Abstract
Background
Chloroplasts of most plants are responsible for photosynthesis and contain a conserved set of about 110 genes that encode components of housekeeping gene expression machinery and photosynthesis-related functions. Heterotrophic plants obtaining nutrients from other organisms and their plastid genomes represent model systems in which to study the effects of relaxed selective pressure on photosynthetic function. The most evident is a reduction in the size and gene content of the plastome, which correlates with the loss of genes encoding photosynthetic machinery which become unnecessary. Transition to a non-photosynthetic lifestyle is expected also to relax the selective pressure on photosynthetic machinery in the nuclear genome, however, the corresponding changes are less known.
Results
Here we report the complete sequence of the plastid genome of Monotropa hypopitys, an achlorophyllous obligately mycoheterotrophic plant belonging to the family Ericaceae. The plastome of M. hypopitys is greatly reduced in size (35,336 bp) and lacks the typical quadripartite structure with two single-copy regions and an inverted repeat. Only 45 genes remained presumably intact– those encoding ribosomal proteins, ribosomal and transfer RNA and housekeeping genes infA, matK, accD and clpP. The clpP and accD genes probably remain functional, although their sequences are highly diverged. The sets of genes for ribosomal protein and transfer RNA are incomplete relative to chloroplasts of a photosynthetic plant. Comparison of the plastid genomes of two subspecies-level isolates of M. hypopitys revealed major structural rearrangements associated with repeat-driven recombination and the presence of isolate-specific tRNA genes. Analysis of the M. hypopitys transcriptome by RNA-Seq showed the absence of expression of nuclear-encoded components of photosystem I and II reaction center proteins, components of cytochrome b6f complex, ATP synthase, ribulose bisphosphate carboxylase components, as well as chlorophyll from protoporphyrin IX biosynthesis pathway.
Conclusions
With the complete loss of genes related to photosynthesis, NADH dehydrogenase, plastid-encoded RNA polymerase and ATP synthase, the M. hypopitys plastid genome is among the most functionally reduced ones characteristic of obligate non-photosynthetic parasitic species. Analysis of the M. hypopitys transcriptome revealed coordinated evolution of the nuclear and plastome genomes and the loss of photosynthesis-related functions in both genomes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0929-7) contains supplementary material, which is available to authorized users.
Keywords: Chloroplast genome, Parasitic plant, Mycoheterotrophy, Photosynthesis, Gene loss, Transcriptome
Background
Heterotrophic plants obtaining nutrients from other organisms, either directly from other plants (direct parasites) or through mycorrhizal fungi (mycoheterotrophs) with which they associate, often display a reduction in morphological and genomic features. Some heterotrophic plants retain photosynthetic capabilities being hemiparasites, while others completely lose the ability to carry out photosynthesis (i.e. holoparasites). Plastid genomes of parasitic plants represent model systems in which to study the effects of relaxed selective pressure on photosynthetic function. The most evident is a reduction in the size and gene content of the plastid genome, which correlates with the loss of genes encoding photosynthetic machinery which becomes unnecessary [1–3]. Nearly all heterotrophic plants demonstrate a reduction of their plastomes compared with photosynthetic relatives, ranging from minimal in some “early” parasites such as Corallorhiza striata [4] and Cuscuta sp. [5], to extreme in some endoparasitic species like Pilostyles aethiopica [6], and possibly even the complete loss of the plastome in Rafflesia lagascae [7].
Transition to a non-photosynthetic lifestyle is expected to relax the selective pressure on photosynthetic machinery encoded not only in the chloroplast, but also in the nuclear genome. However, the corresponding changes in the nuclear genome and transcriptome of parasitic plants are less known and limited to few studies (e.g. [8, 9]). For example, transcriptome analysis of a holoparasitic plant Phelipanche aegyptiaca of the family Orobanchaceae revealed an expected loss of expression of photosynthesis-related genes and surprising conservation and transcription of the chlorophyll biosynthesis pathway [10].
Mycoheterotrophy evolved independently in at least 50 plant lineages comprising approximately 400 species [11]. Most of the full mycoheterotrophs are monocots, primarily representing the Orchidaceae family [12, 13]. Relatively few complete plastid genomes of mycoheterotrophic plants have been sequenced, including the orchid species Corallorhiza striata, Epigogium roseum, Epigogium aphyllum, Neottia nidus-avis and Rhizanthella gardneri [4, 14–16], Petrosavia stellaris [17], Sciaphila densiflora [18], and the liverwort Aneura mirabilis [19]. Among eudicots, mycoheterotrophs were found in the family Ericaceae [11], which belongs to the order Ericales of Asterids [20]. Many species of Ericaeae forms associations with mycorrhizal fungi and obtain nutrients from them. It is supposed that mycoheterotrophic plants have evolved from photosynthetic mycorrhizal lineages under low-light conditions , where they loss photosynthetic capabilities and established tight association with fungi, which penetrate the roots of green trees and provided the sufficient amounts of carbon and nutrients to its mycoheterotrophic partner [12, 21].
Monotropa hypopitys (pinesap) is an achlorophyllous obligately mycoheterotrophic plant of the family Ericaceae, subfamily Monotropoideae (reviewed in [22]). The above-ground part of M. hypopitys plant is 5–35 cm tall unbranched adventitious raceme-like inflorescence with 2 to 11 flowers on the top and scale-like bracts which cover most of the inflorescence (Additional file 1: Figure S1). Short roots are invested by a net of sheathing mycorrhizas forming haustorium-like structures used to attach the fungal partner. Recently we sequenced the plastid genome of M. hypopitys isolate MON2-KALR [23], the first reported plastome of mycogheterotrophic eudicots. The plastid genome is only 34800-bp long; it is highly rearranged relative to the gene order typical of most the other plastomes of angiosperms, lacking the quadripartite structure with two inverted repeats regions. The reduction in size correlates with the loss of genes encoding NADH dehydrogenase, photosynthesis-related proteins, the plastid-encoded RNA polymerase and some other functions [23]. The loss of photosynthesis-related genes in mycoheterotrophic Ericaceae was also previously shown by Braukmann and Stefanovic [24] using a slot-blot Southern hybridization approach.
Our previous studies revealed the substantial genetic diversity within M. hypopitys and the possible existence of two distinct subspecies or varieties [25, 26]. Taking into account the accelerated evolution of the chloroplast genomes in parasitic plants, existence of such close but distinct subspecies provides an opportunity for a comparative analysis of the molecular changes that mark the progression of plastid genome degradation. Here we report the full plastome sequence of the second subspecies isolate of M. hypopitys, MON-1VOLR and present a comparative analysis of plastomes of both subspecies. In addition, in this study we analysed the transcriptome of M. hypopitys to assess the changes in the nuclear genome associated with a transition to mycoheterotrophy and the loss of the ability to perform photosynthesis.
Methods
Plastid genome sequencing and annotation
A single Monotropa hypopitys plant was collected from the Vologda region, Russia (58°35’14” N, 37°59’20” E). The specimen is stored under accession number MON-1VOLR. Total genomic DNA was extracted from fresh leaves of a single individual and sequenced with a Roche Genome Sequencer GS FLX (Roche, Switzerland) using the Titanium XL+ protocol for a shotgun genome library. About 86 Mb of sequences with an average read length of 356 bp were generated. De novo assembly was performed with Newbler Assembler v. 2.9 (454 Life Sciences, Branford, USA), which yielded three chloroplast DNA contigs with 18-fold coverage. These contigs were identified based on sequence similarity to chloroplast genomes of related species (Ericales) and high coverage. The complete plastid genome sequence was obtained upon the generation of appropriate PCR fragments covering the gaps between the contigs and their sequencing by Sanger method on ABI PRISM 3730 (Applied Biosystems). To verify the correct assembly of the reconstructed plastid genome, raw reads were mapped against the reconstructed sequence with GS Reference mapper (454 Life Sciences, Branford, USA). Plastid genome annotation was performed using DOGMA [27] with further manual correction. Circular map of the plastome was drawn using OrganellarGenomeDRAW tools [28].
The sequence of the plastid genome of M. hypopitys MON-1VOLR was submitted to GenBank under accession number KU640957.
Transcriptome sequencing and assembly
Leaves (flower bracts) and flowers of two individual Monotropa hypopitys plants (not the same specimen used for chloroplast genome sequencing) and two pooled samples of roots with haustoria were collected for transcriptome analysis. Total RNA was extracted from ~0.3 g tissue for each six samples using the RNeasy Plant Mini kit (Qiagen, Valencia, CA). RNA samples were sequenced using the Illumina HiSeq2500 platform (100-bp reads) according to the manufacturer’s instructions (Illumina Inc., USA). RNA-seq read data has been deposited in the NCBI SRA database under accession SRP069226.
M. hypopitys transcriptome sequencing resulted in a total of 103 million high quality sequencing reads after primer and quality trimming with Cutadapt [29] and Sickle (https://github.com/najoshi/sickle), respectively. Assembly of the transcriptome from the combined six RNA-seq datasets was carried out using the Trinity platform [30]. The transcriptome was assembled into 98350 unigenes ranging from 201 to 12993 bp in length with N50 of 1342 bp. Coding region identification in Trinity assembly was done using TransDecoder (http://transdecoder.github.io). Trinotate (https://trinotate.github.io/) was used to assign hits from TrEMBL and Swiss-Prot databases (http://www.uniprot.org/uniprot/), and to assign GO terms and pfam domains (accessed 15/02/2016). 37977 unigenes were annotated in the TrEMBL protein database using predicted protein sequences and 38419 unigenes were annotated in the Swiss-Prot database using BLASTX). Protein-coding genes were assigned with the KEGG orthology identifiers using web-based KAAS server [31].
The levels of transcription of protein-coding genes in the plastid genome were quantitated employing RSEM [32] and Bowtie 2 program [33]. Transcription levels were expressed as fragments per kilobase of exon per million fragments mapped (FPKM) values.
Results and discussion
Plastome size, gene content and expression
The plastid chromosome of Monotropa hypopitys MON1-VOLR is only 35,336 bp in length and lacks a typical quadripartite structure with two single-copy regions and an inverted repeat (Fig. 1). This is one of the most highly reduced plastid genome known from nonphotosynthetic angiosperms after those of Pilostyles sp (11–15 kb, [6]), Sciaphila densiflora (21 kb, [18]), and Epigogium sp. (19–31 kb, [16]). The M. hypopitys MON1 plastome is predicted to contain 45 presumably intact genes (Table 1). Most of these genes are involved in protein synthesis: four ribosomal RNA genes, 19 transfer RNA genes, nine genes code for small subunit and nine for large subunit ribosomal proteins. The remaining genes are infA (translation initiation factor), matK (the maturase for splicing of group IIA introns), accD and clpP.
Fig. 1.
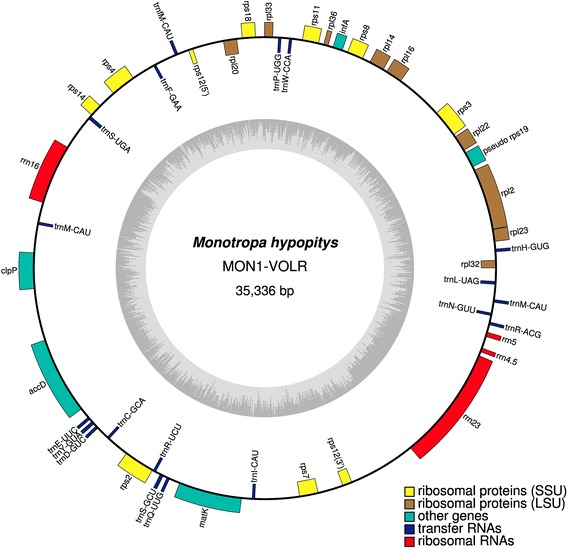
Circular map of the plastid genome of M. hypopitys MON1-VOLR. Genes shown inside the circle are transcribed clockwise, those outside the circle are transcribed counterclockwise. Asterisks indicate intron-containing genes, dark gray bars inside the inner circle indicate guanine-cytosine content
Table 1.
Summary of genes retained in the M. hypopitys plastome
| Function | Genes |
|---|---|
| Ribosomal proteins (large subunit) | rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Ribosomal proteins (small subunit) | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps18 |
| rRNAs | rrn4.5, rrn5, rrn16, rrn23 |
| tRNAs | trnH-GUG, trnW-CCA, trnP-UGG, trnfM-CAU a , trnF-GAA, trnS-UGA, trnM-CAU b , trnE-UUC, trnY-GUA, trnD-GUC, trnC-GCA, trnR-UCU, trnS-GCU, trnQ-UUG, trnI-CAU, trnR-ACG, trnN-GUU, trnL-UAG, trnF-AAA c |
| Other protein-coding genes | infA, matK, clpP, accD d |
a two copies in MON1 and one copy in MON2 plastome
b missing in MON2 plastome
c missing in MON1 plastome
d functionality is unclear
The functionality of the accD gene coding for the β-subunit of the acetyl-CoA carboxylase is unclear since its deduced protein product is very diverged in the N-terminal region and has long repeat-containing internal insertion relative to typical AccD proteins (e.g. from Nicotiana tabacum, Fig. 2). However, the insertions do not interrupt the open reading frame. AccD is involved in fatty-acid synthesis and leaf development [34] and is supposed to be essential to plastome maintenance [35]. Most plastomes of parasitic plants contain a functional copy of the accD gene; however, in Phelipanche it is also highly diverged, containing long indels and appears to lack a standard start codon [36].
Fig. 2.

Alignment of predicted amino acid sequences of AccD proteins from M. hypopitys MON1-VOLR (MON1-accD), M. hypopitys MON2-KALR (MON2-accD) and Nicotiana tabacum (Ntav_accD). Repeats in the MON1-accD sequence are shown by arrows. The AccD conserved domain (COG0777) is boxed. Note that it is interrupted by in-frame insertions in both M. hypopitys plastomes
Like accD, the clpP gene is supposed to be essential for plastid maintenance and is present even in strongly reduced plastomes of parasitic plants [14]. clpP encodes a proteolytic subunit of Clp protease involved in the import of proteins into the plastid [35]. M. hypopitys MON1 plastome the clpP gene lacks both introns and has a very diverged protein sequence with an N-terminal extension, although the whole functional domain (pfam00574) is present (Additional file 2: Figure S2). The closest relative of this protein is ClpP from the chloroplast of the eudicot Asclepias nivea with only 33 % amino acid sequence identity. It should be noted that accelerated evolution of ClpP and intron losses was observed in several parasitic and photosynthetic angiosperm lineages (for example, [36, 37]).
The rps19 is likely a pseudogene judging from its 5’-terminal truncation and the lack of start codon associated with an accumulation of short tandem repeats in this region (Additional file 3: Figure S3). The functionality of rpl22 is unclear since the predicted protein product of this gene is shorter than its homologs in related species due to C-terminal truncation (130 a.a. vs 154–159 a.a.), although the full Rpl22 conservative domain (pfam00237/CHL00034) is retained. Genes for ribosomal proteins Rps15 and Rps16 are missing like in many other plastomes of parasitic plants [28, 36].
Consistently with the obligate mycoheterotrophy of M. hypopitys, its plastome lacks all genes coding for the thylakoid NADH-dehydrogenase complex, photosynthesis-related proteins (photosystems I and II, cytochrome b 6 f complex, rubisco large subunit and ATP synthase). The genes for plastid-encoded RNA polymerase, photosynthesis-related gene ycf1 [38] and conserved genes ycf2, ycf3 and ycf4 are also missing.
Reduction of the size and gene content of the M. hypopitys plastome is also reflected in the loss of introns. Only a single intron was found in the rpl2 gene, while introns in rpl16 and clpP are missing. The rps12 is a trans-splicing gene as in most other angiosperms, but it consists of only two rather than three exons indicating the lack of one intron. The presence of matK maturase in spite of strong reduction of the plastome genome could be determined by the retention of the group IIA intron in the rpl2 gene which requires maturase activity for processing [39].
We analyzed expression of predicted protein-coding genes in the M. hypopitys plastome using RNA-seq data from above-ground part of a single plant. The reads were mapped to the plastid genome and the number of reads corresponding to protein-coding genes (exons) were calculated and normalized per kilobase of the gene length. A total of 33,659 reads were mapped to the protein coding genes. Expression was observed for all genes (Table 2) and, interestingly, the most highly expressed genes were accD and clpP with FPKM value >100,000. In contrast, matK and rpl32 showed lower expression (FPKM value <10,000).
Table 2.
Transcription of protein coding genes in the M. hypopitys MON1-VOLR plastid genome
| Gene | Position (start-end) | FPKM value |
|---|---|---|
| rpl32 | 195-1 | 4424 |
| rpl23 | 650-949 | 36165 |
| rpl2 | 954-1349, 2032-2460a | 66036 |
| rpl22 | 3007-3399 | 71151 |
| rps3 | 3465-4136 | 92022 |
| rpl16 | 5308-5718 | 98751 |
| rpl14 | 5817-6185 | 86138 |
| rps8 | 6383-6781 | 75817 |
| infA | 6916-7155 | 83289 |
| rpl36 | 7278-7391 | 105328 |
| rps11 | 7519-7920 | 27030 |
| rpl33 | 8642-8845 | 29301 |
| rps18 | 9060-9386 | 36275 |
| rpl20 | 9855-9517 | 29237 |
| rps12 | 28675-28433, 10730-10617b | 97942 |
| rps4 | 12232-12843 | 56329 |
| rps14 | 13291-13593 | 89745 |
| clpP | 17294-18169 | 102855 |
| accD | 19448-21352 | 261840 |
| rps2 | 22894-23646 | 37404 |
| matK | 24380-25930 | 4868 |
| rps7 | 27814-27347 | 155291 |
a two exons
b two exons, trans-splicing
Comparison of two plastid genomes of M. hypopitys revealed extensive rearrangements
The availability of the second complete plastome genome of M. hypopitys, obtained from another subspecies-range sample, termed MON2-KALR [23], allowed us to study evolutionary events at a short time scale by comparative analysis. Surprisingly for a single species, these two plastid genomes not only differ slightly in length (35,336 bp for MON1 vs 34,800 bp for MON2 genome), but also show differences in gene order and content (Fig. 3).
Fig. 3.
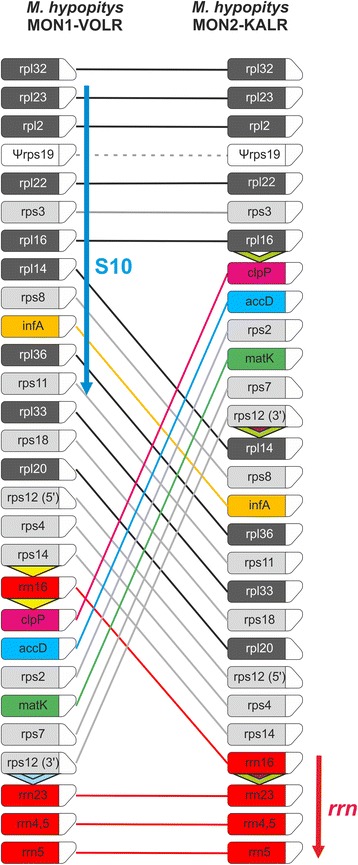
Schematic gene order comparison maps between the plastid genomes of M. hypopitys MON1-VOLR and M. hypopitys MON2-KALR. The linear representation of the circular mapping genomes using rpl32 genes as the starting point does not reflect the actual gene size or spacing between the coding regions. Boxes represent protein-encoding and rRNA genes with the direction of transcription shown by the chamfered top corner. Triangles in M. hypopitys maps indicate repeat sequences. The S10 and rrn operons are shown by arrows. Note that comparison of the structures of plastid genomes of M. hypopitys and Nicotiana tabacum is shown in Additional file 4: Figure S4
The genomes differ in a single major rearrangement and several local minor indels. The major difference is an exchange of positions of genome regions comprising, according to the M. hypopitys MON1 plastome map, genes rps14-rrn16 and clpP- rps12 (5’-exon). In both genomes gene order significantly deviates from the order conserved in photosynthetic angiosperms (Additional file 4: Figure S4) and none could be considered as an “ancestral” form. The first genome, MON1, retains an intact structure of the highly conserved S10 operon (rps11, rpl36, infA, rps8, rpl14, rpl16, rps3, rpl22, rps19, rpl2, rpl23) which is broken in the second genome by the rearrangement. In contrast, the M. hypopitys MON2 plastome contains rrn16-rrn23-rrn4.5-rrn5 gene cluster, which is split in the MON1 genome (Fig. 3). The structures of the S10 and ribosomal RNA operons are highly conserved in the chloroplast genome since it ensured co-expression of the components required for assembly of the ribosome. Despite numerous rearrangements in the course of transition to the non-photosynthetic lifestyle, the gene order in the S10 and rrn operons is conserved even in such highly reduced genomes as in Sciaphila and Epigogium sp. [16, 18].
The rearranged regions are flanked by a repeat sequence that could facilitate recombination events [40, 41]. Three such repeats were found in the MON2 plastome at the “junction” points marking the rearranged regions (Fig. 3); an exchange of the fragment located between the first and the second repeat with one between the second and the third in the MON2 plastome would produce the observed MON1 gene plastome structure.
The content of protein-coding genes is the same in both M. hypopitys plastomes, although some genes whose functionality is questionable are quite variable. The predicted protein products of rpl22 differ in length and both are shorter than in related Ericales species. The deduced products of accD both have N-terminal extensions and internal insertions of different size and low similarity (Fig. 2). The predicted clpP gene in the MON2 plastome contain tandem 72-bp long imperfect duplications in its 5’ region (Additional file 2: Figure S2), but the common parts of the encoded proteins are highly similar (87 % identity) suggesting conservation of functionality. While this article was in review, a report describing the plastid genomes of Monotropa uniflora and another isolate of M. hypopitys was published [42]. It confirmed our previous findings regarding the reduction of the M. hypopitys plastid genome [23], and provided evidences that clpP and accD are intact in these species.
Surprisingly, the two plastid genomes encode a similar but not identical inventory of tRNAs. The MON1 plastome encodes 19 tRNA species for 15 amino acids, and the second plastome encodes 16 tRNA for 14 amino acids. Two tRNA genes are present only in MON1 and one only in the MON2 plastome (Table 1). Interestingly, the second plastome encodes trnF-AAA which is unusual for plant chloroplast genomes and has been reported previously in the parasitic plant E. roseum [16].
Recently Barrett and Davis proposed a model of chloroplast genome degradation in course of transition to heterotrophy [4]. According to this model plastid genes are lost in the following order: NADH dehydrogenase, genes responsible for photosynthesis (psa, psb, pet, rbcL), the plastid-encoded RNA polymerase and ATP synthase genes. Genes encoding ribosomal RNAs and ribosomal proteins, transfer RNAs and several other essential genes (accD, clpP, matK, ycf1, ycf2) are the last to be lost. The gene content of the M. hypopitys plastid genome indicates that this plastome is close to the end of the reduction process since it lacks even some housekeeping genes (ycf1, ycf2, some tRNAs and ribosomal proteins). The notable exceptions are clpP and accD genes which probably remains functional, although their sequences are highly diverged. High levels of transcription of these genes in the plastome also support their functionality. The AccD and ClpP are the components of multi-subunit protein complexes that must be assembled within the chloroplast and thus the clpP and accD genes are retained in the plastomes [14]. The presence of genes coding for other subunits of Clp protease in the M. hypopitys transcriptome (to be published elsewhere) further supports this hypothesis.
The loss of nuclear photosynthesis-related genes, as revealed by transcriptome analysis
The structure and gene content of the M. hypopitys plastid genome clearly shows the complete loss of genes related to photosynthesis. Therefore, we analysed whether the loss of photosynthesis function is accompanied by the loss of the corresponding genes encoded at the nuclear genome. To answer this question we analysed the presence of the photosynthesis-related genes in the M. hypopitys transcriptome using the Arabidopsis reference pathways as a query [10]. Among 63 genes of the KEGG pathway “Photosynthesis” only three were detected in the transcriptome (Additional file 5: Table S1). Of the photosystem I and II reaction center proteins, components of cytochrome b 6 f complex, ATP synthase and ribulose bisphosphate carboxylase components, none was detected in the transcriptome, indicating gene losses and/or absence of expression. Sequencing and analysis of the nuclear genome of M. hypopitys will answer the question whether these genes were lost or only silenced.
Unlike the components of the photosynthetic reaction centers, expression of the chlorophyll biosynthesis genes was previously detected in non-photosynthetic parasitic plant P. aegyptiaca [10]. However, analysis of the M. hypopitys transcriptome revealed that among the chlorophyll biosynthesis pathway components [43] only a single gene, divinyl chlorophyllide a 8-vinyl-reductase, was expressed at a low level and only in flowers, while protoporphyrin IX Mg-chelatase, protoporphyrin IX methyltransferase, Mg-protoporphyrin IX monomethyl ester oxidative cyclase, protochlorophyllide oxidoreductase and chlorophyll synthase were absent from the transcriptome. These findings are consistent with the lack of chlorophyll in M. hypopitys [44]. The upstream biosynthetic pathway generating protoporphyrin IX from aminolevulinic acid is present and expressed, as expected since these steps are common for both heme and chlorophyll synthesis.
Conclusions
With the complete loss of genes related to photosynthesis, namely NAD(P)H dehydrogenase, plastid-encoded RNA polymerase and ATP synthase, the M. hypopitys plastid genome is among the most functionally reduced ones among obligate non-photosynthetic parasitic species. Gene loss and genome reduction were associated with rearrangements, loss of the typical quadripartite structure of the plastid genome and also the loss of introns. The advanced process of plastid genome degradation also involved some housekeeping functions as evidenced by the loss of some ribosomal protein and transfer RNA genes. Comparison of the plastid genomes of two subspecies-level isolates of M. hypopitys revealed major structural rearrangements associated with repeat-driven recombination and the presence of isolate-specific tRNA genes. Analysis of the M. hypopitys transcriptome revealed the coordinated evolution of the nuclear and plastid genomes and the loss of photosynthesis-related functions in both genomes.
Acknowledgements
We are grateful to two anonymous reviewers for their constructive comments, which helped us to improve the manuscript. This work was performed using the scientific equipment of Core Research Facility ‘Bioengineering’.
Declarations
This article has been published as part of BMC Plant Biology Volume 16 Supplement 3, 2016: Selected articles from BGRS\SB-2016: plant biology. The full contents of the supplement are available online at http://bmcplantbiol.biomedcentral.com/articles/supplements/volume-16-supplement-3.
Funding
This work was supported by Russian Science Foundation grant 14-24-00175 (transcriptome and MON2-KALR plastome sequencing) and Russian Foundation for Basic Research grant 14-14-00749 (MON1-VOLR plastome sequencing). The publication costs were funded by Russian Science Foundation.
Authors’ contributions
NVR conceived the study, analysed the data and drafted the manuscript. EVG, AVB and VVK analysed the genome and transcriptome data. AMM, EBP and AVM performed sequencing. MAF and EZK collected the plant samples and isolated nucleic acids. KGS conceived the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Figure S1. M. hypopitys plants in its natural environment. (PDF 436 kb)
Figure S2. ClpP coding regions. (PDF 97 kb)
Figure S3. Nucleotide sequences of putative rps19 pseudogene and upstream intergenic regions. (PDF 68 kb)
Figure S4. Schematic gene order comparison maps between the plastid genomes of M. hypopitys MON1-VOLR, M. hypopitys MON2-KALR and Nicotiana tabacum. (PDF 238 kb)
Table S1. Analysis of the presence of genes of the KEGG pathway “Photosynthesis” in the transcriptome. (XLS 29 kb)
Contributor Information
Nikolai V. Ravin, Email: nravin@biengi.ac.ru, Email: nravin@mail.ru
Eugeny V. Gruzdev, Email: gruevg@ya.ru
Alexey V. Beletsky, Email: mortu@yandex.ru
Alexander M. Mazur, Email: mazur.am@gmail.com
Egor B. Prokhortchouk, Email: prokhortchouk@biengi.ac.ru
Mikhail A. Filyushin, Email: michel7753@mail.ru
Elena Z. Kochieva, Email: kochieva@biengi.ac.ru
Vitaly V. Kadnikov, Email: vkadnikov@bk.ru
Andrey V. Mardanov, Email: mardanov@biengi.ac.ru
Konstantin G. Skryabin, Email: skryabin@biengi.ac.ru
References
- 1.dePamphilis CW, Palmer JD. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990;348:337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 2014;31:3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- 4.Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 2012;99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- 5.Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellot S, Renner SS. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol Evol. 2015;8(1):189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres MJ, Tomilov AA, Tomilova N, Reagan RL, Yoder JI. Pscroph, a parasitic plant EST database enriched for parasite associated transcripts. BMC Plant Biol. 2005;5:24. doi: 10.1186/1471-2229-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida S, Ishida JK, Kamal NM, Ali AM, Namba S, Shirasu K. A full-length enriched cDNA library and expressed sequence tag analysis of the parasitic weed, Striga hermonthica. BMC Plant Biol. 2010;10:55. doi: 10.1186/1471-2229-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickett NJ, Honaas LA, Wafula EK, Das M, Huang K, Wu BA, Landherr L, Timko MP, Yoder J, Westwood JH, et al. Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol. 2011;21:2098–2104. doi: 10.1016/j.cub.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Merckx V, Freudenstein JV. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 2010;185:605–609. doi: 10.1111/j.1469-8137.2009.03155.x. [DOI] [PubMed] [Google Scholar]
- 12.Bidartondo MI. The evolutionary ecology of myco-heterotrophy. New Phytol. 2005;167(2):335–352. doi: 10.1111/j.1469-8137.2005.01429.x. [DOI] [PubMed] [Google Scholar]
- 13.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Delannoy E, Fujii S, des Francs-Small CC, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logacheva MD, Schelkunov MI, Penin AA. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse MA, Penin AA, Logacheva MD. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epigogium aphyllum and Epigogium roseum. Genome Biol Evol. 2015;7:1179–1191. doi: 10.1093/gbe/evv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 2014;6:238. doi: 10.1093/gbe/evu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam VK, Soto Gomez M, Graham SW. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol Evol. 2015;7(8):2220–2236. doi: 10.1093/gbe/evv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, Plock SA, Wolf PG, DePamphilis CW, Boore JL, Goffinet B. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 2008;25:393–401. doi: 10.1093/molbev/msm267. [DOI] [PubMed] [Google Scholar]
- 20.Anderberg AA, Rydin C, Kallersjo M. Phylogenetic relationships in the order Ericales s.l.: analyses of molecular data from five genes from the plastid and mitochondrial genomes. Am J Bot. 2002;89:677–687. doi: 10.3732/ajb.89.4.677. [DOI] [PubMed] [Google Scholar]
- 21.Motomura H, Selosse MA, Martos F, Kagawa A, Yukawa T. Mycoheterotrophy evolved from mixotrophic ancestors: evidence in Cymbidium (Orchidaceae). Ann. Bot. 2010;106(4):573–81. [DOI] [PMC free article] [PubMed]
- 22.Kron KA, Judd WS, Stevens PF, Crayn DM, Anderberg AA, Gadek PA, Quinn CJ, Luteyn JL. Phylogenetic classification of Ericaceae: Molecular and morphological evidence. Bot Rev. 2002;68(3):335–423. doi: 10.1663/0006-8101(2002)068[0335:PCOEMA]2.0.CO;2. [DOI] [Google Scholar]
- 23.Gruzdev EV, Mardanov AV, Beletsky AV, Kochieva EZ, Ravin NV, Skryabin KG. The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: extensive gene losses and size reduction. Mitochondrial DNA Part B: Resources. 2016;1(1):212–213. doi: 10.1080/23802359.2016.1155090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braukmann T, Stefanović S. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol Biol. 2012;79:5–20. doi: 10.1007/s11103-012-9884-3. [DOI] [PubMed] [Google Scholar]
- 25.Filyushin MA, Reshetnikova NM, Kochieva EZ, Skryabin KG. Intraspecific variability of ITS sequences in the parasitic plant Monotropa hypopitys L. from the European Russian populations. Genetika. 2015;51(11):1330–1334. [PubMed] [Google Scholar]
- 26.Filyushin MA, Reshetnikova NM, Kochieva EZ, Skryabin KG. Polymorphism of sequences and the secondary structure of b/c intron of mitochondrial gene nad1 in Monotropa hypopitys and related Ericaceae species. Biol Bull. 2016;43(3):271–275. doi: 10.1134/S1062359016030043. [DOI] [Google Scholar]
- 27.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 28.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW--a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41(Web Server issue):W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 30.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kode V, Mudd EA, Iamtham S, Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44:237–244. doi: 10.1111/j.1365-313X.2005.02533.x. [DOI] [PubMed] [Google Scholar]
- 35.Krause K. Plastid genomes of parasitic plants: a trail of reductions and losses. In: Bullerwell CE, editor. Organelle genetics. Berlin, Heidelberg: Springer; 2012. pp. 79–103. [Google Scholar]
- 36.Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25(10):3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae) Mol Phylogenet Evol. 2014;72:82–89. doi: 10.1016/j.ympev.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339:571–574. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]
- 39.Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogihara Y, Terachi T, Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short directrepeat sequences in wheat species. Proc Natl Acad Sci U S A. 1988;85:8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- 42.Logacheva MD, Schelkunov MI, Shtratnikova VY, Matveeva MV, Penin AA. Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives. Sci Rep. 2016;6:30042. doi: 10.1038/srep30042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beale SI. Enzymes of chlorophyll biosynthesis. Photosynth Res. 1999;60:43–73. doi: 10.1023/A:1006297731456. [DOI] [Google Scholar]
- 44.Leake JR. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994;127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]


