig. 2.
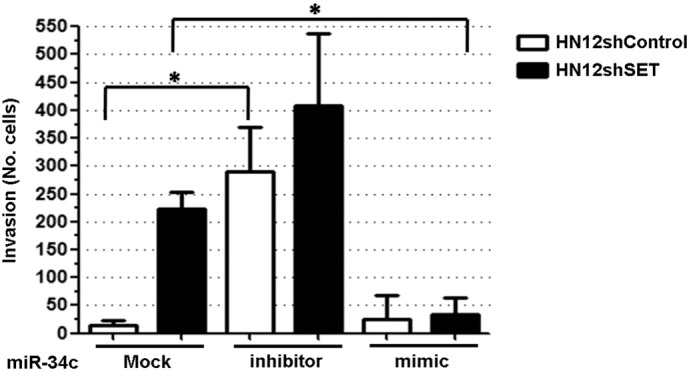
MiRNA-34c inhibits invasion in HNSCC cells. HN12shControl and HN12shSET cells were transfected with miR-34c inhibitor or miR-34c mimic, submitted to transwell® chamber (8.0 μm pore size membrane) with Matrigel™, and their invasion capacity was determined. Values are reported as mean and standard deviation. Statistical significance was assessed by Student's t-test in GraphPad Prism 5.01. *Means p < 0.05.
4. Discussion
Lymph node invasion (LNI) is one of the most important indicators of distant metastasis and survival in the majority of cancer types [21]. LNI means that tumor cells have acquired the ability to penetrate blood vessels and possibly survive in the vascular environment, to circulate through the body, and to initiate new tumors at secondary sites [22].
The present study is the first to report the association between miR-15a, miR-34c and miR-199b levels and lymph node metastasis in HNSCC. In tumors from patients with lymph node metastasis (N +), the levels of these three miRNAs were lower when compared to those of patients without LNI (N0). MiR-15a is a tumor suppressor miR that targets the 3′-UTR of anti-apoptotic protein Bcl2, which is decreased in chronic lymphocytic leukemia (CLL) [23]. Li et al. [14] reported that overexpression of miR-34c in nasopharyngeal carcinoma suppressed tumor growth and metastasis by targeting the proto-oncogene MET. Chao et al. [20] reported that, when the level of miR-199b is reduced, the SET level is higher and cells have more proliferative capacity. Therefore, the decreased levels of these miRs may confer an advantage concerning migration/invasion and proliferation of tumor cells.
Perineural invasion (PNI) is the process of neoplastic invasion of nerves, reported first in head and neck cancers, that exhibit a predilection for growth along nerves (reviewed by [27]). PNI can be a source of distant tumor spread and can be associated with a poor prognosis in distinct cancer types such as pancreatic [24], gastric [25], rectal [26] and prostate [27] cancers.
Reduced levels of miR-199b in HNSCC are reported here for the first time. Chao et al. [20] reported that SET protein is a miR-199b target. The authors described decreased miR-199b levels in human choriocarcinoma, while SET protein levels and cell proliferation capacity were increased compared to normal cells. This evidence suggests proliferative advantages for HNSCC with PNI, since the reduced miR-199b levels induce increased SET protein levels and consequently lead to more proliferation [20].
The present study showed higher miR-21 levels in HNSCC samples compared to the resection margins. MiR-21 has been reported to simultaneously regulate multiple cellular programs that enhance cell proliferation, apoptosis and tumor invasiveness by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma [13].
We also observed lower miR-100 and miR-125b levels in HNSCC samples compared to resection margins (Table 3), in agreement with data reported by Henson et al. [28] regarding reduced miR-100 and miR-125b levels in oral squamous cell cancer (OSCC) lines. After transfection of these two miRs in OSCC lines, Henson et al. [28] reported that they reduced cell proliferation by reducing the levels of many mRNAs, including those that code for MMP13, EGFR3 (miR-100) and KLF13 (miR-125b) proteins. On the basis of these reports, our results suggest that reduced miR-100 and miR-125b levels are important for cell proliferation capacity.
In the present study, we demonstrated that miR-21 is up-regulated in HNSCC tissues when compared to non-tumor resection margin. In Fig. 1, the tumor and resection margin tissues were obtained from the same patients and tumor tissues with higher miR-21 levels presented low PTEN score, while margin tissues with lower miR-21 levels presented high PTEN score. This is a new evidence that miR-21 is an important miR in HNSCC, and miR-21 increase is associated with low PTEN. MiR-21 was also higher in laryngeal and hypopharyngeal cancer, and its levels were reversely correlated with tumor suppressor “PTEN” protein levels [29].
Another protein assessed in the present study by immunohistochemistry was SET (I2PP2A). Leopoldino et al. [30] reported that SET is accumulated in HNSCC and contributes to cell survival and antioxidant defense. Almeida et al. [31] reported that SET interacts with ribonucleoprotein hnRNPK and enhances cell proliferation in OSCC lines. We demonstrated higher SET scores and lower miR-199b in matched HNSCC samples compared to their resection margins. These results are consistent with a role of miR-199b in the control of SET levels and suggest the participation of miR-199b in the control of cell proliferation by down-regulation of SET protein in HNSCC.
A strategy to confirm the biological relevance of a specific miRNA is the ectopic expression or inhibition [14]. In order to validate miR-34c as a metastasis suppressor in HNSCC, we transfected the miR-34c inhibitor and miR-34c mimic in both HN12shControl and HN12shSET cells. The transfection of miR-34c inhibitor significantly raised the number of invasive HN12shControl cells, whereas the transfection of miR-34c mimic significantly reduced the number of invasive HN12shSET cells. Since miR-34c was reduced in patients with lymph node invasion, this result stresses the participation of this miRNA in the regulation of cancer cell invasion and metastasis. Moreover, our findings suggest that SET protein is regulating invasion by target regulated by miR-34c.
5. Study limitations
Limitations of the current study: small sample size and cancer-specific survival data.
6. Conclusions
The results of the present study demonstrate that miRs participate in the malignant transformation in HNSCC. The association of clinical-pathological parameters observed for miR-15a, miR-34C and miR-199b opens a window regarding molecules to be investigated in depth as circulating molecules, as therapeutic targets and/or biomarkers for early HNSCC diagnosis. The increased miR-21 levels, the reduced miR-100 and miR-125b levels, and the higher SET protein levels confer a higher proliferative capacity and lower cell death levels to HNSCC. The increase of miR-21 and the reduction of PTEN levels contribute to cancer growth and spread. The reduced miR-199b levels observed in HNSCC tumors, in association with increased SET levels, have been reported to provide proliferation advantages for choriocarcinoma cells [12], [21].
Briefly, the present study shows that deregulation of miRNAs, with consequent impact on the levels of proteins controlling cancer hallmarks such as cell proliferation, apoptosis or migration/invasion, forms a panel that establishes favorable conditions for generation and maintenance of HNSCC. Therefore, these miRs and proteins are strong candidates as potential targets for cancer diagnosis and/or treatment.
The following is the supplementary data related to this article.
Funding
This work was supported by the São Paulo Research Foundation (FAPESP, grant no. 2013/10898 and by a FAPESP/CEPID grant no. 2013/08135-2).
Disclosure of potential conflicts of interest
No potential conflicts of interest were reported.
Transparency document
Transparency document.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. Dec 3 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. Oct 26 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 3.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedländer M.R., Lizano E., Houben A.J.S., Bezdan D., Báñez-Coronel M., Kudla G. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. Apr 7 2014;15:R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23(5):243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Yao R., Ma Y.-L., Liang W., Li H.H., Ma Z.J., Yu X. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon R.A., Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S. MicroRNA-34a regulates cardiac ageing and function. Nature. Mar 7 2013;495(7439):107. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B.G., Li J.F., Yu B.Q., Zhu Z.G., Liu B.Y., Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting P TEN. Oncol. Rep. 2012;27(4):1019–1026. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summerer I., Unger K., Braselmann H., Schuettrumpf L., Maihoefer C., Baumeister P. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br. J. Cancer. Jun 30 2015;113(1):76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. Mar 4 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharjya S., Nath S., Ghose J., Maiti G.P., Biswas N., Bandyopadhyay S. miR-125b promotes cell death by targeting spindle assembly checkpoint gene MAD1 and modulating mitotic progression. Cell Death Differ. Mar 2013;20(3):430–442. doi: 10.1038/cdd.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., Zheng X., Li X., Fesler A., Hu W., Chen L. Reduction of gastric cancer proliferation and invasion by miR-15a mediated suppression of Bmi-1 translation. Oncotarget. Mar 22 2016;7(12):14522–14536. doi: 10.18632/oncotarget.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L.F., Wu Z.P., Chen Y., Zhu Q.S., Hamidi S., Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. Aug 1 2014;9(8) doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y.Q., Ren X.Y., He Q.M., Xu Y.F., Tang X.R., Sun Y. MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. Jan 22 2015;6 doi: 10.1038/cddis.2014.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobral L.M., Sousa L.O., Coletta R.D., Cabral H., Greene L.J., Tajara E.H. Stable SET knockdown in head and neck squamous cell carcinoma promotes cell invasion and the mesenchymal-like phenotype in vitro, as well as necrosis, cisplatin sensitivity and lymph node metastasis in xenograft tumor models. Mol. Cancer. Feb 20 2014;13:32. doi: 10.1186/1476-4598-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milella M., Falcone I., Conciatori F., Cesta Incani U., Del Curatolo A., Inzerilli N. PTEN: multiple functions in human malignant tumors. Front. Oncol. Feb 16 2015;5:24. doi: 10.3389/fonc.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Squarize C.H., Castilho R.M., Santos Pinto D., Jr. Immunohistochemical evidence of PTEN in oral squamous cell carcinoma and its correlation with the histological malignancy grading system. J. Oral Pathol. Med. 2002;31(7):379–384. doi: 10.1034/j.1600-0714.2002.00142.x. [DOI] [PubMed] [Google Scholar]
- 18.Metindir J., Dilek G.B., Pak I. Staining characterization by immunohistochemistry of tumor cancer antigen in patients with endometrial cancer. Eur. J. Gynaecol. Oncol. 2008;29(5):489–492. [PubMed] [Google Scholar]
- 19.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao A., Tsai C.L., Wei C.P., Hsueh S., Chao A.S., Wang C.J. Decreased expression of microRNA-199b increases protein levels of SET (protein phosphatase 2A inhibitor) in human choriocarcinoma. Cancer Lett. May 1 2010;291(1):99–107. doi: 10.1016/j.canlet.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Fidler I.J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen D.X., Massagué J. Genetic determinants of cancer metastasis. Nat. Rev. Genet. 2007;8(5):341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 23.Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. Sep 27 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozaki H., Hiraoka T., Mizumoto R., Matsuno S., Matsumoto Y., Nakayama T. The prognostic significance of lymph node metastasis and intrapancreatic perineural invasion in pancreatic cancer after curative resection. Surg. Today. 1999;29(1):16–22. doi: 10.1007/BF02482964. [DOI] [PubMed] [Google Scholar]
- 25.Duraker N., Sisman S., Can G. The significance of perineural invasion as a prognostic factor in patients with gastric carcinoma. Surg. Today. 2003;33(2):95–100. doi: 10.1007/s005950300020. [DOI] [PubMed] [Google Scholar]
- 26.Law W.L., Chu K.W. Anterior resection for rectal cancer with mesorectal excision: a prospective evaluation of 622 patients. Ann. Surg. 2004;240(2):260–268. doi: 10.1097/01.sla.0000133185.23514.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard C.J., Chen M.H., Cote K., Loffredo M., Renshaw A.A., Hurwitz M. Perineural invasion is associated with increased relapse after external beam radiotherapy for men with low-risk prostate cancer and may be a marker for occult, high-grade cancer. Int. J. Radiat. Oncol. Biol. Phys. Jan 1 2004;58(1):19–24. doi: 10.1016/s0360-3016(03)01433-0. [DOI] [PubMed] [Google Scholar]
- 28.Henson B.J., Bhattacharjee S., O'Dee D.M., Feingold E., Gollin S.M. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosom. Cancer. 2009;48(7):569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Lei D.P., Jin T., Zhao X.N., Li G., Pan X.L. Altered expression of miR-21 and PTEN in human laryngeal and hypopharyngeal squamous cell carcinomas. Asian Pac. J. Cancer Prev. 2011;12(10):2653–2657. [PubMed] [Google Scholar]
- 30.Leopoldino A.M., Squarize C.H., Garcia C.B., Almeida L.O., Pestana C.R., Sobral L.M. SET protein accumulates in HNSCC and contributes to cell survival: antioxidant defense, Akt phosphorylation and AVOs acidification. Oral Oncol. 2012;48(11):1106–1113. doi: 10.1016/j.oraloncology.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Almeida L.O., Garcia C.B., Matos-Silva F.A., Curti C., Leopoldino A.M. Accumulated SET protein up-regulates and interacts with hnRNPK, increasing its binding to nucleic acids, the Bcl-xS repression, and cellular proliferation. Biochem. Biophys. Res. Commun. Feb 28 2014;445(1):196–202. doi: 10.1016/j.bbrc.2014.01.175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.