Abstract
Observations of distributions of microorganisms and their differences in community composition across habitats provide evidence of biogeographical patterns. However, little is known about the processes controlling transfers across habitat gradients. By analysing the overall microbial community composition (bacteria, fungi, archaea) across a terrestrial-freshwater gradient, the aim of this study was to understand the spatial distribution patterns of populations and identify taxa capable of crossing biome borders. Barcoded 454 pyrosequencing of taxonomic gene markers was used to describe the microbial communities in adjacent soil, freshwater and sediment samples and study the role of biotic and spatial factors in shaping their composition. Few habitat generalists but a high number of specialists were detected indicating that microbial community composition was mainly regulated by species sorting and niche partitioning. Biotic interactions within microbial groups based on an association network underlined the importance of Actinobacteria, Sordariomycetes, Agaricomycetes and Nitrososphaerales in connecting among biomes. Even if dispersion seemed limited, the shore of the lake represented a transition area, allowing populations to cross the biome boundaries. In finding few broadly distributed populations, our study points to biome specialization within microbial communities with limited potential for dispersal and colonization of new habitats along the terrestrial-freshwater continuum.
Microorganisms, comprising much of the biodiversity on Earth, play major roles in functions and processes in a wide range of ecosystems such as soils or freshwaters1. Bacterial communities are central to most biogeochemical cycles in both soils and aquatic ecosystems2,3,4. Fungi are ubiquitous and diverse. They play a dominant role in decomposition and nutrient cycling in soil through their saprotrophic activities, take part in mycorrhizal associations in soil and can be abundant in freshwaters3,5,6,7,8,9,10. Archaea are also widespread and phylogenetically diverse in terrestrial and freshwater ecosystems, the latter being considered as the largest reservoirs of archaeal genetic diversity11,12,13. Archaea are actively participating in ecosystem processes such as methanogenesis and nitrogen cycle14. Whereas ecosystem functioning can be affected by the identity and relative abundance of microbial taxa15,16,17, an understanding of the local distribution of microorganisms within the microbial metacommunity is needed to improve predictions of ecosystem responses to global change.
Distributions and co-occurrence patterns of microorganisms as well as differences in community composition across habitats (beta diversity) provide evidence of biogeographical patterns for microorganisms18,19,20,21. Both deterministic and stochastic processes have been used to explain microbial community assembly22,23. Deterministic processes assume that community composition is driven by environmental factors leading to species sorting24. Species interactions and competition may further affect the composition of microbial communities and drive their distribution patterns25,26,27,28. Stochastic processes on the other hand control community composition by dispersal limitation, mass effect and random birth/death events29. Microbial community composition seems to be governed by both deterministic and stochastic processes30. However, depending on the habitat, the spatial scale and the community considered the relative influence of these processes will differ23,31. It has been shown that habitat generalists, showing broad environmental tolerances, and habitat specialists, having more restricted habitat ranges, are assembled by different mechanisms leading to complex metacommunity dynamics32,33.
Most studies on distribution of microorganisms, as well as structure and turn-over of microbial communities focus on one biome and little is known about the processes mediating and controlling transfers across a landscape gradient involving terrestrial and aquatic biomes, and the resulting biogeographical patterns of microbial community distribution. However, there is evidence of microbial community linkages between soil and adjacent freshwater systems34,35. Therefore, the aim of the present study was to understand the spatial distribution patterns of microbial populations and identify taxa capable of crossing the terrestrial-freshwater biome border (habitat generalists) as well as those specific to a particular habitat (habitat specialists). Moreover, we explored the co-occurrence patterns between microorganisms across the terrestrial-freshwater gradient using correlation network analysis. In this way, potential interactions among microorganisms can be dissected20. We analysed all three microbial domains of life, focusing on bacteria, archaea and fungi as they are all key players in ecosystem functioning. Five adjacent soils presenting different aboveground vegetation types, land managements or environmental properties were sampled around lake Erken in central Sweden, and freshwater and sediment samples were collected at different depths, including the shore line (Fig. 1). To our knowledge this is the first study to comprehensively investigate the bacterial, fungal and archaeal communities across a terrestrial-freshwater gradient.
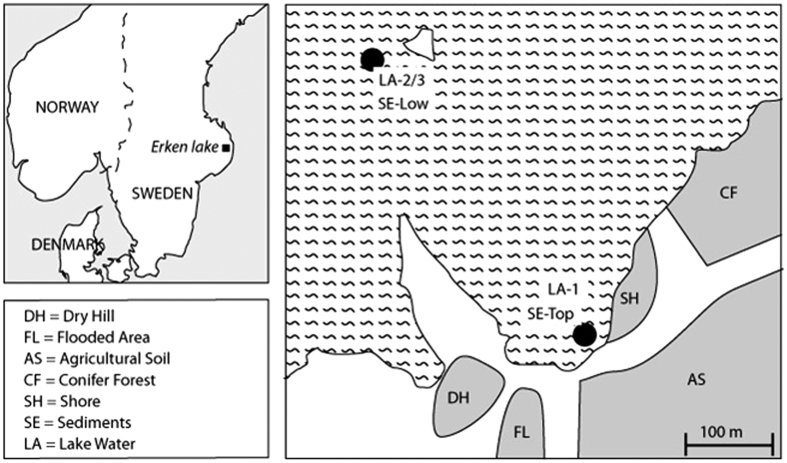
Figure 1. Location and description of sampling sites.
The figure was composed using Adobe Illustrator CS3 software (www.adobe.com) based on Google Earth (Map data: Google, DigitalGlobe).
Results
Microbial community distribution across the terrestrial-freshwater continuum
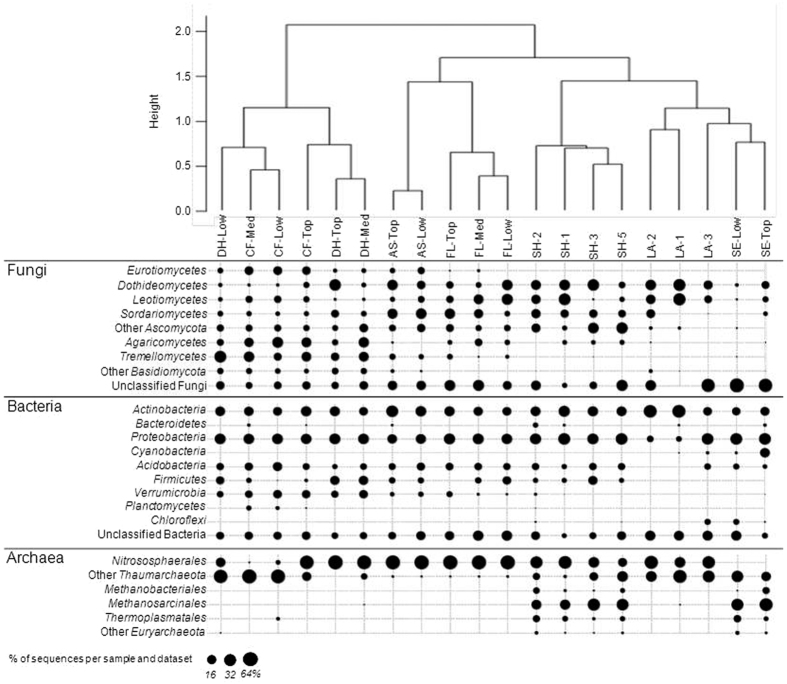
The different samples from the terrestrial-freshwater gradient were clustered according to the similarity of the community composition based on the main OTUs across the environmental gradient (Fig. 2). Two main clusters were evident; one including the dry hill (DH) and conifer forest (CF) site samples and the other with the rest of the sites. One sub-group within the latter was formed by the agricultural soil (AS) and flooded area (FL) samples, while the shore (SH), lake (LA) and sediment (SE) samples made up a remaining sub-group. This indicates a gradient of increasing similarity in the microbial communities from forested sites to the lake. The composition of the microbial communities within each unique site was very similar since the samples from each particular site grouped together regardless of sampling depth, which suggests habitat specificity. The exception was the samples from the dry hill (DH) and the coniferous forest (CF). These sites clustered together apart from the other sites, but differed within each of the two sites according to depth. This difference was mainly due to a lower relative abundance of Nitrososphaerales in the lower depths of the forest and dry hill samples, whereas the top samples displayed higher abundances (Fig. 2).
Figure 2. Similarity of community composition as revealed by hierarchical clustering of the distribution of the main OTUs with a dot plot indicating the relative abundance of the main microbial taxa at different depths in the environmental samples: Dry hill (DH), Conifer forest (CF), Flooded area (FL), Shore (SH), Agricultural soil (AS), lake side (LA-1), Lake surface (LA-2), 10 m depth lake (LA-3), Sediments at the lake side (SE-Top) and in the middle of the lake (SE-Low).
Each sample corresponds to the averages of the biological triplicates (Fig. S2).
The microbial communities from the shore were more similar to the microbial communities from the lake water and sediments than to those from the other samples. In the freshwater sampled at the lake surface (LA-1 and LA-2), the most common fungal, bacterial and archaeal taxa were Ascomycota, Actinobacteria and Thaumarchaeota, respectively. In the deep water (LA-3), the microbial community composition was more similar to the ones observed in the sediments than to those in the littoral and surface water (LA-1 and LA-2) with lower relative abundances of Actinobacteria and higher relative abundance of Proteobacteria, respectively, as well as a higher presence of Acidobacteria (Fig. 2).
The distribution of the fungal taxa across the environmental gradient showed that the Basidiomycota (Agaricomycetes and Tremellomycetes) were dominant in the DH and CF samples, whereas the Ascomycota (mainly the Dothideomycetes, Leotiomycetes and Sordariomycetes) were more abundant in the other sites (Fig. 3). Regarding the bacterial phyla, the Actinobacteria and the Proteobacteria were present in all environmental samples, and the Acidobacteria in nearly all samples (Fig. 2). The Firmicutes and Verrucomicrobia were typically found in terrestrial samples, with the latter mainly at the driest sites, whereas the Chloroflexi were mainly detected in the deep lake samples (LA-3 and SE-Low) and the Cyanobacteria in the sediments from the littoral zone (SE-Top). Archaea were represented at all the sites across the environmental gradient, especially the Thaumarchaeota that were detected in all samples. The Euryarchaeota, which include methanogenic archaea (Methanobacteriales and Methanosarcinales) were predominant in the shore and sediment samples (Fig. 2).
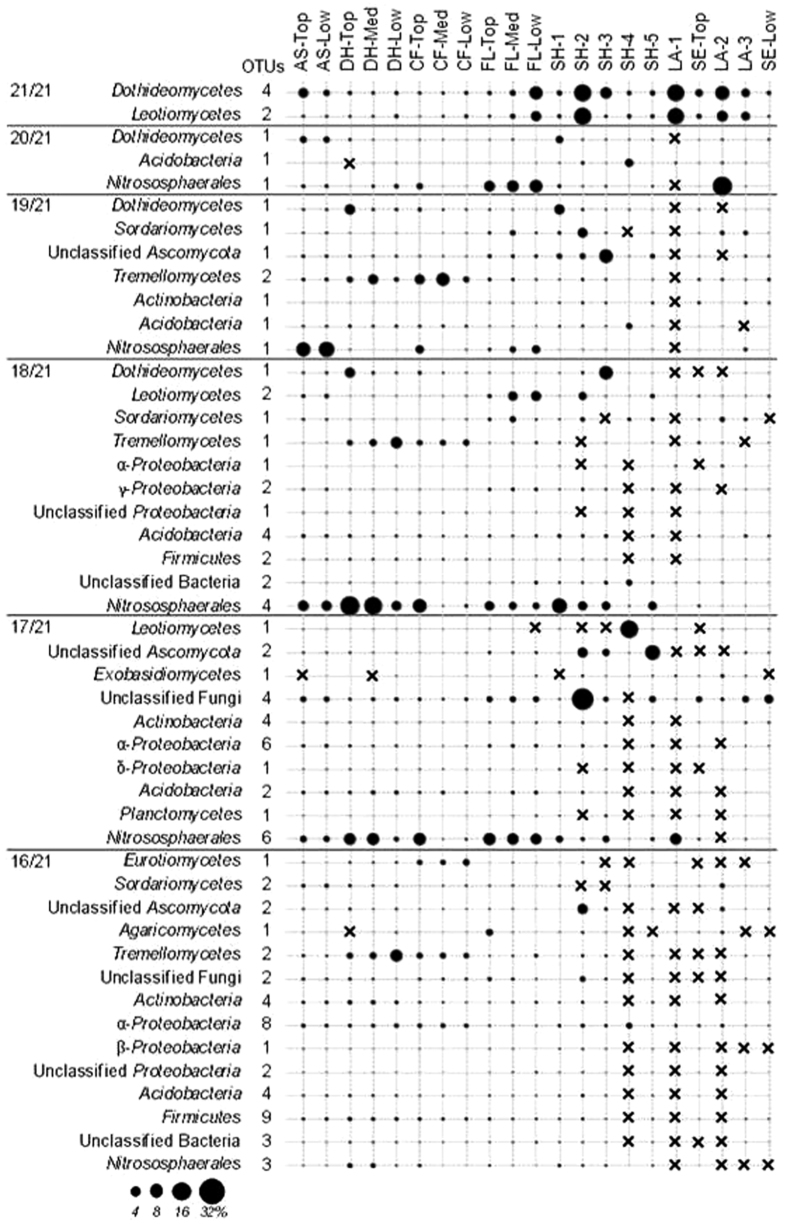
Figure 3. Dot plot of the relative abundance of the habitat generalists detected in all (occurrence 21/21) and almost all the sampling sites (occurrence <21/21).
Each sample corresponds to the averages of the biological triplicates. Crosses indicate no detection.
Ubiquity and specificity of microorganisms along the gradient
Along the environmental gradient, only a few OTUs were habitat generalists, being ubiquitously distributed all along the terrestrial-freshwater gradient, whereas the majority was more narrowly distributed and specific to a unique site or even site depth, indicating environmental niche specialization (habitat specialists). Among the habitat generalists, only six fungal OTUs belonging to the Dothideomycetes and the Leotiomycetes were detected at all sites and all depths (Fig. 3). Among these taxa, Cladosporium sp., Pilidium concavum and two Capnodiales were identified and they were most abundant in freshwater and mineral soils (FL-Low and SH-2; Fig. 3). Some Tremellomycetes affiliated to Trichosporon sp. and Cryptococcus sp. were detected in almost all samples with the exception of one freshwater sample (LA-1), and they were found in higher relative abundance in the dry hill (DH) and conifer forest (CF) soils (Fig. 3). For the bacteria, only one OTU belonging to the subgroup Gp16 of the Acidobacteria was widely distributed and detected in all samples but the ones from the upper layer of the dry hill (DH-Top), with higher relative abundance in the lower mineral layer of the shore site (SH-4; Fig. 3). Regarding the archaeal OTUs, we only detected Nitrososphaerales as habitat generalists and one OTU was present in all samples, except for the freshwater sample from the littoral zone (LA-1). The relative abundances of the Nitrosphaerales generalist OTUs were the lowest in the deeper layers at several sites: conifer forest soil (CF-Med and -Low), mineral layer from the shore (SH-4), sediments and freshwater (LA-3; Fig. 3).
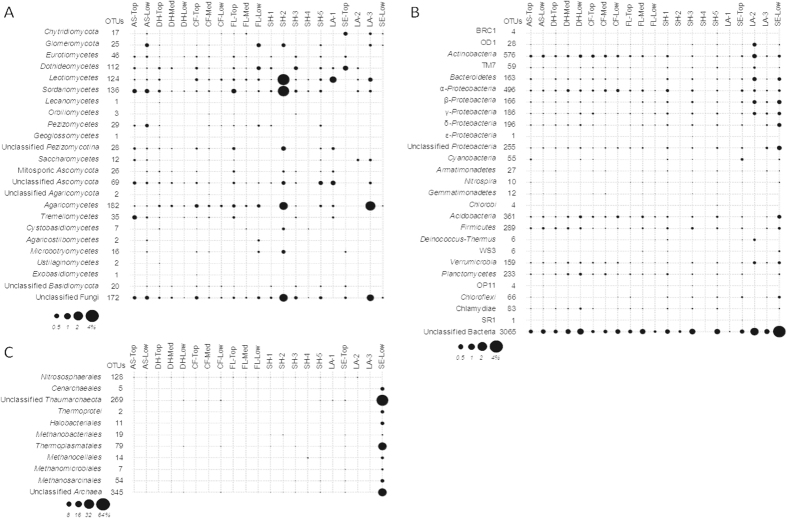
Several microbial OTUs distributed across a wide range of different habitats were not detected in the surface freshwater sampled in the littoral zone (LA-1), and in the middle of the lake (LA-2) or within the deepest mineral layer from the shore (SH-4). This could be attributed to the lower bacterial abundances detected in these habitats (Fig. S1). These sites might potentially correspond to unique and particularly challenging habitats for bacteria. Many microbial OTUs were in fact habitat specialists, being detected at only one site and depth (Fig. 4). Within the fungal domain, the highest specificity was observed for the top mineral layer of the shore site (SH-2; Fig. 4A) with a high relative abundance of Leotiomycetes, Sordariomycetes and Agaricomycetes. This habitat specificity towards the SH-2 sample was not observed for bacteria nor archaea (Fig. 4B,C). Instead, habitat specialists among the bacterial OTUs were found for most of the sites, but not for the mineral soil layers of the shore (SH-2 and SH-4; Fig. 4B). In the surface lake sample (LA-2) and the deep sediments (SE-Low), high proportions of specific bacteria were observed within the Actinobacteria, Bacteroidetes and Proteobacteria (Fig. 4B). Except for the Nitrososphaerales, the deep sediment of the lake was a highly specific habitat for archaea (Fig. 4C).
Figure 4.
Dot plot of the relative abundance of the habitat specialists (OTUs within microbial taxa) detected in only one site and depth along the terrestrial – freshwater biomes taxa ((A) Fungi; (B) Bacteria; (C) Archaea).
Co-occurrence patterns of microbial communities
The microbial co-occurrence patterns were explored using network inference based on strong and significant Spearman’s correlations (ρ > 0.06; P < 0.001) and using the main OTUs (relative abundance higher than 1%; Fig. 5). The resulting microbial network consisted of 710 nodes (OTUs) and 10 812 edges, representing the microbial associations, with an average degree of 15.2 node connectivity that corresponds to the number of connections to other nodes in the network. Among the total microorganisms, the fungal OTUs were the most represented within the network with 470 nodes, whereas the bacterial and archaeal OTUs were represented by 93 and 147 nodes, respectively. Five bacterial OTUs (three unclassified bacteria and two Actinobacteria) had the highest degree of association (degree = 54) with correlation coefficients ranging from 0.6 to 1. Within the fungal and archaeal domains, a single Sordariomycetes and some Thaumarchaeota (Nitrososphaerales and unclassified) had the highest degrees of association (37 and 36, respectively).
Figure 5. Co-occurrence network of the main OTUs based on correlation analysis.
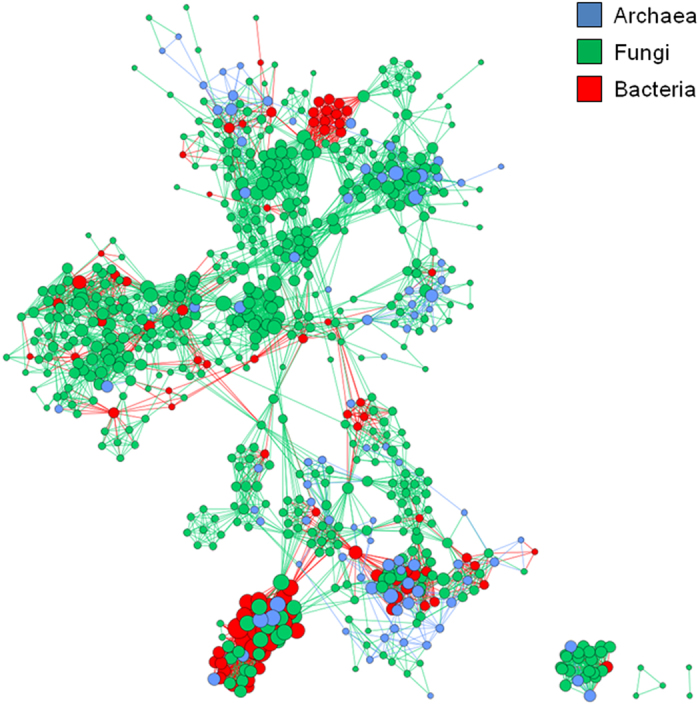
Nodes correspond to microbial OTUs and edges to the microbial associations. A connection stands for a strong (Spearman’s ρ > 0.6) and highly significant (P < 0.001) correlation. The size of each node is proportional to the number of degree.
Within the bacterial domain, the Actinobacteria and the Proteobacteria were the most represented phyla with 30 and 27 nodes and an average degree of 29.9 and 17.2 node connectivity, respectively. Among the archaea, an unclassified Thaumarchaeota and a Nitrososphaerales were the most represented phyla with 53 and 52 nodes and an average degree of 15.4 and 13.9 node connectivity, respectively. Finally, in the fungal domain, the Sordariomycetes was the most represented phyla with 88 nodes and an average degree of 14.1 node connectivity. Some co-occurrence patterns, involving the most frequently connected microbial taxa were observed, e.g. the co-occurrence of two Actinobacteria (153 degrees), two Sordariomycetes (144 degrees), some Nitrososphaerales and Sordariomycetes (128 degrees) and between two Agaricomycetes (127 degrees). Focusing on the six fungal habitat generalists identified previously, only the one identified as Pilidium concavum did not have significant microbial association, whereas the other OTUs had degrees of association varying from 7 to 18.
Discussion
By analysing the fungal, bacterial and archaeal community composition along a terrestrial-freshwater gradient, we identified the dominant habitat specialists and generalists within the three domains of life across the habitats along the environmental gradient. Habitat specialists were defined as OTUs detected at only one site. These OTUs could possibly be present also at other sites but at lower abundances that escape detection in the pyrosequencing assay used. However, by pooling the three sample replicates, we further increased the probability of detecting a specific OTU at a particular site even if it is rare. We thus assume that OTUs detected at only one site can be considered as habitat specialists.
Across the biomes that were sampled, different spatial patterns were observed for the different domains. As expected, members of Actinobacteria and Proteobacteria were detected along the entire gradient, which can be explained by their known diversity and dominance in the environment36,37. The Euryarchaeota, especially the methanogenic Methanosarcinales, were more abundant in the sediments and the shore samples compared to the terrestrial sites, which agrees with previous observations13,38. At the OTU level, only a few habitat generalists, but a high number of specialists were detected. Only six fungal OTUs appear to cross the biome boundaries with occurrences in all samples. Similarly, Barberán et al.39 only found 2% of all bacterial OTUs in more than 300 of the 596 soil samples they collected in Central Park. Moreover, they detected 58% of the OTUs in less than 10 of the soils analysed. The ubiquity of microorganisms can be explained by either better dispersion, higher adaptation and competition abilities or less resource needs, which all would increase their niche breadth along the terrestrial-freshwater gradient in our study32. Considering the significant co-occurrence patterns observed for five of the six identified fungal habitat generalists, these organisms should be both strong competitors and dispersers, whereas a trade-off between competition and colonization potentially occurs for the OTU that did not feature any significant co-occurrence pattern40. Slow growing oligotrophy, such as often observed for Acidobacteria41, could also be an advantage for the ubiquity of microorganisms that are more resistant to nutrient resource changes within their habitat and thus, making them able to sustain a viable population in heterogeneous environments. Acidobacteria were indeed widely distributed and especially subgroup 16, a common member of acidobacterial communities42. In the archaeal domain, only OTUs from the Nitrososphaerales were identified as habitat generalists being distributed along the entire gradient except in the freshwater samples from the littoral zone, corroborating previous results showing the wide range of habitats in which these ammonia-oxidizing archaea have been detected43. Some specific habitats did not harbour habitat generalists that were otherwise widely distributed across the sites. These habitats (the surface freshwater from the littoral zone and from the middle of the lake and the deepest mineral layer from the shore site) harbored the lowest bacterial abundances in agreement with previous work reporting lower microbial biomass in freshwater than in soil44. The surface freshwater from the littoral zone and from the middle of the lake along with the deepest mineral layer from the shore site may be less hospitable or more favorable to competitive non-generalist microorganisms, constituting particular niches where few habitat generalists could establish. As previously observed, niche partitioning can strongly structure microbial communities45. Habitat specialists are known to have a limited niche, but the highest fitness in their optimal habitat40. However, due to habitat specialisation, specialists are much more susceptible to changes in environmental conditions and thereby to extinction as compared to the generalists32. For example, even though generalist bacteria were detected in the mineral soil layer in the shore samples (Acidobacteria, Actinobacteria and Proteobacteria), none of these phyla and only a few unclassified bacterial OTUs were identified as habitat specialist in these samples. Moreover, except for OTUs belonging to the Nitrososphaerales, many archaea were specific to the sediments sampled at the deepest point of the lake. By using the ecological concept of ‘indicator species’ for archaeal lineages and analysing sequences from various environmental samples (freshwater and marine sediments and plankton, hydrothermal vents, soils), Auguet et al.13 concluded that the Methanomicrobiales could be considered as an indicator group of freshwater sediments. In our study, the Methanomicrobiales did not constitute the most important specialist archaeal taxa in the sediments. Instead, the sediments were a more specific habitat for Thaumarchaeota and Thermoplasmatales, indicating that the concept of ‘indicator species’ is context dependent and therefore difficult to apply globally.
Interestingly, the upper mineral soil layer from the shore harboured a high proportion of habitat-specific fungi. Whereas the relative abundance of the habitat specialists Leotiomycetes and Sordariomycetes coincided with overall fungal abundances across the sites this was not the case for the habitat specialist Agaricomycetes, which were relatively more abundant locally than the total Agaricomycetes community. This indicates that the habitat specialist distribution across an environmental gradient does not necessarily reflect the one at the metacommunity level and that specialist taxa are not always rare taxa as previously hypothesized33. Habitat specialist could be considered as species banks for dispersal and colonization of new and favorable habitats facilitated by production of resting cysts or spores which can survive long periods and disperse through air and water46,47. In the present study, the shore site, through its physical localisation between the soil, lake water and sediments, could represent a transitional dispersion area for microbial populations between these different environments. This site is subjected to flooding events which would allow inoculation of the lake by soil microorganisms and vice versa due to the physical exchange, as proposed by Crump et al.35. Vertical dispersal could also explain the similarity observed between the deep lake water and the sediments, which both harboured similar relative abundances of Actinobacteria, Proteobacteria, Acidobacteria and Chloroflexi. The exchange might be driven by both sedimentation in the water column and by the nutrient fluxes at the water-sediment interface48.
Contrasting community assembly mechanisms have been proposed for habitat generalists and specialists, either through species sorting or spatial factors32,33. However, considering that these two groups would respond differently to environmental and spatial factors, the spatial turnover of microbial communities will be modified according to their relative importance. In the present study, we observed that microbial communities were more similar within habitats than across habitats, most likely due to environmental filtering as suggested by the few habitat generalists detected. Species sorting could also result from biotic interactions between microorganisms such as competition, mutualistic or syntrophic relationships. These interactions were analysed and visualized through the co-occurrence analyses and association network, which may correspond to microorganisms performing similar or complementary functions and/or sharing similar preferred environmental conditions, but not necessarily having physical interactions27,49,50. Interestingly, while the fungal taxa were the most represented within the network, bacteria, especially two actinobacterial OTUs, had the highest number of associations. This suggests that they have a large impact on the microbial community structure along the environmental gradient. The network was supported by both microbial intra-and inter-phyla associations and underlined the importance of Actinobacteria, Sordariomycetes, Agaricomycetes and Nitrososphaerales in the complex ecological interactions shaping the microbial community structure across the biomes studied.
Conclusion
By analysing bacterial, fungal and archaeal community composition across a terrestrial-freshwater gradient, we identified only a few microbial OTUs as habitat generalists able to cross biome boundaries. Many habitat specialists were detected at each site, which suggests that environmental filtering was an important assembly mechanism for microorganisms within all three domains and that dispersal was limited. This low ubiquity of microorganisms shows the susceptibility of microbial communities to changing environments. However, areas allowing dispersal, such as the shore that links soil and freshwater, were likely important to maintain diversity across the environmental gradient.
Materials and Methods
Environmental samples
To create a terrestrial-freshwater gradient, soil, sediment and water were sampled in adjacent sites and within lake Erken in central Sweden (59°51′N, 18°36′E) in October 2009 (Fig. 1). Soil cores (22 cm depth × 3 cm diameter) were sampled at five different sites differing in their vegetation, characterised and denoted as ‘Dry hill’ (DH) dominated by Quercus robur in the tree layer, ‘Flooded area’ (FL) dominated by Alnus glutinosa in the tree layer, ‘Agricultural soil’ (AS) dominated by annual cereal crops, ‘Conifer forest’ (CF) dominated by Picea abies in the tree layer, and ‘Shore’ (SH) mainly covered by Salix in the bush layer. For each site, three replicates were sampled. According to the soil profile, each soil core was subsampled either into i) a top (0–5 cm depth), a medium (8–13 cm depth) and a lower (17–22 cm depth) layer (DH, CF and FL -Top/-Med/-Low) or ii) a top (0–5 cm depth) and a lower (17–22 cm depth) layer (AS -Top/-Low). The FL-Low layer was visually a clay-rich mineral soil. The SH cores were divided into five equal layers since they were composed of alternated organic (SH -1/-3/-5) and mineral-sandy layers (SH-2/-4). Each subsample was homogenised by sieving (2 mm mesh size).
Lakewater samples were obtained from the littoral zone (LA-1), and from both surface (1 m depth; LA-2) and 10 m depth (LA-3) at the deepest point of the lake. Three replicates of 500 mL of water were immediately filtered through 0.22 μm pore size polycarbonate filters. Sediment samples (50 mL) were collected from the littoral zone (SE-Top) and from the central part of the lake (21 m depth; SE-Low) in three replicates from each site. All samples (soil cores, filters and sediments) were stored at −20 °C for subsequent molecular analysis.
DNA extraction
DNA from soil and pelletized sediments was extracted from 4 × 0.5 g aliquots. DNA from water samples was obtained by dividing the filters into four pieces and extracting the DNA from each filter piece individually in order to improve the extraction yield. The Griffiths protocol51 was used with the modifications described by Monard et al.52. DNA quality and quantity were checked at 260 nm (NanoDrop Technologies). All 4 extraction-replicates were pooled and stored at −70 °C.
PCR amplification and pyrosequencing
The bacterial 16 S rRNA gene amplifications were performed using the 341 F and 805 R primers53 and the fungal ITS amplifications using the ITS1F54 and ITS455 primers. For the Archaea, nested PCR amplifications of the 16 S rRNA gene were performed because some samples were not successfully amplified with the direct archaeal-specific arch344f56-806R57 primer set. The arch21F-958R primer set58 was used for the first round of amplifications, followed by the arch344f-806R primer set for the second round of amplifications. The 805 R, ITS1F, ITS4 and arch806R primers contained a unique additional 6 bp length barcode used to tag each PCR product according to the original environmental sample and the bacterial and archaeal primer sets were supplemented with the 454 FLX adaptors ‘A’ and ‘B’.
All PCR amplifications were performed in duplicates using 4 μL of DNA diluted 100 and 1000 times as template. The PCR reaction further contained 2.5 units of DreamTaq green DNA polymerase (Fermentas), 1 X PCR buffer supplied by the manufacturer, 1.6 mM MgCl2, 80 μM of dNTP, 1.6 μg of BSA, 0.4 μM of each primer and H2O to a final volume of 40 μL. The PCR conditions were: 94 °C for 5 min, followed by between 26 and 31 cycles of denaturation (94 °C; 30 sec), annealing (53, 55 and 50 °C for the bacteria, fungi and archaea, respectively; 40 sec) and extension (72 °C; 30 sec); followed by the final elongation (72 °C; 7 min). The number of PCR cycles was determined according to previous qPCRs performed on our DNA samples (Fig. S1). All diluted DNA extracts were amplified in duplicates.
Each PCR sample was purified using the GeneJet purification kit (Fermentas) following the manufacturer’s instructions and quantified using a Qubit Fluorometer (Invitrogen) and an equal amount of DNA (25 ng) from each sample and each DNA dilution was pooled. To remove potential primer dimers, the pooled DNA was finally gel purified using the Qiaquick gel extraction kit (Qiagen). The final samples were sent to LGC Genomics and ligation of the 454 FLX sequencing adaptors ‘A’ and ‘B’ was performed on the fungal amplicons. The bacterial, fungal and archaeal amplicons were then sequenced on a 454 Genome Sequencer FLX (Roche) machine from the 805 R, both ITS1F and ITS4 and the arch806R sides, respectively. The fasta and quality files from each sample have been submitted to the Sequence Read Archive (www.ncbi.nlm.nih.gov/sra; accession SRX981059).
Processing of pyrosequencing data
The two fungal sequence datasets (ITS1 and ITS2) were processed using the SCATA pipeline (http://scata.mykopat.slu.se refs 52,59) whereas the pipeline provided by the Ribosomal Database Project60 was used for both bacterial and archaeal sequences. Adaptor and primer sequences were trimmed, and sequences of low quality (exponential quality score lower than 10) or shorter than 200 bp were removed. The 6 bp DNA barcode attached to the primers was used to assign sequences to samples. Operational taxonomic units (OTUs) were defined using single linkage clustering at the 98.5% identity level for the fungal sequences according to Wallander et al.61 and Blaalid et al.62. For bacterial and archaeal sequences, OTUs were defined using complete linkage clustering at the 97% identity level63. After removing all the singletons, a total of 641 230, 182 365 and 624 321 sequences were obtained for the bacterial, fungal and archaeal communities, respectively. These corresponded to 41 418, 2 295, 2 062 and 3 497 bacterial, fungal (ITS1 and ITS2) and archaeal OTUs respectively, distributed across the 21 environmental samples replicated three times. The OTUs were taxonomically identified using the RDPII taxonomy tool63 and the SILVA-ARB database64 for the bacterial and archaeal OTUs, respectively (see Table S1 for details). The fungal ITS sequences were phylogenetically assigned using MEGAN v.4. (MEtaGenome Analyzer, Center for bioinformatics, Tübingen, Germany65) according to their best matches from the GenBank database66.
Community and statistical analysis
After checking that the Bray-Curtis distance within the three replicates were significantly lower than between depths or sites (vegdist function in the Vegan library program implemented in R (http://www.r-project.org/); P < 0.001, t-test; Fig. S2), data from the different sample replicates were pooled to increase the amount of sequences and the representativeness of each sample. The pooled data were then clustered according to the dissimilarity (Bray-Curtis) of their microbial community composition. The main OTUs (relative abundance per sample higher than 1‰ for each dataset – fungi, bacteria and archaea) in the different environmental samples, the habitat generalists detected in at least 16 of the 21 samples and the habitat specialists detected in only one sample type were visualised using the Perl script bubble.pl (available at http://hallam.microbiology.ubc.ca/).
To explore co-occurrence patterns, a network analysis was performed in R by calculating all possible Spearman’s rank correlations between OTUs with a relative abundance per sample higher than 1%. This filtering step removed poorly represented OTUs and reduced network complexity. A valid co-occurrence event was considered to be a robust correlation if the Spearman’s correlation coefficient (ρ) was both >0.6 and highly statistically significant (P < 0.001). Global descriptors of the network were obtained using Network Analyzer in Cytoscape v3.0.167 and the network of coexisting microorganisms was visualised using the Gephi v0.8.2-beta software68.
Additional Information
How to cite this article: Monard, C. et al. Habitat generalists and specialists in microbial communities across a terrestrial-freshwater gradient. Sci. Rep. 6, 37719; doi: 10.1038/srep37719 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Uppsala Microbiomics Centre (UMC) funded by the Swedish Research Council FORMAS.
Footnotes
Author Contributions C.M., S.G., S.B. S.H. and J.S. designed research. C.M. and S.G. performed research. C.M. analysed the data and wrote the manuscript. All authors commented on the manuscript at all stages.
References
- Torsvik V., Ovreas L. & Thingstad T. F. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science. 296, 1064–1066 (2002). [DOI] [PubMed] [Google Scholar]
- Brussaard L. Biodiversity and ecosystem functioning in soil. Ambio 26, 563–570 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heijden M. G. A., Bardgett R. D. & Van Straalen N. M. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008). [DOI] [PubMed] [Google Scholar]
- Cotner J. B. & Biddanda B. A. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5, 105–121 (2002). [Google Scholar]
- Bailey V. L., Smith J. L. & Bolton H. Jr Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol. Biochem. 34, 997–1007 (2002). [Google Scholar]
- Lindahl B. D. et al. Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 173, 611–20 (2007). [DOI] [PubMed] [Google Scholar]
- Peay K. G., Kennedy P. G. & Bruns T. D. Fungal community ecology: a hybrid beast with a molecular master. Bioscience 58, 799–810 (2008). [Google Scholar]
- Buée M. et al. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184, 449–456 (2009). [DOI] [PubMed] [Google Scholar]
- Krauss G.-J. et al. Fungi in freshwaters: ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 35, 620–51 (2011). [DOI] [PubMed] [Google Scholar]
- Clemmensen K. E. et al. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339, 1615–8 (2013). [DOI] [PubMed] [Google Scholar]
- Oline D. K., Schmidt S. K. & Grant M. C. Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microb. Ecol. 52, 480–490 (2006). [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T., Selezi D., Quaiser A., Bonch-Osmolovskaya L. & Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5, 787–797 (2003). [DOI] [PubMed] [Google Scholar]
- Auguet J.-C., Barberan A. & Casamayor E. O. Global ecological patterns in uncultured Archaea. ISME J. 4, 182–90 (2010). [DOI] [PubMed] [Google Scholar]
- Leininger S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 (2006). [DOI] [PubMed] [Google Scholar]
- Reed H. E. & Martiny J. B. H. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol. Ecol. 62, 161–170 (2007). [DOI] [PubMed] [Google Scholar]
- Strickland M. S., Lauber C., Fierer N. & Bradford M. A. Testing the functional significance of microbial community composition. Ecology 90, 441–451 (2009). [DOI] [PubMed] [Google Scholar]
- Reed H. E. & Martiny J. B. H. Microbial composition affects the functioning of estuarine sediments. ISME J. 7, 868–879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. L. et al. Spatial scaling of microbial eukaryote diversity. Nature 432, 747–750 (2004). [DOI] [PubMed] [Google Scholar]
- Horner-Devine M. C., Lage M., Hughes J. B. & Bohannan B. J. M. A taxa-area relationship for bacteria. Nature 432, 750–753 (2004). [DOI] [PubMed] [Google Scholar]
- Barberán A., Bates S. T., Casamayor E. O. & Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein L. M. & Blackwood C. B. The spatial scaling of saprotrophic fungal beta diversity in decomposing leaves. Mol. Ecol. 22, 1171–84 (2013). [DOI] [PubMed] [Google Scholar]
- Besemer K. et al. Unraveling assembly of stream biofilm communities. ISME J. 6, 1459–1468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J. 7, 1310–1321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas-Becking L. G. M. Geobiologie of inleiding tot de milieukunde. (1934).
- Fierer N. & Jackson R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed D. J. & Martiny J. B. H. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 5, 379–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. A. et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J. M. et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. USA 111, 6341–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell S. P. The unified neutral theory of biodiversity and biogeography. (2001). [DOI] [PubMed]
- Langenheder S. & Székely A. J. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 5, 1086–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-M., Cao P., Fu B., Hughes J. M. & He J.-Z. Ecological drivers of biogeographic patterns of soil archaeal community. PLoS One 8, e63375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S. N., Kolasa J. & Cottenie K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 90, 2253–2262 (2009). [DOI] [PubMed] [Google Scholar]
- Székely A. J. & Langenheder S. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol. Ecol. 87, 102–112 (2014). [DOI] [PubMed] [Google Scholar]
- Bergström A. & Jansson M. Bacterioplankton production in humic lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microb. Ecol. 39, 101–115 (2000). [DOI] [PubMed] [Google Scholar]
- Crump B. C., Amaral-Zettler L. A. & Kling G. W. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J. 6, 1629–1639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner F. O. et al. Comparative 16S rRNA Analysis of Lake Bacterioplankton Reveals Globally Distributed Phylogenetic Clusters Including an Abundant Group of Actinobacteria. Appl. Environ. Microbiol. 66, 5053–5065 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P. H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl. Environ. Microbiol. 72, 1719–1728 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller L. et al. Composition of bacterial and archaeal communities in freshwater sediments with different contamination levels (Lake Geneva, Switzerland). Water Res. 45, 1213–28 (2011). [DOI] [PubMed] [Google Scholar]
- Barberán A. et al. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802 (2014). [DOI] [PubMed] [Google Scholar]
- Kneitel J. M. & Chase J. M. Trade-offs in community ecology: Linking spatial scales and species coexistence. Ecol. Lett. 7, 69–80 (2004). [Google Scholar]
- Fierer N., Bradford M. A. & Jackson R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007). [DOI] [PubMed] [Google Scholar]
- Jones R. T. et al. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A. & de la Torre J. R. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101 (2012). [DOI] [PubMed] [Google Scholar]
- Beier S. et al. Pronounced seasonal dynamics of freshwater chitinase genes and chitin processing. Environ. Microbiol. 14, 2467–79 (2012). [DOI] [PubMed] [Google Scholar]
- Oakley B. B., Carbonero F., Dowd S. E., Hawkins R. J. & Purdy K. J. Contrasting patterns of niche partitioning between two anaerobic terminal oxidizers of organic matter. ISME J. 6, 905–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. J. Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002). [DOI] [PubMed] [Google Scholar]
- Hanson C. A., Fuhrman J. A., Horner-Devine M. C. & Martiny J. B. H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012). [DOI] [PubMed] [Google Scholar]
- Heinen E. A. & McManus J. Carbon and nutrient cycling at the sediment-water boundary in western lake superior. J. Great Lakes Res. 30, 113–132 (2004). [Google Scholar]
- Fuhrman J. A. Microbial community structure and its functional implications. Nature 459, 193–199 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou J., Deng Y., Luo F., He Z. & Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio 2, e00122–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. I., Whiteley A. S., O’Donnell A. G. & Bailey M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monard C., Gantner S. & Stenlid J. Utilizing ITS1 and ITS2 to study environmental fungal diversity using pyrosequencing. FEMS Microbiol. Ecol. 84, 165–175 (2013). [DOI] [PubMed] [Google Scholar]
- Herlemann D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M. & Bruns T. D. ITS primers with enhanced specificity for Basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993). [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T. D., Lee S. B. & Taylor J. W. In PCR Protoc. - a Guid. to methods Appl . (eds. Innis M. A., Gelfand D. H., Sninsky J. J. & White T. J.) 315–322 (Academic Press, 1990). [Google Scholar]
- Casamayor E. O. et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4, 338–348 (2002). [DOI] [PubMed] [Google Scholar]
- Takai K. & Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. 89, 5685–5689 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl B. D. et al. Fungal community analysis by high-throughput sequencing of amplified markers-a user’s guide. New Phytol. 199, 288–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander H., Johansson U., Sterkenburg E., Brandström Durling M. & Lindahl B. D. Production of ectomycorrhizal mycelium peaks during canopy closure in Norway spruce forests. New Phytol. 187, 1124–1134 (2010). [DOI] [PubMed] [Google Scholar]
- Blaalid R. et al. Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol. Ecol. 21, 1897–1908 (2012). [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R. & Amann R. The species concept for prokaryotes. FEMS Microbiol. Rev. 25, 39–67 (2001). [DOI] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S. & Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Third Int. ICWSM Conf. 361–362 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.