Abstract
Canonical signal transduction via heterotrimeric G proteins is spatially and temporally restricted, i.e., triggered exclusively at the plasma membrane (PM), only by agonist activation of G-protein-coupled receptors (GPCRs) via a process that completes within a few hundred milliseconds. Recently, a rapidly emerging paradigm has revealed a non-canonical pathway for activation of trimeric G proteins by the non-receptor guanidine-nucleotide exchange factor (GEF), GIV/Girdin. This pathway has distinctive temporal and spatial features and an unusual profile of receptor engagement: Diverse classes of receptors, not just GPCRs can engage with GIV-GEF to trigger such activation. Such activation is spatially and temporally unrestricted, i.e., can occur both at the PM and on internal membranes discontinuous with the PM, and can continue for prolonged periods of time. Here we review the molecular mechanisms that govern non-canonical G protein activation by GIV-GEF and the relevance of this new paradigm in health and disease.
INTRODUCTION
Heterotrimeric (henceforth trimeric) G proteins work as molecular switches that control the flow of information from extracellular cues perceived at the cell surface to a wide array of intracellular effector proteins that control cell behavior[19,20]. Canonical G protein signaling is initiated when inactive trimers (i.e., GDP-bound) Gα subunits in complex with Gβγ are activated by ligand-occupied G Protein Coupled Receptors (GPCRs), which are Guanine nucleotide Exchange Factors (GEFs) and promote the exchange of GDP for GTP on the α subunit[19] (Figure 1). Signaling is terminated by the intrinsic GTPase activity of the Gα subunit, leading to re-association of Gα with Gβγ. This sequence of "on" and "off" events regulate the so-called “G protein cycle”, and represents the core components and events of signal transduction via GPCRs. The importance of canonical signal transduction via GPCRs/G proteins in modern medicine is unparalleled by any other. For example, extensive work during the past decades has revealed how dysregulation of G protein signaling influences the pathogenesis in a myriad of human diseases, from cancer, thru fibrosis, neurodegeneration, diabetes and cardiovascular diseases, to name a few. More importantly, attempts to develop therapies targeting this pathway has also been rewarded with unparalleled success; today canonical G protein signaling by GPCRs represent the target for ~40% of marketed drugs[21].
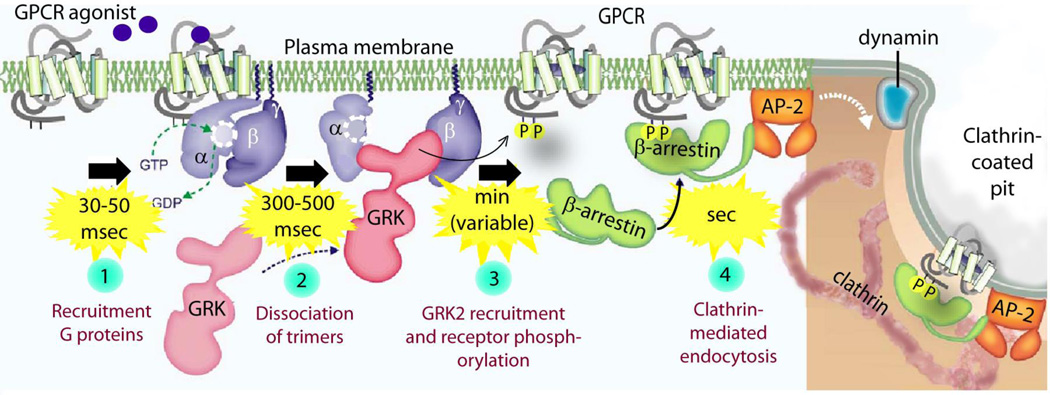
Figure 1. Canonical G proteins signaling is restricted in time and space.
Schematic shows the steps in GPCR signaling. Kinetics of various steps in receptor/G protein signaling chain as determined by FRET and BRET assays in intact cells are displayed [42]. Therefore, these values represent estimates of half-lives (in msec) for each step at maximal agonist concentration and overexpressed protein levels. Specific values cited here reflects the events during activation of β1-adrenoreceptor/Gαs/cAMP cascade. Similar values were reported also for the α2A-adrenoreceptors/Gαi/o and for the A2A-adenosine/Gαi pathways.
G protein signaling that is initiated by GPCRs is further fine-tuned by a heterogeneous set of “accessory proteins” capable of modulating the activity of G proteins by various mechanisms (Figure 2). These accessory proteins include GTPase activating proteins (GAPs), guanine nucleotide dissociation inhibitors (GDIs) and non-receptor GEFs[22–26]. The characterization of the biological role of these proteins has been largely driven by the discovery of structurally well-defined conserved signature motifs or domains, e.g., the “GoLoco/ GPR motif” (~20–30 aa) [27,28] and the “RGS box” (~120 aa) [29–32] that are necessary and sufficient to exert the GDI or GAP enzymatic activity, respectively, on Gα subunits.
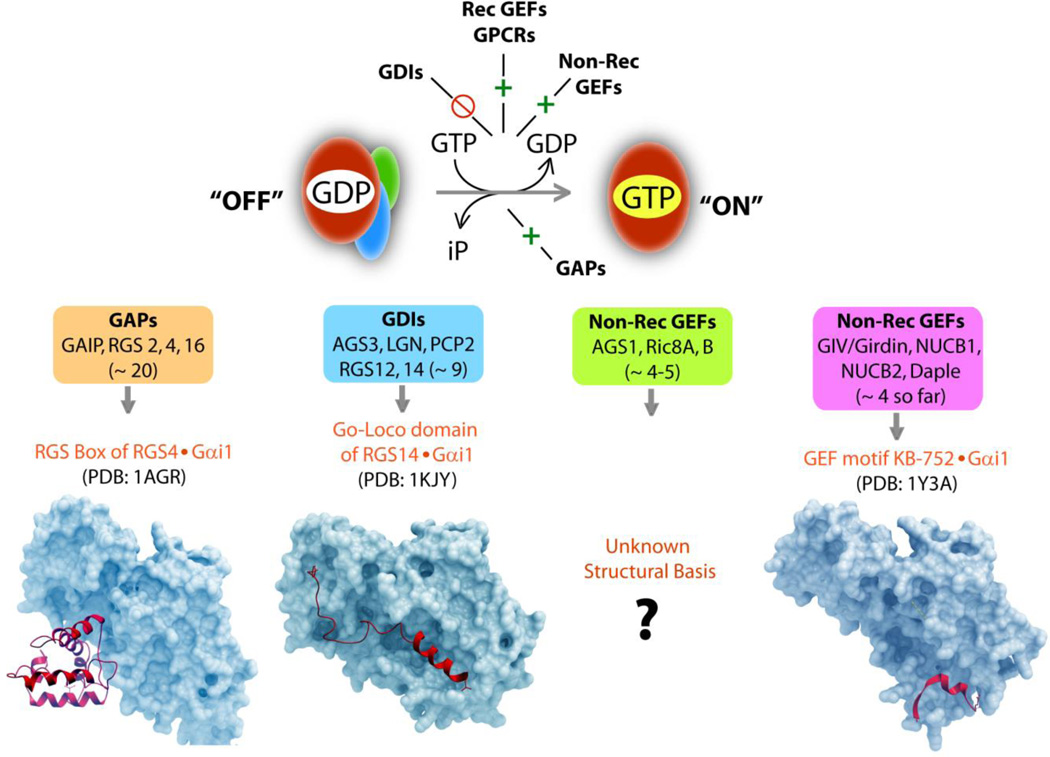
Figure 2. Network of regulatory proteins coordinately function to maintain finiteness of G protein signaling.
Upper panel: Schematic of the G protein cycle is shown. Gα-subunits in GDP-bound "OFF" bind βγ-heterodimers and exist as inactive trimers, until nucleotide exchange and cycle of activation is triggered (green  ) by chance encounter with GEFs, which could be either GPCRs, i.e., receptor GEFs or non-receptor GEFs (such as GIV/Girdin). Such nucleotide exchange is inhibited (red
) by chance encounter with GEFs, which could be either GPCRs, i.e., receptor GEFs or non-receptor GEFs (such as GIV/Girdin). Such nucleotide exchange is inhibited (red  ) by GDIs. Once GTP-bound, Gα is active, dissociates from Gβγ-dimers until the GTP is hydrolyzed, inorganic phosphate (iP) is released and Gα returns to inactive state. This step of GTP hydrolysis is sped up by GAPs. Lower panel: The structural modules (red ribbons) which impart enzymatic properties to GAPs, GDIs and GEFs is shown in complex with Gα-subunit (in blue): (from left to right) GAPs accelerate GTP hydrolysis via a RGS box domain, as shown here in the case of RGS4 bound to an active conformation of Gαi1. GDIs inhibit nucleotide exchange via a Go-Loco/GPR domain, as shown here in the case of RGS14 bound to an inactive conformation of Gαi1. In the case of non-receptor GEFs, while some work via unknown module(s), a newly emerging subfamily uses a short stretch of amino acids to trigger nucleotide exchange using similar structural basis as shown here in the case of KB-752 synthetic peptide bound to an inactive conformation of Gαi1.
) by GDIs. Once GTP-bound, Gα is active, dissociates from Gβγ-dimers until the GTP is hydrolyzed, inorganic phosphate (iP) is released and Gα returns to inactive state. This step of GTP hydrolysis is sped up by GAPs. Lower panel: The structural modules (red ribbons) which impart enzymatic properties to GAPs, GDIs and GEFs is shown in complex with Gα-subunit (in blue): (from left to right) GAPs accelerate GTP hydrolysis via a RGS box domain, as shown here in the case of RGS4 bound to an active conformation of Gαi1. GDIs inhibit nucleotide exchange via a Go-Loco/GPR domain, as shown here in the case of RGS14 bound to an inactive conformation of Gαi1. In the case of non-receptor GEFs, while some work via unknown module(s), a newly emerging subfamily uses a short stretch of amino acids to trigger nucleotide exchange using similar structural basis as shown here in the case of KB-752 synthetic peptide bound to an inactive conformation of Gαi1.
Although the discovery of GoLoco/ GPR or RGS domains propelled the biological characterization of GAPs and GDIs, and paved the way for rationale designing of therapeutics[33,34], the lack of similar structurally well-defined motifs or domains pose a serious limitation to unlocking the biological relevance of the group of accessory proteins called non-receptor GEFs. Non-receptor GEFs represent a heterogeneous group of proteins, such as AGS1 [35], Ric-8A [36], Ric-8B [37], Arr4 [38] or CSPα [39], and some others [22] which serve as GEFs for different Gα subunits. The lack of understanding what motif/domain imparts the non-receptor GEFs their intrinsic enzymatic activity had serious consequences; rationale designing of selective GEF-deficient mutants or tools could not happen, and therefore, selective interrogation of the GEF function without altering the functions of other domains/modules remained impossible [38,40,41]. Consequently, an in-depth understanding of their biological roles or spatiotemporal features of non-canonical, (i.e., GPCR-independent) activation of G proteins is lacking in the case of most members of this family. Regardless, what is clear is that canonical activation of G proteins by GPCRs is tightly regulated by the network of accessory modulators [23]. These accessory modulators, several kinases and phosphatases, and adaptors that mediate clathrin-mediated endocytosis function coordinately to maintain finiteness of signal transduction via G proteins[20] by ensuring that their activation is spatially and temporally restricted, i.e., triggered exclusively at the plasma membrane (PM) by agonist activation of GPCRs via a process that is terminated within a few hundred milliseconds [42] (Figure 1).
Breaking the rules of engagement
The discovery of the first well-defined GEF motif in Gα-Interacting Vesicle associated protein (GIV; a.k.a Girdin)[10] has provided a unique opportunity to understand non-receptor GEFs in a way that has never been possible. GIV is a multi-modular signal transducer and a non-receptor GEF for Gαi[10]. Unlike the canonical GPCR/G protein pathway, in which the G proteins engage exclusively with ligand-activated GPCR-GEFs, GIV, on the other hand, either enhances, or suppresses a diverse range of signaling pathways by engaging G proteins with an equally diverse variety of receptors (Figure 3), all via its ability to bind and activate Gαi in the close proximity of these ligand-activated receptors. Multiple studies [summarized in[12,43]] employing a selective GEF-deficient GIV mutant (F1685A) have demonstrated that the signaling network triggered in cells with wild-type GIV is a mirror image of the network in cells expressing a GEF-deficient mutant GIV; signals that are enhanced in cells that are GEF-proficient are suppressed in cells that are GEF-deficient, and vice versa. It is because cells can alter (increase or decrease) the levels of GIV mRNA/protein or selectively modulate GIV's GEF activity to modulate growth factor signaling pathways across a range of intensities [3], we likened GIV to a cellular “rheostat” for signal transduction [2]. Consistent with its ability to integrate signals downstream of multiple receptors, GIV modulates diverse cellular processes (Table 1), and GIV-dependent signaling has been implicated in a number of pathophysiologic conditions (Table 2).
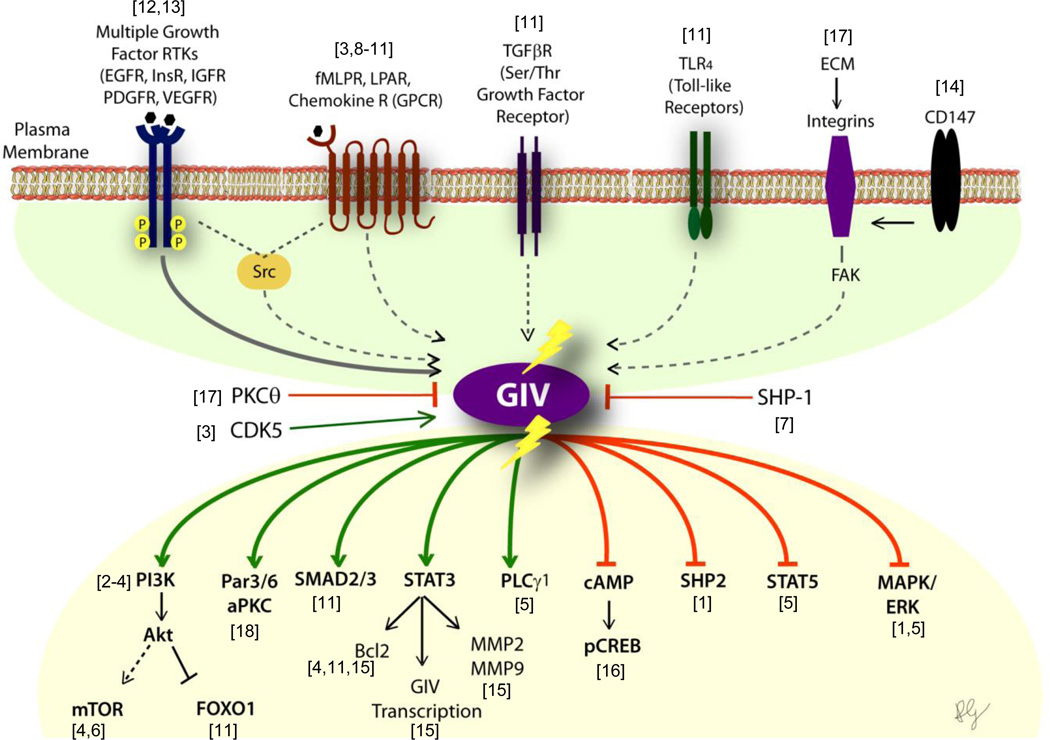
Figure 3. Activation of G proteins by GIV-GEF modulates multi-receptor signaling and broadly impacts diverse pathways within the post-receptor signaling network.
Upper parts of the schematic shows multiple classes of receptors, some that sense a variety of chemical stimuli and others that sense mechanical signals, all of which utilize GIV-GEF to transactivate G proteins. The solid arrow connecting RTKs and GIV indicates that the mechanism for such engagement is well understood. The broken arrows linking all other receptors and GIV indicate that little or nothing is known as to how GIV may engage with those receptors. Lower part of the schematic shows the consequence of such activation (when GIV-GEF is turned "ON") on diverse signaling pathway within the signaling network. Green = enhancement; Red = suppression. This profile of signaling is reversed when GIV-GEF is switched "OFF", i.e., enhanced signals are suppressed and vice versa. Shown in the middle are three known ways to inhibit GIV-dependent signaling (PKCθ selectively phosphoinhibits GIV-GEF[3]; SHP-1 dephosphorylates tyrosine-phosphorylated GIV [7]) or activate (CDK5 phosphoactivates GIV-GEF [17]).
Table 1.
GIV is essential for a variety of physiologic cellular processes.
| Cellular Processes | Effect of GIV's GEF function | Receptor(s) studied | Citation |
|---|---|---|---|
| Cell Migration | “ON” = Enhances; “OFF” = Inhibits | EGFR, PDGFR, VEGFR, IGF1R, LPAR1 |
[2–5,8–10,44] |
|
Golgi Structure, Secretory Function |
“ON” = Preserves/enhances; “OFF” = Disrupts, delays |
n/a | [70] |
| Autophagy | “ON” = Halt/rescue; “OFF” = Initiate/promote |
InsR | [6] |
| Endosome Maturation | “ON” = Rapid; “OFF” = Slowed | EGFR | [87] |
| Cell Survival | “ON” = Survive; “OFF” = Apoptosis | PDGFR, VEGFR, EGFR | [4,11] |
| Cell polarity*** | “ON” = Polarity achieved; “OFF” = Loss of polarity |
Serum (multiple receptors) | [18,88] |
| Cell Division | Not examined | n/a | [89] |
| Endocytosis | Not examined | EGFR, TfR | [90] |
|
Cell-cell junctions, permeability*** |
Not examined | VEGFR | [91] |
|
Neuron migration, differentiation*** |
Not examined | n/a | [92,93] |
| Macrophage chemotaxis | Not examined | fMLPR | [8] |
| Growth, Cell Size*** | Not examined | n/a | [94] |
= In vivo evidence is available.
Table 2.
Modulation of G protein signaling by GIV affects diverse pathophysiologic states.
| Disease/Pathology Investigated |
Effect of GIV's GEF function |
Receptor(s) Studied |
Citation | |
|---|---|---|---|---|
|
Cancer Progression |
Migration/Invasion | “ON” = Enhances “OFF” = Inhibits |
IGF1R, EGFR, β1 integrin, Multi-receptor* |
[15– 17,95,96] |
| Stemness | Not examined | -- | [97] | |
| Chemoresistance | Not examined | -- | [98] | |
| Tumor-Stroma Interactions | Not examined | PDGFR, TGFβR, CXCR4 |
[99] | |
| Tumor angiogenesis | Not examined | VEGFR | [100] | |
|
Organ Fibrosis (Liver) |
Myofibroblast transdifferentiation, collagen production, chemotaxis, mitosis, anti-apoptotic signaling |
“ON” = Enhances “OFF” = Inhibits |
PDGFR, CCR1, TGFβR | [11] |
|
Dermal Wound Healing |
Wound closure | “ON” = Enhances “OFF” = Inhibits |
Multi-receptor* | [17] |
|
Nephrotic Syndrome |
Podocyte survival after glomerular injury |
“ON” = Enhances survival “OFF” = Inhibits survival |
VEGFR | [4] |
|
Insulin Resistance, Type II Diabetes |
Metabolic insulin response in the skeletal muscle |
“ON” = Enhances “OFF” = Inhibits |
InsR | [101,102] |
|
Disorders of Blood Vessels |
Neonatal vascular development; Pathologic neovascularization; vein repair; vein graft |
Not examined | PDGF, Angiotensin II, VEGF |
[103–106] |
|
Neuronal Plasticity, Memory formation |
Synaptic plasticity | Not examined | NMDA | [107] |
The molecular mechanisms that govern how GIV influences a diverse range of pathophysiologic processes and how it may couple activation of G protein to multiple receptors have only recently come to light [44], and best understood in the context of a numerous RTKs that signal via GIV (Figure 4). RTKs, much like GPCR/G proteins are a major signaling hub in eukaryotes. For several decades the RTK and GPCR/G protein pathways were believed to operate in a discrete mode by transducing signals through their respective downstream intermediates; upon ligand stimulation RTKs propagate the signals to the interior of the cell via adaptor proteins that are recruited to phosphotyrosines on the receptor tail [45], whereas GPCRs recruit and activate G proteins by triggering the exchange of GDP with GTP nucleotide [19]. However, mounting evidence over time has unfolded a complex array of cross-talk between these two pathways-- such that activated receptors from one pathway transactivate the other pathway either by directly activating the receptors [46] or by indirectly activating the downstream adaptor proteins [47]. For example, transactivation of RTKs by GPCRs via scaffolding proteins such as β-arrestins [48] is a well-documented and widely-accepted phenomenon. However, the reverse concept, i.e., transactivation of trimeric G proteins by RTKs remains controversial. Despite numerous clues that support the concept that growth factors trigger activation of heterotrimeric G proteins [49], the fundamental question as to how that occurs in cells remained poorly understood and the concept itself was met with skepticism. Why? Largely because there was no evidence that G proteins and ligand-activated RTKs come within close proximity in cells, nor that RTKs, or any member of the growing family of signal transducing adaptors used by RTKs can serve as GEFs.
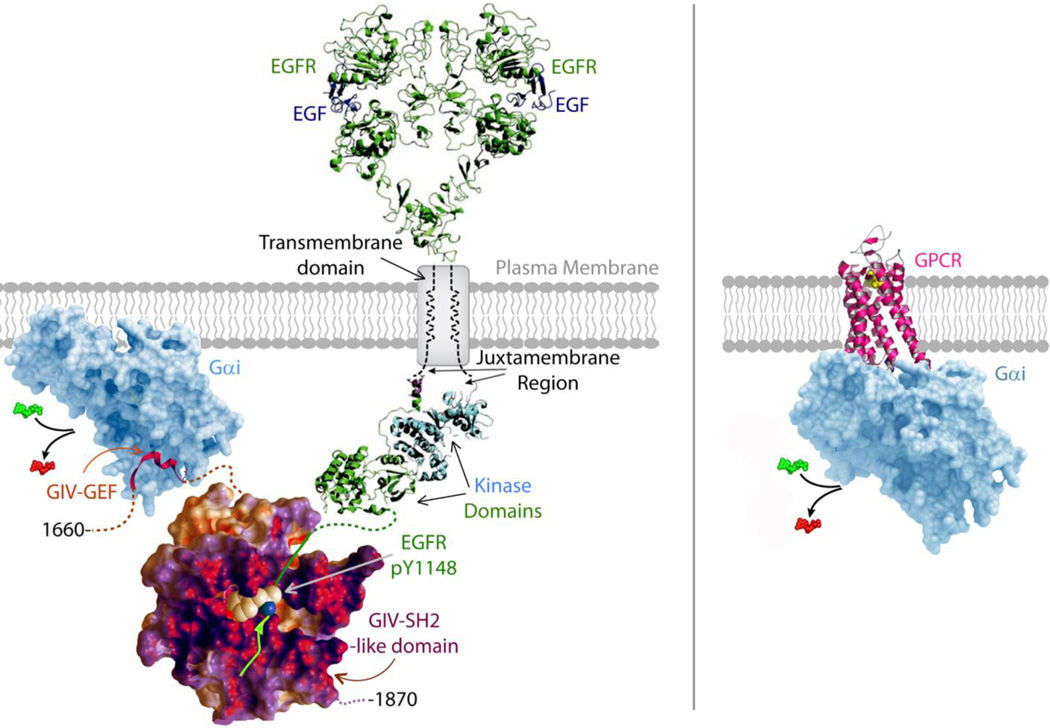
Figure 4. GIV plus RTKs, equals GPCRs.
Schematic on the left shows the modular makeup of G-protein coupled growth factor RTKs. Ligand stimulation of growth factor receptor tyrosine kinases, e.g., EGFR (shown here) leads to receptor dimerization and autophosphorylation of its cytoplasmic tail (green interrupted line). Within ~3–5 minutes after growth factor stimulation, a ~110 aa long intrinsically disordered stretch within GIV's C-terminus (aa 1660–1870) recognizes and folds into a SH2-like module (red-purple-white) and directly docks onto the phosphotyrosine ligand (pTyr1148) presented by the cytoplasmic tail of ligand-activated RTKs. The mechanism of phosphotyrosine recognition and the kinetics of recruitment of GIV's SH2-like module to the RTK tail mimics that of other SH2 adaptors, e.g., Grb2. Just upstream and adjacent to the SH2-like module is GIV's GEF module (red ribbon) which binds and triggers nucleotide exchange (GTP, green for GDP, red), thereby activating Gαi subunits (light blue) in the vicinity of ligand-activated RTKs. It is the unique coexistence of two (GEF and SH2-like) modules that is key, because their collaboration is necessary and sufficient to assemble RTK-GIV-Gαi complexes at the PM within 5 minutes after ligand stimulation. One of the major consequences of the assembly of such complexes is transactivation of Gi and suppression of cellular cAMP after growth factor stimulation. In doing so, GIV's C-terminus enables the assembly of G-protein coupled RTKs, which subsequently leads to the non-canonical transactivation of G proteins. Schematic on the right shown canonical activation of G protein by ligand-activated receptor GEFs, i.e., GPCRs which can directly trigger nucleotide exchange.
A lot of these unanswered questions got clarified by the discovery and characterization of the unique modular make-up of GIV, which allows it to bind both ligand-activated RTKs and G proteins with high degrees of specificity and affinity (Figure 4). More specifically, GIV-dependent growth factor signaling relies heavily on the multi-modular nature of its C-terminus (CT), within which two unlikely domains coexist-- 1) a previously defined GEF motif via which GIV binds and activates Gi [10] and 2) adjacent to this GEF motif, a newly defined ~110 aa stretch which folds into a SH2-like domain in the presence of phosphotyrosine ligands; the latter is necessary and sufficient to recognize and bind specific sites of autophosphorylation on the receptor tail [5,50]. There are several unique features of GIV's SH2-like module. No conventional programs predict its existence, mostly because GIV's SH2-like module, unlike the remaining ~140 or more SH2 domains, is intrinsically disordered or partially structured at resting state. By that token, GIV joined the rank of numerous examples of eukaryotic proteins that are intrinsically disordered or partially structured under physiological conditions and fold into functional modules, especially in the context of signal transduction [51–54]; in many of these cases, binding and folding are coupled [55]. Much like those intrinsically disordered proteins, GIV’s SH2-like folded structure is induced upon binding to its biological target, that is, activated RTKs. Upon encountering phosphotyrosine ligands, e.g., autophosphorylated tyrosines on cytoplasmic tails of ligand-activated RTKs, the ~110 aa stretch within GIV's CT stably folds into a functional SH2-like module to assemble the RTK•GIV signaling interface. GIV's CT serves as a platform that links RTKs to G proteins within RTK-GIV-Gαi ternary complexes only when both its GEF and SH2-like modules are intact[1]. In the absence of either of these modules, ligand-activated RTKs and Gαi are uncoupled, and the recruitment of Gαi to RTKs and subsequent activation of G proteins is impaired[1].
These findings have fundamentally enriched our knowledge of GIV's unique C-terminal stretch and provide many clues into what might be the molecular mechanism(s) behind GIV's ability to engage, directly or indirectly, with multiple upstream receptors/pathways (Figure 3). The fact that GIV-CT has two-states, one that is intrinsically disordered and another that is folded, perhaps provide the biggest clue. Intrinsically disordered proteins, while structurally poor, are functionally rich by virtue of the flexibility of their modular structures, as recently described in the case of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) [56–58]. Post-translational modifications, burial or exposure of conserved linear motifs and molecular recognition features present in the CT of PTEN during folded- vs disordered states directly regulate PTEN's interactions with other proteins, which are required for executing diverse cellular functions. It has also been shown that PTEN's disordered and folded-state interactomes are further enriched in proteins that are intrinsically disordered, revealing how PTEN functions are regulated by the disordered CT to nucleate flexible network-hubs and orchestrate 'on-demand' modes of signaling. Based on the diversity of pathways and receptors that GIV modulates, we suspect that the evolution of structural plasticity in the intrinsically disordered GIV-CT is likely to assemble distinct interactomes in the disordered versus SH2-folded states, thereby contributing to functional enrichment. Because GIV’s C-terminus is enriched in Ser/Thr residues, some of which are heavily phosphorylated (www.phosphosite.org), it is possible that additional phosphoevents on GIV’s C-terminus trigger and/or regulate folding of GIV’s SH2-like domain and binding to RTKs. Further biochemical, structural and biophysical studies are essential to understand how plasticity of the GIV-CT influences when and where it may engage receptors/proteins within signaling pathways, depending on whether it is in folded-vs disordered state, to nucleate distinct signaling hubs.
Breaking the rules of time
Although the discovery of coexisting SH2-like and GEF modules in-tandem within GIV-CT supported the idea that GIV's C-terminus may be sufficient to link multiple RTKs to G proteins, it was not until recently, when FRET studies using genetically encoded fluorescent proteins unraveled the unique temporal features of non-canonical G protein signaling via GIV-GEF (Table 3). FRET is a principal method of choice to study dynamic protein-protein interactions because it extends the resolution limitation (~250 nm) of confocal microscopy to ~10 nm and serves as a widely accepted tool for estimating the proximity of macromolecules in living cells[59]. First, biochemical and FRET studies using GIV-derived biosensors confirmed that the evolutionarily conserved C-terminus of GIV indeed represents the smallest, functionally autonomous unit that retains most key signaling properties of full length GIV [16] : 1) They retain the properties of ligand-dependent receptor recruitment. FRET and coimmunoprecipitation studies were in agreement that the timing of recruitment of GIV to EGFR or insulin receptor (InsR) at the PM is ~3–5 min, which resembles the kinetics demonstrated in the case of multiple other SH2 adaptor proteins, e.g., Grb2[60]. Additionally, bimolecular fluorescent complementation (BiFC) studies using SH2-deficient mutants showed that the GIV-biosensors require a functionally intact SH2-like domain for recruitment to ligand-activated RTKs[50]. 2) They can bind Gαi, assemble GIV-Gαi complexes at the PM in a GEF dependent manner within ~5 to ~15 min, and undergo disassembly by ~ 30 min.: FRET and GST-pulldown assays were in agreement that although some GIV:Gαi complexes are pre-formed (i.e., in serum starved cells), a large fraction of them assemble exclusively after growth factor stimulation, only to disassemble later. A recently concluded work[17] has now revealed the molecular basis for such enhanced association after growth factor stimulation. Turns out, targeting of GIV to the receptor at the PM is insufficient for maximal activation of the GIV-Gαi pathway. Instead, an additional step of phosphoactivation of GIV-GEF (at Ser 1674) by CDK5 is essential for GIV to initiate non-canonical G protein signaling downstream of growth factor RTKs. Because CDK5 is activated within seconds after growth factor stimulation[61], it is likely that once activated, CDK5 can promptly phosphoactivate GIV-GEF before or during the latter’s recruitment to the activated receptor at the PM, ensuring subsequent maximal coupling to and activation of Gαi. As for the mechanism of disassembly of most of the GIV-Gαi complexes at ~30 min after ligand stimulation, it is likely to be brought on by a negative feedback loop initiated by kinases like PKCθ which phosphoinhibits GIV's GEF motif (at Ser1689) and selectively terminates GIV's ability to bind or activate Gαi[3]. It is possible that kinases other than PKCθ can also accomplish this goal. 3) The combined synergy of BiFC and FRET studies further confirmed that GIV-CT probes serve as bona fide platforms for the assembly of RTK-GIV-Gαi complexes at the PM in response to growth factors. The complexes are not assembled if GIV and Gαi cannot bind each other. 4) They retain the signaling properties and fulfill the phenotypic functions characteristic of full length GIV, i.e., enhances the PI3K-Akt pathway and triggers cell migration/invasion through basement membrane matrix. Thus, comprised of the essential modules (GEF and SH2-like domains), GIV's CT is necessary and sufficient for linking G proteins to RTKs in the vicinity of ligand-activated RTKs (see legend for Figure 4).
Table 3.
Temporal dynamics of signaling via GPCTKs revealed by FRET-based assays in live cells.
| Major Steps in GPCTK Signaling | Kinetics | Reference |
|---|---|---|
| Dimerization of ligand-activated RTKs | ~15 sec | [108] |
| Lateral mobility of ligand-activated RTKs | 0.17 µm2/s for monomers; 0.08 µm2/s for dimers | [109] |
| Recruitment of GIV and other SH2-adaptors | ~1–6 min | [16,60,110] |
| RTK-G protein interaction | 5 min | [16,85,111] |
| Transactivation of G proteins by growth factors | 5 min | [16,85] |
| Growth factors induced cAMP changes | ~ 6–10 min | [16,85] |
Second, use of fluorescent G proteins [Gαi3-YFP (internal tag), CFP-Gβ1 (N-terminal tag) and Gγ2] in FRET assays, as originally developed and validated by the groups led by Gilman and Bunemann[62,63] and extensively used thereafter to study the canonical GPCR/G protein pathway by others[64,65] accomplished three goals: 1) They leveled the ground and allowed, for the first time, a head-to-head comparison of non-canonical G protein signaling with the canonical pathway; 2) They revealed the unusual temporal dynamics of transactivation of G proteins by growth factors; and 3) They confirmed the obligate need for GIV to trigger such transactivation. Such comparative analysis revealed that although the extent of Gi activation downstream of RTKs (EGFR; current work) and GPCRs (α2 AR; [62]) appear similar, the temporal dynamics of non-canonical G protein activation by GIV represent a clear deviation from the dynamics of canonical G protein signaling that is triggered by GPCRs (Tables 3 and 4). For example, multiple FRET- and BRET-based studies have revealed that canonical signal transduction via trimeric G proteins is spatially and temporally restricted [42,66]. Agonist activation of GPCRs trigger activation of G proteins exclusively at the PM, that activation completes within a few hundred milliseconds [67] (summarized in Figure 1, Table 4), and that such finiteness is imposed upon by a network of regulatory proteins, and more definitively via clathrin-mediated endocytosis of the ligand-activated GPCRs; the latter effectively sequesters the receptor from its agonists/ligands in the extracellular space. By contrast, transactivation of Gi and dissociation of the trimer in response to growth factors starts at ~5 min and lasts several minutes (Table 3, 4), and depletion of GIV abrogates such growth factor-responsive signaling. Consistent with the contrasting temporal patterns of canonical vs non-canonical Gi activation, suppression of cAMP by Gi-coupled GPCRs within the canonical pathway occurs rapidly (i.e., within seconds) [68], but non-canonical transactivation of Gi by RTKs leads to a delayed suppression of cAMP (i.e., several minutes). The fundamental molecular basis that governs such delayed activation of Gi and suppression of cAMP is the dynamics of binding of GIV's SH2-like domain to ligand-activated RTKs; the latter is a prerequisite for facilitating the proximity between G proteins and RTKs (Figure 3). These findings challenge a fundamental long-held tenet in canonical G protein signaling, i.e., activation of G proteins is triggered exclusively by GPCRs, and that RTKs do not have the wherewithal to trigger such activation. These studies also helped nucleate a new paradigm, in which RTKs access and activate G proteins in living cells, utilizing GIV as a platform for cross-talk. Further studies using a combination of, but not limited to, mathematical modeling, conformational antibodies (nanobodies), FRET and BRET studies are essential to dissect the unique temporal aspects of GIV-dependent G protein signaling.
Table 4.
Striking differences between canonical (GPCR-triggered) and RTK-triggered G protein activation via GIV-GEF
| Canonical G protein Signaling by G protein coupled Receptor GEFs (GPCRs) |
Non-Canonical G Protein Signaling by RTKs via GIV-GEF (GPCTK pathway) |
|
|---|---|---|
|
Input Signal |
Exclusivity of GPCRs (Receptor GEFs) | Diverse classes of receptors that engage in tyrosine-based signaling converge on GIV to activate G proteins [13,43,112] |
|
Temporal Dynamics |
Finite (activation of G proteins by 300–500 ms; receptor endocytosis by minutes) [42] |
Extended periods of time (several min) [16,85] |
|
Spatial Dynamics |
Primarily at the PM; to some extent on early signaling endosomes during endocytosis [69] |
Internal membranes noncontiguous with the PM, e.g., autophagosomes and the Golgi membranes [6,70] |
Breaking the rules of space
The spatial pattern of non-canonical G protein activation by GIV also provide a stark contrast with that of canonical signaling (Table 4). Canonical G protein activation is largely limited to the PM. Although recent studies using nanobodies have revealed that some signaling continues also within endosomes[69], to date, no such activation on internal membranes that are discontinuous with the PM has ever been observed. By contrast, GIV-dependent signaling has been described at multiple intracellular compartments (summarized in Table 1), including autophagosomes[6], and more recently, on Golgi membranes[70]. The specific role of activation of G proteins by GIV-GEF has been investigated in the context of autophagy[6], secretory functions of the Golgi[70], and during the establishment of cell polarity[18], and these 3 studies have accomplished 3 key goals: 1) they prove that G proteins are active at internal locations; 2) that such activation can be brought on by cytosolic non-receptor GEF, GIV; and finally, 3) they provide valuable clues into how the same GEF, i.e., GIV may coordinate G protein signaling at the PM and on internal membranes.
In the context of autophagy, it had been known for decades that constitutively active mutants of Gαi3 inhibit whereas inactive mutants promote autophagy[71,72]. However, who activates Gαi3 on autophagosomes and how such activation may be linked to the availability of nutrients and growth factors was unknown. We showed that GIV rescues/reverses autophagy via activation of Gαi at the PM and on autophagosomes in response to growth factors [6]. Poised at the cross-roads of G protein and growth factor signaling pathways, GIV enhances the metabolic insulin signaling at the PM and alters autophagosome-resident G protein complexes, primarily by binding and activating Gαi at both sites. On the autophagosome membranes, the key mechanism is reversible regulation of Gαi3 by GIV and AGS3 in response to growth factors-- Upon starvation, Gαi3 localizes to the autophagosomes and preferentially interacts with its inhibitor, AGS3, whereas upon growth factor stimulation Gαi3 preferentially interacts with its activator, GIV, which activates and releases Gαi3 from the autophagosome and redistributes it to the PM. This shift from AGS3-Gi to GIV-Gi complexes occurs exclusively in the presence of a functional GEF motif in GIV, and is mediated by the competitive binding of GEF and the GDI to an overlapping site on Gαi. The yin-yang effect of GIV and AGS3 on Gαi3 correlates with their respective roles in reversibly inhibiting and promoting autophagy [71,72]. These findings not only defined a new paradigm in yin-yang reversible regulation of Gα activity by paired modulators, AGS3 and GIV, but also pinpointed the hitherto elusive link between growth factors, the G protein, modulators of G protein and the autophagosome-resident molecular machinery.
As for the Golgi, heterotrimeric G proteins were detected at this location over two decades ago [73,74], and numerous studies have provided clues that they may regulate membrane traffic and maintain the structural integrity of the Golgi [75]. However, the concept of G protein activation at the Golgi and the potential impact of such activation were met with skepticism, primarily due to the lack of direct proof of G protein activation. Some argued that G proteins shuttle between the PM and Golgi membranes [76] only to acquire palmitoylation by Golgi-resident palmitoyl transferases [77]. Others claimed that KDEL receptors are predicted to fold like GPCRs and couple to Gαq/11 [78] but did not provide direct evidence or mechanism for activation of Gα subunits. Some evidence existed for the role of 'free' Gβγ in regulation of trans-Golgi network (TGN)-to-PM trafficking via its effector, PKD [79]. Using a combination of FRET imaging (Figure 5A), conformational antibodies and specific mutant GIV and/or G proteins (e.g., GEF-deficient GIV and GIV-insensitive G protein), we have recently defined GIV as a bona fide activator of the trimeric G protein Gαi at the Golgi and an effector of active Arf1[70]. Activation of Gi by GIV serves the fundamental role of ensuring finiteness of Arf1 signaling at the Golgi[70]. By virtue of its ability to modulate Arf1 signaling, GIV regulates both structure and function of the Golgi, two closely intertwined processes regulated by Arf1. Our results provide mechanistic insights into how GIV orchestrates a two-pronged mechanism to suppress Arf1 signaling when COPI vesicles carrying cargo proteins from the ER/ERGIC dock on the Golgi membranes: First, activation of Gαi by GIV at the Golgi complex releases 'free' Gβγ which in turn inhibits Arf1 signaling. Second, GIV interacts with both β-COP and ArfGAP2/3 and facilitates the recruitment of the coat-dependent GAP protein onto the Golgi and COPI vesicles. Once recruited, the catalytic activity of ArfGAP2/3 is enhanced, presumably at/near the target Golgi membranes, and consequently Arf1 signaling is efficiently terminated and COPI vesicles uncoat prior to fusion. The first event, i.e., activation of Gi by GIV and release of Gβγ (GIV→Gβγ pathway) on Golgi membranes, is a hierarchically dominant step because without a functional GEF motif in GIV, ArfGAP3 localized to Golgi/COPI vesicles normally, but Arf1 activity remained elevated, vesicle uncoating was impaired, trafficking was delayed and Golgi ribbons were dispersed. Activation of Gi and release of 'free' Gβγ dimers is also a central mechanism by which GIV terminates Arf1 at/near acceptor (i.e., Golgi) membranes, and is consistent with prior work demonstrating the inhibitory role of Gβγ in Arf signaling [80,81]. Thus, this work elucidated some of the well-known but poorly understood functions of G proteins on the Golgi and illuminated the role of trimeric G protein signaling in the secretory pathway. Because G proteins and the other essential components (Arf1, ArfGAP3 and GIV) are also present on other membranes, it is possible that the fundamental mechanisms we define here also facilitate vesicular trafficking from the TGN to the PM and/or endolysosomal system, as shown in the case of ArfGAP3 [82].
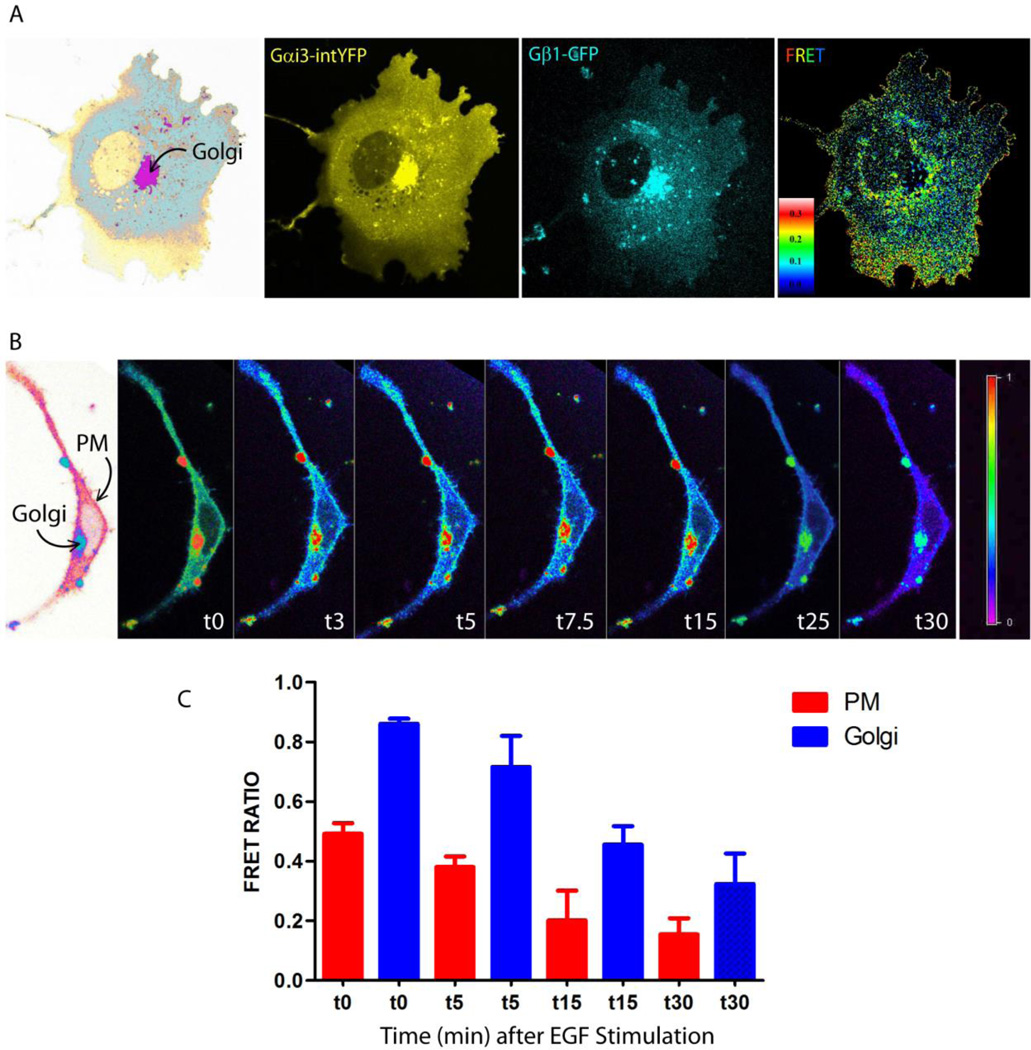
Figure 5. G proteins are active at the Golgi and on other internal membranes; such activation can be triggered in response to growth factors.
A. Cos7 cell co-expressing Gαi1-YFP (internal tag) and CFP-Gβ1 and Gγ2 (untagged) were analyzed at steady-state by FRET imaging. Individual YFP and CFP channels show concentrated localization of G proteins on a perinuclear compartment, confirmed to be Golgi in a prior study [70]. FRET scale is shown as an inset. FRET is observed at the PM (yellow pixels), indicative of the formation of inactive trimers. By contrast, at steady-state, little or no FRET was seen at the Golgi, indicative of dissociated trimers. Activation of G protein and dissociation of trimers at the Golgi is abolished in GIV-depleted cells [70]. B–C. HeLa cell coexpressing Gαi1-YFP (internal tag) and CFP-Gβ1 and Gγ2 (untagged) as in A, were serum starved in 0.2% FBS and then stimulated with 50 nM EGF and analyzed by FRET imaging for dissociation of Gαβγ trimers. Briefly, ratiometric FRET imaging (FRET/CFP) was carried out using a Nikon Ti Eclipse which allows for perfect focus that locks the field and plane of image designed for long term live cell imaging. To reduce bleaching, only 5% of 405 laser was used. Upon growth factor stimulation, FRET is diminished both at the PM and at the Golgi. Bar graphs in C display the quantification of change in FRET ratio (as determined using 5–8 ROIs / cell for the PM and 1–2 ROIs for the Golgi region), in 3 cells, during 3 independent experiments.
In the context of epithelial cell polarity, it has been known for some time that the formation of a complex among atypical protein kinase C (aPKC), PAR-3 and PAR-6 is critical. Once again, using specific mutant GIV and/or G proteins (e.g., GEF-deficient GIV and GIV-insensitive G protein) Ohno and colleagues recently elucidated the links between PAR–aPKC complexes and GIV-dependent G protein signaling[18] and firmed the vital role of GIV-GEF in establishing cell polarity. In the normal polarized epithelium, the PAR-3–aPKC complex is required for GIV-GEF transcription and maintenance of GIV protein levels via an AP-2α-dependent pathway. Additionally, Par3 also physically interacts with GIV, and GIV-dependent activation of Gαi3 is critical for the formation of tight junction formation, development of the apical domain and actin organization. Depletion of GIV disrupts actin organisation, delays in the formation of tight junctions and generates an aberrant number of vacuolar apical compartments, indicating a problem with the formation of apical domains. When GIV or Gαi3 cannot bind each other, and GIV-dependent activation of Gi is abolished, severely deformed cysts are formed in 3D cyst morphogenesis assays, the latter is a widely used method to study cell polarity.
It is noteworthy that while all three aforementioned studies investigated the fundamental mechanisms of how GIV-GEF may affect G protein activity on specific intracellular locations, they revealed the complexity and variation of the interactome that allows GIV-GEF to function at the given location. For example, GIV straddles both PAR-3 and G proteins signaling during the establishment of cell polarity, somewhat analogous to the way it straddles active Arf1 and G protein signaling on Golgi membranes, or AGS3/G proteins and LC3/Atg8 on autophagosomes. Because GIV serves many other roles at a variety of other intracellular sites (Table 1), it is tempting to speculate that those roles are, at least in part, regulated by GIV's ability to activate trimeric G proteins also at those locations and will involve novel protein-protein interactions unique to those locations. Finally, our understanding of how GIV-GEF localizes to various places within a cell and coordinately activate G proteins at the PM as well as on internal membranes is lacking. In the case of GIV's role during autophagy, we speculated that signaling programs that are initiated by growth factor receptors could trigger post-translational modifications such as phosphorylation and/or altered localization of Gαi3 and/or GIV, and thereby shift the composition of Gαi3-bound complexes on the autophagosome membrane. Because GIV’s GEF function modulates growth factor signaling at the PM, it is possible that one or more of those signaling intermediates/pathways (e.g., calcium, cAMP, kinases, phosphatases) establish rapid loops of feed-back regulation which impacts GIV's ability to bind and activate Gi on internal membranes. Similarly, in the case of the Golgi, it is possible that the previously observed cross-talk between growth factor signals initiated at the PM and functions of the Golgi [83,84] may be orchestrated in part via the GIV platform. Such cross-talk may coordinately trigger secretion in response to growth factors when the GIV-GEF is activated and/or coordinate the dispersal of Golgi in response to mitogenic signals when the GIV-GEF is disabled. Consistent with that notion, we find that growth factors can indeed trigger the activation of G proteins at the PM as well as on internal membranes, with variable kinetics (Figure 5B, C). It is possible that the cross-talk GIV sets up between trimeric G proteins at two locations within the cell may represent an evolutionary advantage that allows for regulation of Golgi (or for that matter any other internal location) functions by external environmental cues such as growth factors. What is clear is that meaningful G protein activation on internal membranes is achieved by GIV, and so far the evidence supports the notion that such non-canonical activation is essential for making internal organelles responsive to growth factors. Further studies are underway to dissect how signals triggered by the RTK-GIV-Gi axis at the PM travel in time and space to coordinately trigger the GIV-Gi axis on the internal membranes.
Therapeutic potential of non-canonical G protein signaling
With the emergence of a new paradigm that broadly impacts a variety of disease states (summarized in Table 2), the next hurdle is to devise a way to target it. The canonical GPCR/G protein pathway has long been the target of small molecule therapeutics accounting for 30–40% of the launched drug targets. But the unusual spatiotemporal features of GPCTK signaling (Tables 3, 4) poses a unique set of advantages as well as challenges. For example, it is far more complex to target or exogenously modulate a pathway that appears to be ubiquitously expressed, serves as a point of convergence downstream of multiple receptors [13], performs a broad array of physiologic functions, and is frequently deregulated in multiple pathologic states (Tables 1, 2). Other challenges include the absence of high-resolution structures; in the absence of such structural information, computational modeling has provided some clues into the 'druggability' of the GIV•Gαi interface. Computational homology modeling approaches have been used to model the GEF motif sequence bound to Gαi3 and such model has been validated experimentally[9,10]. The model also revealed that GIV’s binding site on Gαi does not overlap with the binding site of GPCRs, raising hopes that GIV's interface with G protein may be selectively targetable without affecting their canonical activation by GPCRs. The GIV•Gαi3 interface has also been extensively characterized using biochemistry and enzymology to characterize multiple G protein mutants[9,10], all supportive of the notion that it is theoretically possible to selectively abolish GIV binding. Based on the broad range of receptor-initiated signals that converge on GIV and the variety of signaling pathways within 'disease networks' that are modulated via GIV's GEF function (Figure 3), it is predicted that disrupting the GIV•Gαi interface will be effective and specific for inhibiting non-canonical G protein signaling that is initiated by multiple receptors via GIV-GEF. Thus, one major advantage of targeting the GIV•Gαi interface is that such approach circumvents the need to target individual receptors in diseases that are driven by multiple receptors, and may even bypass the need to pinpoint which upstream or downstream pathways are involved. By the same token, inhibition of the GIV•Gαi interface is expected to have the tremendous advantage of allowing 'network-based therapy' irrespective of the receptor of origin[43]. Recently, we showed just that in a proof-of-concept study using cell-permeable peptides[85]. Selective modulation of the GIV•Gαi interface using cell-permeable GIV-CT peptides fused to a TAT-peptide transduction domain (TAT-PTD) containing the minimal modular elements of GIV that are necessary and sufficient for activation of Gi downstream of RTKs can effectively engineer signaling networks and alter cell behavior[85]. In the presence of an intact GEF motif, TAT-GIV-CT peptides enhanced diverse processes in which GIV's GEF function has previously been implicated; e.g., 2D cell migration after scratch-wounding, invasion of cancer cells, and finally, myofibroblast activation and collagen production. Furthermore, topical application of TAT-GIV-CT peptides enhanced wound repair in mice in a GEF-dependent manner. The impact of these findings is two-fold. First, the findings described here using TAT-GIV-CT peptides represent a significant advancement in our ability to access, interrogate and manipulate that platform, and thereby, modulate the cross-talk it facilitates. Second, G-proteins are an ideal target for therapeutic intervention because they serve as signal amplification switches, and potent and pathway-selective activators/inhibitors of a G protein can serve multiple purposes ranging from being a research tool to pharmacologic probe for use in experimental and clinical therapeutics[86]. The cell-permeable peptides allow for exogenous manipulation of the RTK-GIV-Gαi pathway by enhancing or suppressing coupling of G protein with RTKs and their subsequent transactivation, in a dose dependent manner while minimizing the risk of tampering with other physiologic functions/interactions of G proteins/or other components within the network of modulators of G protein signaling (Figure 2).
The therapeutic advantages of using these cell-permeable GIV-CT peptides for activation/inactivation of Gαi proteins are also many-fold. First, this approach circumvents the need to target individual receptors in diseases that are driven by multiple receptors. Second, GIV's SH2 like domain can directly bind multiple ligand-activated RTKs and re-wire several components of downstream signaling, and therefore, these peptides offer a versatile tool to simultaneously modulate multiple pathways downstream of many RTKs (i.e., broad), even in diseases/processes where upstream and downstream events are incompletely understood (i.e., circumvents the limitations of unknown). Third, because GIV binds preferentially to Gi subfamily members but can discriminate within this subfamily by binding to Gαi subunits but not to the close homologue Gαo (~75% overall similarity to Gαi1/2/3 subunits)[10], TAT-GIV-CT peptides selectively affect the activation of Gαi1/2/3, but not Gαo (i.e., specific). Fourth, these peptides circumvent the limitation that no promising 'druggable' pockets have been identified within GIV's C-terminus, and that small molecules that can selectively block this platform are yet to be identified. Last, these GIV-CT peptides are predicted to directly address the upstream component of RTK-related signaling in cases of mutations, polymorphisms, and expression-related defects often seen in disease.
Although cell-permeable peptides allowed exogenous modulation of the fundamental function of GIV, i.e., activate Gi downstream of growth factor RTKs, it is unlikely that these TAT-appended peptides will serve as marketable pharmacologic agents. But the lessons we learned are invaluable because it appears that GIV-CT peptides may be optimal for potential gene therapy applications to manipulate Gαi activation downstream of multiple growth factors in different cell types and in a diverse array of pathophysiologic conditions. The therapeutic potential of these peptides is expected to only grow with the rapidly growing list of pathophysiologic processes that GIV modulates. We speculate that these peptides will also modulate other pathophysiologic conditions in which GIV is implicated, but the role of its GEF function is yet to be interrogated (see Tables 1, 2).
Conclusions and Future perspectives
The insights gained just within the past half-decade has shaped a paradigm of GIV-dependent non-canonical G protein signaling by receptors that are typically not believed to signal via G proteins. Despite these insights, it is clear that a lot remains unknown. For example, although we have some understanding of how RTKs transactivate G proteins via GIV, how other classes of receptors, such as GPCRs, β1 integrins, Toll-like receptors (TLRs), Transforming growth factor (TGFβ) receptors also do the same remains unclear. Knowing how GIV engages receptors is of utmost importance because an in-depth insight into that mechanism(s) will fundamentally revolutionize our understanding of the new rules of engagement of non-canonical G protein signaling via GIV. Because GIV's C-terminus offers structural/conformational plasticity, which should directly impact protein-protein interactions, it is possible that such structural plasticity provides context-dependent engagement with a variety of receptors, some directly and some others indirectly.
As for the newly revealed temporal and spatial features of non-canonical G protein signaling, several interesting questions remain unsolved. One such unanswered question is how does non-canonical G protein activation at the PM by GIV-GEF coordinately trigger the same on internal membranes. Last, but not least, although homology modeling has proved insightful thus far, obtaining structural insights into how GIV engages with ligand-activated RTKs and Gα-subunits is an urgent and an unmet need. Such insights are expected to greatly facilitate the development of small molecules that can selectively target the GIV:RTK and/or GIV:Gαi interfaces. Thus, it is clear that there are many unanswered questions and it is expected that solving these questions will encounter many conceptual, technical, and logical problems. Such challenges can only be overcome by engagement of more groups in the scientific community to systematically dissect this emerging paradigm of non-canonical G protein signaling from the atomic level to pathway modeling.
Acknowledgments
This work was funded by NIH (R01CA160911 and DK099226) to P.G.
REFERENCES
- 1.Lin C, Ear J, Midde K, Lopez-Sanchez I, et al. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol Biol Cell. 2014;25:3654–3671. doi: 10.1091/mbc.E14-05-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh P, Garcia-Marcos M, Farquhar MG. GIV/Girdin is a rheostat that fine-tunes growth factor signals during tumor progression. Cell Adh Migr. 2011;5:237–248. doi: 10.4161/cam.5.3.15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Sanchez I, Garcia-Marcos M, Mittal Y, Aznar N, et al. Protein kinase C-theta (PKCtheta) phosphorylates and inhibits the guanine exchange factor, GIV/Girdin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5510–5515. doi: 10.1073/pnas.1303392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Misaki T, Taupin V, Eguchi A, et al. GIV/girdin links vascular endothelial growth factor signaling to Akt survival signaling in podocytes independent of nephrin. J Am Soc Nephrol. 2015;26:314–327. doi: 10.1681/ASN.2013090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh P, Beas AO, Bornheimer SJ, Garcia-Marcos M, et al. A G{alpha}i-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol Biol Cell. 2010;21:2338–2354. doi: 10.1091/mbc.E10-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal Y, Pavlova Y, Garcia-Marcos M, Ghosh P. Src homology domain 2-containing protein-tyrosine phosphatase-1 (SHP-1) binds and dephosphorylates G(alpha)-interacting, vesicle-associated protein (GIV)/Girdin and attenuates the GIV-phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway. The Journal of biological chemistry. 2011;286:32404–32415. doi: 10.1074/jbc.M111.275685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. The Journal of biological chemistry. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Sanchez I, Dunkel Y, Roh YS, Mittal Y, et al. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat Commun. 2014;5:4451. doi: 10.1038/ncomms5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh P. G protein-Coupled Growth Factor Receptor Tyrosine Kinases: No Longer an Oxymoron. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1066538. (Invited, Peer Reviewed Review; In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J Biol Chem. 2015;290:6697–6704. doi: 10.1074/jbc.R114.613414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Yuan L, Yang XM, Wei D, et al. A chimeric antibody targeting CD147 inhibits hepatocellular carcinoma cell motility via FAK-PI3K-Akt-Girdin signaling pathway. Clin Exp Metastasis. 2015;32:39–53. doi: 10.1007/s10585-014-9689-7. [DOI] [PubMed] [Google Scholar]
- 15.Dunkel Y, Ong A, Notani D, Mittal Y, et al. STAT3 protein up-regulates Galpha-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. The Journal of biological chemistry. 2012;287:41667–41683. doi: 10.1074/jbc.M112.390781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Midde KK, Aznar N, Laederich MB, Ma GS, et al. Multimodular biosensors reveal a novel platform for activation of G proteins by growth factor receptors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E937–E946. doi: 10.1073/pnas.1420140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhandari DL-SI, To A, Niesman I, Gupta V, Leyme AC, Lo I, Aznar N, Garcia-Marcos M, Farquhar MG, Ghosh P. Cyclin Dependent Kinase 5 Phosphorylates and Activates Guanidine Exchange Factor GIV/Girdin. Proceedings National Academy of Sciences. 2015 doi: 10.1073/pnas.1514157112. (Accepted, In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, Kakuwa T, Akimoto K, Koga H, et al. Regulation of epithelial cell polarity by PAR-3 depends on Girdin transcription and Girdin-Galphai3 signaling. J Cell Sci. 2015 doi: 10.1242/jcs.160879. [DOI] [PubMed] [Google Scholar]
- 19.Gilman AG. G proteins: transducers of receptor-generated signals. Annual review of biochemistry. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 20.Morris AJ, Malbon CC. Physiological regulation of G protein-linked signaling. Physiol Rev. 1999;79:1373–1430. doi: 10.1152/physrev.1999.79.4.1373. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins AL, Groom CR. The druggable genome. Nature reviews. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 22.Sato M, Blumer JB, Simon V, Lanier SM. Accessory proteins for G proteins: partners in signaling. Annual review of pharmacology and toxicology. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 23.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annual review of biochemistry. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 25.Blumer JB, Oner SS, Lanier SM. Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta physiologica (Oxford, England) 2012;204:202–218. doi: 10.1111/j.1748-1716.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 26.De Vries L, Zheng B, Fischer T, Elenko E, et al. The regulator of G protein signaling family. Annual review of pharmacology and toxicology. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 27.Kimple RJ, Kimple ME, Betts L, Sondek J, et al. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- 28.Peterson YK, Hazard S, 3rd, Graber SG, Lanier SM. Identification of structural features in the G-protein regulatory motif required for regulation of heterotrimeric G-proteins. The Journal of biological chemistry. 2002;277:6767–6770. doi: 10.1074/jbc.C100699200. [DOI] [PubMed] [Google Scholar]
- 29.De Vries L, Mousli M, Wurmser A, Farquhar MG. GAIP, a protein that specifically interacts with the trimeric G protein G alpha i3, is a member of a protein family with a highly conserved core domain. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasa SP, Watson N, Overton MC, Blumer KJ. Mechanism of RGS4, a GTPase-activating protein for G protein alpha subunits. The Journal of biological chemistry. 1998;273:1529–1533. doi: 10.1074/jbc.273.3.1529. [DOI] [PubMed] [Google Scholar]
- 32.Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 33.Blazer LL, Roman DL, Chung A, Larsen MJ, et al. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Molecular pharmacology. 2011;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimple AJ, Yasgar A, Hughes M, Jadhav A, et al. A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-alpha interaction. Combinatorial chemistry & high throughput screening. 2008;11:396–409. doi: 10.2174/138620708784534770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cismowski MJ, Ma C, Ribas C, Xie X, et al. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. The Journal of biological chemistry. 2000;275:23421–23424. doi: 10.1074/jbc.C000322200. [DOI] [PubMed] [Google Scholar]
- 36.Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. The Journal of biological chemistry. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 37.Chan P, Gabay M, Wright FA, Tall GG. Ric-8B is a GTP-dependent G protein alphas guanine nucleotide exchange factor. The Journal of biological chemistry. 2011;286:19932–19942. doi: 10.1074/jbc.M110.163675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MJ, Dohlman HG. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol. 2008;18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natochin M, Campbell TN, Barren B, Miller LC, et al. Characterization of the G alpha(s) regulator cysteine string protein. The Journal of biological chemistry. 2005;280:30236–30241. doi: 10.1074/jbc.M500722200. [DOI] [PubMed] [Google Scholar]
- 40.Vaidyanathan G, Cismowski MJ, Wang G, Vincent TS, et al. The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene. 2004;23:5858–5863. doi: 10.1038/sj.onc.1207774. [DOI] [PubMed] [Google Scholar]
- 41.Tonissoo T, Lulla S, Meier R, Saare M, et al. Nucleotide exchange factor RIC-8 is indispensable in mammalian early development. Dev Dyn. 2012;239:3404–3415. doi: 10.1002/dvdy.22480. [DOI] [PubMed] [Google Scholar]
- 42.Lohse MJ, Hein P, Hoffmann C, Nikolaev VO, et al. Kinetics of G-protein-coupled receptor signals in intact cells. Br J Pharmacol. 2008;153(Suppl 1):S125–S132. doi: 10.1038/sj.bjp.0707656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh P. G proteins as Emerging Targets for Network Based Therapy in Cancer: End of a long futile campaign striking heads of a Hydra. Aging. 2015 doi: 10.18632/aging.100781. (Invited Editorial; In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/ Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.R114.613414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlessinger J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 47.Natarajan K, Berk BC. Crosstalk coregulation mechanisms of G protein-coupled receptors and receptor tyrosine kinases. Methods in molecular biology. 2006;332:51–77. doi: 10.1385/1-59745-048-0:51. [DOI] [PubMed] [Google Scholar]
- 48.Pierce KL, Luttrell LM, Lefkowitz RJ. New mechanisms in heptahelical receptor signaling to mitogen activated protein kinase cascades. Oncogene. 2001;20:1532–1539. doi: 10.1038/sj.onc.1204184. [DOI] [PubMed] [Google Scholar]
- 49.Marty C, Ye RD. Heterotrimeric G protein signaling outside the realm of seven transmembrane domain receptors. Molecular pharmacology. 2010;78:12–18. doi: 10.1124/mol.110.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin CEJ, Midde KK, Pavlova Y, Garcia-Marcos M, Kufareva I, Abagyan R, Ghosh P. Structural basis for multi-receptor signal enhancement via the metastasis-related protein GIV/Girdin. Structure-Cell. 2014 [Google Scholar]
- 51.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 52.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 53.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, et al. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 54.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, et al. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 55.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 56.Malaney P, Uversky VN, Dave V. Identification of intrinsically disordered regions in PTEN and delineation of its function via a network approach. Methods. 2015;77–78:69–74. doi: 10.1016/j.ymeth.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Malaney P, Uversky VN, Dave V. The PTEN Long N-tail is intrinsically disordered: increased viability for PTEN therapy. Mol Biosyst. 2013;9:2877–2888. doi: 10.1039/c3mb70267g. [DOI] [PubMed] [Google Scholar]
- 58.Malaney P, Pathak RR, Xue B, Uversky VN, et al. Intrinsic disorder in PTEN and its interactome confers structural plasticity and functional versatility. Sci Rep. 2013;3:2035. doi: 10.1038/srep02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szollosi J, Damjanovich S, Nagy P, Vereb G, et al. Principles of resonance energy transfer. Curr Protoc Cytom. 2006;Chapter 1(Unit1 12) doi: 10.1002/0471142956.cy0112s38. [DOI] [PubMed] [Google Scholar]
- 60.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 61.Lee HY, Jung H, Jang IH, Suh PG, et al. Cdk5 phosphorylates PLD2 to mediate EGF-dependent insulin secretion. Cell Signal. 2008;20:1787–1794. doi: 10.1016/j.cellsig.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Gibson SK, Gilman AG. Gialpha and Gbeta subunits both define selectivity of G protein activation by alpha2-adrenergic receptors. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:212–217. doi: 10.1073/pnas.0509763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 65.Yi TM, Kitano H, Simon MI. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10764–10769. doi: 10.1073/pnas.1834247100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lohse MJ, Hoffmann C, Nikolaev VO, Vilardaga JP, et al. Kinetic analysis of G protein-coupled receptor signaling using fluorescence resonance energy transfer in living cells. Adv Protein Chem. 2007;74:167–188. doi: 10.1016/S0065-3233(07)74005-6. [DOI] [PubMed] [Google Scholar]
- 67.Ross EM. Coordinating speed and amplitude in G-protein signaling. Curr Biol. 2008;18:R777–R783. doi: 10.1016/j.cub.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ponsioen B, Zhao J, Riedl J, Zwartkruis F, et al. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO reports. 2004;5:1176–1180. doi: 10.1038/sj.embor.7400290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lo IC, Gupta V, Midde KK, Taupin V, et al. Activation of Galphai at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling. Dev Cell. 2015;33:189–203. doi: 10.1016/j.devcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. The Journal of biological chemistry. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 72.Ogier-Denis E, Petiot A, Bauvy C, Codogno P. Control of the expression and activity of the Galpha-interacting protein (GAIP) in human intestinal cells. The Journal of biological chemistry. 1997;272:24599–24603. doi: 10.1074/jbc.272.39.24599. [DOI] [PubMed] [Google Scholar]
- 73.Stow JL, de Almeida JB, Narula N, Holtzman EJ, et al. A heterotrimeric G protein, G alpha i-3, on Golgi membranes regulates the secretion of a heparan sulfate proteoglycan in LLC-PK1 epithelial cells. J Cell Biol. 1991;114:1113–1124. doi: 10.1083/jcb.114.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barr FA, Leyte A, Huttner WB. Trimeric G proteins and vesicle formation. Trends in cell biology. 1992;2:91–94. doi: 10.1016/0962-8924(92)90001-4. [DOI] [PubMed] [Google Scholar]
- 75.Cancino J, Luini A. Signaling circuits on the Golgi complex. Traffic. 2013;14:121–134. doi: 10.1111/tra.12022. [DOI] [PubMed] [Google Scholar]
- 76.Chisari M, Saini DK, Kalyanaraman V, Gautam N. Shuttling of G protein subunits between the plasma membrane and intracellular membranes. The Journal of biological chemistry. 2007;282:24092–24098. doi: 10.1074/jbc.M704246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, et al. Identification of G protein alpha subunit-palmitoylating enzyme. Molecular and cellular biology. 2009;29:435–447. doi: 10.1128/MCB.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giannotta M, Ruggiero C, Grossi M, Cancino J, et al. The KDEL receptor couples to Galphaq/11 to activate Src kinases and regulate transport through the Golgi. The EMBO journal. 2012;31:2869–2881. doi: 10.1038/emboj.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jamora C, Yamanouye N, Van Lint J, Laudenslager J, et al. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell. 1999;98:59–68. doi: 10.1016/S0092-8674(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 80.Colombo MI, Inglese J, D'Souza-Schorey C, Beron W, et al. Heterotrimeric G proteins interact with the small GTPase ARF. Possibilities for the regulation of vesicular traffic. The Journal of biological chemistry. 1995;270:24564–24571. doi: 10.1074/jbc.270.41.24564. [DOI] [PubMed] [Google Scholar]
- 81.Cohen LA, Donaldson JG. Analysis of Arf GTP-binding protein function in cells. Current protocols in cell biology / editorial board, Juan S Bonifacino [et al] 2010;Chapter 3(Unit 14):2, 1–7. doi: 10.1002/0471143030.cb1412s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shiba Y, Kametaka S, Waguri S, Presley JF, et al. ArfGAP3 regulates the transport of cation-independent mannose 6-phosphate receptor in the post-Golgi compartment. Curr Biol. 2013;23:1945–1951. doi: 10.1016/j.cub.2013.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, et al. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weller SG, Capitani M, Cao H, Micaroni M, et al. Src kinase regulates the integrity and function of the Golgi apparatus via activation of dynamin 2. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5863–5868. doi: 10.1073/pnas.0915123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma GS, Aznar N, Kalogriopoulos N, Midde KK, et al. Therapeutic effects of cell-permeant peptides that activate G proteins downstream of growth factors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E2602–E2610. doi: 10.1073/pnas.1505543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iiri T, Farfel Z, Bourne HR. G-protein diseases furnish a model for the turn-on switch. Nature. 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 87.Beas AO, Taupin V, Teodorof C, Nguyen LT, et al. Galphas promotes EEA1 endosome maturation and shuts down proliferative signaling through interaction with GIV (Girdin) Mol Biol Cell. 2012;23:4623–4634. doi: 10.1091/mbc.E12-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohara K, Enomoto A, Kato T, Hashimoto T, et al. Involvement of Girdin in the determination of cell polarity during cell migration. PloS one. 2012;7:e36681. doi: 10.1371/journal.pone.0036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao JZ, Jiang P, Cui SP, Ren YL, et al. Girdin locates in centrosome and midbody and plays an important role in cell division. Cancer Sci. 2012;103:1780–1787. doi: 10.1111/j.1349-7006.2012.02378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weng L, Enomoto A, Miyoshi H, Takahashi K, et al. Regulation of cargo-selective endocytosis by dynamin 2 GTPase-activating protein girdin. The EMBO journal. 2014;33:2098–2112. doi: 10.15252/embj.201488289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ichimiya H, Maeda K, Enomoto A, Weng L, et al. Girdin/GIV regulates transendothelial permeability by controlling VE-cadherin trafficking through the small GTPase, R-Ras. Biochem Biophys Res Commun. 2015 doi: 10.1016/j.bbrc.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 92.Enomoto A, Asai N, Namba T, Wang Y, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 93.Wang Y, Kaneko N, Asai N, Enomoto A, et al. Girdin is an intrinsic regulator of neuroblast chain migration in the rostral migratory stream of the postnatal brain. J Neurosci. 2011;31:8109–8122. doi: 10.1523/JNEUROSCI.1130-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puseenam A, Yoshioka Y, Nagai R, Hashimoto R, et al. A novel Drosophila Girdin-like protein is involved in Akt pathway control of cell size. Exp Cell Res. 2009;315:3370–3380. doi: 10.1016/j.yexcr.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 95.Enomoto A, Murakami H, Asai N, Morone N, et al. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Jiang P, Enomoto A, Jijiwa M, Kato T, et al. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer research. 2008;68:1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- 97.Natsume A, Kato T, Kinjo S, Enomoto A, et al. Girdin maintains the stemness of glioblastoma stem cells. Oncogene. 2012;31:2715–2724. doi: 10.1038/onc.2011.466. [DOI] [PubMed] [Google Scholar]
- 98.Zhang YJ, Li AJ, Han Y, Yin L, et al. Inhibition of Girdin enhances chemosensitivity of colorectal cancer cells to oxaliplatin. World J Gastroenterol. 2014;20:8229–8236. doi: 10.3748/wjg.v20.i25.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamamura Y, Asai N, Enomoto A, Kato T, et al. Akt-Girdin signaling in cancer-associated fibroblasts contributes to tumor progression. Cancer research. 2015;75:813–823. doi: 10.1158/0008-5472.CAN-14-1317. [DOI] [PubMed] [Google Scholar]
- 100.Kitamura T, Asai N, Enomoto A, Maeda K, et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nature cell biology. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- 101.Hartung A, Ordelheide AM, Staiger H, Melzer M, et al. The Akt substrate Girdin is a regulator of insulin signaling in myoblast cells. Biochim Biophys Acta. 2013;1833:2803–2811. doi: 10.1016/j.bbamcr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 102.Ma GS, Lopez-Sanchez I, Aznar N, Kalogriopoulos N, Pedram S, Midde K, Ciaraldi T, Henry RR, Ghosh P. Activation of G proteins by GIV-GEF is a Pivot Point for Insulin Resistance and Sensitivity. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-08-0553. Accepted, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito T, Komeima K, Yasuma T, Enomoto A, et al. Girdin and its phosphorylation dynamically regulate neonatal vascular development and pathological neovascularization in the retina. Am J Pathol. 2013;182:586–596. doi: 10.1016/j.ajpath.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 104.Miyachi H, Mii S, Enomoto A, Murakumo Y, et al. Role of Girdin in intimal hyperplasia in vein grafts and efficacy of atelocollagen-mediated application of small interfering RNA for vein graft failure. J Vasc Surg. 2014;60:479–489. e5. doi: 10.1016/j.jvs.2013.06.080. [DOI] [PubMed] [Google Scholar]
- 105.Miyake H, Maeda K, Asai N, Shibata R, et al. The actin-binding protein Girdin and its Akt-mediated phosphorylation regulate neointima formation after vascular injury. Circulation research. 2011;108:1170–1179. doi: 10.1161/CIRCRESAHA.110.236174. [DOI] [PubMed] [Google Scholar]
- 106.Miyachi H, Takahashi M, Komori K. A Novel Approach against Vascular Intimal Hyperplasia Through the Suppression of Girdin. Ann Vasc Dis. 2015;8:69–73. doi: 10.3400/avd.ra.14-00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakai T, Nagai T, Tanaka M, Itoh N, et al. Girdin phosphorylation is crucial for synaptic plasticity and memory: a potential role in the interaction of BDNF/TrkB/Akt signaling with NMDA receptor. J Neurosci. 2014;34:14995–15008. doi: 10.1523/JNEUROSCI.2228-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blakely BT, Rossi FM, Tillotson B, Palmer M, et al. Epidermal growth factor receptor dimerization monitored in live cells. Nat Biotechnol. 2000;18:218–222. doi: 10.1038/72686. [DOI] [PubMed] [Google Scholar]
- 109.Chung I, Akita R, Vandlen R, Toomre D, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 110.Schmoll HJ, Van Cutsem E, Stein A, Valentini V, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2012;23:2479–2516. doi: 10.1093/annonc/mds236. [DOI] [PubMed] [Google Scholar]
- 111.Dalle S, Ricketts W, Imamura T, Vollenweider P, et al. Insulin and insulin-like growth factor I receptors utilize different G protein signaling components. The Journal of biological chemistry. 2001;276:15688–15695. doi: 10.1074/jbc.M010884200. [DOI] [PubMed] [Google Scholar]
- 112.Ghosh P. G protein-Coupled Growth Factor Receptor Tyrosine Kinases: No Longer an Oxymoron. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1066538. (Invited Extra View; In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]