Abstract
Background
Microbial antimonite [Sb(III)] oxidation converts toxic Sb(III) into less toxic antimonate [Sb(V)] and plays an important role in the biogeochemical Sb cycle. Currently, little is known about the mechanisms underlying bacterial Sb(III) resistance and oxidation.
Results
In this study, Tn5 transposon mutagenesis was conducted in the Sb(III)-oxidizing strain Pseudomonas stutzeri TS44 to isolate the genes responsible for Sb(III) resistance and oxidation. An insertion mutation into gshA, encoding a glutamate cysteine ligase involved in glutathione biosynthesis, generated a strain called P. stutzeri TS44-gshA540. This mutant strain was complemented with a plasmid carrying gshA to generate strain P. stutzeri TS44-gshA-C. The transcription of gshA, the two superoxide dismutase (SOD)-encoding genes sodB and sodC as well as the catalase-encoding gene katE was monitored because gshA-encoded glutamate cysteine ligase is responsible for the biosynthesis of glutathione (GSH) and involved in the cellular stress defense system as are superoxide dismutase and catalase responsible for the conversion of ROS. In addition, the cellular content of total ROS and in particular H2O2 was analyzed. Compared to the wild type P. stutzeri TS44 and TS44-gshA-C, the mutant P. stutzeri TS44-gshA540 had a lower GSH content and exhibited an increased content of total ROS and H2O2 and increased the Sb(III) oxidation rate. Furthermore, the transcription of sodB, sodC and katE was induced by Sb(III). A positive linear correlation was found between the Sb(III) oxidation rate and the H2O2 content (R 2 = 0.97), indicating that the accumulated H2O2 is correlated to the increased Sb(III) oxidation rate.
Conclusions
Based on the results, we propose that a disruption of the pathway involved in ROS-protection allowed H2O2 to accumulate. In addition to the previously reported enzyme mediated Sb(III) oxidation, the mechanism of bacterial oxidation of Sb(III) to Sb(V) includes a non-enzymatic mediated step using H2O2 as the oxidant.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0902-5) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas stutzeri, Sb(III) oxidation, H2O2, Transposon mutagenesis, gshA, Reactive oxygen species (ROS)
Background
Antimony (Sb) is a metal belonging to group V in the periodic table. It is present in aquatic systems and soil, with stibnite (Sb2S3) representing the most common mineral form [1]. Antimonials have been used to treat leishmaniasis for over 60 years [2, 3]. However, the U.S. Environmental Protection Agency recognized Sb as a priority pollutant [4], and the WHO has proposed 5 μg/L to be the highest acceptable Sb concentration in potable water [5]. Although Sb pollution has gained increased attention in recent decades [6–8], the exact mechanism of Sb toxicity in both mammals and microorganism remains unclear [9, 10].
A comparison of two common inorganic forms of Sb revealed that antimonite [Sb(III)] was more toxic than antimonate [Sb(V)] [7]. The abiotic Sb(III) oxidation in the natural environment is extremely slow [11]. In contrast, Sb(III)-oxidizing bacteria can oxidize Sb(III) at a relatively high rate. Thus, Sb(III) oxidation was proposed to be a microbial detoxification process that could be useful for environmental Sb bioremediation [12–14]. A few Sb(III)-oxidizing bacteria had been reported in earlier literature [15, 16]. Recently, dozens of Sb(III)-oxidizing bacteria have been isolated and identified both by our group and by others [12, 13, 17, 18]. The arsenite oxidase AioAB in Agrobacterium tumefaciens was also found to be able to oxidize Sb(III) [19]. In addition, the oxidoreductase AnoA was shown to be responsible for bacterial Sb(III) oxidation [20]. However, the disruption of both of these genes did not result in a complete loss of Sb oxidation, indicating the existence of other mechanisms responsible for bacterial Sb(III) oxidation.
In general, aerobic respiration is inevitably accompanied by the production of reactive oxygen species (ROS). ROS include superoxide (O2 •-), hydroxyl (OH•), hydroperoxyl (HO2 •), and peroxyl (RO2 •) radicals and other oxidizing agents, such as hydrogen peroxide (H2O2) [21]. Among the various ROS, O2 •- is the primary radical that results from the univalent reduction of molecular oxygen (O2) via the bacterial respiratory chain. Subsequently, O2 •- can be converted into H2O2 and O2 by superoxide dismutase (SOD). Both O2 •- and H2O2 can generate highly reactive hydroxyl radicals via the Haber-Weiss reaction or the Fenton reaction [22]. ROS can damage different types of macromolecules; thus, bacteria have evolved defense mechanisms against ROS and induce specific genes in response to oxidative stress [23]. Glutathione (GSH) is the central substrate involved in cellular protection against ROS and their toxic products in eukaryotic cells [24]. In bacteria GSH plays multiple role in protection against environmental stresses such as osmotic shock and acidity, as well as against toxins and oxidative stress. Indeed, GSH represents the primary low molecular weight thiol in many Gram-negative bacteria [25]. Two enzymes (glutamate cysteine ligase, the rate-limiting enzyme, and GSH synthetase) catalyze the de novo synthesis of GSH. These two enzymes are encoded by distinct genes (gshA and gshB, respectively) in most bacteria [26].
Previously, the aerobic, arsenite-oxidizing bacterium Pseudomonas stutzeri TS44 was isolated from arsenic-contaminated soil [27] and the genome sequence was published (Accession No. AJXE00000000, [28]). In this study, we showed that strain TS44 is able to oxidize Sb(III) to Sb(V). Therefore, in an effort to determine the molecular mechanism underlying bacterial Sb(III) oxidation, more than 3000 Tn5 transposon insertion mutants were generated and screened. We isolated a mutant of gshA (TS44-gshA540) that showed an increase in Sb(III) oxidation rate. The Sb(III) oxidation rates of the wild type, mutant and complemented strain, the gene transcription of gshA, sod and katE, as well as the cellular contents of ROS, GSH and H2O2 were analyzed since gshA is responsible for the biosynthesis of GSH and therefore involved in the cellular stress defense system.
Methods
Bacterial strains, plasmids, primers and culture conditions
The strains, plasmids and primers used in this study are listed in Table 1 and Additional file 1: Table S1. The Pseudomonas stutzeri strains were grown in a chemically defined medium (CDM) [29] containing 0.114 mmol/L phosphate (0.07 mmol/L K2HPO4•3H2O and 0.044 mmol/L KH2PO4) and shaken under aerobic conditions at 28 °C. The Escherichia coli strains were cultured at 37 °C in Luria-Bertani medium [30]. Rifampin (Rif, 50 mg/mL), kanamycin (Kan, 50 mg/mL), tetracycline (Tet, 5 mg/mL) or chloromycetin (Cm, 50 mg/mL) were added when needed.
Table 1.
The strains and plasmids used in this study
| Strain/plasmid | Relevant properties or derivation | Source or reference |
|---|---|---|
| Pseudomonas stutzeri | ||
| TS44 | Wild-type, As(III)-oxidizing phenotype | [27] |
| TS44-gshA540 | Kanr; gshA mutant by Tn5 random transposon mutagenesis | This study |
| TS44-gshA-C | Kanr Cmr; gshA complemented strain | This study |
| TS44 (PgshA) | Kanr; TS44 with pLSP-PgshA | This study |
| TS44 (PsodB) | Kanr; TS44 with pLSP-PsodB | This study |
| TS44 (PsodC) | Kanr; TS44 with pLSP-PsodC | This study |
| TS44-gshA540
(PgshA) |
Kanr; TS44-gshA540 with pLSP-PgshA | This study |
| TS44-gshA540
(PsodB) |
Kanr; TS44-gshA540 with pLSP-PsodB | This study |
| TS44-gshA540
(PsodC) |
Kanr; TS44-gshA540 with pLSP-PsodC | This study |
| Escherichia coli | ||
| DH5α | supE44 lacU169(j80lacZM15) hRDR17 recA1 endA1 gyrA96 thi-1 relA1 |
[35] |
| S17-1 | Pro− Mob+; conjugation donor | [36] |
| pir116 | mcrA, Δ(mrr-hsdRMS-mcrBC) recA1 R6Kγ lacZΔM15 λpir | Epicenter, Madison, WI |
| Plasmids | ||
| pRL27-Kan | Kanr Transposon vector, oriR6K | [37] |
| pCT-Zori | broad host vector, pUC ori, Cmr | [34] |
| pCT-Zori-gshA | gshA complementation vector | This study |
| pLSP-kt2lacZ | Kanr oriV; lacZ fusion vector used for lacZ fusion constructs | T. R. McDermott, MSU |
| pLSP-PgshA | pLSP-kt2lacZ containing gshA promoter region | This study |
| pLSP-PsodB | pLSP-kt2lacZ containing sodB promoter region | This study |
| pLSP-PsodC | pLSP-kt2lacZ containing sodC promoter region | This study |
Isolation of a Sb(III) sensitive transposon mutant and complementation of the mutant strain
Plasmid pRL27-Kan was transferred into P. stutzeri TS44 by conjugation from E. coli strain S17-1 carrying Tn5 as described previously [31]. To obtain Sb(III) sensitive mutants, the colonies of transformants from the mating plates were spread onto LB agar plates containing 50 μg/mL Kan and 50 μg/mL Rif, in the presence or absence of 0.4 mmol/L C8H4K2O12Sb2S3(H2O) [as Sb(III)]. The mutants did not grow in the presence of the indicated concentration of Sb(III). The insertion sequence of the mutant was determined using a plasmid rescue strategy as described by Zheng et al. [31]. The sequences were compared to the draft genome sequence of P. stutzeri TS44 using BLAST [32] and compared to the protein sequence database at GenBank using the BlastX algorithm [33]. Finally, the Sb-sensitive Tn5-insertion mutant TS44-gshA540 was isolated.
The construction of the gshA complementary strain TS44-gshA-C was accomplished using the high-copy broad host range plasmid pCT-Zori containing a Kan resistance determinant ([34], Table 1). Briefly, a 2023 bp DNA fragment containing the complete gshA coding region along with 156 bp upstream and 289 bp downstream sequence was PCR-amplified using the gshA-F/gshA-R primers (Additional file 1: Table S1) and subsequently cloned into HindIII + BamHI-digested pCT-Zori. The resulting plasmid pCT-Zori-gshA was transformed into E. coli S17-1 and conjugated into the mutant TS44-gshA540, yielding the complementary strain TS44-gshA-C. The integrity of the mutant and the complementary strain was confirmed by PCR amplification and subsequent DNA sequencing.
Growth and Sb(III) oxidation tests
Overnight cultures of P. stutzeri strains TS44, TS44-gshA540 and TS44-gshA-C (OD600 = 1.0) were inoculated into 100 mL of CDM medium with or without 0.2 mmol/L Sb(III) and incubated at 28 °C for 48 h with shaking at 170 rpm. Culture samples were collected to determine the OD600 value using a UV spectrophotometer (DU800, Beckman, USA) at the designated time points. Sb(III)/Sb(V) concentrations were monitored using hydride-generation atomic fluorescence spectroscopy combining HPLC (HPLC-HG-AFS, Beijing Titan Instruments Co., Ltd., China) according to the method described by Li et al. [13]. The measured data were analyzed with single factor analysis of variance (one way ANOVA) method in Excel program.
GshA activity and determination of GSH content
To detect GshA activity and GSH content, P. stutzeri strains TS44, TS44-gshA540 and TS44-gshA-C were cultured under aerobic condition with shaking in 100 mL of liquid CDM medium at 28 °C. Sb(III) was added at a final concentration of 0.2 mmol/L when the OD600 reached 0.3. Cultures without the addition of Sb(III) were used as controls. One mililiter of cells was centrifuged at 13,400 × g at 4 °C after 30 min of cultivation. The pellet was washed twice using phosphate-buffered saline (PBS, pH 7.0), then resuspended in 1 mL of PBS and sonicated on ice to dissolve the cell membranes. The total protein content of the sonicate was measured by the Coomassie brilliant blue G-250-staining method [38]. Bovine serum albumin was used as the standard.
The GshA activity and GSH content were determined using 2, 3-naphthalenedicarboxyaldehyde (NDA) (Aladdin Industrial Co., Shanghai, China) as described previously [39]. To measure GshA activity, an equal volume of cell lysate was mixed with the GshA reaction buffer and substrate solution (50 μL each). Fifty microliter of reaction terminator (1 mol/L Na2CO3) was added after 45 min of incubation at 28 °C. The mixture was incubated on ice for 20 min and centrifuged at 8000 × g for 5 min. Then, 20 μL of the supernatants were transferred to wells of black Microlon 96-well plates (Greiner). The samples were mixed with 180 μL of NDA derivatization solution (50 mmol/L Tris–HCl, pH = 10, 0.5 N NaOH, and 10 mmol/L NDA in Me2SO, v/v/v 1.4/0.2/0.2). Fluorescence was measured (472 ex/528 em) with an EnVision® Multimode Plate Reader (Perkin Elmer) after 60 min of incubation at 37 °C and converted to the γ-glutamylcysteine concentration using appropriate calibration standards. The procedure for measuring GSH content was almost the same as the procedure to detect GshA activity, except that the reaction terminator was added immediately when the equal volume of cell lysate was mixed with the GshA reaction buffer and substrate solution (50 μL each).
Determination of the ROS content
The cellular ROS content was tested with the fluorescent probe 2, 7-dichlorofluorescin diacetate (DCFH-DA) (Sigma Chemical Co., St. Louis, MO, USA). The cells were cultivated and collected for processing as described above. Cells from 0.5 mL cultures were washed two times with PBS buffer and resuspended in 0.5 mL of PBS buffer. Then, a 0.2 mL cell suspension was mixed with 5 μL of 0.1 mmol/L DCFH-DA and incubated at 37 °C for 30 min to develop the fluorescent product DCF. The cells were harvested by centrifugation for 3 min at 13,400 × g and washed two times using PBS buffer (pH 7.0) to remove background fluorescence. The fluorescence was measured (488 ex/535 em) on the EnVision® Multimode Plate Reader (Perkin Elmer) [40]. The ROS contents were normalized to the total protein content as described above.
Quantification of gene expression using a lacZ reporter gene fusion and qRT-PCR
Quantitative reverse transcription PCR (qRT-PCR) was employed to test the transcription of gshA, two superoxide dismutase (SOD)-encoding genes (sodB and sodC) and the catalase katE. The strains were inoculated into 100 mL of CDM medium. 0.2 mmol/L of Sb(III) was added (or not) when the OD600 reached 0.3. The cells were harvested after 30 min. Total RNA was extracted using the TRIzol® Reagent (Invitrogen) and treated with DNaseI following the manufacturer’s instructions. The synthesis of cDNA from 300 ng of total RNA was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo) [41]. The resulting cDNA was used as a template for qRT-PCR with the SYBR-Green® qPCR Master Mix (Takara). Primers RT-gshA-F/RT-gshA-R, RT-sodB-F/RT-sodB-R, RT-sodC-F/RT-sodC-R and RT-katE-F/RT-katE-R were used to test the expression of gshA, sodB, sodC and katE, respectively. The RT-PCRs were performed using the AB ViiA 7 RT-PCR system (Life Technologies) following the manufacturer’s recommended protocol. The annealing temperature for gshA and sodB was 55 °C, while for sodC and katE it was 48 °C.
For the lacZ reporter fusion analysis, plasmid pLSP-kt2lacZ was used to construct the lacZ fusions. DNA fragments containing the predicted promoter regions of gshA, sodB and sodC were amplified by PCR using the primers PgshA-F/PgshA-R, PsodB-F/PsodB-R and PsodC-F/PsodC-R, respectively. The DNA fragments were digested using EcoRI and BamHI and ligated into the double-digested pLSP-kt2lacz. The resulting plasmids pLSP-PgshA, pLSP-PsodB, and pLSP-PsodC were separately transferred into strains TS44 and TS44-gshA540 by conjugation employing E. coli S17-1. The resulting strains containing the above constructs were inoculated into 100 mL of CDM medium and cultivated with shaking at 28 °C. 0.2 mmol/L of Sb(III) was added when the OD600 reached 0.3. The samples were collected and the β-galactosidase activity was tested according to the method described by Miller [42] after 30 min of incubation.
Determination of the linear correlation between H2O2 and Sb(III) oxidation rate
To test the H2O2 content, strains TS44, TS44-gshA540 and TS44-gshA-C were incubated as described above. To eliminate the H2O2 content difference caused by the different amount of cell collection, Sb(III) was added when the OD600 reached 0.3, and then 2 mL of cells were harvested after a 30 min incubation with Sb(III). The cells were resuspended in 1 mL of K3PO4 (pH 7.8) after washing twice with 50 mmol/L K3PO4 (pH 7.8) and sonicated on ice. Then, the sonicated cell lysates were centrifuged at 13,400 × g at 4 °C, and 0.1 mL of the supernatant was transferred to wells of black Microlon 96-well plates (Greiner). The samples were mixed with 50 μL of amplex red (AR) (Sigma Chemical Co., St. Louis, MO, USA) and 50 μL of horseradish peroxidase (HRP) (F. Hoffmann-La Roche Ltd, Shanghai, China), then incubated at 37 °C for 15 min. Fluorescence was measured (530 ex/587 em) as described above using the EnVision® Multimode Plate Reader (Perkin Elmer) and converted to the H2O2 concentration using appropriate calibration standards [43].
To detect the dynamic variation of the H2O2 and Sb(III) contents, strains TS44, TS44-gshA540 and TS44-gshA-C were inoculated into 100 mL of CDM medium supplemented with 0.2 mmol/L Sb(III). The cells were harvested and sonicated as described above at the designated time points. The Sb(III) contents were monitored using HPLC-HG-AFS as described above, while the H2O2 contents were determined by the AR/HRP method as described above. A boiled CDM culture of strain TS44-gshA540 was used as a control. To test the effect of H2O2 on bacterial growth, strain TS44 was inoculated onto CDM plates containing different concentrations of H2O2 and cultivated at 28 °C for 48 h.
In vitro oxidation of Sb(III) by H2O2 was performed using an un-inoculated liquid CDM medium containing 0.2 mmol/L of Sb(III) and the media with the addition of 0, 0.02, 0.05, 0.1, 0.3, 0.4 and 0.5 mmol/L of H2O2 and reacted within 10 min.
Results
A Sb(III)-sensitive mutant could be generated by transposon mutagenesis
More than 3000 transposon insertions were isolated and screened for loss of Sb(III) resistance with one mutant displaying lower Sb(III) resistance. This strain, TS44-gshA540, was selected for further characterization (data not shown). The Tn5 transposon had inserted into gshA encoding a putative glutamate-cysteine ligase at nucleotide 540 (Additional file 2: Figure S1A). The gshA encoded glutamate-cysteine ligase is the rate-limiting enzyme in de novo GSH biosynthesis and therefore is involved in conferring resistance to ROS and their toxic products [44]. Additionally, a strain complementing the gshA insertion was constructed and named TS44-gshA-C. Diagnostic PCRs were used to confirm the transposon mutation and the complementation (Additional file 2: Figure S1B-C).
An insertion in gshA led to an increased Sb(III) oxidation rate
Cells of P. stutzeri TS44, TS44-gshA540 and TS44-gshA-C were incubated in CDM medium lacking Sb(III) supplementation after thorough washing. The growth of strain TS44-gshA540 was slightly slower compared to the wild type strain (Fig. 1a). The growth of strain TS44-gshA540 was further delayed by supplementation with 0.2 mmol/L Sb(III), causing strain TS44-gshA540 to need an extra 24 h to reach the lag phase (Fig. 1b). The gshA complementing strain TS44-gshA-C showed no difference in growth compared to the wild type strain regardless of whether Sb(III) was added or not. Although the growth of TS44-gshA540 was delayed, strain TS44-gshA540 oxidized 88% Sb(III) to Sb(V) in 48 h (Fig. 2b). In contrast, strains TS44 and TS44-gshA-C both oxidized only 48% Sb(III) to Sb(V) in 48 h (Fig. 2a and c). The Sb(III)-oxidation rate of TS44-gshA540 significantly increased by 83% in the monitored 48 h time frame (p < 0.01) (Fig. 2).
Fig. 1.
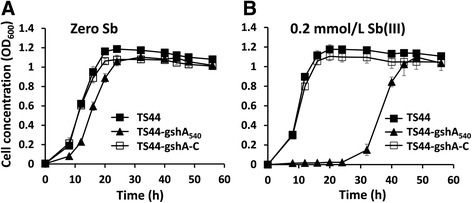
Growth curves of P. stutzeri strains TS44, TS44-gshA540 and TS44-gshA-C without (a) or with (b) the addition of 0.2 mmol/L Sb(III). Error bars correspond to the standard deviations of the means from three independent experiments
Fig. 2.
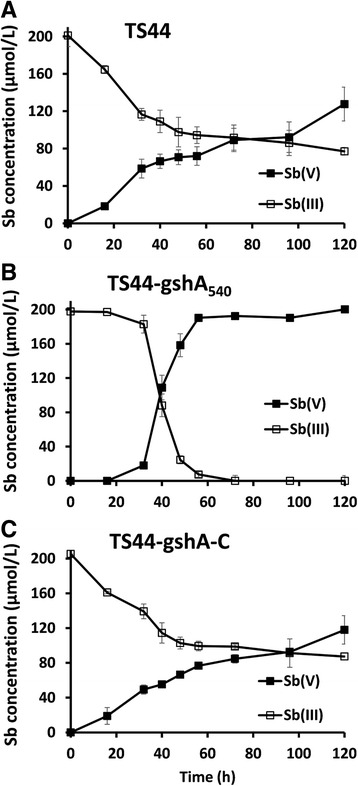
Sb(III) oxidation in P. stutzeri strains TS44, TS44-gshA540 and TS44-gshA-C with the addition of 0.2 mmol/L Sb(III). The amounts of Sb(III) and Sb(V) in culture fluids were calculated based on culture volume to normalize for the total amount of antimony added. Sb(III) and Sb(V) concentrations in the culture fluids were measured using HPLC-HG-AFS. Error bars correspond to the standard deviations of the means from three independent experiments
An insertion in gshA eliminated GshA activity and influenced cellular contents of GSH and ROS
We investigated both GshA activity and the GSH and ROS content to elucidate how the presence of GshA affected Sb(III) oxidation. The GshA activity in strains TS44 and TS44-gshA-C was slightly increased following the addition of Sb(III); however, GshA activity was undetectable in the insertional mutant strain TS44-gshA540 (Fig. 3a), indicating that the transposon insertion functionally disrupted gshA in strain TS44. Consistent with this result, the GSH content was decreased by approximately 66% in the mutant compared to strains TS44 and TS44-gshA-C regardless of whether Sb(III) was provided (Fig. 3b).
Fig. 3.
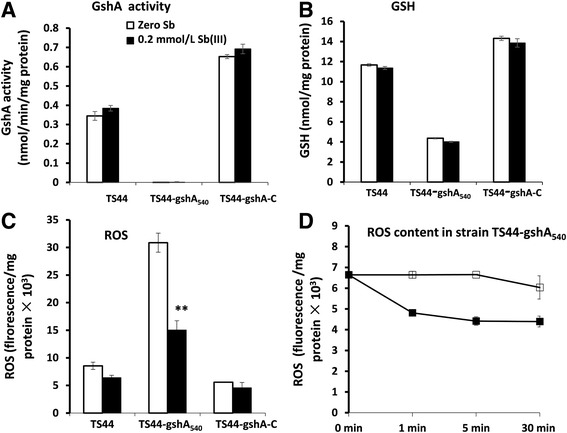
gshA insertion affected GSH content, GshA activity and ROS content. a, GshA activity, (b), GSH content, (c), ROS content and (d) ROS content in strain TS44-gshA540. Data symbols shown in panels (a), (b) and (c) are the same. Data are expressed as the mean ± SD, N = 3. **Indicates a significant difference from the control (p < 0.01)
As a next step, cellular ROS content was determined because GSH is the central substrate for cellular protection against ROS and their toxic products [24]. The cellular ROS content in strain TS44-gshA540 was increased by approximately three-fold compared to strains TS44 and TS44-gshA-C regardless of whether Sb(III) was provided or not. The cellular ROS content was significantly decreased in strain TS44-gshA540 after 30 min of incubation with Sb(III) compared to no Sb(III) addition (Fig. 3b-c), despite the fact that gshA was mutated and the GSH content was obviously decreased (Fig. 3a-b). After addition of Sb(III), the ROS content changed quickly in 30 min, and decreased significantly in the strain TS44-gshA540 (Fig. 3d). In such a short time, the oxidation rate of Sb(III) did not significantly change. The Sb(III) oxidation mainly occurred in the 36–48 h time frame (Fig. 2).
Sb(III) affected transcription of gshA, sodB, sodC and katE differently
The expression of gshA, sodB, sodC and katE was monitored, because GshA, SodB, SodC and KatE are all involved in changes of GSH, ROS or H2O2. There are two distinct genes (sodB and sodC) encoding superoxide dismutase (SOD) on the genome of P. stutzeri TS44 that encode for Fe SOD and Cu-Zn SOD, respectively. These two enzymes shared over 83% and 58% amino acid sequence similarity with the respective enzymes of other species of Pseudomonas, respectively, and can convert O2 •- to H2O2 and O2. In the lacZ reporter fusion analysis, we only transformed the lacZ fusions designated pLSP-PgshA, pLSP-PsodB and pLSP-PsodC into strains TS44 and TS44-gshA540 because the gshA complementary strain TS44-gshA-C carried a plasmid that might be incompatible with the lacZ fusion.
The transcription levels of gshA, sodB, sodC and katE were tested by qRT-PCR in all three strains. Sb(III) had no effect on the transcription of gshA in strains TS44, TS44-gshA540 or TS44-gshA-C (Fig. 4a and d). The gshA::lacZ fusion only showed a low level of background expression levels in the gshA insertional mutation strain TS44-gshA540, (Fig. 4a), which indicated a very low promoter activity of gshA, while qRT-PCR analysis showed no transcription of gshA in strain TS44-gshA540 (Fig. 5d). Moreover, lacZ fusion and qRT-PCR both showed that the sodB gene was strongly induced by Sb(III) in all three strains (Fig. 4b and e). Sb(III) only induced transcription of sodC and katE in the mutant strain TS44-gshA540, but not in strains TS44 and TS44-gshA-C (Fig. 4c, f and g).
Fig. 4.
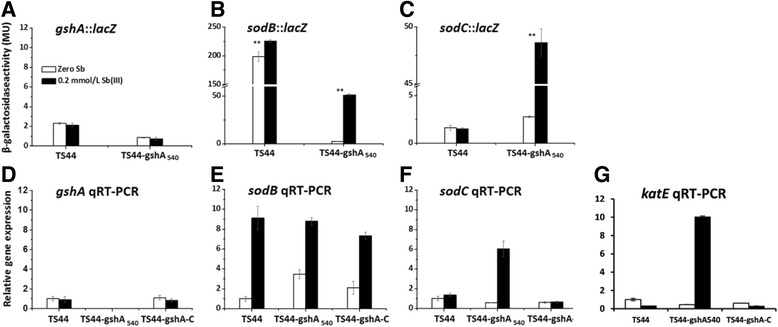
The effect of Sb(III) to the transcription of gshA, sodB, sodC and katE. The transcription of gshA, sodB and sodC was tested by a lacZ reporter fusion and qRT-PCR, while the katE transcription was only tested by qRT-PCR. For lacZ reporter fusion (a, b and c), data are expressed as the mean ± SD, N = 3. **Indicates a significant difference from the control (p < 0.01). For qRT-PCR, error bars correspond to the standard deviations of the means from three biological replicates. Gene expression was normalized to the 16S rRNA gene. The results are presented as the mean gene expression normalized to mRNA levels in Sb(III)-free CDM. Data symbols shown in all panels are the same
Fig. 5.
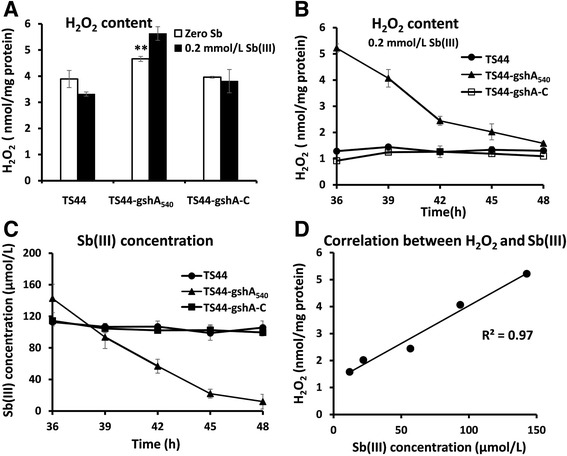
The H2O2 content is correlated with bacterial Sb(III) oxidation. The P. stutzeri strains were cultured as described above. a The H2O2 concentration was tested after 30 min of incubation with Sb(III). **Indicates a significant difference from the control (p < 0.01). b H2O2 content and (c) Sb(III) concentration in strains TS44, TS44-gshA540 and TS44-gshA-C from 36 to 48 h of incubation in cultures supplemented with 0.2 mmol/L Sb(III). Data are expressed as the mean ± SD, N = 3. d Correlation between H2O2 and Sb(III) concentrations in strain TS44-gshA540
Disruption of GSH synthesis allowed accumulation of H2O2
In order to examine whether the ROS-protection system is involved in H2O2 production, we tested the H2O2 content in the presence and absence of Sb(III) in all strains. The H2O2 content was slightly higher in gshA mutant strain TS44-gshA540 without addition of Sb(III) compared to strains TS44 and TS44-gshA-C (Fig. 5a). Notably, after a 30 min incubation with 0.2 mmol/L Sb(III), the H2O2 content was increased in the mutant strain TS44-gshA540, but slightly decreased in strains TS44 and TS44-gshA-C (Fig. 5a), indicating that the disruption of GSH synthesis is related to the accumulation of H2O2. In addition, it appears the conversion of ROS by SOD was more efficient than the H2O2 conversion by KatE and thus would both sod and katE were induced by Sb(III).
H2O2 is responsible for Sb(III) oxidation
H2O2 was shown to be an efficient Sb(III) oxidant in vitro (Additional file 3: Figure S2) as the un-inoculated control showed no Sb(III) oxidation (Additional file 3: Figure S2). A control with dead cells after boiling the culture also showed no oxidation of Sb(III) (data not shown). As the efficiency of Sb(III) oxidation in the mutant strain TS44-gshA540 was shown to be highest from 32 to 48 h (Fig. 2), the H2O2 and Sb(III) content in strains TS44, TS44-gshA540 and TS44-gshA-C were also measured from 36 to 48 h. Interestingly, the H2O2 content in strain TS44-gshA540 was significantly decreased from 5.2 to 1.1 nmol/mg protein, while the H2O2 content was stable at a low level in strains TS44 and TS44-gshA-C (Fig. 5b). Correspondingly, Sb(III) was oxidized to Sb(V) concomitant with a decrease in H2O2 content, while the Sb(III) concentration was almost stable in strains TS44 and TS44-gshA-C (Fig. 5c). The consumed H2O2 content and the oxidized Sb(III) showed a linear correlation with a correlation coefficient of 0.97 (Fig. 5d), indicating that H2O2 is responsible for Sb(III) oxidation. In addition, we could show that growth was inhibited with increasing concentration of H2O2 (Additional file 4: Figure S3), thus Sb(III) oxidation may contribute to the bacterial detoxification of H2O2 and Sb(III).
Discussion
At present, several Sb(III)-oxidizing strains have been reported [13, 14, 18]; some of these strains were also able to oxidize As(III) [13, 15, 19]. The arsenite oxidase AioAB was demonstrated to be capable of oxidizing Sb(III), but the AioAB kinetic rate of the reaction was orders of magnitude higher for As(III) than for Sb(III) [19]. Moreover, AnoA belonging to the SDR superfamily was reported to be able to catalyze Sb(III) oxidation using NADP+ as a cofactor [20]. However, disruption of aioA and anoA in Agrobacterium tumefaciens caused a decrease in Sb(III) oxidation of only about 25% [19] and 27% [20], respectively, indicating the existence of other bacterial Sb(III) oxidation mechanisms. After discovering that the presence of gshA affected the Sb(III) oxidation rate by transposon mutagenesis in strain TS44-gshA540, we proposed that some abiotic cellular components, such as GSH, ROS or H2O2, may have a role in the bacterial oxidation of Sb(III).
Subsequently, we conducted a comprehensive analysis. First, we found that a disruption of gshA caused a decrease in the cellular GSH amount; Second, it is conceivable that the gshA mutant strain resulted in an increase in cellular ROS content compared to the wild type; Third, we could also show a linear correlation between the decrease of H2O2 content and the increase in Sb(III) oxidation rate, indicating that Sb(III) oxidation consumed H2O2 and acts as a detoxification mechanism to counter this cellular stressor. In addition, it appeared that Sb(III) directly caused a disruption of the ROS-protection system in strain TS44-gshA540 and allowed the accumulation of H2O2 instead of affecting GSH levels, since neither the activity of GshA nor the GSH content were influenced by Sb(III). Previously, it was reported that Sb(III) may consume residual GSH (forming a stable Sb(GS)3 complex) in red blood cells [45]. However, we did not observe significant changes in GSH content when Sb(III) was added to wild type strain TS44 and the gshA complemented strain TS44-gshA-C, indicating little of this complex was formed and Sb(III) was mainly oxidized to Sb(V) by H2O2. We therefore propose a new model for Sb(III) oxidation in P. stutzeri TS44. i) the addition of Sb(III) would trigger the ROS-protective system by inducing the transcription of sodB, sodC and katE, with SodB and SodC catalyzing the conversion of ROS to H2O2, and KatE responsible for the depletion of excessive H2O2; ii) the increased cellular H2O2 content enhanced the Sb(III) oxidation rate; and iii) the addition of Sb(III) played a selection role on the characterization of the gshA insertion (Fig. 6); iv) in addition, the accumulated H2O2 was partially consumed by KatE (data not shown), since katE was induced by Sb(III) (Fig. 4 g), and similar result was described recently with transcription of the peroxidase-encoding gene katA being induced by both Sb(III) and H2O2 [46].
Fig. 6.
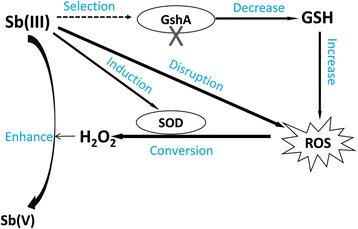
The proposed model for Sb(III) bacterial oxidation in P. steutzeri TS44. In this study, i) the addition of Sb(III) would trigger the ROS-protective system by inducing the transcription of sodB, sodC and katE, with SodB and SodC catalyzing the conversion of ROS to H2O2, while KatE responsible for the degradation of excessive H2O2 (data not shown); ii) the increased cellular H2O2 content enhanced the Sb(III) oxidation rate; and iii) the addition of Sb(III) played a selection role on the characterization of gshA insertion; iv) in addition, the accumulated H2O2 is partially cosumed by the upregulated catalase KatE (data not shown)
Several chemical substances (i.e., amorphous iron, manganese oxyhydroxides and H2O2) have been reported to be capable of mediating Sb(III) oxidation in vitro [47–49]. OH• was the oxidant in acidic solutions and H2O2 was the main oxidant in neutral and alkaline solutions involved in Sb(III) oxidation, with a pH range of 8.1 to 11.7 [48, 49]. Sb(III) was reported to cause cellular oxidative stress in lymphoid tumoral cells [50]. H2O2 is a substantial component of cellular oxidative stress [51] and can inhibit cellular growth [52]. In this study, the pH of the cultures changed from the initial 7.0 to approximately 8.0 following exposure to Sb(III), indicating that H2O2 may correlate with an increase in the Sb(III) oxidation rate. In addition, the in vitro experiment provided direct evidence that H2O2 can oxidize Sb(III) to Sb(V) (Additional file 4: Figure S3) and a correlation between the concentrations of Sb(III) and H2O2 was found in vivo (Fig. 5d). As an moderatedly reactive intermediate, O2 •- may also have an effect on Sb(III) oxidation. However, in the presence of high level of SOD, the steady-state of O2 •- would be no more than 0.1 nmol/L and thus the effect of O2 •- is negligible [53]. Thus, we conclude that H2O2 oxidized Sb(III) to Sb(V).
Conclusions
This study proposed a novel mechanism for microbial antimonite oxidation involving changes in cellular content of components related to oxidative stress (GSH, ROS and H2O2). Although H2O2 was reported to be capable of oxidizing Sb(III) chemically as early as 2005 [11], this is the first study to demonstrate that H2O2 is responsible for Sb(III)-oxidation in a microbe. The results showed that a disruption of bacterial GSH-dependent ROS-protection mechanism allowed H2O2 to build up and thus promote the oxidation of Sb(III) to Sb(V). In addition, the oxidation of Sb(III) to Sb(V) is a detoxification process against the cellular stressor H2O2. We do not exclude the possibility of an additional enzymatic process responsible for Sb(III) oxidation in strain TS44, since genes encoding a putative arsenite oxidase AioBA were found on its genome [28] which may function as a Sb(III) oxidase. Our data and other findings [19, 20, 46] could show that microbial antimonite oxidation contains both biotic and abiotic components.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (31170106) to GW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors’ contributions
DW performed the experiments and wrote the draft of the manuscript, FZ designed and performed the experiments and helped to draft the manuscript. QW and CR revised the draft of the manuscript. PY and JG performed the experiments. GW designed the study and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Primers used in this study. (DOC 35 kb)
The gene cluster containing gshA in strain TS44 and Diagnostic PCR confirming the transposon mutation to create mutant strain TS44-gshA540 and complementation to create TS44-gshA540-C. (A), The transposon insertion site of the gshA mutant is shown by the vertical arrow. (B), PCR used primers R6K-F/R6K-R and TnpA-F/TnpA-R (Additional file 4: Table S1) using genomic DNA of strain TS44-gshA540 as template. Lane 1 is the amplification of R6K fragment, while line 2 represents the TnpA fragment. (C), PCR used primers gshA-F/gshA-F (Additional file 4: Table S1). Lane 1, strain TS44, lane 2, gshA gene insertional inactivation strain TS44-gshA540 and lane 3, the complemented strain TS44-gshA540-C. M, the molecular weight marker (DL 2000 plus). Amplicon identities were confirmed by DNA sequencing. (TIF 104 kb)
In vitro oxidation of Sb(III) by H2O2. (A) Sb(III) was added into uninoculated liquid CDM medium to a final concentration of 0.2 mmol/L. (B) CDM containing 0.2 mmol/L of Sb(III) and with the addition of 0, 0.02, 0.05, 0.1, 0.3, 0.4 and 0.5 mmol/L of H2O2, respectively. (TIF 59 kb)
The tolerance of P. stutzeri strains TS44, TS44-gshA540 and TS44-gshA-C to H2O2. The strains were grown in liquid CDM medium until reaching an OD600 of 1.0 and then serially diluted 10-fold. The 100, 10−1, 10−3, and 10−5 dilutions were spotted onto agar plates with different H2O2 concentrations. (TIF 426 kb)
References
- 1.Ehrlich HL. Newman DK. Geomicrobiology: CRC press; 2008. [Google Scholar]
- 2.Goyeneche-Patino DA, Valderrama L, Walker J, Saravia NG. Antimony resistance and trypanothione in experimentally selected and clinical strains of Leishmania panamensis. Antimicrob Agents Ch. 2008;52(12):4503–6. doi: 10.1128/AAC.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liarte DB, Murta SM. Selection and phenotype characterization of potassium antimony tartrate-resistant populations of four New World Leishmania species. Parasitol Res. 2010;107(1):205–12. doi: 10.1007/s00436-010-1852-8. [DOI] [PubMed] [Google Scholar]
- 4.Callahan MA. Water-related environmental fate of 129 priority pollutants. Office of Water Planning and Standards. Office of Water and Waste Management, US Environmental Protection Agency; Washington. 1979.
- 5.WHO. Antimony in Drinking-water-Background. Document for Development of WHO Guidelines for Drinking Water Quality. Geneva. 2003.
- 6.Filella M, Belzile N, Chen Y-W. Antimony in the environment: a review focused on natural waters: I. Occurrence. Earth-Sci Rev. 2002;57(1):125–76. doi: 10.1016/S0012-8252(01)00070-8. [DOI] [Google Scholar]
- 7.Filella M, Belzile N, Chen Y-W. 2002b. Antimony in the environment: a review focused on natural waters: II. Relevant solution chemistry. Earth-Sci Rev. 2002;59(1):265–85. doi: 10.1016/S0012-8252(02)00089-2. [DOI] [Google Scholar]
- 8.Wilson SC, Lockwood PV, Ashley PM, Tighe M. The chemistry and behaviour of antimony in the soil environment with comparisons to arsenic: a critical review. Environ Pollut. 2010;158(5):1169–81. doi: 10.1016/j.envpol.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 9.Gebel T. Arsenic and antimony: comparative approach on mechanistic toxicology. Chem-bio interact. 1997;107(3):131–44. doi: 10.1016/S0009-2797(97)00087-2. [DOI] [PubMed] [Google Scholar]
- 10.De Boeck M, Kirsch-Volders M, Lison D. Cobalt and antimony: genotoxicity and carcinogenicity. Mutat Res-fund Mol M. 2003;533(1):135–52. doi: 10.1016/j.mrfmmm.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Leuz A-K, Johnson CA. Oxidation of Sb III to Sb V by O2 and H2O2 in aqueous solutions. Geochim Cosmochim Ac. 2005;69(5):1165–72. doi: 10.1016/j.gca.2004.08.019. [DOI] [Google Scholar]
- 12.Smichowski P. Antimony in the environment as a global pollutant: a review on analytical methodologies for its determination in atmospheric aerosols. Talanta. 2008;75(1):2–14. doi: 10.1016/j.talanta.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Wang Q, Zhang SZ, Qin D, Wang GJ. Phylogenetic and genome analyses of antimony-oxidizing bacteria isolated from antimony mined soil. Int Biodet Biodegr. 2013;76:76–80. doi: 10.1016/j.ibiod.2012.06.009. [DOI] [Google Scholar]
- 14.Shi Z, Cao Z, Qin D, Zhu W, Wang Q, Li M, et al. Correlation models between environmental factors and bacterial resistance to antimony and copper. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehr CR, Kashyap DR, McDermott TR. New insights into microbial oxidation of antimony and arsenic. Appl Environ Microb. 2007;73(7):2386–9. doi: 10.1128/AEM.02789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lialikova N. Stibiobacter senarmontii--a new microorganism oxidizing antimony. Mikrobiologiia. 1974;43(6):941. [PubMed] [Google Scholar]
- 17.Hamamura N, Fukushima K, Itai T. Identification of antimony- and arsenic-oxidizing bacteria associated with antimony mine tailing. Microbes Environ/JSME. 2013;28(2):257–63. doi: 10.1264/jsme2.ME12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen VK, Lee J-U. Antimony-oxidizing bacteria isolated from antimony-contaminated sediment–a phylogenetic study. Geomicrobiol J. 2015;32(1):50–8. doi: 10.1080/01490451.2014.925009. [DOI] [Google Scholar]
- 19.Wang Q, Warelow TP, Kang Y-S, Romano C, Osborne TH, Lehr CR, et al. Arsenite oxidase also functions as an antimonite oxidase. Appl Environ Microb. 2015: AEM. 02981–14 [DOI] [PMC free article] [PubMed]
- 20.Li JX, Wang Q, Li MS, Yang BR, Shi MM, Guo W, et al. Proteomics and genetics for identification of a bacterial antimonite oxidase in Agrobacterium tumefaciens. Environ Sci Technol. 2015 doi: 10.1021/es506318b. [DOI] [PubMed] [Google Scholar]
- 21.Halliwell B, Gutteridge JM. Free radicals in biology and medicine. Clarendon: Oxford University Press; 1999. [Google Scholar]
- 22.Bayr H. Reactive oxygen species. Crit Care Med. 2005;33(12):S498–501. doi: 10.1097/01.CCM.0000186787.64500.12. [DOI] [PubMed] [Google Scholar]
- 23.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2010;3(1):3–8. [PubMed] [Google Scholar]
- 24.Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radical Res. 1999;31(4):273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 25.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Sign. 2006;8(5–6):753–62. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 26.Janowiak BE, Griffith OW. Glutathione Synthesis in Streptococcus agalactiae one protein accounts for γ-glutamylcysteine synthetase and glutathione synthetase activities. J Biol Chem. 2005;280(12):11829–39. doi: 10.1074/jbc.M414326200. [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Rensing C, Li X, Wang G. Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers: Achromobacter sp. SY8 and Pseudomonas sp. TS44. Appl Microbiol Biot. 2009;83(4):715–25. doi: 10.1007/s00253-009-1929-4. [DOI] [PubMed] [Google Scholar]
- 28.Li XY, Gong J, Hu Y, Cai L, Johnstone L, Grass G, et al. Genome sequence of the moderately halotolerant, arsenite-oxidizing bacterium Pseudomonas stutzeri TS44. J Bacteriol. 2012;194(16):4473–4. doi: 10.1128/JB.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeger W, Lievremont D, Perret M, Lagarde F, Hubert J-C, Leroy M, et al. Oxidation of arsenite to arsenate by a bacterium isolated from an aquatic environment. Biometals. 1999;12(2):141–9. doi: 10.1023/A:1009255012328. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Zheng S, Su J, Wang L, Yao R, Wang D, Deng Y, et al. Selenite reduction by the obligate aerobic bacterium Comamonas testosteroni S44 isolated from a metal-contaminated soil. BMC Microbiol. 2014;14(1):204. doi: 10.1186/s12866-014-0204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology RAST. Nucleic Acids Res. 2014;42(D1):D206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Chen F, Cao Y, Wei S, Li Y, Li X, Wang Q, Wang G. Regulation of Arsenite Oxidation by the Phosphate Two-Component System PhoBR in Halomonas sp. HAL1. Front Microbiol. 2015;6:923. doi: 10.3389/fmicb.2015.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–80. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1(9):784–91. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 37.Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178(3):193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.White CC, Viernes H, Krejsa CM, Botta D, Kavanagh TJ. Fluorescence-based microtiter plate assay for glutamate–cysteine ligase activity. Anal Biochem. 2003;318(2):175–80. doi: 10.1016/S0003-2697(03)00143-X. [DOI] [PubMed] [Google Scholar]
- 40.Guo FF, Yang W, Jiang W, Geng S, Peng T, Li JL. Magnetosomes eliminate intracellular reactive oxygen species in Magnetospirillum gryphiswaldense MSR-1. Environ Microbiol. 2012;14(7):1722–9. doi: 10.1111/j.1462-2920.2012.02707.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Lei Y, Xu X, Wang G, Chen L-L. Theoretical prediction and experimental verification of protein-coding genes in plant pathogen genome Agrobacterium tumefaciens strain C58. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0043176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JH. Assay of β-galactosidase. In: Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press; 1972. p. 352–55.
- 43.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183(24):7173–81. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murata K, Kimura A. Cloning of a gene responsible for the biosynthesis of glutathione in Escherichia coli B. Appl Environ Microb. 1982;44(6):1444–8. doi: 10.1128/aem.44.6.1444-1448.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun H, Yan SC, Cheng WS. Interaction of antimony tartrate with the tripeptide glutathione. Eur J Biochem. 2000;267(17):5450–7. doi: 10.1046/j.1432-1327.2000.01605.x. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Wang Q, Oremland R S, Kulp T R, Rensing C, Wang G. Microbial antimony biogeochemistry-enzymes, regulation and related metabolic pathways. Appl Environ Microb. 2016: AEM. 01375–16. [DOI] [PMC free article] [PubMed]
- 47.Belzile N, Chen Y-W, Wang Z. Oxidation of antimony III by amorphous iron and manganese oxyhydroxides. Chem Geol. 2001;174(4):379–87. doi: 10.1016/S0009-2541(00)00287-4. [DOI] [Google Scholar]
- 48.Leuz A-K, Hug SJ, Wehrli B, Johnson CA. Iron-mediated oxidation of antimony III by oxygen and hydrogen peroxide compared to arsenic III oxidation. Environ Sci Technol. 2006;40(8):2565–71. doi: 10.1021/es052059h. [DOI] [PubMed] [Google Scholar]
- 49.Kong L, Hu X, He M. Mechanisms of Sb III oxidation by pyrite-induced hydroxyl radicals and hydrogen peroxide. Environ Sci Technol. 2015;49:3499–505. doi: 10.1021/es505584r. [DOI] [PubMed] [Google Scholar]
- 50.Lecureur V, Le Thiec A, Le Meur A, Amiot L, Drenou B, Bernard M, et al. Potassium antimonyl tartrate induces caspase‐and reactive oxygen species‐dependent apoptosis in lymphoid tumoral cells. Brit J haematol. 2002;119(3):608–15. doi: 10.1046/j.1365-2141.2002.03863.x. [DOI] [PubMed] [Google Scholar]
- 51.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240(4852):640–2. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 52.McLeod JW, Gordon J. Production of hydrogen peroxide by bacteria. Biochem J. 1922;16(4):499. doi: 10.1042/bj0160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266(11):6957–65. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


