Abstract
A complete understanding of the function of Ten-eleven translocation (TET) family of dioxygenase-mediated DNA demethylation requires new methods to quantitatively map oxidized 5-methylcytosine (5mC) bases at high-resolution. We have recently developed a methylase-assisted bisulfite sequencing (MAB-seq) method that allows base-resolution mapping of 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), two oxidized 5mC bases indicative of active DNA demethylation events. In standard bisulfite sequencing (BS-seq), unmodified C, 5fC and 5caC are read as thymine. Thus 5fC/5caC cannot be distinguished from C; in MAB-seq, unmodified C is enzymatically converted to 5mC, allowing direct mapping of rare modifications such as 5fC and 5caC. By combining MAB-seq with chemical reduction of 5fC to 5hmC, we also developed caMAB-seq, a method for direct 5caC mapping. Compared to subtraction-based mapping methods, MAB-seq and caMAB-seq require less sequencing efforts and enable robust statistical calling of 5fC/5caC. MAB-seq and caMAB-seq can be adapted to map 5fC/5caC at the scale of whole genome (WG-MAB-seq), within specific genomic regions enriched for enhancer-marking histone modifications (ChIP-MAB-seq), or at CpG-rich sequences (reduce-reprepsentation (RR)-MAB-seq) such as gene promoters. The full protocol, including DNA preparation, enzymatic treatment, library preparation and sequencing, can be completed within 6-8 d.
Keywords: Epigenetics, active DNA demethylation, Ten-eleven translocation (TET) proteins, Thymine DNA glycosylase (TDG), 5-formylcytosine (5fC), 5-carboxymethylcytosine (5caC)
INTRODUCTION
DNA cytosine methylation is an evolutionarily conserved epigenetic modification and is indispensable for normal mammalian development 1, 2. Enzyme-catalyzed active DNA demethylation contributes substantially to dynamic regulation of DNA methylome during development and in diseases. In mammals, active DNA demethylation (converting 5mC back to C) is primarily initiated by the enzymatic activity of ten-eleven translocation (TET) family of 5mC dioxygenases 3–5. TET proteins convert 5mC into 5-hydroxymethylcytosine (5hmC) 6–8. Further oxidative modification of 5hmC by TET results in 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) 9, 10, both of which can be efficiently excised by Thymine DNA glycosylase (TDG) and restored to unmodified cytosines through base excision repair (BER) pathway (Figure 1) 5, 9, 11. Genetic analyses of mutant mice deficient in TET proteins indicated that these enzymes play essential roles in a wide range of biological processes, including gene regulation 12–18, embryonic development 19–21, stem cell differentiation 8, 22, 23, meiotic gene control 24, erasure of genomic imprinting 19, 25, learning and memory 26–28, and cancer 29, 30.
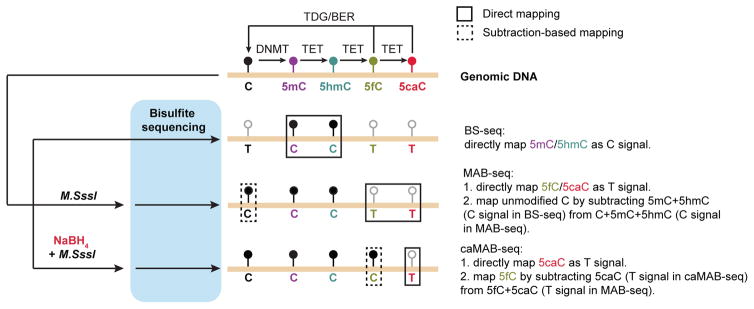
Figure 1. Schematic diagram of BS-seq, MAB-seq and caMAB-seq.
In mammalian cells, de novo and maintenance DNA methyltransferases (DNMTs) methylate unmodified cytosines (C) to generate 5-methylcytosines (5mC). Ten-eleven translocation (TET) family of DNA dioxygenase (TET1–3) is capable of iteratively oxidizing 5mC and its derivatives to generate three oxidized methylcytosines, i.e. 5hmC, 5fC and 5caC. Highly oxidized cytosine bases, 5fC and 5caC, are enzymatically excised by Thymine DNA glycosylase (TDG), and resulting abasic sites are repaired by the base-excision repair (BER) pathway to regenerate unmodified C, completing the DNA demethylation process (5mC to C). In standard bisulfite sequencing (BS-seq), 5mC and 5hmC are resistant to sodium bisulfite-mediated deamination and read as C in subsequent sequencing, whereas unmodified C, 5fC and 5caC are read as T. M.SssI exhibits robust methylase activity toward unmodified cytosines within CpGs. In MAB-seq, only 5fC and 5caC are read as T after genomic DNA is treated with M.SssI. In caMAB-seq, genomic DNA is first treated with sodium borohydride (NaBH4) to reduce 5fC back to 5hmC, enabling 5caC to be directly mapped as T.
The TET/TDG-dependent active demethylation pathway involves generation and excision repair of 5fC/5caC and may occur in both proliferating and post-mitotic cells. Besides acting as intermediates of active DNA demethylation pathway, oxidized methylcytosines may also possess unique regulatory functions 31, 32. Recent studies have identified potential reader proteins for oxidized methylcytosines, many of which are transcription factors, chromatin modifying enzymes or proteins linked to DNA repair process 33–35. Furthermore, 5fC and 5caC accumulated within gene bodies may have a regulatory role in decreasing the elongation rate of RNA polymerase II 36, 37. Finally, clustered 5fCpG sites have recently been shown to affect base-pairing and DNA structure in vitro 38, 39 and may thus impact DNA-templated processes by directly modulating DNA conformation. A better mechanistic understanding of these potential regulatory roles of 5fC and 5caC requires the ability to systematically map the genomic position and determine absolute levels of these oxidized methylcytosines in the mammalian genome. Here we present detailed protocols of the MAB-seq method that we recently developed for single-base resolution mapping of 5fC and 5caC 40. We also present the procedure for caMAB-seq, a modified version of MAB-seq for direct 5caC mapping 40.
Development of MAB-seq
Identification of cytosines that are committed to active DNA demethylation requires quantitative measurement of TDG-mediated excision of 5fC/5caC at high resolution. In addition, because both 5fC and 5caC are excised by TDG, strand-specific preference of active DNA demethylation activity requires simultaneous mapping of 5fC and 5caC in a single experiment. Using either modification-specific antibodies or chemical tagging, recent studies have shown that genomic regions enriched for cytosines undergoing TET/TDG-dependent active demethylation can be identified by analyzing ectopic accumulation of 5fC/5caC in Tdg-depleted cells 41, 42. However, 5fC and 5caC maps generated by affinity-enrichment methods are of limited resolution (a few hundred base-pairs). In addition, these 5fC and 5caC profiles represent only relative enrichment and lacks strand distribution information. To circumvent these limitations, we have developed MAB-seq, a modified BS-seq strategy that allows simultaneous and quantitative mapping of both 5fC and 5caC at single-base resolution 40, 43. In traditional BS-seq, C/5fC/5caC are efficiently deaminated through sodium bisulfite treatment and all read as thymine (T) in subsequent sequencing experiments. In contrast, 5mC and 5hmC are resistant to this chemical conversion and are read as C. In MAB-seq, genomic DNA is first treated with the bacterial DNA CpG methyltransferase M.SssI, an enzyme (originally isolated from Spiroplasma sp. strain MQ1) that efficiently methylates cytosines within CpG dinucleotides. Bisulfite conversion of M.SssI-treated DNA only deaminates 5fC and 5caC; originally unmodified C within CpGs becomes resistant to bisulfite conversion due to its conversion to 5mC. Subsequent sequencing would identify 5fC and 5caC as T, whereas C/5mC/5hmC is read as C (Figure 1). We have further developed a method termed caMAB-seq (5caC methylase-assisted bisulfite sequencing) to directly map 5caC at single-base resolution. In this modified version of MAB-seq, 5fC is first reduced by NaBH4 to 5hmC. By combining NaBH4 reduction with M.SssI treatment, 5caC is sequenced as T after bisulfite conversion whereas C/5mC/5hmC/5fC are read as C.
Application and limitations of MAB-seq
A major advantage of the MAB-seq method is that it allows for direct and simultaneous mapping of 5fC and 5caC, which requires less sequencing efforts and enables quantitative detection of all TET/TDG-dependent DNA demethylation events in a single experiment at single-base resolution. MAB-seq is also amenable to both genome-scale and locus-specific analysis (Figure 2). For instance, MAB-seq has been applied to study both early embryos and mouse embryonic stem cells (ESCs), and generated whole genome map of 5fC/5caC as well as quantitative profile of 5fC/5caC at specific loci 40, 43, 44. The detection limit of MAB-seq is governed by several factors, including the error rate of the M.SssI methylase, efficiency of bisulfite conversion of 5fC/5caC, the abundance of 5fC/5caC at the modified base, and overall sequencing depth. With the protocol described here, highly efficient conversion of C to 5mC and deamination of 5fC/5caC in genomic DNA can be achieved. Thus, sequencing depth is a major factor for sensitive and specific detection of 5fC/5caC by MAB-seq. However, MAB-seq is unable to distinguish 5fC/5caC from unmodified C within a non-CpG context due to the poor methylase activity of M.SssI toward C outside CpG dinucleotides. This limitation does not greatly affect the application of this technique due to the following two reasons. First, whole-genome base-resolution mapping indicates that 5hmC is found almost exclusively in the CpG context (>99% in CpGs), even in mouse ESCs and neurons where non-CpG methylation is prevalent 45, 46; Second, TET proteins have a strong preference for 5mC within CpG context compared to that in non-CpG context 47, 48. Thus, 5mC within CpG dinucleotides is the primary target for TET proteins, and MAB-seq allows quantitative measurement of the abundance of 5fC/5caC within the CpG context.
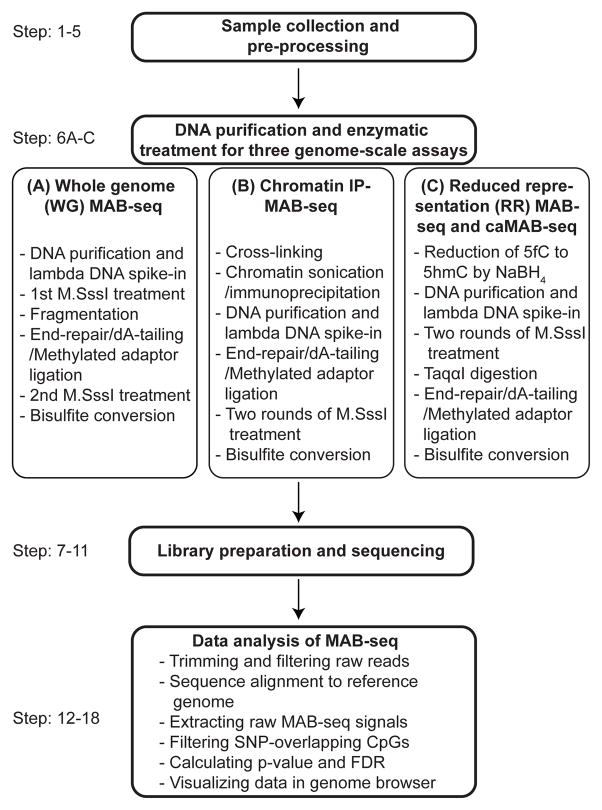
Figure 2.
Diagrammatic presentation of the work-flow of MAB/caMAB-seq.
Comparison of MAB-seq with other base-resolution 5fC or 5caC methods
Both bisulfite-based and bisulfite-free mapping methods have been developed for mapping oxidized methylcytosines at single-base resolution (Figure 3) 32. Bisulfite-free base-resolution mapping methods, such as Pvu-Seal-seq 49 and fC-CET 50, currently provide only relative enrichment of cytosines marked by 5fC. Compared to bisulfite-free methods, a major advantage of bisulfite-based mapping methods (including MAB-seq and caMAB-seq) is their ability to determine both base-resolution location and absolute levels of 5fC or 5caC. Prior to MAB-seq 40, 43, several bisulfite-based mapping methods have been developed by utilizing specific chemical treatment to protect 5fC (fCAB-seq 41 and redBS-seq 51) or 5caC (caCAB-seq 52) from bisulfite-induced deamination. However, these methods require subtracting the C signals between standard BS-seq and those of modified BS-seq to determine the position and abundance of 5fC or 5caC, therefore doubling required sequencing efforts. In addition, MAB-seq is the only method capable of simultaneously detecting 5fC and 5caC, which is required for base-resolution analysis of strand-specificity of TET/TDG-mediated active DNA demethylation events. Moreover, MAB-seq and caMAB-seq can be integrated together to map 5fC/5caC together (MAB-seq), 5fC individually (subtract caMAB-seq from MAB-seq), and 5caC individually (caMAB-seq) at single-base resolution. Thus, with two genome-scale mapping experiments (MAB-seq plus caMAB-seq), this integrated approach not only provides quantitative base-resolution mapping of active DNA demethylation events (generation and excision of 5fC/5caC), but also distinguishes 5fC from 5caC at base-resolution.
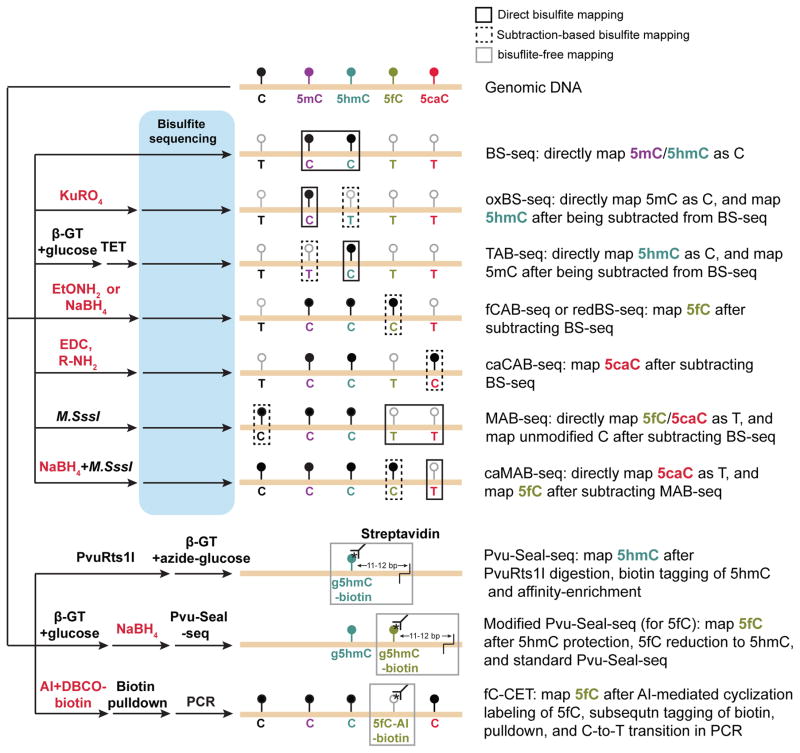
Figure 3. Schematic diagram of base-resolution mapping methods for oxidized methylcytosines.
Coupled with various chemical (in red) and enzymatic (in black) treatments, bisulfite sequencing (BS-seq) based (upper panels) or bisulfite-free (lower panels) base-resolution mapping methods have been developed to profile 5fC and 5caC. In direct bisulfite mapping (targeting modification highlighted in black line boxes), position and abundance of specific oxidized methylcytosines (e.g. mapping of 5hmC by MAB-seq 46) can be directly determined. In subtraction-based bisulfite mapping (targeting modification highlighted in dash line boxes), subtracting signals between conventional BS-seq and those of modified BS-seq (e.g. mapping of 5hmC by subtracting signals of oxBS-seq 67 from those of standard BS-seq) is required to indirectly determine the position and abundance of oxidized methylcytosines. In two bisulfite-free mapping strategies (targeting modification highlighted in gray line boxes), modification-sensitive restriction enzyme (5hmC-specific endonuclease PvuRts1l for Pvu-Seal-seq) or chemical (AI [azido derivative of 1,3-indandione] for fC-CET) assisted tagging of oxidized methylcytosines is utilized to both enrich the genomic fragments containing methylcytosines (giving relative abundance) and to identify the position of these cytosine variants.
Experimental design
Enzymatic treatment (Step 6A (iv–v) and (xi–xii), Step 6C (v–vi))
Because incomplete methylation of unmodified CpGs will result in false-positive 5fC/5caC signals, the conditions of the enzymatic treatment with M.SssI is critical to the success of MAB-seq. S-adenosylmethionine (SAM) is unstable at 37 °C and is sensitive to degradation at elevated pH (i.e. pH 7.5). Moreover, S-adenosylhomocysteine (SAH), the byproduct of the methylation reaction binds more tightly to M.SssI than does SAM and greatly reduces the methylation reaction rate as time passes. Thus, it is important to ensure that only fresh SAM is used as co-factors in the enzymatic reaction, and adding more SAM after two hours can increase the methylation rate. Because high concentration of input DNA leads to accumulation of high SAH levels, we recommend that the concentration of input DNA does not exceed 20 ng/μL. M.SssI is more processive in the absence of magnesium and more distributive in the supplied reaction buffer (NEBuffer2 contains 10 mM MgCl2). To maximize the methylation rate, we recommend treating genomic DNA in Mg2+-free buffer for one round and in the supplied reaction buffer for another rounds.
NaBH4 reduction of 5fC to 5hmC (Step 6C (ii–iii))
Several studies have shown that NaBH4 can efficiently reduces 5fC to 5hmC, which changes the behavior of 5fC during bisulfite sequencing and offers a strategy for 5fC mapping 40, 41, 49, 51. In caMAB-seq, a step of NaBH4 reduction is added before M.SssI treatment. In our hands, this reduction step is compatible with M.SssI treatment and library preparation based on reduced representation bisulfite sequencing (RRBS) strategy 53. We termed this genome-scale direct mapping method of 5caC as RR-caMAB-seq (Step 6C).
Bisulfite conversion of 5fC/5caC (Step 6A (xiv–xvi), 6C (xiii))
Unlike unmodified C and 5caC (deamination rate >99%), converting 5fC to uracil by bisulfite treatment is less efficient 51. We have optimized the bisulfite conversion conditions using different concentrations of sodium bisulfite (Epitect Fast kit provides a much higher concentration of sodium bisulfite reagent than does the standard Epitect kit) and various incubation times. With the optimized bisulfite conversion protocol described in Table 1 (standard Epitect + 10 h treatment), ~85% of 5fC can be deaminated with minimal effect on the conversion of 5mC or 5hmC (measured by methylated lambda DNA or modification-specific synthetic oligonucleotides).
Table 1.
Comparison of different bisulfite conversion kits and protocols.
| Bisulfite conversion kit | Time | 5mC conversion rate | 5hmC conversion rate | 5fC conversion rate | 5caC conversion rate |
|---|---|---|---|---|---|
| Epitect | 10 h | 2.2% | 3.2% | 83.8% | 99.6% |
| Epitect | 10 h + 5 h | 3.5% | 3.7% | N.A. | N.A. |
| Epitect Fast | 20 min × 2 | 2.8% | 3.3% | 59.9% | 99.5% |
| Epitect Fast | 20 min × 4 | 5.1% | 4.4% | 76.5% | N.A. |
| Epitect Fast | 20 min × 6 | 7.5% | 5.5% | 84.4% | N.A. |
| Epitect Fast | 40 min × 4 | 7.6% | 5.5% | 84.2% | N.A. |
Note 1:
Numbers shown are % of T sequenced as C after bisulfite conversion.
Note 2:
“Epitect, 10h” refers to the recommended protocol (the standard 5-hour thermal program is performed twice).
“Epitect 10h+5h” refers to purifying the DNA after the 10-hour recommended protocol and performing another round of bisulfite conversion using a standard 5-hour thermal program.
For Epitect Fast kit, the standard thermal program is to perform two cycles of 95 °C 5 minutes, 60 °C 10 minutes. All four tested protocols are either elongating the 10 minutes at 60 °C to 20 minutes / 40 minutes or doing four / six cycles instead of two.
Note 3:
There is a clear trend that harsher conditions lead to more side reactions such as unintended deamination of 5mC and 5hmC. 5mC conversion rate is assessed using methylated lambda DNA. 5hmC/5fC/5caC conversion rate is assessed using synthetic 38-mer oligos. The actual conversion rate may slightly differ from the measured conversion rate, as the methylated lambda DNA and synthetic oligos may not be perfect.
Quality controls (Box 1 and 2)
Box 1. Quality control of MAB-seq and caMAB-seq experiment.
Lambda DNA spike-in for assessingM.SssImethylation and bisulfite conversion
-
1
Before starting M.SssI treatment, spike in 0.25% of unmethylated lambda DNA (wt/wt) into sample genomic DNA.
-
2
Perform MAB-seq using the selected protocol (H3K4me1-MAB-seq, WG-MAB-seq, RR-MAB-seq or RR-caMAB-seq).
-
3
Align sequencing reads to lambda DNA genome. CpG sites are expected to be fully methylated (97–99%) while non-CpG sites are expected not to be efficiently methylated (0–1.5%).
Tet1/2/3triple-knockout (Tet TKO) cells orDnmt1/3a/3btriple-knockout (Dnmt TKO) cells as negative controls for assessing false discovery rate (FDR)
-
4
For each set of experiments, include one sample using Tet TKO or Dnmt TKO cells as a negative control and perform the experiments in parallel with samples of interest. When these two cell lines are not available, other samples with low 5fC/5caC levels can serve as a negative control.
-
5
During data analysis, FDR for a sample of interest is calculated as (# of 5fC/5caC-modified CpG called in negative control) / (# of 5fC/5caC-modified CpG called in the sample of interest).
Box 2. 5hmC/5fC/5caC-modified oligo spike-in for validating experimental conditions.
Combine 250 pg modified oligo with 250 ng carrier genomic DNA.
When testing caMAB-seq, perform Step 6C (ii-ix). When testing MAB-seq, perform Step 6C (v-ix).
Perform bisulfite conversion following the protocol in Step 6A (xiv-xvi).
Amplify the bisulfite-converted oligo using NEBNext multiplex oligo for Illumina, purify the amplified DNA and perform Illumina sequencing. A few thousand reads are sufficient for the purpose of quality control.
After sequencing data is obtained, align reads to oligo template sequence. Non-CpG sites are expected not to be methylated, while 5hmC/5fC/5caC-modified CpG sites should behave as expected (Table 1).
To assess the methylation rate within the CpG context, we use unmethylated lambda DNA as spike-in controls in both genome-wide and locus-specific MAB-seq experiments (Box 1). To assess the behavior of oxidized bases (5hmC/5fC/5caC) in experiments for optimizing/validating reagents and conditions, we use modification-specific 38-mer synthetic double-stranded oligonucleotides containing 9 CpG sites (Box 2). Validation of bisulfite treatment reagents by measuring deamination rate of unmodified cytosine can be performed along with MAB-seq experiments, and the bisulfite conversion rate of unmodified cytosine is ~99.5% under optimal experimental conditions.
Enrichment strategy (Step 6B, 6C and Box 3)
Box 3. Locus-specific MAB-seq and caMAB-seq experiment.
-
1
Spike in 0.25% lambda DNA (wt/wt) into genomic DNA of interest.
-
2
When testing caMAB-seq, perform Step 6C (ii-ix). When testing MAB-seq, perform Step 6C (v-ix).
-
3
Perform bisulfite conversion following the protocol in Step 6A (xiv-xvi).
-
4
Amplify regions of interest by PCR using primers designed for bisulfite converted genomic DNA. Programs such as methprimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) can facilitate the BS-PCR primer design. We recommend choosing primers that generate PCR amplicons between 200 and 500bp. In addition, amplify a region from lambda DNA for spike-in controls. For each pair of primers, set up a 15-μL reaction using 2× KAPA HiFi Uracil+ HotStart ReadyMix. Using properly designed primers and sufficient starting material (we typically start with 250 ng genomic DNA and use 1/20 of the bisulfite-converted DNA as template for each BS-PCR reaction), 35 to 40 cycles of amplification should generate sufficient products for visualization on agarose gel.
-
5
Perform downstream analysis using either Sanger sequencing (option A) or Illumina sequencing (option B), depending on the experimental purpose and service availability.
(A) Sanger sequencing
Clone individual PCR product with the Zero Blunt TOPO PCR cloning kit, following manufacturer’s instructions.
For each amplified locus of a sample, pick individual colonies (>30 clones) for Sanger sequencing.
(B) Illumina sequencing
Cast a 2% (wt/vol) agarose gel. Run 1 μL of BS-PCR products (diluted in 9 μL of water with 2 μL of 6x loading dye added) at 20 V cm−1 for 20 min[AU: Please specify conditions i.e. loading dye, run time and voltage]. Include in one lane 10 μL of the quantitative DNA ladder. Image the gel using a standard gel imaging system. By comparing with ladder with known DNA mass, quantify the concentration of all BS-PCR amplicons based on signal intensity using ImageJ or other gel quantification software.
Adjust the concentration of purified PCR products by adding nuclease-free water so that each PCR amplicons has the same molar concentration. Based on the concentrations of BS-PCR products, pool comparable amounts of PCR products for multiple genomic loci (from one sample) together.
Sonicate the mixture to a range between 100 and 200bp. If the PCR amplicons are relatively small (from 200 to 350 bp), it is difficult to completely shear PCR products to smaller fragments. In this case, if more than 20% of the total PCR amplicons appear to be sonicated, proceed to the next step.
Purify the sonicated PCR products using Qiagen MinElute PCR purification kit and prepare sequencing libraries using NEBNext DNA Library Prep Master Mix Set for Illumina
Perform Illumina sequencing using HiSeq 2500 or equivalent sequencers.
-
6
Align sequencing results to amplicon sequences to quantify 5fC/5caC signals.
Whole genome (WG) maps of 5fC and 5caC can be generated by applying MAB-seq analysis to unenriched genomic DNA (termed WG-MAB-seq, Step 6A). However, WG-MAB-seq is sequencing-intensive and can be cost prohibitive for samples with relatively low levels of 5fC/5caC. To reduce sequencing efforts, enrichment strategies can be integrated with the MAB-seq workflow to generate genome-scale maps. By combining chromatin immunoprecipitation (ChIP) with MAB-seq (termed ChIP-MAB-seq), 5fC/5caC abundance can be examined within a fraction of the genome where 5fC/5caC marks tend to enrich (Step 6B) 40. In addition to histone antibodies, DNA immunoprecipitation using an antibody against 5fC and/or 5caC could also be used to first enrich DNA fragments containing these rare modified bases before MAB-seq analysis (unpublished observations). Coupling restriction digestion-based reduced representation (RR) strategy with MAB-seq (termed as RR-MAB-seq) allows 5fC/5caC mapping within genomic regions containing CpG-rich sequences (mostly gene promoters and some repeat sequences) (Step 6C) 43. It is important to note that the MspI enzyme used in standard RRBS only partially digests 5fC-containing C^CGG sites and completely fails to cut 5caCpGs, which will lead to underestimation of 5fC/5caC levels at the digestion sites 10. Therefore, it is essential to choose a restriction enzyme (e.g. TaqαI) that efficiently cut 5fC and 5caC-modified CpGs. Finally, locus-specific MAB/caMAB-seq analysis of regions of interest can be performed using appropriately designed PCR primers (Box 3).
Data analysis (Step 12–18 and Box 4)
Box 4. Identification of 5fC/5caC-enriched genomic intervals.
-
Segment the genome into non-overlapping 100 bp genomic bins. The bin information can be stored in bed format:
chr1 0 99 chr1 100 199 … Identify the CpG sites covered for at least five times in both the sample of interest and the negative control (common 5× CpG sites). Store the information in bed format.
- Identify genomic bins with more than two common 5× CpG sites. Example command using coverageBed (from BEDTools):
/path-to-bedtools/coverageBed -a <common 5× CpG bed file> -b <100 bp bin bed file> | awk -v OFS=‘\t’ ‘{if ($4>=2) print $1, $2, $3, $4;}’ > <bin with at least two 5× CpG sites> For each genomic bin with more than two common 5× CpG sites, sum up the C and T counts from all the common 5× CpG sites in this bin. MAB-seq or caMAB-seq signals of this bin is calculated as [Sum(NT) /(Sum(NT)+ Sum(NC))].
Identify 5fC/5caC-modified bins and assess empirical FDR using the approach described in Step 18.
For MAB-seq analysis, raw signals were calculated as % of T/(C+T) at each CpG dinucleotide. For BS-seq analysis, raw signals were calculated as % of C/(C+T) at each CpG site. For each CpG dinucleotide, we counted the number of “T” bases from MAB-seq reads as 5fC/5caC (denoted NT) and the number of “C” bases as other forms of cytosines (C/5mC/5hmC; denoted NC).
There are potentially two major sources of false positive signals: 1) unmethylated CpG sites that failed to be methylated by M.SssI; 2) bisulfite treatment-induced deamination of 5mC and 5hmC. To statistically account for both sources of false positive signals, we modeled these presumably stochastic events with a binomial distribution, X ~ B(N, p) (N as the sequencing coverage (NT + NC) at a given CpG site and p as the probability of detecting false positive signals, which is the sum of both failure rate of M.SssI and deamination rate of 5mC) 40. Using unmethylated lambda DNA as an internal spike-in control, we can experimentally determine p for every experiment (in our published study, we used 2.04% as p, which was averaged from multiple genome-scale MAB-seq experiments 40) and calculate the probability of whether raw MAB-seq signals [NT/(NC+NT)] are significantly higher than expected by chance. A notable caveat for this statistical filtering strategy is that deamination of 5hmC cannot be directly accounted for. However, given that 5hmC modification level within CpGs is generally ranging from 10–30% and deamination rate of 5hmC in our optimized bisulfite conversion condition is quite small (3.2%), the chance of detecting deaminated 5hmC as 5fC/5caC is quite low (0.3% to 1%) at a given CpG site 40, 46. Indeed, when raw MAB-seq signals for called CpGs in Tdg-depleted cells are compared with those in wild-type cells (where 5hmC is present at similar levels), global MAB-seq signals in Tdg mutant cells are 2–3 fold higher than in wild-type within 5fC/5caC-enriched genomic regions 40. In addition, we have shown that >90% of 5hmC-modified CpGs are non-overlapping with 5fC/5caC-modified sites in mouse ES cells 40. Together, these observations indicate that 5hmC is unlikely to contribute significantly to the false positive signals in MAB-seq.
To estimate empirical false discovery rate (FDR) of calling 5fC/5caC-modified CpGs, the steps above are repeated on MAB-seq signals of negative control sample. For instance, we have successfully used genomic DNAs extracted from Dnmt1/3a/3b / or Tet1/2/3 / mouse ESCs as negative control samples, since these cells do not contain any 5fC or 5caC modifications (Box 1). The FDR for a given P-value cutoff is the number of called CpG sites (false positive signals) in negative controls divided by the number of called CpG sites (potential true signals) in the sample of interest. Please note that it is ideal to have a negative control processed side-by-side with the samples of interest for each batch of experiments, because this allows the estimation of FDR based on parallel experiments.
MATERIALS
REAGENTS
Wild-type feeder-independent mouse ESC line E14TG2a (ATCC, cat. no. ATCC® CRL-1821)
UltraPure™ DNase/RNase-free distilled water (Life Technologies, cat. no. 10977-023)
DPBS (Life Technologies, cat. no. 14190-250)
-
Ethanol, absolute (200 Proof) (Fisher Scientific, cat. no. BP28184)
! CAUTION Ethanol is highly flammable. Handle it in a fume hood.
Glycine (Sigma-Aldrich, cat. no. 50046)
-
Formaldehyde (37%; Fisher Scientific, cat. no. BP531)
! CAUTION Formaldehyde is toxic if absorbed through skin, swallowed or inhaled. It is also flammable. Handle it in a fume hood with appropriate equipment.
-
Triton X-100 (Sigma-Aldrich, cat.no.T8787)
! CAUTION Triton X-100 can cause skin and eye irritation. Handle it with caution.
HEPES buffer solution (1 M; Sigma-Aldrich, cat. no. 83264)
-
KOH (Sigma-Aldrich, cat. no. 484016)
! CAUTION KOH is corrosive if swallowed and causes skin and eye burn. Wear laboratory clothing (gloves, lab coat and goggle) when handling it.
-
Sodium deoxycholate (Sigma-Aldrich, cat. no. D6750)
! CAUTION Sodium deoxycholate is harmful if swallowed and can lead to respiratory irritation. Handle it with caution.
NP-40 Surfact-Amps™ detergent solution (10% w/v; Life Technologies, cat. no. 28324)
N-Lauroylsarcosine sodium salt solution (20%; Sigma-Aldrich, L7414)
LiCl (Sigma-Aldrich, cat. no. L9650)
NaCl (5 M; Life Technologies, cat. no. AM9760G)
-
NaBH4 (Sigma-Aldrich, cat. no. 480886)
! CAUTION NaBH4 is corrosive. Hydrogen gas is produced when NaBH4 reacts with water. Handle it with suitable equipment.
Sodium acetate buffer solution (3 M, pH 5.2±0.1; Sigma-Aldrich, cat. no. S7899)
EDTA (0.5 M, pH8.0; Life Technologies, cat. no. 15575-020)
EGTA (0.5 M, pH8.0; prepared from Sigma-Aldrich, cat. no. 03777)
RNase A (10 mg/mL; Life Technologies, cat. no. EN0531)
Anti-H3K4me1 (10 μg/mL; Abcam, cat. no. ab8895)
TaqαI (20 U/μL; New England Biolabs, cat. no. R0149L)
Klenow fragment (exo–, 5 U/μL; Thermo Scientific, cat. no. EP0422)
T4 DNA ligase (2000 U/μL; New England Biolabs, cat. no. M0202M)
CutSmart buffer (10×; New England Biolabs, cat. no. B7204S)
ATP (100 mM; Thermo Scientific, cat. no. R0441)
dNTP set 100 mM solutions (Life Technologies, cat. no. R0181)
SPRIselect reagent kit (Beckman Coulter, cat. no. B23318)
DNeasy blood & tissue kit (Qiagen, cat. no. 69504)
Qiagen EpiTect DNA bisulfite kit (Qiagen, cat. no. 59104)
QIAquick nucleotide removal kit (Qiagen, cat. no. 28304)
MinElute PCR purification kit (Qiagen, cat. no. 28004)
M.SssI (20 U/μL, New England Biolabs, M0226M)
NEBuffer 2 (10x; cat. no. B7002S)
-
S-adenosylmethionine (SAM) (32 mM; New England Biolabs, cat. no. B9003S)
CRITICAL Always use SAM before its expiration date and make aliquots to avoid multiple cycles of freeze / thaw.
KAPA HiFi Uracil+ HotStart ReadyMix (Kapa Biosystems, KK2801)
NEBNext Multiplex Oligos for Illumina (Index Primers Set 1; New England Biolabs, cat. no. E7335L)
NEBNext Multiplex Oligos for Illumina (Index Primers Set 2; New England Biolabs, cat. no. E7500L)
NEBNext ultra DNA Library Prep Kit for Illumina (New England Biolabs, cat. no. E7370S)
NEBNext DNA Library Prep Master Mix Set for Illumina (New England Biolabs, cat. no. E6040S)
-
Custom methylated adapters (asterisk denotes phosphorothioate bond, all cytosines are modified as 5mC; Integrated DNA Technologies):
Forward: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′
Reverse: 5′-/5Phos/GATCGGAAGAGCACACGTCTGAACTCCAGTC-3′
-
Custom 5hmC/5fC/5caC-modified oligo (X: 5hmC, 5fC or 5caC):
Forward: 5′-AGCCXGXGCXGXGCXGGTXGAGXGGCXGCTCCXGCAGC-3′,
Reverse: 5′-GCTGXGGGAGXGGCXGCTXGACXGGXGXGGXGXGGGCT-3′
CRITICAL Only CpG site of these oligos are modified, which is different from the methylated adaptor in which all cytosines are modified as 5mC.
Unmethylated Lambda DNA (Promega, cat. no. D1521)
Agilent High Sensitivity DNA Kit (Agilent Technologies, cat. no. 5067-4626) cOmplete EDTA-free proteinase inhibitor (Roche, cat. no. 11873580001)
Dynabeads Protein G for Immunoprecipitation (ThermoFisher Scientific, cat. no. 10004D)
RNase A (Life Technologies, cat. no. 12091-201)
Proteinase K (New England Biolabs, cat. no. P8107S)
Glycogen (Roche Life Science, cat. no. 10901393001)
5 PRIME Phase Lock Gel heavy 2 mL (Fisher Scientific, cat. no. FP2302830)
-
Phenol - chloroform - isoamyl alcohol mixture (Sigma-Aldrich, cat. no. 77617)
! CAUTION Phenol can cause skin and eye burn. Wear laboratory clothing (gloves, lab coat and goggle) and handle it with caution.
Qubit dsDNA BR Assay Kit (Life Technologies, cat. no. Q32853)
Qubit dsDNA HS Assay Kit (Life Technologies, cat. no. Q32854)
Zero Blunt TOPO PCR cloning kit (Life Technologies, cat. no. K2800-20).
Illumina sequencing reagent for HiSeq 2500 or equivalent sequencers
EQUIPMENT
Eppendorf 0.2 mL PCR tube strips (Fisher Scientific, cat. no. E0030124286)
Eppendorf RNA/DNA LoBind microcentrifuge tubes (1.5 mL, Sigma-Aldrich, cat. no. Z666548)
Corning™ CentriStar™ centrifuge tubes (15 mL; Fisher Scientific, cat. no. 0553859A)
Corning™ CentriStar™ centrifuge tubes (50 mL; Fisher Scientific, cat. no.0553860)
Agilent 2100 Bioanalyzer (Agilent, cat. no. G2939AA)
Qubit 2.0 Fluorometer (Life Technologies, cat. no. Q32866)
Qubit assay tubes (Life Technologies, cat. no. Q32856)
Branson Sonifier 450 (Branson Ultrasonics)
M220 Focused-ultrasonicator (Covaris)
microTUBE-50 AFA Fiber Screw-Cap (Covaris, cat. No. 5201660)
Filter tips (10, 20, 200, 1000 μL; Genesee Scientific)
Serological pipet (1, 5, 10, 25 mL; Genesee Scientific)
C1000 thermal cycler (Biorad, cat. no. 185-1096)
Eppendorf Thermomixer R (Fisher Scientific, cat. no. 05-400-203)
Vortex-genie 2 (Scientific Industries, cat. no. SI-0286)
Illumina sequencer (HiSeq 2500 or equivalent models)
Software
Trim galore (version 0.3.7): http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
FastQC (version 0.10.1): http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bismark (version 0.12.5) 54: http://www.bioinformatics.babraham.ac.uk/projects/bismark/
Bowtie2 (version 2.2.2) 55: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
Picard (version 1.91): http://broadinstitute.github.io/picard/
Samtools (version 0.1.19) 56: http://samtools.sourceforge.net
IGV genome browser 57: download from https://www.broadinstitute.org/igv/
BEDTools (version 2.19.0) 58: http://bedtools.readthedocs.org/en/latest/
BisSNP (version 0.82.2) 59: http://people.csail.mit.edu/dnaase/bissnp2011/
REAGENT SETUP
Cross-linking buffer (11×)
ChIP cross-linking buffer consists of 50 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 11% (vol/vol) Formaldehyde. This solution is freshly prepared.
Blocking solution
Mix 0.5% (wt/vol) BSA in 1×DPBS. This solution is freshly prepared.
ChIP Lysis buffer
Combine 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% (wt/vol) Sodium deoxycholate, 0.5% (wt/vol) N-lauroylsarcosine, 0.1% (wt/vol) SDS. This solution can be stored at room temperature (22–25 °C) for 1 year.
RIPA buffer
Make ChIP RIPA washing buffer with the following composition: 50 mM HEPES-KOH (pH7.5), 500 mM LiCl, 1 mM EDTA, 1% (wt/vol) NP-40, 0.7% (wt/vol) Na-Deoxycholate. This solution can be stored at 4 °C for 1 year.
TE buffer
This buffer consists of 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. This solution can be stored at room temperature for 1 year.
Elution buffer
The ChIP elution buffer consists of 50 mM Tris-HCl, pH8.0, 10 mM EDTA, 1% SDS. This solution can be stored at room temperature for 1 year.
Mg2+-free M.SssI buffer (10×)
Make 10× home-made M.SssI buffer with the following composition: 100 mM Tris-HCl (pH 8.0), 500 mM NaCl, 100 mM EDTA. Dilute to 1× final concentration when used in reaction. Compared with the supplied NEBuffer 2, this Mg2+-free buffer may help M.SssI work in a more processive manner. This buffer can be stored at 4 °C for at least six months.
dNTP mix for RR-MAB-seq and RR-caMAB-seq
Make dNTP mix stock solution with the following composition: 10 mM dATP, 1 mM dGTP, 1 mM dCTP. Dilute the stock solution 10 times in nuclease-free water to working solution (1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP) immediately before use and add the indicated volume to achieve final concentration. The stock solution can be stored at −20 °C for at least six months.
CRITICAL TaqαI digestion generates 5′ CG overhang (5′-T^CGA-3′, ‘^’ denotes cutting site), therefore dCTP and dGTP are required for end repair. The high concentration of dATP is for 3′ dA-tailing after end repair (both processes are taking place in a single reaction). However, when other restriction enzymes generating different overhangs are used, the dNTP composition needs to be adjusted accordingly. Make working solution each time before use and do not store it.
Methylated adaptors
Dilute the forward and reverse oligos to 100 μM with nuclease-free water. Set up the following reaction to anneal the forward and reverse oligos: 10 μL forward oligo, 10 μL reverse oligo, 2.5 μL water, 2.5 μL NEBuffer 2. The thermal cycle for annealing is: 94 °C 10 min; slowly ramp down to 60 °C and hold for 10 min (1% ramp if using an Eppendorf thermal cycler, or similar rate (for example 0.1 °C per second) if using other thermal cyclers); slowly ramp down to 4 °C and hold (same ramp rate as the previous step). This will generate 40 μM annealed adaptor. Dilute the annealed adaptor to 15 μM, make aliquots and stock at −20 °C. For RR-MAB-seq or RR-caMAB-seq adaptor ligation, dilute the adaptor stock (15 μM) 20 times with water to working solution (0.75 μM) and add the indicated volume. CRITICAL Use LoBind tubes for all procedures to avoid loss of oligos during the preparation and storage, and do not stock diluted oligos.
5hmC/5fC/5caC-modified oligo
Dilute the forward and reverse modified oligos (ssDNA) with nuclease-free water to 1 μg/μL. Set up the following reaction to anneal the forward and reverse oligos: 4 μL forward oligo, 4 μL reverse oligo, 1 μL water, 1 μL NEBuffer 2. Perform oligo annealing using the same thermal program for annealing methylated adaptor oligos described above (the annealing reaction can be scaled down proportionally), which will give rise to 0.8 μg/μL 38-bp dsDNA. Take 2 μL (1.6 μg) for end repair and dA-tailing by NEBNext DNA Library Prep Master Mix Set for Illumina following the manufacturer’s instruction, with two modifications: reaction is scaled down to 60% (60 μL for end repair and 30 μL for dA-tailings); purification after end repair and dA-tailings are performed using QIAquick nucleotide removal kit. Use Qubit dsDNA HS kit to measure the concentration of the recovered 38-bp dsDNA and ligate it to the methylated adaptor with a DNA:adaptor molar ratio of 1:10 (the ligation reaction is set up in 30 μL using the quick T4 ligase provided by the library preparation kit). Purify the adaptor-ligated oligos with QIAquick nucleotide removal kit and adjust the concentration to 1 ng/μL for storage at −20 °C.
CRITICAL Use LoBind tubes for all procedures to avoid loss of oligos during the preparation and storage, and do not stock diluted oligos.
Sodium borohydride aqueous solution for caMAB-seq
Before each experiment, make 1M sodium borohydride solution by dissolving sodium borohydride in nuclease-free water. CRITICAL Sodium borohydride is unstable in aqueous solution and should be made freshly.
PROCEDURE
Cell culture and vitamin C treatment. TIMING 4–5 d
CRITICAL Steps 1–5 can be readily adapted for cell types other than mouse ESCs.
-
1|
Culture wild-type (e.g. E14TG2a), Tdg-depleted 42, and negative control (Tet1/2/3 triple knockout 60 or Dnmt1/3a/3b triple knockout 61) mouse ESCs in feeder-free conditions on 0.1% gelatin coated 6-well plates or 10-cm dishes. Passage the cells every 2–3 days and change culture medium daily.
-
2|
To better mimic metabolic milieu in vivo, maintain mouse ES cells in culture medium containing 100 μg/mL (final concentration) of vitamin C (VC) for 60 hours.
CRITICAL STEP Because VC is present at a relatively high level in both embryonic and adult mouse tissues 62 and VC positively regulates catalytic activity of TET enzymes 63, 64, it is recommended that a physiologically relevant amount of VC is supplemented to cell culture medium either transiently or for long-term.
-
3|
At approximately 70% confluency, aspirate the medium and wash the cells with 1×DPBS. Aspirate DPBS and add sufficient 0.05% (wt/vol) typsin to the culture dish (3 ml for each 10-cm dish). Incubate at 37 °C for 5 min.
-
4|
After the cells have detached, add DMEM with 20% (vol/vol) FBS to inactivate the trypsin (6 ml of 10-cm dish), gently pipette to dissociate cells and collect the cell suspension into a 15-mL Falcon conical tube.
-
5|
Centrifuge the cell suspension at 500 x g for 5 min at room temperature and aspirate the supernatant and discard.
DNA purification and M.SssI treatment
-
6|
Prepare genomic DNA for different scale of analysis and perform enzymatic treatment using CpG DNA methyltransferase M.SssI to protect unmodified CpGs in the genome from bisulfite conversion. Below are protocols for base-resolution mapping of 5fC/5caC using whole genomic DNA [option A: whole genome (WG) MAB-seq], chromatin immunoprecipitated DNA (option B: ChIP-MAB-seq), enzyme digested genomic DNA (option C: reduced representation (RR)-MAB-seq). WG-MAB-seq (option A) provides an unbiased view of 5fC/5caC distribution across the entire genome. ChIP-MAB-seq (option B) using antibody against specific histone marks (e.g. histone 3 lysine 4 monomethylation (H3K4me1) or H3K27me3) can preferentially enriches genomic regions containing relatively high levels of 5fC/5caC (e.g. poised promoters and active enhancers), thus requires less sequencing efforts compared to WG-MAB-seq. Enzymatic digestion-based RR-MAB-seq (option C) provides an approach to enrich CpG-rich promoters and repeat sequences. In option C, we also describe procedures combining caMAB-seq with the enzymatic digestion-based strategy for genome-scale 5caC mapping (RR-caMAB-seq).
(A) Whole genome (WG) MAB-seq TIMING 3 d
-
Wash the cell pellet with 1 mL of 1×DPBS, centrifuge it at 1000 rpm for 5 min at room temperature and aspirate the supernatant and discard.
PAUSE POINT Snap-freeze the cell pellet in liquid nitrogen and the pellet may be stored at −80 °C freezer for at least one month.
Extract genomic DNA from frozen cell pellets with the DNeasy Blood & Tissue Kit and elute purified genomic DNA in 100 μL of nuclease-free water by following the manufacturer’s instructions.
Add 2.5 ng (0.25%, wt/wt) of unmethylated lambda DNA to 1 μg genomic DNA.
-
Set up the first round of M.SssI treatment reaction as follows. Add each component in order of list below. Mix well and incubate the reaction at 37 °C for 4 h.
Component Volume (μL) Final concentration Genomic DNA in Nuclease-free water 41.5 Mg2+-free M.SssI buffer (10×) 5.0 1× S-adenosylmethionine (32 mM) 1.0 0.64 mM M.SssI methylase (20 U μL−1) 2.0 0.8 U μL−1 Total 50 CRITICAL STEP Use fresh S-adenosylmethionine.
TROUBLESHOOTING
Add additional 0.5 μL M.SssI and 1 μL S-adenosylmethionine to the 50 μL reaction. Incubate at 37°C for another 4 h.
Fragment M.SssI-treated genomic DNA (in 50 μL) to an average size of 300–600 bp with Covaris M220 (20% duty factor, 200 cycles per burst, 80 s × 2).
-
Purify sheared DNA with SPRI beads (1.2×). Specifically, vortex the SPRI beads until they are well dispersed. Add 60 μL of SPRI beads to 50 μL of sheared genomic DNA and mix well by repeated pipetting. Incubate the mixture at room temperature for 10 min, and then place the tubes on the magnetic rack until the solution becomes clear (at least 5 min). Remove and discard the supernatant carefully without disturbing the beads. Add 200 μL of freshly prepared 80% (vol/vol) ethanol and wait for 30 seconds; carefully remove the supernatant. Rpeat the 80% ethanol wash step once more. Let the tubes stand at room temperature for approximately 10 min with the lids open until the beads become dry. Resuspend the beads with 60 μL nuclease-free water, mix well by repeated pipetting and incubate the mixture at room temperature for 2 min. Place the tube on the magnetic rack and wait for at least 5 min until the solution becomes clear. Transfer the supernatant to new tubes carefully to avoid disturbing the beads.
CRITICAL STEP The size range of sheared genomic DNA can be examined by Agilent Bioanalyzer and should be 150–800 bp.
-
Perform end-repair/dA-tailing of sheared DNA with the NEBNext Ultra DNA Library Prep kit from Illumina. Set up the end-repair/dA-tailing reaction as follows and perform on the thermocycler: first at 20 °C for 30 min, then at 65 °C for 30 min and finally at 4 °C for temporary storage.
Component Volume (μL) Purified DNA from Step 6A(vii) 55.5 End repair reaction buffer (10×) 6.5 End prep enzyme mix 3 Total 65 -
Perform adaptor ligation reaction with the NEBNext Ultra DNA Library Prep kit from Illumina. Set up the ligation reaction as follows and perform on the thermocycler: first at 20 °C for 15 min, then at 4 °C.
Component Volume (μL) End-repaired/dA-tailed DNA from Step 6A(viii) 65 Blunt/TA ligase mix 15 Ligation enhancer 1 Methylated adaptors (15 μM) 2.5 Total 83.5 Purify methylated-adaptor-ligated DNA with SPRI beads (1.0×) and elute with 44 μL nuclease-free water.
-
Set up the second round of M.SssI treatment reaction as follows and add each component in order of list below. Mix well and incubate the reaction at 37 °C for 4 h.
Component Volume (μL) Final concentration Genomic DNA in Nuclease-free water from Step 6A(x) 41.5 NEBuffer 2 (10×) 5.0 1× S-adenosylmethionine (32 mM) 1.0 0.64 mM M.SssI methylase (20 U μL−1) 2.0 0.8 U μL−1 Total 50 Add additional 0.5 μL M.SssI and 1 μL S-adenosylmethionine to the 50 μL reaction. Incubate at 37°C for another 4 h.
Purify sheared DNA with SPRI beads (1.2×) and elute with 22 μL nuclease-free water.
-
Perform bisulfite conversion of the purified DNA with the Qiagen EpiTect bisulfite kit. Prepare the bisulfite reactions in 200-μL PCR tubes according to the instruction below and add each component in the order listed.
Component Volume (μL) M.SssI-treated DNA 20 Bisulfite conversion mix 85 DNA protection buffer 35 Total 140 TROUBLESHOOTING
-
Perform the bisulfite conversion reaction on a thermal cycler and run the following thermal cycling program.
Step Temperature (°C) Time (min) 1. Denaturation 95 5 2. Incubation 60 25 3. Denaturation 95 5 4. Incubation 60 85 5. Denaturation 95 5 6. Incubation 60 175 Repeat step 1–6 one more time (two cycles of step 1–6 in total) - - 7. Hold 20 Indefinite -
Purify bisulfite converted DNA according to the Qiagen EpiTect bisulfite kit manufacturer’s instructions and elute the DNA with 20 μL EB buffer.
CRITICAL STEP Add carrier RNA provided by the Qiagen EpiTect bisulfite kit to the supplied BL buffer to enhance the recovery rate.
CRITICAL STEP To prepare the carrier RNA solution, add 310 μL RNase-free water to the lyophilized carrier RNA (310 μg per vial) to obtain a 1 μg/μL solution. Dissolve the carrier RNA thoroughly by vortexing.
CRITICAL STEP Elute with pre-warmed (50 °C) EB buffer to increase yield.
TROUBLESHOOTING
(B) Chromatin immunoprecipitation for genome-scale MAB-seq TIMING 5 d
CRITICAL Step 6B (i-xiii) describes the protocol of chromatin immunoprecipitation used for the published H3K4me1-MAB-seq analysis 40. The protocol may need to be optimized for other antibodies.
-
Resuspend the cell pellet (approximately 10 million cells from Step 5) in 10 mL of cold 1×DPBS. Add 1 mL cross-linking buffer and incubate for 10 min at room temperature on a rotating platform. Quench the cross-linking reaction with 125 mM glycine for 5 min.
CAUTION Formaldehyde is harmful if inhaled or absorbed through skin. Waste should be disposed of according to local regulation.
-
After two washes with 10 mL cold 1×DPBS, centrifuge the cell suspension at 1000×g for 5 min.
PAUSE POINT Snap-freeze the cell pellet in liquid nitrogen and the pellet may be stored at −80 °C freezer for at least one month.
Prepare 100 μL protein G Dynabeads for each ChIP sample. Wash beads with 1 mL Blocking solution by inverting the tubes several times and then place the tubes on magnetic rack to collect beads. Resuspend Dynabeads in 250 μL Blocking solution and add 10 μg anti-H3K4me1 antibodies. Immobilize the antibody with beads for at least 6 h at 4 °C. After incubation, wash beads with 1 mL Blocking solution twice and resuspend beads in 50 μL Blocking solution.
-
Lyse cell pellet from Step 6Bii with 1 mL ChIP lysis buffer and incubate on ice for 10 min.
CRITICAL STEP Add freshly prepared proteinase inhibitor (50x stock) to ChIP lysis buffer.
-
Sonicate chromatin using a microtip in a container of ice/water mixture. We use the Branson sonifier 450 with 30 cycles of 10 sec ON/30 sec OFF pulses at 20% power amplitude. Pellet cell debris at 16,000 ×g at 4 °C for 10 min. Transfer the supernatant to a new tube and discard the pellet.
CAUTION Always wear ear protection when operating the sonicator.
CRITICAL STEP Size range sheared fragments should be 200–1,000 bp in length. We recommend optimizing the sonication conditions before the actual ChIP experiment. It should be noted that different sonifiers and cell types require separate optimizations. To experimentally estimate the fragment size range, add 150 μL of elution buffer to 50 μL of sheared chromatin and reverse cross-link the sample by incubating the tube at 65 °C overnight. Purify the DNA (see Step 6B (ix-xiv)) and examine the DNA fragment size by standard Agarose gel electrophoresis or Agilent Bioanalyzer.
Measure chromatin concentration using Nanodrop (concentration [μg/μL] is estimated by OD260×50×dilution factor/1000). Dilute the chromatin to the concentration of 0.5 mg/mL in ChIP lysis buffer and aliquot 500 μg of chromatin (in 1 mL) for each ChIP sample.
-
Add 100 μL of 10% (vol/vol) Triton-X 100 to 1 mL of 500 μg chromatin immediately before IP. Perform Immunoprecipitation overnight at 4 °C by adding 50 μL antibody-conjugated Dynabeads from Step 6B (iii).
CRITICAL STEP Optimal immunoprecipitation conditions need to be separately determined for other antibodies.
Wash DNA/protein complexes with 1 mL of RIPA buffer for five times and with 1 mL of TE/50 mM NaCl once. To wash the sample, first invert several times and then place the sample tubes on a rocking platform for 2 min. Place tube on magnetic rack for 1 min to collect beads each time (until the solution becomes clear). After the final wash, spin at 1000×g for 3 min at 4 °C to pellet the magnetic beads and discard TE/50 mM NaCl buffer.
Add 210 μL of elution buffer to the bead pellet and incubate at 65 °C for 15 min on a Thermomixer (shaking at speed of 1000×rpm). After incubation, spin down the beads at 16,000×g for 1 min at room temperature. Transfer 200 μL of supernatant and reverse cross-link DNA/protein complexes by incubating the sample at 65 °C overnight on a Thermomixer.
Add 200 μL of TE to each tube of reverse cross-linked IP sample. Add 8 μL of 10 mg/mL RNaseA (0.2 mg/mL final concentration) and incubate at 37 °C for 2 h. Add 7 μL of 300 mM CaCl2 and 4 μL of 20 mg/mL proteinase K (0.2 mg/mL final concentration) sequentially and incubate at 55 °C for 30 min.
-
Add 400 μL of phenol:chloroform:isoamyl (25:24:1) alcohol to each tube and mix the sample on a vortex mixer. Prepare one Phase Lock Gel tube for each ChIP sample by spinning tube at 16,000×g at room temperature for 30 sec. Transfer 800 μL of sample to the Phase Lock tube. Spin the sample in a centrifuge at 16,000×g for 5 min at room temperature.
CAUTION Phenol and chloroform are both toxic and should only be used in a fume hood.
Transfer the aqueous layer (~400 μL) to a new 1.5 mL LoBind microcentrifuge tube. Add 16 μL of 5M NaCl (200 mM final concentration), 1.5 μL of 20 μg/μL glycogen (30 μg total), and 880 μL EtOH. Mix and cool the mixture for 30 min at −80 °C.
Spin the mixture at 16,000×g for 10 min at 4 °C, discard supernatant and wash the pellet with 1 mL of cold 70% EtOH.
-
Dry the pellet at 37 °C for 5–10 min until residual EtOH is completely removed. Thoroughly resuspend each pellet in 30 μL of EB buffer.
PAUSE POINT Eluate can be stored at −20 °C for at least 1 month.
-
Measure the concentration of ChIP DNA with Qubit dsDNA HS Assay Kit and add 0.25% (wt/wt) of unmethylated lambda DNA to ChIP DNA.
CRITICAL STEP The expected yield from the H3K4me1 ChIP experiment described here is 100–200 ng, thus 0.25–0.5 ng of unmethylated lambda DNA should be added to H3K4me1 ChIP DNA as internal controls for M.SssI treatment.
End-repair/dA-tailing immunprecipitated DNA as in Step 6A (viii).
-
Perform methylated adaptor ligation as in Step 6A (ix–x).
CRITICAL STEP The concentration of the methylated adaptor should be adjusted according to the approximate concentration of ChIP DNA. For instance, we use 10-fold less adaptor for H3K4me1 ChIP DNA in this step (compared to WG-MAB-seq).
Perform the first round of M.SssI treatment as in Step 6A (iv–v).
Purify sheared DNA with SPRI beads (1.2×) and elute with 44 μL nuclease-free water.
Perform the second round of M.SssI treatment as in Step 6A (xi–xii).
Purify sheared DNA with SPRI beads (1.2×) and elute with 22 μL nuclease-free water.
Perform bisulfite conversion of M.SssI-treated, methylated adaptor-ligated ChIP DNA using Qiagen Epitect Bisulfite kit as in Step 6A (xiv–xvi).
(C) Enzymatic digestion-based caMAB-seq and MAB-seq TIMING 3 d
CRITICAL Step 6C (ii-iv) is specifically for caMAB-seq. For standard MAB-seq method, skip step 6C (ii-iv) and start from Step 6C (v) using 250 ng genomic DNA mixed with 0.625 ng unmethylated lambda DNA (0.25% w/w spike-in).
-
Purify genomic DNA from cultured cells as in Step 6A (i–ii).
CRITICAL STEP For RR-MAB-seq based simultaneous mapping of 5fC/5caC, jump to Step 6C (v).
Add 5 μL freshly prepared sodium borohydride aqueous solution (1 M) to 250 ng genomic DNA mixed with 0.625ng unmethylated lambda DNA (0.25% w/w spike-in) in 15 μL nuclease-free water. Incubate the reaction at room temperature in darkness for 1 h. During the 1 h incubation, briefly spin the reaction tube and open the lid to release the pressure every 15 min..
-
Quench the reaction by slowly adding (dorp-by-drop) 10 μL sodium acetate (0.75 M, pH=5) to the 20 μL reaction from Step 6C (ii). Incubate until no bubbles are observed (~1.5 h).
CRITICAL STEP The reaction needs to be quenched completely to prevent any interference with downstream steps. Check every 15 min until no more gas is released.
Add 370 μL of nuclease-free water to 30 μL of sample from Step 6C (iii). Purify the DNA by phenol:chloroform:isoamyl (25:24:1) alcohol extraction and ethanol precipitation following the procedure described in Step 6B (xi-xiii). Dry the pellet at 37 °C for 5–10 min until residual EtOH is completely removed. Thoroughly resuspend each pellet in 45 μL of EB buffer.
-
Set up a 50 μL M.SssI treatment reaction as follows and incubate at 37 °C for 4 h.
Component Volume (μL) Final concentration DNA in Nuclease-free water from Step 6C (i) or 6C (iv) 43 Mg2+-free M.SssI buffer (10×) 5 1× S-adenosylmethionine (32 mM) 1 0.64 mM M.SssI methylase (20 U μL−1) 1 0.4 U μL−1 Total 50 CRITICAL STEP Step 6C (v-xiii) are common steps for RR-MAB-seq and RR-caMAB-seq.
CRITICAL STEP Use fresh S-adenosylmethionine.
Add additional 1 μL M.SssI and 1 μL S-adenosylmethionine to the 50 μL reaction. Incubate at 37 °C for another 4 h.
Heat inactivate the reaction at 65 °C for 20 min.
Add 350 μL of nuclease-free water to ~50 μL of sample from Step 6C (vii). Purify the DNA by phenol:chloroform:isoamyl (25:24:1) alcohol extraction and ethanol precipitation following the procedure described in Step 6C (iv). Dissolve the DNA pellet in 45 μL nuclease-free water.
Repeat the procedure described in Step 6C (v-viii) for the second round of M.SssI treatment except replacing Mg2+-free M.SssI buffer with NEBuffer 2 (10×).
-
Set up the TaqαI digestion reaction as follows using 1 ng of purified M.SssI-treated DNA. Incubate at 65 °C for 3 h followed by heat inactivation at 80 °C for 20 min.
Component Volume (μL) Final amount/concentration DNA + water 15.7 1 ng NEB Cutsmart buffer 1.8 1× TaqαI (20 U μL−1) 0.5 0.556 U μL−1 Total 18 -
Set up an end-repair reaction by adding 2 μL end-prep mix (prepared as follows) to the 18 μL mixture from Step 6C (x). Incubate at 37 °C for 40 min followed by heat inactivation at 75 °C for 15 min.
End-prep mix (2 μL per reaction) Volume (μL) Final concentration Klenow fragment exo-(5 U μL−1) 1.0 0.25 U μL−1 NEB Cutsmart buffer 0.2 1× dNTP mix for RRBS (1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP) 0.8 40 μM dATP, 4 μM dCTP, 4 μM dGTP (in 20 μL) Total 2 μL (total reaction volume: 20 μL) CRITICAL STEP Dilute dNTP mix stock (10 mM dATP, 1 mM dCTP, 1 mM dGTP) 10 times in nuclease-free water to working concentration immediately before use.
-
Set up an adaptor-ligation reaction by first adding all components of ligation mix except T4 ligase (4 μL) to the 20 μL mixture from Step 6C (xi). Mix the reaction by gentle tapping the tube. Then add 1 μL of T4 ligase and immediately mix the reaction by gentle tapping. Incubate at room temperature for 10 min, 16 °C for 3 h and 4 °C overnight followed by heat inactivation at 65 °C for 20 min.
Ligation mix (5 μL per reaction) Volume (μL) Final concentration Nuclease-free water 2.25 T4 ligase (2000 U μL−1) 1.0 80 U μL−1 NEB Cutsmart buffer 0.5 1× ATP (100 mM) 0.25 1 mM (in 25 μL) Methylated adaptor (0.75 μM) 1.0 30 nM (in 25 μL) Total 5 (total reaction volume: 25 μL) CRITICAL STEP Dilute methylated adaptor stock solution (15 μM) 20 times in nuclease-free water to working solution (0.75 μM) immediately before use. Discard the remaining working solution.
CRITICAL STEP To avoid the potential ligation between methylated adaptors (which leads to unwanted adaptor dimers in the final library), we recommend adding the ligation mix in a stepwise manner as described above and mixing the reaction immediately after the T4 ligase is added.
-
Add 10 μL nuclease-free water to the 25 μL ligation reaction mixture from Step 6C (xii).
Perform bisulfite conversion and DNA purification using Qiagen Epitect Bisulfite kit as in Step 6A (xiv–xvi) with minor modifications in reaction composition as below:
Component Volume (μL) M.SssI-treated DNA + water 35 Bisulfite conversion mix 85 DNA protection buffer 20 Total 140
Library preparation and quality control TIMING 2–3 d
-
7|
Prepare the library amplification PCR reaction as follows. Add each component in the order listed.
Component Volume (μL) Nuclease-free water 3.5 Bisulfite converted DNA 19.0 NEBNext Universal primer (10 μM) 1.25 NEBNext Index primer (10 μM) 1.25 KAPA HiFi Uracil+ ReadyMix (2×) 25 Total 50 CRITICAL STEP KAPA HiFi Uracil+ ReadyMix must be used in this PCR reaction because this enzyme is insensitive to the presence of uracil in the DNA template (uracil usually causes DNA polymerase to stall). Alternatively, PfuTurbo Cx hotstart DNA polymerase (Agilent), which is also resistant to uracil stalling, can be used in this step.
CRITICAL STEP NEBNext Universal and Index primers are from the NEBNext Multiplex Oligos for Illumina kits. Depends on the version of the kits, the primer concentration can be either 25 μM (older version) or 10 μM (lastest version). Our PCR reaction composition is calculated using the latest version of the kits. Adjust the composition accordingly if 25 μM primers are to be used.
TROUBLESHOOTING
-
8|
Perform library preparative PCR using the thermal cycling program below.
CRITICAL STEP The total number of PCR cycle needs to be adjusted according to initial DNA amount. WG-MAB-seq (from Step 6A) typically needs 5–6 cycles of PCR amplification; H3K4me1-MAB-seq (from Step 6B) usually requires 10–12 cycles; enzymatic digestion-based MAB-seq (from Step 6C) generally requires 15–17 cycles.
Step Temperature (°C) Time 1. Denature 95 2 min 2. Denature 98 20 sec 3. Anneal 63 30 sec 4. Extend 72 1 min Repeat step 2–4 for 5–17 cycles - - 5. Extend 72 5 min 6. Hold 4 Indefinite TROUBLESHOOTING
-
9|
Purify library preparative PCR products with SPRI beads, using option A for DNA from WG-MAB-Seq (Step 6A) and H3K4me1-MAB-Seq (Step 6B) or option B for DNA from RR-MAB-Seq or RR-caMAB-seq (Step 6C).
(A) WG-MAB-seq or H3K4me1-MAB-seq library
Purify the amplified library with 60 μL SPRI beads (1.2×) following the procedure described in Step 6A (vii) and elute with 15 μL nuclease-free water.
(B) RR-MAB-seq or RR-caMAB-seq library
Perform size selection of amplified library with SPRI beads using double size selection strategy. First, add 30 μL (0.60×) SPRI beads to 50 ul PCR reactions. Incubate the mixture at room temperature for 10 min, and then place the tubes on the magnetic rack for 10 min (until the solution becomes clear). Transfer the supernatant to a new tube and discard the beads. Then, add 40 μL (1.4×) SPRI beads to 80 μL supernatant and place the tubes on the magnetic rack for 10 min. Discard the supernatant and wash the beads with 200 μL of freshly prepared 80% ethanol twice. Let the beads air dry at room temperature for 10 min. Elute size-selected final libraries with 15 μL nuclease-free water.
-
10|
Quantify the concentration of final libraries (from Step 9) with a Qubit fluorometer and the dsDNA HS assay kit. Assess the size distribution of final libraries with Agilent Bioanalyzer 2100 and high-sensitivity DNA kit (Figure 4). Calculate the molar concentration (nM) of final libraries based on the average size (in base pairs, measured by Bioanalyzer) and library concentration (in ng/μL, measured by Qubit) using the following formula.
Sample concentration (nM) = (library concentration (ng/μL) / 1000) / (Average fragment size (bp) × 660) × 109
TROUBLESHOOTING
-
11|
Sequence the libraries using Illumina HiSeq 2000/2500 or NextSeq 500 sequencers with a final concentration at ~80% of that used in regular sequencing experiments.
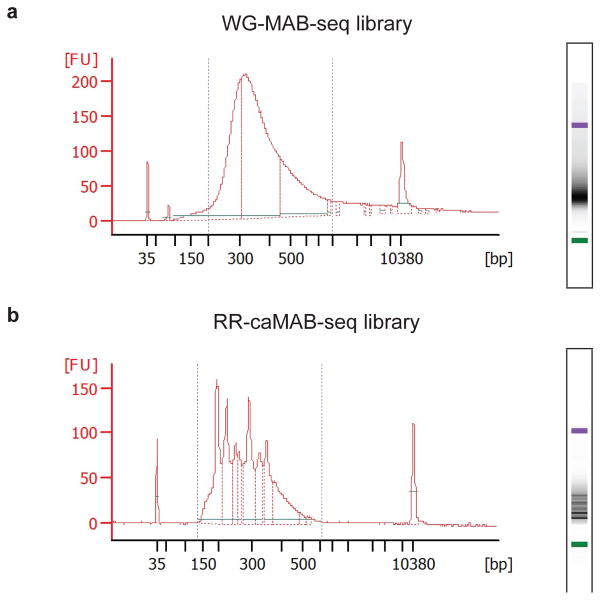
Figure 4. Bioanalyzer electropherogram of sequencing libraries of MAB/caMAB-seq.
(a) Representative example of final library size distribution obtained from whole-genome MAB-seq experiments.
(b) Representative example of final library size distribution obtained from TaqαI-digested RR-caMAB-seq experiments. Sharp peaks are from digestion of repetitive elements.
Data analysis of MAB-seq Timing 2d
-
12|
Analyze data using option A for WG-MAB-seq or ChIP-MAB-seq, or using option B for RR-MAB-seq or RR-caMAB-seq
(A) WG-MAB-seq and H3K4me1-MAB-seq
Trim low-quality bases and contaminating adaptor sequences using the Trim Galore program. Example command:
trim_galore --fastqc --gzip --length 36 <raw.fastq.gz>
(B) RR-MAB-seq or RR-caMAB-seq
Trim raw sequencing reads for Illumina adaptor sequences, low-quality bases and experimentally-introduced 3′ CG using Trim Galore. Example command:
trim_galore --fastqc --gzip --three_prime_clip_R1 2 --length 36 <raw.fastq.gz>
CRITICAL STEP During the end repair following TaqαI digestion, an unmodified CG is experimentally introduced at 3′ end of the fragments and will be sequenced as TG after bisulfite conversion. To ensure that this artificial CG does not interfere with the analysis, after adaptors and low-quality bases are trimmed, an additional two bases at 3′ end are trimmed from all reads.
CRITICAL STEP In this case, we only keep reads no less than 36bp in length after trimming. This cutoff can be adjusted for sequencing libraries with different read length.
-
13|Align trimmed reads to the bisulfite-converted reference genome using the Bismark program. The reference genome sequences (e.g. mm9.fasta plus lambda.fasta) need to be first bisulfite converted and indexed in silico. Example command:
/path-to-bismark/bismark_genome_preparation --bowtie2 -path_to_bowtie <bowtie2 folder> --verbose <genome folder>
Once the bisulfite-converted reference genome index for bismark/bowtie2 is generated, use the following command line to map trimmed reads:/path-to-bismark/bismark --bowtie2 --path_to_bowtie <bowtie2_folder> - - samtools_path <samtools folder> --fastq -p 8 --gzip --bam <genome folder> <trimmed.fastq.gz>
This command will generate a BAM file containing all the alignment output.
-
14|Use SortSam.jar and MarkDuplicate.jar in the Picard toolkit to sort the mapped reads and remove PCR duplicates, respectively. Example command:
java -Xmx32g -jar /path-to-picard-tools-1.91/SortSam.jar \ INPUT=<bismark-aligned.bam> \ OUTPUT=<sorted.bam> \ SORT_ORDER=coordinate CREATE_INDEX=true java -Xmx32g -jar /path-to-picard-tools-1.91/MarkDuplicates.jar ASSUME_SORTED=true REMOVE_DUPLICATES=true \ INPUT=<sorted.bam> \ OUTPUT=<deduplicated.bam> \ METRICS_FILE=MarkDuplicates.metrics.txt \ VALIDATION_STRINGENCY=SILENT CREATE_INDEX=true
These commands will generate a sorted BAM file containing only monoclonal reads.
CRITICAL STEP Because restriction digestion instead of random shearing is used for fragmenting genomic DNA in RRBS-based experiments, skip this step for RR-MAB-seq or RR-caMAB-seq analysis.
-
15|Use Bismark Methylation Extractor program provided by the Bismark program to extract 5fC/5caC modification information for each CpG site. For MAB-seq, 5fC+5caC level at a CpG site equals to NT /(NT + NC). For caMAB-seq, 5caC level at a CpG site equals to NT /(NT + NC). Example command:
/path-to-bismark/bismark_methylation_extractor --single-end --gzip -- bedGraph --counts --genome_folder <genome sequence folder> --output <output folder> <deduplicated.bam>
This commands will generate a compressed bedgraph file, <bismark.cov.gz>, which can be used to extract raw MAB-seq signals and sequencing coverage information for each CpG site in the genome. This bedgraph file contains six columns: <chromosome> <start position> <end position> <percentage of C> <number of C> <number of T>.
CRITICAL STEP Because the samples are spiked in with unmethylated lambda DNA, the M.SssI methylase efficiency can be estimated by the methylation level within CpG dinucleotides of lambda DNA.CRITICAL STEP For downstream analysis, we recommend using CpG sites with at least 5× coverage, because some low-coverage CpG sites arise from wrong alignment or other unwanted situations.
-
16|Visualize raw MAB-seq signals in the IGV genome brower. Example command:
zcat <bismark.cov.gz> | awk ‘BEGIN{OFS=“\t”} {coverage=$5+$6; if (coverage >= 5) print $1,$2,$3,100-$4}’ | gzip > <bismark.bedgraph.gz> igvtools tile <bismark.bedgraph.gz> <bismark.bedgraph.tdf> mm9This command will generate a .tdf file for visualizing raw MAB-seq signals in the IGV browser (Figure 5) 40.
-
17|Mouse strain or human tissue/cell-line-specific single nucleotide polymorphisms (SNPs; e.g. C-to-T mutations) overlapping with CpG dinucleotides in the reference genome can affect the calculation of MAB-seq signals [NT /(NT + NC)] by introducing false positive or false negative signals. Known SNPs can be used to exclude the affected CpG sites from downstream analysis. Obtain SNP information from Sanger Institute (https://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1303)65 and create a bed file summarizing all the genomic CpG sites containing SNP (at C, G or both). Exclude these SNP-containing CpG sites using the intersectBed command from the BEDTools. Example commands:
/path-to-bedtools/intersectBed -a <Unfiltered bed file> -b <SNP- containing CpG bed file> -v > <Filtered bed file>
CRITICAL STEP If WG-MAB-seq or ChIP-MAB-seq data sets are of sufficient sequencing depth (e.g. at least 10 reads per strand), de novo SNPs overlapping with annotated CpG sites in the reference genome can be identified and removed by using the BisSNP program. In the case of RR-MAB-seq and RR-caMAB-seq, only a small proportion of the CpG sites are covered in both positive and negative strands. Thus, de novo SNP detection which relies on sequencing information obtained from both strands will not remove all the SNPs.
CRITICAL STEP In addition to SNP filtering, we recommend removing CpG sites with unusually high levels (for instance, CpGs in >= 80% reads are sequenced as TpGs) of raw MAB-seq signals [NT /(NT + NC)] in both the negative control (e.g. DNMT1/3a/3b mouse ES cells) and the sample of interest. It is possible that these annotated CpG sites present in reference genome could be mutated (e.g. C-to-T) in the sample.
-
18|To identify CpG sites modified by significant level of 5fC/5caC, we employ the binomial distribution (N as the sequencing coverage (NT + NC) and p as the probability of detecting false-positive signals (the sum of M.SssI methylase failure rate and deamination rate of 5mC) to assess the probability of observing NT or greater by chance. Example R command:
apply(df, 1, function(x) diff(pbinom(c(x[1],x[2]), size=x[2], prob=p)))
df: a data frame (df) contains two columns - the first one (x[1]) is the number of T reads and the second one (x[2]) is the total number of reads [C+T]).
p: the probability of detecting false-positive signals (we used 2.04% in the published study).
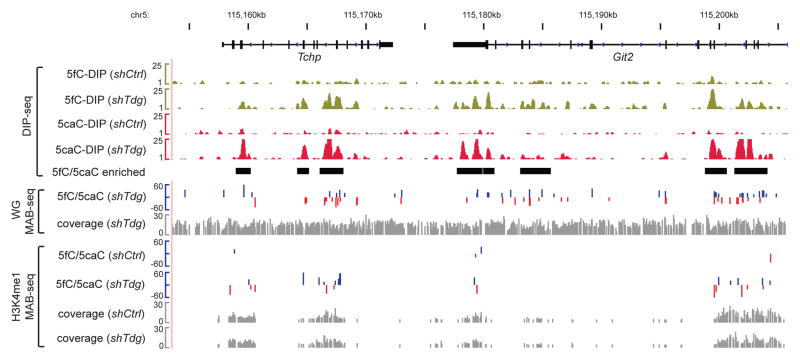
Figure 5. Base-resolution 5fC/5caC maps and affinity-enrichment-based 5fC/5caC maps at the Tchp-Git2 locus in mouse ES cells.
For WG-MAB-seq or H3K4me1-MAB-seq datasets, positive values (blue) indicate CpGs on the Watson strand, whereas negative values (red) indicate CpGs on the Crick strand (vertical axis limits are −60% to +60%). Only CpGs sequenced to depth 5 and associated with statistically significant level of 5fC/5caC (FDR=5%) are shown. Sequencing coverage for WG-MAB-seq or H3K4me1-MAB-seq is shown in gray in separate tracks (vertical axis limits are 0 to 30 reads). Highlighted by black horizontal bars are 5fC/5caC-enriched genomic regions identified by DNA immunoprecipitation following by sequencing (DIP-seq). The vertical axis limits for DIP-seq datasets are 1 to 25 (in rp10m: reads per 10 million reads). shCtrl, control knockdown mouse ES cells; shTdg, Tdg knockdown mouse ES cells.
To estimate empirical FDR of calling 5fC/5caC-modified CpGs, calculate the above statistics at each common CpGs in negative control samples (Dnmt1/3a/3b or Tet1-3 deficient cells) in which true 5fC/5caC signals are absent. The empirical FDR for a given P-value cutoff is the number of called CpGs in negative controls divided by the number detected in the sample of interest. For RR-MAB-seq and RR-caMAB-seq analysis, 5fC/5caC-modified sites identified using the binomial distribution-based p-value cut-off approach can be further filtered by a numeric cut-off (for example NT /(NT + NC) should be at least 10%). In our hands, applying this additional numeric filter to RR-MAB-seq and RR-caMAB-seq datasets allows comparatively low FDR without compromising the detection sensitivity. For data sets with low sequencing depth or of cell-types with relatively low levels of 5fC/5caC, analyzing genomic bins (divide genome into 100bp bins) instead of single nucleotides may identify 5fC/5caC-marked regions with increased accuracy and sensitivity (Box 4). This is probably because true 5fC/5caC signals tend to cluster together while false positive signals tend to disperse randomly in the genome (at least in mouse ESCs). Identification of 5fC/5caC-enriched genomic regions by binning strategy is conceptually similar to identification of differentially methylated regions (DMRs) in WGBS or RRBS analysis. Therefore, other tests (e.g. Fisher exact test) can be used as alternatives to binomial distribution-based test.
TIMING
Steps 1–5, cell culture and sample harvest: 4–5 d
Step 6A, DNA purification and enzymatic treatment of WG-MAB-seq: 3 d
Step 6B, DNA purification and enzymatic treatment of ChIP-MAB-seq: 5 d
Step 6C, DNA purification and enzymatic treatment of RR-MAB-seq: 3 d
Steps 7–10, library preparation PCR and purification of final library: 1 d
Steps 10–11, quality control and high-throughput DNA sequencing: 2 d
Steps 12–18, data analysis for MAB-seq data: 2 d
Box 1, quality control of MAB-seq and caMAB-seq experiment: 1 d
Box 2, oxidized methylcytosine synthetic oligo spike-in experiment: 2 d
Box 3, locus-specific MAB-seq and caMAB-seq experiment: 3 d
Box 4, Identification of 5fC/5caC-enriched genomic intervals 1 d
TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 6A (iv) | Low M.SssI methylation efficiency | Poor quality reagents (e.g. expired SAM) Too much DNA is used |
Use fresh SAM and make SAM aliquots stored at −20 °C to avoid excessive rounds of freezing / thawing. Methyl transfer reaction generates SAH, a potent inhibitor of M.SssI. Therefore, reducing the amount of input DNA will help to reduce the generation of SAH and increase M.SssI efficiency. Additionally, when input DNA is sufficient, perform multiple rounds of M.SssI treatment (with DNA purification through phenol:chloroform:isoamyl (25:24:1) alcohol extraction and ethanol precipitation in between; following the procedure described in Step 6B (xi–xiii).). |
| 6A (xiv–xvi) | Incomplete / excessive bisulfite conversion | Poor quality reagents Non-optimal protocols when using other bisulfite conversion kits |
If Epitect bisulfite kit is used, the dissolved bisulfite mix can be stored at −20 °C and used within a month, but avoid freezing / thawing more than once. DNA protection buffer and buffer BD should be stored at 4 °C and, ideally, replaced every 6 months. When using kits other than Qiagen Epitect bisulfite kit, test the protocol first using synthetic oligos and lambda DNA to make sure that 5fC is efficiently converted while 5mC and 5hmC are not wrongly deaminated. |
| 6A (xiv–xvi) | Poor yield of DNA after bisulfite conversion | DNA degradation during bisulfite conversion DNA loss during purification |
Purify the DNA shortly (within 6 hours) after the thermal program is completed to avoid any potential DNA degradation. Use fresh DNA protection buffer (green color and less than 6 months old). Carrier RNA should be added freshly to buffer BL before use to facilitate DNA recovery, and dissolved carrier RNA should be stored at −20 °C. Use EB warmed to 50 °C to facilitate elution from the column. |
| 7–8 | Low PCR amplification efficiency of the library | Poor DNA yield / quality before PCR amplification Poor quality of PCR reagents |
Use LoBind tubes when dealing with small amounts of DNA and avoid unnecessary pipetting. Use glycogen to facilitate DNA precipitation during PCI purification. Once start, try to avoid pausing for a long time (>12 hours) in between steps. Make aliquots of PCR reagents and avoid excessive cycles of freezing / thawing. |
| 10 | Abnormal size / shape of library after PCR amplification | Insufficient / excessive sonication of DNA (for WG-MAB-seq and H3K4me1-MAB-seq) Poor DNA yield / quality (for RR-MAB-seq) |
Adjust sonication conditions correspondingly for different amounts of input DNA / different equipment / different enrichment methods. For RR-MAB-seq library preparation, avoid pausing for a long time (>6 hours) once end preparation is started. |
ANTICIPATED RESULTS
Sequencing library preparation
For H3K4me1-MAB-seq and WG-MAB-seq, the expected yield after SPRI bead purification is 2–5 ng/μL in 15 μL, if the PCR conditions are optimized. For RRBS-MAB-seq and RRBS-caMAB-seq, the expected yield after SPRIselect purification is 3–5 ng/μL in 15 μL. The expected size range of an ideal library is 200–500 bp for H3K4me1-MAB-seq and for WG-MAB-seq, with an average size of 350–400 bp (Figure 4a). For RR-MAB-seq and RR-caMAB-seq based on TaqαI digestion, the expected size range of an ideal library is 150–650 bp (this range can be different if other enzymes are used), with an average size of 250–350 bp (Figure 4b).
Sequencing coverage for different genome-scale MAB-seq experiments
For WG-MAB-seq analysis of mouse genome, we have sequenced approximately 300–400 million reads (100–150 bp single-end) per library on HiSeq 2500 or NextSeq 500. After trimming adaptor sequences and low quality bases, roughly 80% of reads can be uniquely aligned to the mouse genome. By combining two biologically independent sequencing libraries (>500 million uniquely mapped and monoclonal reads), >95% of CpG dyads (out of a total of ~21 million CpG dyads) in the mouse genome can be covered with an average of 28x coverage per CpG dyad (combine both strands). Under this sequencing coverage, ~34 million CpGs (~80% of a total of ~42 million CpGs) can be covered by at least 5 times per strand. For H3K4me1-MAB-seq, we typically sequence roughly 100 million reads (100 bp single-end) per sample. Approximately 80% of reads can be uniquely mapped to mouse genome after quality filtering. Under this sequencing depth, 3–4 millions of CpG dyads can be covered by at least 5 times per CpG dyad. For RR-MAB-seq or RR-caMAB-seq analysis, we usually sequence around 30 million reads (100 bp single-end) per library. After adaptor trimming and filtering low-quality / short reads, around 45% of the reads can be mapped uniquely to the reference genome using the parameters specified in the data analysis part, giving rise to around 12 million uniquely mapped reads. Under this sequencing depth, around one million CpG sites are covered for at least 5 times per CpG (on each strand).
Statistical calling of 5fC/5caC-modified CpGs
In our published study 40, WG-MAB-seq analysis of Tdg-depleted and VC-treated (60 h) mouse ESCs identified 675,325 5fC/5caC-modified CpGs (2.7% out of 24,872,637 CpGs with coverage 10, using a stringent p-value cut-off leading to an FDR<5%). With a similar FDR cutoff (<5%) and sequencing depth filtering ( 10 per CpG dyads), H3K4me1-MAB-seq analysis identified 127,576 (7.6% out of 1,670,036 CpG dyads with N 10) 5fC/5caC-modified CpG dyads in VC-treated, Tdg-depleted mouse ESCs (shTdg+VC). The higher percentage of 5fC/5caC-modified CpG identified in H3K4me1-marked genomic domains is expected as 5fC and 5caC are preferentially enriched in these gene regulatory regions. Choosing the p-value cutoff partly depends on the purpose of the downstream analyses. For a biological sample with much less 5fC/5caC (e.g. wild-type mouse ES cells), it requires much higher sequencing depth to resolve true 5fC/5caC signals from background. To partially circumvent the requirement of ultra-high sequencing depth, analysis of MAB-seq datasets by combining individual CpG sites into genomic bins (e.g. 100 bp intervals) and by statistical calling with a looser FDR cutoff (e.g. 20%) may provide a lower-resolution map of 5fC/5caC-modified CpGs in these cell-types with lower levels of 5fC/5caC. Similarly, RRBS-MAB-seq data sets tend to generate a higher FDR at a given p-value due to following two reasons. First, PCR duplicates cannot be removed when using standard RRBS strategy, increasing the probability that random background signals are overly amplified. Second, RRBS preferentially cover GC-rich regions that are generally depleted of 5fC/5caC (at least in mouse ES cells). These potential problems can be in part solved by adopting unique molecule identifier (UMI)-based RRBS approach 66 to remove PCR duplicates, and by using other restriction enzymes that cover more 5fC/5caC-modified regions.
EDITORIAL SUMMARY: This protocol for methylase-assisted bisulfite sequencing (MAB-Seq) and caMAB-Seq methods allows the analysis of active DNA demethylation, directly mapping 5fC and 5caC at single base resolution.
Acknowledgments
H.W. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and is currently supported by the National Human Genome Research Institute (R00HG007982). X.W. was supported by the China Scholarship Council. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS: H.W. and X.W. performed experiments and carried out data analysis. H.W., X.W. and Y.Z. wrote the manuscript.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests. Reprints
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011 doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10 doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang J, et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci U S A. 2015;112:E4236–4245. doi: 10.1073/pnas.1510510112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011 doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 22.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi S, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaas GA, et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET Family Proteins and Their Role in Stem Cell Differentiation and Transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song CX, He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem Sci. 2013 doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Zhang Y. Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol. 2015;22:656–661. doi: 10.1038/nsmb.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Iurlaro M, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015 doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raiber EA, et al. 5-Formylcytosine alters the structure of the DNA double helix. Nat Struct Mol Biol. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szulik MW, et al. Differential stabilities and sequence-dependent base pair opening dynamics of Watson-Crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine. Biochemistry. 2015;54:1294–1305. doi: 10.1021/bi501534x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Wu X, Shen L, Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song CX, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neri F, et al. Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15:447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Lister R, et al. Global epigenomic reconfiguration during Mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu L, et al. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Z, et al. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol Cell. 2015;57:750–761. doi: 10.1016/j.molcel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 50.Xia B, et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nature methods. 2015 doi: 10.1038/nmeth.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nature Chemistry. 2014 doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu X, et al. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13:R61. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsumura A, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 62.Sotiriou S, et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nature medicine. 2002;8:514–517. doi: 10.1038/0502-514. [DOI] [PubMed] [Google Scholar]
- 63.Blaschke K, et al. Vitamin_C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013:1–7. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin R, et al. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. Journal of the American Chemical Society. 2013 doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 65.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K, et al. Q-RRBS: a quantitative reduced representation bisulfite sequencing method for single-cell methylome analyses. Epigenetics. 2015;10:775–783. doi: 10.1080/15592294.2015.1075690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]