Abstract
With the possibility of large-scale terrorist attacks around the world, the need for modeling and development of new medical countermeasures for potential future chemical, biological, radiological and nuclear (CBRN) has been well established. Project Bioshield, initiated in 2004, provided a framework to develop and expedite research in the field of CBRN exposures. To respond to large-scale population exposures from a nuclear event or radiation dispersal device (RDD), new methods for determining received dose using biological modeling became necessary. The field of biodosimetry has advanced significantly beyond this original initiative, with expansion into the fields of genomics, proteomics, metabolomics and transcriptomics. Studies are ongoing to evaluate the use of lymphocyte kinetics for dose assessment, as well as the development of field-deployable EPR technology. In addition, expansion of traditional cytogenetic assessment methods through the use of automated platforms and the development of laboratory surge capacity networks have helped to advance our biodefense preparedness. In this review of the latest advances in the field of biodosimetry we evaluate our progress and identify areas that still need to be addressed to achieve true field-deployment readiness.
INTRODUCTION
Radiation exposure is a continuing threat both from potential “dirty bomb” terrorist events and industrial accidents involving nuclear power or misplaced radioactive sources. Disasters involving radiological materials require specialized planning and preparedness to ensure the safety of first responders, for evacuation and medical treatment for potentially contaminated victims and for general crisis management. In the case of a radiological event, mass screenings of large sections of the relevant population will be required to separate exposed from nonexposed individuals and to determine the severity of the received dose in those determined to have been exposed (1, 2). The identification of potential diagnostic biomarkers for use as radiation biodosimeters is critical for enabling physicians to formulate effective medical treatment in a mass screening scenario (3). It is also exceedingly difficult to determine the level of exposure, due to variations in the radiation field, heterogeneity of exposure pattern, and uncertainties of total-body vs. partial-body irradiation, inhaled airborne particles or ingested products from ground contamination (4, 5). Psychosomatical-induced symptoms also complicate triage during mass casualty events, particularly where potential radiation exposure is a concern, further highlighting the need for independent physiological biomarkers of radiation exposure (6).
The ability to mitigate uncertainty of radiation exposure through biodosimetry applications will substantially improve medical management of casualties complicated by radiation exposure or contamination and provide additional resources to enhance confidence in communication with the affected public (7–10). As experiences from the recent Fukushima disaster illustrate, the need for deployable mass screening radiation biodosimetry methodologies is a relevant and current one.
A radiation biodosimeter can be characterized as a physiologic molecule with an expression pattern that is quantitatively altered when exposed to ionizing radiation. This comprises a very large field of disparate classes of molecules and technology platforms, including DNA, RNA and protein expression, chromosomal aberration, blood count changes and even metabolomics profiles. These potential markers are usually evaluated in blood, plasma, urine or saliva, sample types that are readily obtainable in a field setting, although some markers have also been explored in samples taken from both tooth enamel and nail and hair clippings.
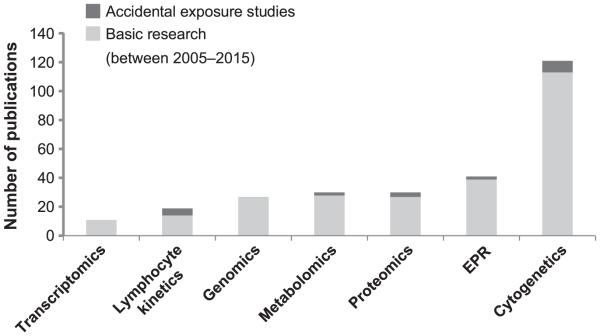
This review of the most recent advances in radiation biodosimetry science addresses how far this field has progressed towards development of viable biodosimetry methodologies for field application through a discussion of the major findings in each area of research and through the use of a scoring assessment based on the phase of development (Table 1). The maturity of each field is also evaluated by the number of publications over the last decade (Fig. 1). Here we present the field of biodosimetry as representing a diverse and growing body of radiation biology research with immediate relevance to public health and security.
TABLE 1.
Scoring Rubric for Biodosimetry Field Phase of Development
| Phase | Score | Description |
|---|---|---|
| Investigative phase | 1 | In vitro/ex vivo radiation model |
| 2 | Murine in vivo model | |
| Early development phase | 3 | NHP in vivo model |
| Late development phase | 4A | Human clinical trials |
| 4B | Human accidental exposure study | |
| Deployment ready | 5 | FDA approved |
Notes. This table provides a scoring rubric for phases of development based on deployment readiness according to model type and recent application in documented clinical care of accidental human exposures. As indicated in the text, an additional assessment at each level includes the accessibility of either an automated platform (X) or laboratory surge capacity (Y). An automated platform is comprised of either high-throughput robotics or clinical laboratory processing capability. Laboratory surge capacity represents established networks for work-intensive co-laboratory scoring.
FIG. 1.
Aggregate number of publications for each field of biodosimetry was determined for the time interval between the years 2005–2015. Search terms in PubMed were used for each field, such as “genomics AND radiation biodosimetry”, in addition to manually curated searches for each subfield. Only primary research articles were counted for this figure, and reviews were not included. The data is split to identify primary research articles and case reports from studies of actual accidental human exposures. The authors made every attempt to identify all biodosimetry related publications for each field.
GENOMICS
Use of recombinant DNA technologies and bioinformatics to investigate changes in global genome expression constitutes the field of genomics. This powerful set of tools used to analyze changes in genomic expression specific to disease state and environmental factors is now being utilized to study radiation-induced changes in gene expression. For example, in one study utilizing qPCR, mRNA levels in blood taken from 0.5–10 Gy total-body-irradiated (TBI) mice collected at 12 h to 7 days after exposure were found to be highly predictive of radiation dose. Using mRNA levels from a relatively few number of genes, highly correlative accuracies between actual and predicted dose were established with Cdkn1a as the strongest predictor of dose assessment (11). In a study using blood from pediatric cancer patients after 2 Gy myeloablative radiation therapy, several radioresponsive genes were identified, which correlated with increased expression in a TBI mouse model (12).
In several studies, microarray analysis using blood samples from TBI mice, ex vivo irradiated human peripheral blood and TBI clinical patients was performed to identify gene expression signatures capable of separating irradiated from nonirradiated samples (13, 14). When these model systems were compared, one study found that gene expression profiles generated from the ex vivo irradiated human samples and clinically irradiated patient samples were correlative, while gene expression signatures from the TBI murine model were not. This study also identified an 18-gene cohort that was highly accurate at predicting exposed radiation dose in both ex vivo and clinical TBI samples (14)
In another study, gene expression profiles in the peripheral blood of partial-body-irradiated mice were examined at exposures of 0.5, 2 and 10 Gy, and were able to distinguish exposed from nonexposed animals, although gene signatures from TBI were not predictive of partial-body irradiation, nor were different partial-body irradiation predictive of each other as each type generated its own unique gene expression pattern (15).
In a large study using peripheral blood irradiated ex vivo between 0.5 and 10 Gy, it was reported that a small group of genes, ASTN2, CDKN1A, GDF15 and ATM, were found to be highly predictive of exposed dose. Expression levels of these genes were predictive of exposures up to 6 Gy, with correlation accuracy slightly diminished at higher doses of 8 and 10 Gy due to saturation (16). In one study using gene expression profiles identified from microarray gene expression analysis of human blood irradiated ex vivo, these identified genes were applied to samples taken from clinical patients who were total-body irradiated with either a single 1.25 Gy dose or an accumulated fractionated dose of 3.75 Gy. This gene cohort had a success rate of 96% for distinguishing between the respective radiation exposures of these clinical samples (17). This same set of genes identified from human blood irradiated ex vivo has been used to develop a quantitative nuclease protection assay (qNPA) capable of generating expression profiles, using a platform that is more accessible for field deployment and requiring only extremely small volumes of blood. This method, which was tested on blood from healthy volunteers that was irradiated ex vivo, successfully differentiated irradiated and nonirradiated samples in a dose-dependent manner (18).
The effect of radiation dose rate on gene expression has also been investigated in a mouse and human blood irradiated ex vivo model using either an acute exposure of 1.03 Gy/min or a low-dose-rate exposure of 3.1 mGy/min. These exposure patterns resulted in unique gene expression patterns at 24 h after exposure and were capable of separating acute and low-dose-rate exposures (19, 20). These studies illustrate the possible utility of gene expression profiling to determine the level of radiation exposure as well as the type of radiation exposure and the dose rate.
While the predominant model for radiation exposure utilizes external TBI from X-ray and gamma sources, alternative models of exposure using internal emitters are now being investigated. Alterations to gene expression profiles were examined in mice using cesium-137 chloride (137CsCl) as the radiation source. Genomic microarray analysis yielded different panels of upregulated genes depending on the time point after 137CsCl injection. A comparison of expression levels of a relevant set of genes was performed using a single 2.8 Gy external radiation dose and the same accumulated whole-body dose of 2.8 Gy internally from a single intraperitoneal 137CsCl injection, and yielded a substantial increase in gene expression with the internal exposure (21). As all of these factors have relevance in the medical management of radiological casualties, genomic biodosimetry has the potential to be a powerful tool in medical triage of radiologic events.
Further studies examining alterations in gene expression due to inherent variance in population, disease state, psychological stress and combined injury for radiation exposure and expanded dose assessment profiles, as well as application of this technology to available human samples from accidental exposures will validate this platform as a viable methodology for radiation biodosimetry. Advances in genomic assessment of radiation response also have relevant clinical applications for patients undergoing radiation therapy. Such information may advance personalized care, and development of organ-specific signatures of radiation response could help mitigate normal tissue toxicity. Establishing a standardized panel of genomic signatures relative to dose and time of exposure will help advance genomics for practical field applications. For current phase of development, the field of genomics for biodosimetry assessment is ranked at 4A (Table 1).
PROTEOMICS
Changes in cytokine, chemokine and other proteomic profiles have been explored for potential application in estimating unknown radiation exposures. C-reactive protein (CRP) and serum amylase were among the first identified protein biomarkers of radiation exposure. Dose-dependent increases of serum amylase have been observed in patients undergoing total- and partial-body radiotherapy, and elevation of serum amylase after radiation exposure was also observed in victims of the Tokai-mura criticality accident (22–24). The relationship between radiation injury and activation of the acute-phase response is also well established with correlations between CRP elevation and onset and severity of acute radiation syndrome (ARS) (4). Elevation of acute-phase proteins such as CRP and serum amyloid A (SAA) were also reported to increase in a dose-dependent manner after irradiation (25, 26).
Efforts to discover additional radiation-responsive proteins are ongoing. One study using irradiated human cell lines of hematological origin screened 161 individual proteins using Western blotting and identified 55 radiation-responsive proteins. The feasibility of applying these in vitro determined targets to a TBI canine model was also demonstrated, with a similar trend between in vitro and in vivo responses to radiation for an example target protein (27). Expression profiles of several individual protein biomarkers have been examined for their relationship to received radiation dose and exposure pattern. In a TBI nonhuman primate (NHP) model, dose-dependent increases in CRP, SAA, FIT3 Ligand (FL), Interleukin-6 (IL6) and amylase were observed in the peripheral blood (28–30). Similar changes in expression were seen in plasma for SAA, FL, IL6 and G-CSF in TBI mice models (31–33). Partial-body exposures have also been shown to correlate with plasma expression of SAA, FL and G-CSF using a murine model (34, 35). These studies were able to show relative increases in expression of plasma biomarkers correlating to an increasing area of partial-body exposure (26, 36).
Investigations into radiation-induced changes to the proteome relevant to partial exposures are also being explored. In one study, changes to the urine proteome by LC-MS/MS were evaluated after 10 Gy TBI or 10 Gy dose locally administered to the kidney in rats. Interestingly, among the many changes in protein content observed after irradiation, the decline in urinary albumin was greater with TBI than with a local acute exposure to the kidney alone (37). This study highlights the relevance of exposure type on the resulting proteomic profile.
A study to examine changes of the glycosylation state of serum proteins in acute partial-body exposures in mice noted changes in MALDI-MD generated glycomic profiles of irradiated animals, in addition to increased expression of IL1B, IL6 and TNFa after exposure. Using 2D-DIGE analysis in this study, similar trends were also observed in the serum proteome profiles of locally irradiated mice and one human patient who accidently received an acute partial-body radiation exposure (38). In one recent study of human saliva, the salivary proteome of single- and accumulated-fraction TBI patients was screened, and IL8, MCP-1 and ICAM-1 were identified as radiation-responsive proteins with elevated expression after exposure, which correlated with accumulated dose; and in another study evaluating cytokine levels in saliva from patients undergoing head and neck radiotherapy, a dose-response relationship was reported for expression of IL4, IL6, IL8, EGF, VEGF, MCP-1 and TNF-a (39, 40). The utility of proteomic markers of radiation response in a variety of models has been demonstrated, and the evaluation of FL, citrulline, oxysterols and other protein markers has even been incorporated into the medical management of recent radiological incidents (41, 42).
Proteomics based approaches to biodosimetry have potential as a useful, deployable point-of-care diagnostic due to technological advances in the methodology allowing ease of use and high-throughput capability and stability of the analyte under field conditions. In addition, studies have shown a highly correlative relationship of expression with received dose. For example, several studies have shown that expression of cytokine FL, exhibits a direct relationship to the amount of radiation and has been used previously in clinical assessment of accidental exposures. (34, 36, 43). While a single marker or small panel of proteomic markers to determine dose has great utility for dose assessment, further proteomics studies are needed to evaluate internal exposures, other types of radiation exposure and preexisting clinical confounders. With the imminence of field-applicable technology and high-throughput capability, there is still no field-deployable platform for proteomic assessment of radiation dose. The current ranking for proteomics for phase of development is 4B(X) based on the use of proteomics in accidental case studies (Table 1). Radiation-responsive proteins represent a new and functional methodology for determining radiation exposure and show promise as one of the leading emerging platforms for radiation biodosimetry.
METABOLOMICS
The field of metabolomics comprises the study of the metabolome and the investigation of changes in the global population, of the metabolite products in the body under different experimental conditions. Metabolite profiles or the sum changes in chemical byproducts of cellular processes are now being studied for their application in radiation biodosimetry (44). Plasma citrulline has been established as a marker of radiation-induced damage to the small bowel, and several clinical studies have shown elevated plasma citrulline to be a predictor of intestinal toxicity in patients undergoing radiation therapy. (45, 46). The relationship between citrulline and gross histological tissue damage to the gastrointestinal system has been well validated in murine and other animal models, and there are ongoing efforts to further characterize this biomarker for radiation exposure (45, 47).
Novel metabolite profiles that correlate with dose response were recently found in a TBI NHP study in urine where a unique panel of 13 metabolites was identified at day 7 postirradiation using ultra-performance liquid chromatography quadrupole time-of-flight mass spectroscopy (UPLC-QTOF-MS). These metabolites evidenced dose-dependent expression between 2 and 10 Gy indicative of metabolic changes to tryptophan and taurine metabolism, steroid hormone biosynthesis, purine catabolism and fatty acid b oxidation. Sex-linked trends within the metabolite profiles were also observed in NHPs (48). Changes in metabolite profiles relative to exposure were also seen at earlier time points between 12 and 84 h postirradiation in NHPs that received 1–8.5 Gy TBI (49).
Distinct metabolite signatures were observed in a human study using urine from TBI cancer patients. Samples were taken after either a single dose of 1.25 Gy TBI at 6 h postirradiation or after an accumulated dose of 3 fractions of 1.25 Gy. Global metabolomics profiling was achieved using UPLC-TOFMS resulting in identification of seven metabolites that demonstrated markedly different patterns of expression after irradiation. The identified metabolites represent changes to metabolic pathways including fatty acid b oxidation and purine catabolism. Differences in metabolite expression patterns were also observed between male and female patients in this study (50). In global metabolic profile analysis, was also able to separate urine samples taken from partial-body irradiated prostate cancer patients receiving fractionated radiotherapy from corresponding nonirradiated samples (44). A review of radiation-sensitive metabolite markers in urine across species revealed increased expression of taurine and xanthine in mice, NHPs and humans (48).
Studies in mice to examine the effects of internal 137Cs exposure on metabolomic and lipidomic profiles in serum and urine have shown radiation-induced changes to metabolites associated with fatty acid and amino acid metabolism. At an accumulated internal 137Cs dose of 2 Gy, metabolomic profiles in serum clearly differentiated irradiated from nonirradiated mice (51, 52). Acute external exposures of 8 Gy TBI in mice have revealed key changes to metabolite signatures involved in amino acid, carnitine and lipid metabolism (53). Dose-dependent changes in metabolite expression were also seen using GC-TOFMS in serum from a TBI rat model receiving gamma exposures between 0.75 and 8 Gy. This study identified nine metabolites with expression that directly correlated with received dose (54). In another study of a rat model, a metabolomics approach was used to determine the effects of chronic low-dose internal exposures from ingested natural uranium or 137Cs. In this system, LC-MS detected metabolomic profiles in serum and urine unique to the internal exposure (55, 56).
Metabolomics has proven to be a powerful tool in the identifying radiation-induced changes to metabolic processes after both external and internal radiation exposures. While the technology platform is not currently field deployable, establishment of surge capacity laboratory resources could make this analysis a valuable diagnostic addition to medical management of radiologic casualties. Metabolomics is ranked 4B(X) within the late development phase category based on its proof of concept, use in select human accidental exposure studies and accessibility of plasma citrulline assessment through clinical laboratory support (Table 1).
TRANSCRIPTOMICS
Transcriptomics is the study of the complete body of RNA products encoded by the genome, including mRNA, siRNA and miRNA, and is currently under investigation for potential applications in the field of biodosimetry. miRNA expression is known to be radiosensitive, as these molecules are integral to cellular responses to radiation-induced oxidative stress (57). One recent set of experiments, utilizing miRNA signatures in serum taken from a TBI mouse model, identified miRNA-200b, miRNA-762 and miRNA-150 as responsive to radiation with expression levels that correlated with dose at exposures between 1 and 8 Gy at 24–48 h postirradiation. Their relative trends of expression remained similar with a clinically relevant fractionated dosing regimen up to 12 Gy. The authors report that miRNA-150 is a strong contender for use as a biomarker for radiation exposure, because its expression negatively correlates with increasing radiation dose, it is expressed in lymphocytes and it is a known regulator of hematopoiesis. miRNA-150 expression also correlates with lymphocyte depletion kinetics. These recent studies of miRNA-150 serum expression in mice suggest it may have potential use as a biomarker of radiation exposure and radiation effect to the hematopoietic system (58). In a murine model analyzing changes in miRNA expression using plasma from 0.5, 2 and 10 Gy TBI animals, distinct miRNA profiles were also expressed in a dose- and time-dependent manner. In addition to the exposure-driven profiles, miR34a upregulation was reported in all irradiated samples in this study (59).
In another study evaluating serum miRNA signatures in C57BL6 and humanized TBI mice given sublethal or lethal radiation doses, miRNA profiles, which were able to differentiate eventual lethality at 24 h postirradiation, were determined. These serum miRNA signatures were found to correlate with residual hematopoietic stem cell status and demonstrate the potential for miRNA to serve as a biomarker for radiation injury in addition to dose (60).
Evaluating the effect of combined injury in the form of TBI concurrent with hemorrhage, a trauma state predicted to be common in a radiological event, one study using a mouse model determined that combined injury alters expression of miRNA in the kidney with changes of miR-29b, miR-30e and let-7e expression. Ingenuity pathway analysis associated these miRNAs with hematopoiesis and inflammatory function (61).
Correlation of miRNA expression has also been explored in a comparison of different radiation types, including low- and high-linear-energy transfer radiation. In a study comparing TBI exposures of c-ray or iron-56 (56Fe) ions, miRNA expression profiles were examined using whole blood from irradiated mice. Under these conditions 31 differentially expressed miRNAs, associated with the various irradiation profiles, were identified. Using Geneontology (GO) analysis these miRNAs were found to be primarily involved in regulation of transcription, nucleic acid metabolism and development. Further, specific miRNA expression profiles were found to be dose- and time dependent as well as specific to radiation type. These miRNA signatures serve as a proof-of-concept illustration of the utility of miRNA as a biomarker of both radiation quantity and quality (62).
To our knowledge, there are only a few reported studies examining radiation-induced miRNA expression relative to dose in human systems. In one of these studies, using an ex vivo irradiated human blood model, a few changes were noted in miRNA expression at doses of 0.5, 2.5 and 5 Gy, with significant changes at 1 Gy (63). Given the highly conserved homology between human and mouse miRNA, there is potential for applicability of mouse model miRNA expression signatures to human physiology. Studies that reveal the radiation bystander effect on miRNA profiles, including persistent alteration of the miRNA population in the spleen after cranial irradiation, demonstrate the potential use of miRNA profiling for partial-body exposures, as well (57). miRNA profiling has relevant future applications as a biomarker of radiation exposure and radiation injury. With the addition of future studies that expand on existing dose-threshold modeling and further analysis of clinical samples, the miRNA platform may serve as a valuable tool in the field of biodosimetry. Within the scoring rubric for phase of development, transcriptomics generally ranks at 2, since the majority of studies have used murine models; however, since there is at least one clinical study, the field qualifies for a score of 4A (Table 1). This field of biodosimetry, however, still requires substantial development based on its minimal number of publications over the last decade (Fig. 1). Development of a cohesive panel of miRNA signatures associated with dose assessment profiles and further studies, including accidental human exposures, should provide the additional validation needed to advance this field of biodosimetry.
CYTOGENETICS
Cytogenetic techniques have historically been used for estimation of unknown radiation exposures to determine biological dose assessments. These techniques include scoring of chromosomal aberrations with the dicentric chromosome assay (DCA), the micronucleus assay or cytokinesis-block micronucleus assay (CBMN), the premature chromosome condensation assay (PCC), fluorescence in situ hybridization (FISH) and the recent addition of c-H2AX scoring as a cytogenetic tool. While each of these assays has specific advantages, DCA remains the “gold standard” among these techniques for estimation of radiation exposure (64). The feasibility of using cytogenetic analysis for total- and partial-body estimates of radiation exposure has been demonstrated using the DCA in cancer patients treated with different dose rates and different radiation fields to the same area of the body (65). Measurement of c-H2AX foci has been shown to be an effective method of estimating dose for both total- and partial-body exposures using human ex vivo and TBI NHP models (66–69)
Mathematical modeling of cytogenetic data to predict unknown whole-body dose has classically used a linear-quadratic model for assessment of dose prediction and is freely available in application software such as CABAS. However, there is currently growing acceptance of Bayesian methodologies, since they allow inclusion of previous information about the circumstances of exposure (70–72). More user-friendly software platforms based on Bayesian modeling have been developed to estimate radiation exposures including CytobayesJ and radir, a cytogenetic biodosimetry dose estimation application tailored for use in the R software platform (73, 74). Application of Bayesian modeling has also been developed both for estimation of partial-body exposure amount and to identify the fraction of the body irradiated (75). MULTIBIODOSE is one example of an application that serves as a software interface designed to coordinate biodosimetry efforts with a laboratory network combining information from multiple assay techniques, both biological and physical, to deliver a single dose assessment for triage decision (76).
While cytogenetic techniques are well established for unknown dose estimation, they are not well suited for mass casualty scenarios because they require extended incubation times and highly trained technical interpretation. To adapt these techniques and address such situations, attempts have been made to establish surge capacity networks of laboratories capable of meeting the need for processing large numbers of samples and to develop high-throughput technology for decreasing output times. In a recent NATO exercise, the ability of a network of 11 institutions to provide dose estimates from ex vivo irradiated blood samples was tested, using DCA, CBMN, c-H2AX or gene expression assays. The results of this study revealed that DCA provided dose estimates with the highest degree of accuracy among the different assays tested and a wide level of variability was observed among the network laboratories, perhaps due to each site using their own particular protocols for each of the cytogenetic assays tested (77). This exercise shows the need for developing a uniform protocol for use in mass dose assessment scenarios. Other studies comparing variance among laboratories have shown the feasibility of co-laboratory efforts for dose-exposure screening, using the DCA, c-H2AX or gene expression assays, including a MULTIBIODOSE exercise where DCA, CBMN and c-H2AX were used with successful correlation of dose estimates (78–83).
Another effort to make the DCA assay more applicable to mass screening is to create an international network of laboratories to participate in a web-based platform for collaborative scoring of DCA images. This step is the most labor intensive in the DCA process, utilizing the expertise of scientists experienced in dicentric scoring from around the world, and represents a surge capacity resource for dose estimation. Such a study, done through the European MULTIBIODOSE project, revealed statistically comparable results among the respective laboratories, with reliable dose estimates (84). Considerable work has also been done to create global biodosimetry networks as a resource for radiological emergencies, including the WHO-sponsored BioDoseNet, the IAEA RANET and the European-based RENEB, as well as networks in North and South America and Japan (64, 85).
The development of automatic platforms capable of supporting cytogenetic assays has been recognized as a necessary step towards the application of cytogenetic methodologies for large scale screening. Within the framework of the European MULTIBIODOSE network, automatic scoring of dicentrics has been tested in a laboratory intercomparison study with promising results (86). In another study using automated detection of dicentrics for partial- and whole-body irradiation, the automated detection method was found to be 33 faster than manual scoring while yielding similar dose estimates (87, 88). The Columbia Center for High-Throughput Minimally-Invasive Biodosimetry (Columbia University Medical Center, New York, NY) has developed an automated cytogenetics system called the Rapid Automated Biodosimetry Tool (RABiT), which is a high-throughput robotics platform capable of performing CBMN, DCA, c-H2AX and chromosome banding (mBand) assays and is compatible with a multi-well plate format. The platform is currently functional and the technology is being optimized for used with commercially available robotic platforms (89).
Although this technology is not currently applicable as a point-of-care diagnostic, it is the most historically relevant assay for biological dosimetry, as reflected in cytogenetics possessing the greatest number of publications compared to other fields in biodosimetry within the last ten years (Fig. 1). With the establishment of surge capacity laboratory resources and development of high-throughput automation of cytogenetic assays, this classic methodology is given a modern application for future radiological events. The field assessment score for cytogenetics is a 5(X, Y). It remains to be seen whether new biological assessment platforms for radiation dosimetry will replace this classic technique.
LYMPHOCYTE KINETICS/CLINICAL EXAM
Lymphocyte depletion kinetics, time to emesis and monitoring of other clinical signs and symptoms represent a potent methodology for determining degree of radiation exposure. Basic clinical signs and symptoms associated with prodromal ARS such as nausea, vomiting, diarrhea and hypertension can be used in basic triage dose assessment. Time to emesis after exposure has been shown to correlate with dose (6). Hematological changes observed in the immediate timeframe after irradiation, such as lymphocyte depletion, an abortive rise in neutrophils and an increase in the ratio of neutrophils to lymphocytes can provide early confirmation of significant radiation exposure (29). The Biodosimetry Assessment Tool (BAT) software platform, developed by the Biological Dosimetry Research Program at the Armed Forces Radiobiology Research Institute (Bethesda, MD), incorporates clinical symptoms with lymphocyte depletion kinetics to provide dose estimates in a user-friendly interface. The Radiation Event Medical Management (REMM) web portal also allows an easy interface to predict radiation exposures either by lymphocyte depletion, time to emesis or DCA data (90).
Multivariate approaches utilizing the integration of protein biomarker assessment concurrent with lymphocyte depletion kinetics and traditional evaluation of clinical signs and symptoms is now being used to determine biological dose prediction (4). Compared to traditional cytogenetic techniques, hematological markers are faster and utilize existing high-throughput methodologies. In several studies, it has been reported that a combined approach using analysis of blood cell populations with concurrent measurement of serum-soluble protein biomarkers shows a strong correlation between predicated and actual received dose in mouse and NHP models (28, 29, 31, 33, 91). In addition, successful use of a multivariate approach in the clinical management of victims of the Dakar radiation accident was reported, in which 63 individuals were screened to determine the level of radiation exposure using a combination of classic cytogenetic biodosimetry, evaluation of blood cell population and measurement of FL. This combined approach helped in prioritization of treatment, and a correlation of dose assessment between the classical cytogenetic analysis and combined blood cell population profile and FL status was observed (43).
HemoDose is a dose prediction software platform capable of estimating such exposures either shortly after a radiological event, or up to four weeks postirradiation, by combining counts from four cell types in a compartmental modeling approach. The use of this modeling platform potentially overcomes previous confounders to using blood cell depletion as a measure of radiation exposure, as it allows dose estimates to be rapidly obtained using blood cell counts alone (92). It has been suggested that dose estimation based on lymphocyte depletion alone is not suitable for triage biodosimetry, due to potentially confounding effects from inherent biological variability in addition to the complication of combined injury. There are also practical concerns; for example, the most informative initial lymphocyte counts must be taken shortly after the event, which may not be possible, and monitoring of lymphocyte counts must be done serially, which is not conducive to mass clinical care (93).
Although screening serial lymphocyte counts in a large field may not be feasible, this methodology might be used as a secondary assessment tool in a clinical setting once radiation exposure has been confirmed in the field. Evaluation of basic symptoms, such as time to emesis and onset of diarrhea, is conducive to field screening and might be used synergistically with other biodosimetry platforms. While the development of designer biomarkers of radiation exposure is both appealing and necessary, it is important to remember the utility of basic dose estimates using standard clinical management techniques. Lymphocyte kinetics is ranked in the late development phase, 4B(X), based on available clinical support and its use in multiple accidental exposure studies.
ELECTRON PARAMAGNETIC RESONANCE (EPR) BIODOSIMETRY
Electron paramagnetic resonance is a promising and unique methodology for radiation biodosimetry, which uses a form of magnetic resonance spectroscopy to measure radiation-induced free radical formation in materials such as hydroxyapatite found in tooth enamel and bone and in keratin found in nails. Unlike biological biodosimetry, EPR is noninvasive and free of potential biological confounders such as preexisting disease states, confounding trauma response, psychosomatic variables and interindividual radiosensitivity (94, 95). Using a modified system developed for noninvasive measurements and field application, EPR can now be used to take in vivo measurements of radiation exposure directly from the tooth enamel surface. Advantages of the EPR system include the ability to obtain immediate dose-exposure estimates on site without further processing and the ability to take repeated measurements, as the process is nondestructive and does not alter the inherent signal. In addition, EPR is not sensitive to dose rate or fractionation, since it estimates the total accumulated exposure up to the time of measurement. While incidental background ultraviolet (UV) radiation exposure has been suggested as a potential confounder of EPR dose estimates, published studies thus far have concluded these small exposures, while relevant from an epidemiological viewpoint, show little relevance in a triage setting (96). Furthermore, the benefits of EPR include a lack of time sensitivity, since the stability of radiation-induced EPR signal is immediate and exceeds the average human lifespan (97).
Electron paramagnetic resonance can be used to assess partial-body exposures through measurements taken from tooth enamel and nails from different limbs, although with heterogeneous exposure patterns EPR might not detect localized exposures to other parts of the body. EPR dose estimation using fingernail samples has also been recently utilized in accidental exposures involving the hands. As many accidental exposures include localized irradiation of the hands, obtaining an effective dose assessment for this specific partial-body exposure is very useful (98, 99)
Electron paramagnetic resonance has a dose-response linearity of up to 30 Gy, and is sensitive to free radical formation from charged particles, X rays, gamma rays and internal contamination (96, 100). EPR can detect neutron radiation over a range of energies but at a reduced sensitivity compared to gamma radiation, which can potentially be used to differentiate gamma from neutron exposures (101, 102). EPR has been used previously to reconstruct dose estimates using tissue samples taken from survivors of Nagasaki and Hiroshima, as well as exposed individuals from Chernobyl and the Techa River region (103–106). For application in a radiological event setting, a field-deployable prototype has been developed and tested successfully in mock simulations in the field using normal human volunteers and previously irradiated clinical patients (100)
The process is relatively efficient with read times per individual of approximately 5 min, allowing a single EPR unit to potentially screen up to 275 people in one day. It is also accurate, with the current field unit capable of obtaining dose assessments within 0.5 Gy of the actual exposure, allowing identification of exposures above the target threshold of 2 Gy with relative accuracy (94). Because EPR technology is insensitive to biological processes, it should be used in conjunction with other biodosimetry tools. Nevertheless, its utility as a first-responder screening tool to separate exposed individuals from the unexposed population makes it a valuable addition to the radiation biodosimetry field. Further development of fingernail dosimetry will advance the ease of use of EPR technology for mass casualty screening. Although EPR is not automated, it does have surge capacity capability through the use of multiple units, and field-deployable systems have been tested. EPR is ranked 4B(Y) based on these criteria and its recent use in several accidental exposure dosimetry assessments.
NASCENT MODELS OF BIODOSIMETRY
Electron spin resonance (ESR) spectroscopy was used to evaluate the applicability of human hair assessment for dosimetric analysis. In this study we used human hair samples irradiated ex vivo that received doses between 5 and 50 Gy and ultrahigh exposures up to 750 Gy, and differences in ESR baseline profiles were noted based on hair follicle color. Although the study demonstrated a dose-response relationship to .5 Gy, this magnitude of exposure required for detection is not practical for the triage setting (107). Plucked hairs from TBI NHPs have successfully been used to demonstrate dose prediction with c-H2AX foci analysis and illustrate a potential application as a method for partial-body dosimetry (66, 67). Evaluation of fecal microbiota by microarray analysis demonstrated alteration in the fecal microbiota profile in rats after 10 and 18 Gy TBI with single or multiple fractions. Several of the observed altered floras are present in both human and rat feces, serving as a potential methodology for dose assessment (108). These model systems represent emerging methods for determining radiation exposure and are considered in the investigative phase of development based on their novel status.
FUKUSHIMA AND BEYOND
Development of biological dosimetry models estimating internal exposures from radionuclides is highly relevant, since a majority of projected exposures from a radiological event will be from radionuclide dispersal (10). After the events of Fukushima, the major source of concern for radioactive exposures was from external or internal contamination of iodine-131 (131I), 124Cs and 137Cs (109). Whole-body counter scintillation detectors were used to screen hundreds of thousands of people for radionuclide exposure (110, 111). Screening to determine contamination was accomplished primarily with the use of hand-held scanners and whole-body counter technologies. Surveillance of radionuclide contamination in the food supply and the environment was used and continues to be used to model predicted biological exposures in the population (112–114). While there is an increasing body of evidence that the exposure levels received from radionuclide fallout from Fukushima were too low to increase cancer risk, there is significant unresolved psychosocial and mental stress in the population experiencing the event and over 350,000 individuals are currently being monitored for possible effects of low-dose radiation over their lifespans (115). In all areas of management of this recent radiological event, a method for biological dose assessment would have been useful. Such a method might have alleviated the significant psychological concerns of the “worried well”, provided personalized dose effect estimates to individuals with internal radionuclide contamination and provided a means for monitoring real biological dose effects for contaminated food consumption and environmental exposures. Biomarkers for dose assessment have significant potential and broad applicability to the many aspects of these scenarios.
The field of radiation biodosimetry has greatly advanced in the last decade, with the development of novel methods for evaluating dose effect in biological systems in the areas of metabolomics, transcriptomics and gene expression. Cytogenetic methodologies are being developed with the future promise of high-throughput capacity, and EPR has been modified for easy field deployment. Lymphocyte depletion kinetics used in combination with expression levels of plasma proteins has already been applied to the clinical management of accidental exposures, and the development of new models of radiation biodosimetry continues.
Future directions for the field of biodosimetry involve the application of these new biomarker dose assessment methodologies to exposure scenarios more closely modeling actual field exposures, including internal contamination from ingestion or inhalation of radionuclides, accumulated fractionated exposures and different qualities of radiation, including neutron radiation. This is highly relevant, since the majority of the existing biodosimetry models are used to evaluate acute gamma or X-ray exposures, which is not applicable to the triage scenario in an actual radiological event where most exposures will be fractionated and from a mixture of radiation qualities. Dose estimates using these new biomarkers also need to be tested in models using combined injury, as the inflammatory response and other biological pathways affected by trauma will complicate evaluation of biomarker expression for dose. Use of these emerging methodologies for evaluating radiation dose should be applied further to understand not only received radiation dose but radiation effect and the complex interaction of biological processes involved in multi-organ failure in ARS. This will address the prevailing thought in the field of biodosimetry that these models of received dose are not really measuring radiation “dose” but radiation “effect” or correlating the biological effect of radiation exposure in a living system to a physical radiation dose in a quantifiable way.
The use of radiation biomarkers to predict level of exposure must also address inherent variance in radiosensitivities across a population. Segregating a population for treatment based on the established 2 Gy threshold may not be the most efficient approach; there will likely be some highly sensitive individuals manifesting biological effects similar to that of a 3 Gy exposure upon receiving a 1.5 Gy exposure, and others not needing immediate medical intervention at doses between 2 and 3 Gy. Application of these biodosimetry assessment tools needs to be further modeled in special populations to include preexisting disease states, as well as the elderly and pediatric populations. Adaptation of this biological science to developing field use technology needs to continue with attention to ease of use, low processing times and rugged engineering, and these future clinical diagnostics need to be integrated with existing emergency management concepts of operations for radiological event planning. This includes practical considerations such as how to label and track samples linked to patients in a disaster setting, building on previous efforts to develop sample tracking systems for biodosimetry assessment (116). How biodosimetry data will integrate with triage systems and how this information will affect subsequent medical management, countermeasure administration and scarce resource allocation also requires consideration.
The most promising and potentially immediately useful platforms for biodosimetry include cytogenetic assessment using surge capacity laboratory networks, proteomics- and genomics-based technologies, which are being developed for field application and EPR. Lymphocyte depletion kinetics has the advantage in its use of an established clinical assay, but requires serial collection of samples and may not be suited to mass casualty scenarios. Transcriptomics and metabolomics remain alternative useful methodologies for biodosimetry assessment, however, they are not readily applicable in triage settings at this time.
In a recent concept-of-operations (CONOPS) assessment of various biodosimetry platforms, similar findings were reported for cytogenetics and blood cell counting as the only biodosimetry diagnostics with immediate capacity for operational use. For this assessment, biodosimetry technologies were ranked based on their technology readiness level score and their suitability to support medical management of radiological exposure scenarios within the military medical support system (117).
As appropriate human models of radiation exposure are not available to validate emerging biodosimetry techniques, greater collaboration between basic research laboratories and clinical management of accidental human exposures needs to occur. While limited, these accidental partial exposures occur regularly in industry, as well as due to lost sources in many countries. Increased efforts are needed to utilize and share human samples from these exposures, not only to advance the field of biodosimetry, but to elevate the level of clinical care for exposed victims (118).
With the accumulated body of biodosimetry research currently available and the application of this science to high-throughput technologies underway, there have been great strides in efforts to prepare for a large scale radiological event, although there is still more work to be done. Greater collaboration within each field of biodosimetry would benefit the development of a standardized panel of biological markers for dosimetry assessment. Assessing the application of radiation biodosimetry in special populations, and development of a rapid assay for assessment of partial-body exposures is needed. Critical organ-specific markers of radiation toxicity also need to be identified and validated. Testing novel prototype technologies in field settings and further investigation and more rigorous modeling of established proof-of-concept research is also needed to make the final step towards practical applications and true emergency readiness.
ACKNOWLEDGMENTS
The authors would like to thank Gregory Koblentz, Director of the Biodefense Graduate Program in the School of Policy Government and International Affairs at George Mason University, Fairfax, VA. This work has been funded by the NIH intramural division project ZIDBC010990.
REFERENCES
- 1.Sullivan JM, Prasanna PGS, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–54. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flood AB, Nicolalde RJ, Demidenko E, Williams BB, Shapiro A, Wiley AL, Jr, et al. A framework for comparative evaluation of dosimetric methods to triage a large population following a radiological event. Radiat Meas. 2011;46:916–22. doi: 10.1016/j.radmeas.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn DF, Goans RE. Nuclear terrorism: triage and medical management of radiation and combined-injury casualties. Surg Clin North Am. 2006;86:601–36. doi: 10.1016/j.suc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Prasanna PG, Blakely WF, Bertho JM, Chute JP, Cohen EP, Goans RE, et al. Synopsis of partial-body radiation diagnostic biomarkers and medical management of radiation injury workshop. Radiat Res. 2010;173:245–53. doi: 10.1667/RR1993.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etherington G, Rothkamm K, Shutt AL, Youngman MJ. Triage, monitoring and dose assessment for people exposed to ionising radiation following a malevolent act. Radiat Prot Dosimetry. 2012;144:534–9. doi: 10.1093/rpd/ncq420. [DOI] [PubMed] [Google Scholar]
- 6.Koenig KL, Goans RE, Hatchett RJ, Mettler FA, Jr., Schumacher TA, Noji EK, et al. Medical treatment of radiological casualties: current concepts. Ann Emerg Med. 2005;45:643–52. doi: 10.1016/j.annemergmed.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Bromet EJ. Emotional consequences of nuclear power plant disasters. Health Phys. 2014;106:206–10. doi: 10.1097/HP.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dainiak N, Skudlarska B, Albanese J. Local, regional and national responses for medical management of a radiological/ nuclear Incident. Dose Response. 2013;11:121–9. doi: 10.2203/dose-response.08-018.Dainiak. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon SL, Coleman CN, Noska MA, Bowman T. Response of the U.S. Department of Health and Human Services in protecting civilian Americans in Japan during the Fukushima nuclear crisis. Health Phys. 2012:102. doi: 10.1097/HP.0b013e31824c79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman CN, Adams S, Adrianopoli C, Ansari A, Bader JL, Buddemeier B, et al. Medical planning and response for a nuclear detonation: a practical guide. Biosecur Bioterror. 2012;10:346–71. doi: 10.1089/bsp.2012.1025. [DOI] [PubMed] [Google Scholar]
- 11.Tucker JD, Divine GW, Grever WE, Thomas RA, Joiner MC, Smolinski JM, et al. Gene expression-based dosimetry by dose and time in mice following acute radiation exposure. PloS One. 2013;8:e83390. doi: 10.1371/journal.pone.0083390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filiano AN, Fathallah-Shaykh HM, Fiveash J, Gage J, Cantor A, Kharbanda S, et al. Gene expression analysis in radiotherapy patients and C57BL/6 mice as a measure of exposure to ionizing radiation. Radiat Res. 2011;176:49–61. doi: 10.1667/RR2419.1. [DOI] [PubMed] [Google Scholar]
- 13.Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, et al. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PloS One. 2008;3:e1912. doi: 10.1371/journal.pone.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas J, Dressman HK, Suchindran S, Nakamura M, Chao NJ, Himburg H, et al. A translatable predictor of human radiation exposure. PloS One. 2014;9:e107897. doi: 10.1371/journal.pone.0107897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, et al. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PloS One. 2010;5:e11535. doi: 10.1371/journal.pone.0011535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucker JD, Joiner MC, Thomas RA, Grever WE, Bakhmutsky MV, Chinkhota CN, et al. Accurate gene expression-based biodosimetry using a minimal set of human gene transcripts. Int J Radiat Oncol Biol Phys. 2014;88:933–9. doi: 10.1016/j.ijrobp.2013.11.248. [DOI] [PubMed] [Google Scholar]
- 17.Paul S, Barker CA, Turner HC, McLane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–65. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brengues M, Paap B, Bittner M, Amundson S, Seligmann B, Korn R, et al. Biodosimetry on small blood volume using gene expression assay. Health Phys. 2010;98:179–85. doi: 10.1097/01.HP.0000346706.44253.5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul S, Smilenov LB, Elliston CD, Amundson SA. Radiation dose-rate effects on gene expression in a mouse biodosimetry model. Radiat Res. 2015;184:24–32. doi: 10.1667/RR14044.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med Genomics. 2015;8:22. doi: 10.1186/s12920-015-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, et al. Gene expression response of mice after a single dose of 137Cs as an internal emitter. Radiat Res. 2014;182:380–9. doi: 10.1667/RR13466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akashi M, Hirama T, Tanosaki S, Kuroiwa N, Nakagawa K, Tsuji H, et al. Initial symptoms of acute radiation syndrome in the JCO criticality accident in Tokai-mura. J Radiat Res. 2001;42:S157–66. doi: 10.1269/jrr.42.s157. [DOI] [PubMed] [Google Scholar]
- 23.Becciolini A, Giannardi G, Cionini L, Porciani S, Fallai C, Pirtoli L. Plasma amylase activity as a biochemical indicator of radiation injury to salivary glands. Acta Radiol Oncol. 1984;23:9–14. doi: 10.3109/02841868409135978. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann R, Schreiber GA, Willich N, Westhaus R, Bogl KW. Increased serum amylase in patients after radiotherapy as a probable bioindicator for radiation exposure. Strahlenther Onkol. 1990;166:688–95. [PubMed] [Google Scholar]
- 25.Ossetrova NI, Sandgren DJ, Blakely WF. C-reactive protein and serum amyloid A as early-phase and prognostic indicators of acute radiation exposure in nonhuman primate total-body irradiation model. Radiat Meas. 2011;46:1019–24. [Google Scholar]
- 26.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Serum amyloid A as a biomarker for radiation exposure. Radiat Res. 2015;184:14–23. doi: 10.1667/RR13927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivey RG, Subramanian O, Lorentzen TD, Paulovich AG. Antibody-based screen for ionizing radiation-dependent changes in the mammalian proteome for use in biodosimetry. Radiat Res. 2009;171:549–61. doi: 10.1667/RR1638.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ossetrova NI, Sandgren DJ, Blakely WF. Protein biomarkers for enhancement of radiation dose and injury assessment in nonhuman primate total-body irradiation model. Radiat Prot Dosimetry. 2014;159:61–76. doi: 10.1093/rpd/ncu165. [DOI] [PubMed] [Google Scholar]
- 29.Blakely WF, Ossetrova NI, Whitnall MH, Sandgren DJ, Krivokrysenko VI, Shakhov A, et al. Multiple parameter radiation injury assessment using a nonhuman primate radiation model-biodosimetry applications. Health Phys. 2010;98:153–9. doi: 10.1097/HP.0b013e3181b0306d. [DOI] [PubMed] [Google Scholar]
- 30.Blakely WF, Ossetrova NI, Manglapus GL, Salter CA, Levine IH, Jackson WE, et al. Amylase and blood cell-count hematological radiation-injury biomarkers in a rhesus monkey radiation model—use of multiparameter and integrated biological dosimetry. Radiat Meas. 2007;42:1164–70. [Google Scholar]
- 31.Ossetrova NI, Sandgren DJ, Gallego S, Blakely WF. Combined approach of hematological biomarkers and plasma protein SAA for improvement of radiation dose assessment triage in biodosimetry applications. Health Phys. 2010;98:204–8. doi: 10.1097/HP.0b013e3181abaabf. [DOI] [PubMed] [Google Scholar]
- 32.Kim D, Marchetti F, Chen Z, Zaric S, Wilson RJ, Hall DA, et al. Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Sci Rep. 2013;3:2234. doi: 10.1038/srep02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ossetrova NI, Condliffe DP, Ney PH, Krasnopolsky K, Hieber KP, Rahman A, et al. Early-response biomarkers for assessment of radiation exposure in a mouse total-body irradiation model. Health Phys. 2014;106:772–86. doi: 10.1097/HP.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 34.Blakely W, Sandgren DJ, Nagy V, Kim SY, Ossetrova NI. Murine partial-body radiation exposure model for biodosimetry studies-preliminary report. Radiat Meas. 2011;46:898–902. [Google Scholar]
- 35.Blakely WF, Sandgren DJ, Nagy V, Kim S-Y, Sigal GB, Ossetrova NI. Further biodosimetry investigations using murine partial-body irradiation model. Radiat Prot Dosimetry. 2014;159:46–51. doi: 10.1093/rpd/ncu127. [DOI] [PubMed] [Google Scholar]
- 36.Sproull M, Avondoglio D, Kramp T, Shankavaram U, Camphausen K. Correlation of plasma FL expression with bone marrow irradiation dose. PloS One. 2013;8:e58558. doi: 10.1371/journal.pone.0058558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma M, Moulder JE. The urine proteome as a radiation biodosimeter. Adv Exp Med Biol. 2013;990:87–100. doi: 10.1007/978-94-007-5896-4_5. [DOI] [PubMed] [Google Scholar]
- 38.Chaze T, Slomianny MC, Milliat F, Tarlet G, Lefebvre-Darroman T, Gourmelon P, et al. Alteration of the serum N-glycome of mice locally exposed to high doses of ionizing radiation. Mol Cell Proteomics: MCP. 2013;12:283–301. doi: 10.1074/mcp.M111.014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Citrin DE, Hitchcock YJ, Chung EJ, Frandsen J, Urick ME, Shield W, et al. Determination of cytokine protein levels in oral secretions in patients undergoing radiotherapy for head and neck malignancies. Radiat Oncol. 2012;7:64. doi: 10.1186/1748-717X-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore HD, Ivey RG, Voytovich UJ, Lin C, Stirewalt DL, Pogosova-Agadjanyan EL, et al. The human salivary proteome is radiation responsive. Radiat Res. 2014;181:521–30. doi: 10.1667/RR13586.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertho JM, Roy L, Souidi M, Benderitter M, Gueguen Y, Lataillade JJ, et al. New biological indicators to evaluate and monitor radiation-induced damage: an accident case report. Radiat Res. 2008;169:543–50. doi: 10.1667/RR1259.1. [DOI] [PubMed] [Google Scholar]
- 42.Gourmelon P, Benderitter M, Bertho JM, Huet C, Gorin NC, De Revel P. European consensus on the medical management of acute radiation syndrome and analysis of the radiation accidents in Belgium and Senegal. Health Phys. 2010;98:825–32. doi: 10.1097/HP.0b013e3181ce64d4. [DOI] [PubMed] [Google Scholar]
- 43.Bertho JM, Roy L. A rapid multiparametric method for victim triage in cases of accidental protracted irradiation or delayed analysis. Br J Radiol. 2009;82:764–70. doi: 10.1259/bjr/49063618. [DOI] [PubMed] [Google Scholar]
- 44.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace AJ. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–23. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onal C, Kotek A, Unal B, Arslan G, Yavuz A, Topkan E, et al. Plasma citrulline levels predict intestinal toxicity in patients treated with pelvic radiotherapy. Acta Oncol. 2011;50:1167–74. doi: 10.3109/0284186X.2011.584557. [DOI] [PubMed] [Google Scholar]
- 46.Lutgens LCHW, Deutz N, Granzier-Peeters M, Beets-Tan R, De Ruysscher D, Gueulette J, et al. Plasma citrulline concentration: a surrogate end point for radiation-induced mucosal atrophy of the small bowel. A feasibility study in 23 patients. Int J Radiat Oncol Biol Phys. 2004;60:275–85. doi: 10.1016/j.ijrobp.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 47.Singh VK, Newman VL, Romaine PL, Hauer-Jensen M, Pollard HB. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Exp Rev Molec Diag. 2016;16:65–81. doi: 10.1586/14737159.2016.1121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJ., Jr. Global metabolomic identification of long-term dose-dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation. Radiat Res. 2015;184:121–33. doi: 10.1667/rr14091.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–40. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–61. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goudarzi M, Weber W, Mak TD, Chung J, Doyle-Eisele M, Melo D, et al. Development of urinary biomarkers for internal exposure by cesium-137 using a metabolomics approach in mice. Radiat Res. 2014;181:54–64. doi: 10.1667/RR13479.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goudarzi M, Weber WM, Mak TD, Chung J, Doyle-Eisele M, Melo DR, et al. Metabolomic and lipidomic analysis of serum from mice exposed to an internal emitter, cesium-137, using a shotgun LC-MS(E) approach. J Proteome Res. 2015;14:374–84. doi: 10.1021/pr500913n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laiakis EC, Strassburg K, Bogumil R, Lai S, Vreeken RJ, Hankemeier T, et al. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014;13:4143–54. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H, Wang Z, Zhang X, Qiao Y, Wu S, Dong F, et al. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat Prot Dosimetry. 2013;154:9–17. doi: 10.1093/rpd/ncs138. [DOI] [PubMed] [Google Scholar]
- 55.Grison S, Martin JC, Grandcolas L, Banzet N, Blanchardon E, Tourlonias E, et al. The metabolomic approach identifies a biological signature of low-dose chronic exposure to cesium 137. J Radiat Res. 2012;53:33–43. doi: 10.1269/jrr.11071. [DOI] [PubMed] [Google Scholar]
- 56.Grison S, Favé G, Maillot M, Manens L, Delissen O, Blanchardon E, et al. Metabolomics identifies a biological response to chronic low-dose natural uranium contamination in urine samples. Metabolomics. 2013;9:1168–80. doi: 10.1007/s11306-013-0544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dickey J, Zemp F, Martin O, Kovalchuk O. The role of miRNA in the direct and indirect effects of ionizing radiation. Radiat Environ Biophys. 2011;50:491–9. doi: 10.1007/s00411-011-0386-5. [DOI] [PubMed] [Google Scholar]
- 58.Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P, et al. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PloS One. 2013;8:e57603. doi: 10.1371/journal.pone.0057603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui W, Ma J, Wang Y, Biswal S. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PloS One. 2011;6:e22988. doi: 10.1371/journal.pone.0022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7:287ra69. doi: 10.1126/scitranslmed.aaa6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiang JG, Anderson MN, Swift JM, Christensen CL, Gupta P, et al. Hemorrhage exacerbates radiation effects on survival, leukocytopenia, thrombopenia, erythropenia, bone marrow cell depletion and hematopoiesis, and inflammation-associated micro-RNAs expression in kidney. PloS One. 2015;10:e0139271. doi: 10.1371/journal.pone.0139271. SJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Templin T, Amundson SA, Brenner DJ, Smilenov LB. Whole mouse blood microRNA as biomarkers for exposure to gammarays and (56)Fe ion. Int J Radiat Biol. 2011;87:653–62. doi: 10.3109/09553002.2010.549537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee K-F, Chen Y-C, Hsu PW-C, Liu IY, Wu LS-H. MicroRNA expression profiling altered by variant dosage of radiation exposure. BioMed Res Int. 2014;2014:10. doi: 10.1155/2014/456323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulka U, Ainsbury L, Atkinson M, Barquinero JF, Barrios L, Beinke C, et al. Realising the European network of biodosimetry (RENEB) Radiat Prot Dosimetry. 2012;151:621–5. doi: 10.1093/rpd/ncs157. [DOI] [PubMed] [Google Scholar]
- 65.Roch-Lefe`vre S, Pouzoulet F, Giraudet AL, Voisin P, Vaurijoux A, Gruel G, et al. Cytogenetic assessment of heterogeneous radiation doses in cancer patients treated with fractionated radiotherapy. Br J Radiol. 2010;83:759–66. doi: 10.1259/bjr/21022597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Redon CE, Nakamura AJ, Gouliaeva K, Rahman A, Blakely WF, Bonner WM. The use of gamma-H2AX as a biodosimeter for total-body radiation exposure in non-human primates. PloS One. 2010;5:e15544. doi: 10.1371/journal.pone.0015544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Redon CE, Nakamura AJ, Gouliaeva K, Rahman A, Blakely WF, Bonner WM. Q(gamma-H2AX), an analysis method for partial-body radiation exposure using gamma-H2AX in nonhuman primate lymphocytes. Radiat Meas. 2011;46:877–81. doi: 10.1016/j.radmeas.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horn S, Barnard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PloS One. 2011;6:e25113. doi: 10.1371/journal.pone.0025113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roch-Lefevre S, Mandina T, Voisin P, Gaetan G, Mesa JE, Valente M, et al. Quantification of gamma-H2AX foci in human lymphocytes: a method for biological dosimetry after ionizing radiation exposure. Radiat Res. 2010;174:185–94. doi: 10.1667/RR1775.1. [DOI] [PubMed] [Google Scholar]
- 70.Deperas J, Szluinska M, Deperas-Kaminska M, Edwards A, Lloyd D, Lindholm C, et al. CABAS: a freely available PC program for fitting calibration curves in chromosome aberration dosimetry. Radiat Prot Dosimetry. 2007;124:115–23. doi: 10.1093/rpd/ncm137. [DOI] [PubMed] [Google Scholar]
- 71.Ainsbury EA, Vinnikov VA, Puig P, Higueras M, Maznyk NA, Lloyd DC, et al. Review of Bayesian statistical analysis methods for cytogenetic radiation biodosimetry, with a practical example. Radiat Prot Dosimetry. 2014;162:185–96. doi: 10.1093/rpd/nct301. [DOI] [PubMed] [Google Scholar]
- 72.Higueras M, Puig P, Ainsbury EA, Rothkamm K. A new inverse regression model applied to radiation biodosimetry. Proc Math Phys Eng Sci. 2015;471:20140588. doi: 10.1098/rspa.2014.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ainsbury EA, Vinnikov V, Puig P, Maznyk N, Rothkamm K, Lloyd DC. CytoBayesJ: software tools for Bayesian analysis of cytogenetic radiation dosimetry data. Mutat Res. 2013;756:184–91. doi: 10.1016/j.mrgentox.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Morina D, Higueras M, Puig P, Ainsbury EA, Rothkamm K. radir package: an R implementation for cytogenetic biodosimetry dose estimation. J Radiat Prot. 2015;35:557–69. doi: 10.1088/0952-4746/35/3/557. [DOI] [PubMed] [Google Scholar]
- 75.Higueras M, Puig P, Ainsbury EA, Vinnikov VA, Rothkamm K. A new Bayesian model applied to cytogenetic partial body irradiation estimation. Radiat Prot Dosimetry. 2015;168:330–6. doi: 10.1093/rpd/ncv356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaworska A, Ainsbury EA, Fattibene P, Lindholm C, Oestreicher U, Rothkamm K, et al. Operational guidance for radiation emergency response organisations in Europe for using biodosimetric tools developed in EU MULTIBIODOSE project. Radiat Prot Dosimetry. 2015;164:165–9. doi: 10.1093/rpd/ncu294. [DOI] [PubMed] [Google Scholar]
- 77.Rothkamm K, Beinke C, Romm H, Badie C, Balagurunathan Y, Barnard S, et al. Comparison of established and emerging biodosimetry assays. Radiat Res. 2013;180:111–9. doi: 10.1667/RR3231.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ainsbury EA, Al-Hafidh J, Bajinskis A, Barnard S, Barquinero JF, Beinke C, et al. Inter- and intra-laboratory comparison of a multibiodosimetric approach to triage in a simulated, large scale radiation emergency. Int J Radiat Biol. 2014;90:193–202. doi: 10.3109/09553002.2014.868616. [DOI] [PubMed] [Google Scholar]
- 79.Romm H, Wilkins RC, Coleman CN, Lillis-Hearne PK, Pellmar TC, Livingston GK, et al. Biological dosimetry by the triage dicentric chromosome assay: potential implications for treatment of acute radiation syndrome in radiological mass casualties. Radiat Res. 2011;175:397–404. doi: 10.1667/RR2321.1. [DOI] [PubMed] [Google Scholar]
- 80.Barnard S, Ainsbury EA, Al-hafidh J, Hadjidekova V, Hristova R, Lindholm C, et al. The first gamma-H2AX biodosimetry intercomparison exercise of the developing European biodosimetry network RENEB. Radiat Prot Dosimetry. 2015;164:265–70. doi: 10.1093/rpd/ncu259. [DOI] [PubMed] [Google Scholar]
- 81.Rothkamm K, Horn S, Scherthan H, Rossler U, De Amicis A, Barnard S, et al. Laboratory intercomparison on the gamma-H2AX foci assay. Radiat Res. 2013;180:149–55. doi: 10.1667/RR3238.1. [DOI] [PubMed] [Google Scholar]
- 82.Beinke C, Barnard S, Boulay-Greene H, De Amicis A, De Sanctis S, Herodin F, et al. Laboratory intercomparison of the dicentric chromosome analysis assay. Radiat Res. 2013;180:129–37. doi: 10.1667/RR3235.1. [DOI] [PubMed] [Google Scholar]
- 83.Badie C, Kabacik S, Balagurunathan Y, Bernard N, Brengues M, Faggioni G, et al. Laboratory intercomparison of gene expression assays. Radiat Res. 2013;180:138–48. doi: 10.1667/RR3236.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Romm H, Ainsbury E, Bajinskis A, Barnard S, Barquinero JF, Barrios L, et al. Web-based scoring of the dicentric assay, a collaborative biodosimetric scoring strategy for population triage in large scale radiation accidents. Radiat Environ Biophys. 2014;53:241–54. doi: 10.1007/s00411-014-0519-8. [DOI] [PubMed] [Google Scholar]
- 85.Blakely WF, Carr Z, Chu MC-M, Dayal-Drager R, Fujimoto K, Hopmeir M, et al. WHO 1st consultation on the development of a global biodosimetry laboratories network for radiation emergencies (BioDoseNet) Radiat Res. 2009;171:127–39. doi: 10.1667/RR1549.1. [DOI] [PubMed] [Google Scholar]
- 86.Romm H, Ainsbury E, Barnard S, Barrios L, Barquinero JF, Beinke C, et al. Automatic scoring of dicentric chromosomes as a tool in large scale radiation accidents. Mutat Res. 2013;756:174–83. doi: 10.1016/j.mrgentox.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 87.Gruel G, Grégoire E, Lecas S, Martin C, Roch-Lefevre S, Vaurijoux A, et al. Biological dosimetry by automated dicentric scoring in a simulated emergency. Radiat Res. 2013;179:557–69. doi: 10.1667/RR3196.1. [DOI] [PubMed] [Google Scholar]
- 88.Vaurijoux A, Gregoire E, Roch-Lefevre S, Voisin P, Martin C, Voisin P, et al. Detection of partial-body exposure to ionizing radiation by the automatic detection of dicentrics. Radiat Res. 2012;178:357–64. doi: 10.1667/rr2728.1. [DOI] [PubMed] [Google Scholar]
- 89.Garty G, Bigelow AW, Repin M, Turner HC, Bian D, Balajee AS, et al. An automated imaging system for radiation biodosimetry. Microsc Res Tech. 2015;78:587–98. doi: 10.1002/jemt.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waller E, Millage K, Blakely WF, Ross JA, Mercier JR, Sandgren DJ, et al. Overview of hazard assessment and emergency planning software of use to RN first responders. Health Phys. 2009;97:145–56. doi: 10.1097/01.HP.0000348464.78396.23. [DOI] [PubMed] [Google Scholar]
- 91.Bolduc DL, Villa V, Sandgren DJ, Ledney GD, Blakely WF, Bunger R. Application of multivariate modeling for radiation injury assessment: a proof of concept. Comput Math Methods Med. 2014;2014:17. doi: 10.1155/2014/685286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu S, Blakely WF, Cucinotta FA. HEMODOSE: a biodosimetry tool based on multi-type blood cell counts. Health Phys. 2015;109:54–68. doi: 10.1097/HP.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ainsbury EA, Bakhanova E, Barquinero JF, Brai M, Chumak V, Correcher V, et al. Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat Prot Dosimetry; 147:573–92. doi: 10.1093/rpd/ncq499. [DOI] [PubMed] [Google Scholar]
- 94.Williams BB, Dong R, Flood AB, Grinberg O, Kmiec M, Lesniewski PN, et al. A deployable in vivo EPR tooth dosimeter for triage after a radiation event involving large populations. Radiat Meas. 2011;46:772–7. doi: 10.1016/j.radmeas.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Swartz H, Williams B, Flood A. Overview of the principles and practice of biodosimetry. Radiat Environ Biophys. 2014;53:221–32. doi: 10.1007/s00411-014-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams BB, Dong R, Nicolalde RJ, Matthews TP, Gladstone DJ, Demidenko E, et al. Physically-based biodosimetry using in vivo EPR of teeth in patients undergoing total body irradiation. Int J Radiat Biol. 2011;87:766–75. doi: 10.3109/09553002.2011.583316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Desrosiers M, Schauer DA. Electron paramagnetic resonance (EPR) biodosimetry. Nucl Instrum Methods Phys Res B. 2001;184:219–28. [Google Scholar]
- 98.Romanyukha A, Trompier F, Reyes RA, Christensen DM, Iddins CJ, Sugarman SL. Electron paramagnetic resonance radiation dose assessment in fingernails of the victim exposed to high dose as result of an accident. Radiat Environ Biophys. 2014;53:755–62. doi: 10.1007/s00411-014-0553-6. [DOI] [PubMed] [Google Scholar]
- 99.Trompier F, Queinnec F, Bey E, De Revel T, Lataillade JJ, Clairand I, et al. EPR retrospective dosimetry with fingernails: report on first application cases. Health Phys. 2014;106:798–805. doi: 10.1097/HP.0000000000000110. [DOI] [PubMed] [Google Scholar]
- 100.Williams B, Flood A, Salikhov I, Kobayashi K, Dong R, Rychert K, et al. In vivo EPR tooth dosimetry for triage after a radiation event involving large populations. Radiat Environ Biophys. 2014;53:335–46. doi: 10.1007/s00411-014-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zdravkova M, Crokart N, Trompier F, Asselineau B, Gallez B, Gaillard-Lecanu E, et al. Retrospective dosimetry after criticality accidents using low-frequency EPR: a study of whole human teeth irradiated in a mixed neutron and gamma-radiation field. Radiat Res. 2003;160:168–73. doi: 10.1667/rr3026. [DOI] [PubMed] [Google Scholar]
- 102.Trompier F, Fattibene P, Tikunov D, Bartolotta A, Carosi A, Doca MC. EPR dosimetry in a mixed neutron and gamma radiation field. Radiat Prot Dosimetry. 2004;110:437–42. doi: 10.1093/rpd/nch225. [DOI] [PubMed] [Google Scholar]
- 103.Ishii H, Ikeya M, Okano M. ESR dosimetry of teeth of residents close to Chernobyl reactor accident. J Nuc Sci Tech. 1990;27:1153–5. [Google Scholar]
- 104.Nakamura N, Miyazawa C, Sawada S, Akiyama M, Awa AA. A close correlation between electron spin resonance (ESR) dosimetry from tooth enamel and cytogenetic dosimetry from lymphocytes of Hiroshima atomic-bomb survivors. Int J Radiat Biol. 1998;73:619–27. doi: 10.1080/095530098141870. [DOI] [PubMed] [Google Scholar]
- 105.Use of electron paramagnetic resonance dosimetry with tooth enamel for retrospective dose assessment. International Atomic Energy Agency; Vienna: 2002. IAEA Report No. IAEA-TECDOC-1331. [Google Scholar]
- 106.Volchkova A, Shishkina EA, Ivanov D, Timofeev Y, Fattibene P, Della Monaca S, et al. Harmonization of dosimetric information obtained by different EPR methods: experience of the Techa river study. Radiat Meas. 2011;46:801–7. [Google Scholar]
- 107.Tepe Çam S, Polat M, Seyhan N. The use of human hair as biodosimeter. Appl Radiat Isot. 2014;94:272–81. doi: 10.1016/j.apradiso.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 108.Lam V, Moulder JE, Salzman NH, Dubinsky EA, Andersen GL, Baker JE. Intestinal microbiota as novel biomarkers of prior radiation exposure. Radiat Res. 2012;177:573–83. doi: 10.1667/rr2691.1. [DOI] [PubMed] [Google Scholar]
- 109.Matsuda N, Kumagai A, Ohtsuru A, Morita N, Miura M, Yoshida M, et al. Assessment of internal exposure doses in Fukushima by a whole body counter within one month after the nuclear power plant accident. Radiat Res. 2013;179:663–8. doi: 10.1667/RR3232.1. [DOI] [PubMed] [Google Scholar]
- 110.Hayano RS, Tsubokura M, Miyazaki M, Satou H, Sato K, Masaki S, et al. Internal radiocesium contamination of adults and children in Fukushima 7 to 20 months after the Fukushima NPP accident as measured by extensive whole-body-counter surveys. Proc Jpn Acad Ser B Phys Biol Sci. 2013;89:157–63. doi: 10.2183/pjab.89.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miyazaki M, Ohtsuru A, Ishikawa T. An overview of internal dose estimation using whole-body counters in Fukushima Prefecture. Fukushima J Med Sci. 2014;60:95–100. doi: 10.5387/fms.2014-10. [DOI] [PubMed] [Google Scholar]
- 112.Yajima K, Kurihara O, Ohmachi Y, Takada M, Omori Y, Akahane K, et al. Estimating annual individual doses for evacuees returning home to areas affected by the Fukushima nuclear accident. Health Phys. 2015;109:122–33. doi: 10.1097/HP.0000000000000308. [DOI] [PubMed] [Google Scholar]
- 113.Kim E, Tani K, Kunishima N, Kurihara O, Sakai K, Akashi M. Estimation of early internal doses to Fukushima residents after the nuclear disaster based on the atmospheric dispersion simulation. Radiat Prot Dosimetry. 2015 doi: 10.1093/rpd/ncv385. http://bit.ly/2cppHHU. [DOI] [PubMed] [Google Scholar]
- 114.Iimoto T, Nunokawa J, Fujii H, Takashima R, Hashimoto M, Fukuhara T, et al. Collaboration of local government and experts responding to increase in environmental radiation level due to the nuclear disaster: focusing on their activities and latest radiological discussion. Radiat Prot Dosimetry. 2015;167:358–64. doi: 10.1093/rpd/ncv279. [DOI] [PubMed] [Google Scholar]
- 115.Clancey G, Chhem R. Hiroshima, Nagasaki, and Fukushima. Lancet. 2015;386:405–6. doi: 10.1016/S0140-6736(15)61414-3. [DOI] [PubMed] [Google Scholar]
- 116.Martin PR, Berdychevski RE, Subramanian U, Blakely WF, Prasanna PGS. Sample tracking in an automated cytogenetic biodosimetry laboratory for radiation mass casualties. Radiat Meas. 2007;42:1119–24. doi: 10.1016/j.radmeas.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Milner EE, Daxon EG, Anastasio MT, Nesler JT, Miller RL, Blakely WF. Concepts of operations (CONOPS) for biodosimetry tools employed in operational environments. Health Phys. 2016;110:370–9. doi: 10.1097/HP.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 118.Chen Y, Trotti A, Coleman CN, Machtay M, Mirimanoff RO, Hay J, et al. Adverse event reporting and developments in radiation biology after normal tissue injury: International Atomic Energy Agency consultation. Int J Radiat Oncol Biol Phys. 2006;64:1442–51. doi: 10.1016/j.ijrobp.2005.10.014. [DOI] [PubMed] [Google Scholar]