Abstract
With the advent of large-scale imaging studies and big health data, and the corresponding growth in analytics, machine learning and computational image analysis methods, there are now exciting opportunities for deepening our understanding of the mechanisms and characteristics of heart disease. Two emerging fields are computational analysis of cardiac remodelling (shape and motion changes due to disease) and computational analysis of physiology and mechanics to estimate biophysical properties from non-invasive imaging. Many large cohort studies now underway around the world have been specifically designed based on non-invasive imaging technologies in order to gain new information about the development of heart disease from asymptomatic to clinical manifestations. These give an unprecedented breadth to the quantification of population variation and disease development. Also, for the individual patient, it is now possible to determine biophysical properties of myocardial tissue in health and disease by interpreting detailed imaging data using computational modelling. For these population and patient-specific computational modelling methods to develop further, we need open benchmarks for algorithm comparison and validation, open sharing of data and algorithms, and demonstration of clinical efficacy in patient management and care. The combination of population and patient-specific modelling will give new insights into the mechanisms of cardiac disease, in particular the development of heart failure, congenital heart disease, myocardial infarction, contractile dysfunction and diastolic dysfunction.
Graphical Abstract
1. Introduction
Heart disease is a leading cause of morbidity and mortality around the world. While substantial advances have been made in the detection and treatment of disease, little is known about the mechanisms and characteristics of disease development. The design of more effective treatments and prevention strategies rely on knowledge of the underlying features of developing disease. For example, patients suffering from heart failure with preserved ejection fraction do not respond well to conventional treatments that work well for other forms of heart failure. It is not known whether these patients exhibit impaired ventricular filling through increased myocardial stiffness or delayed myocyte relaxation. Many heart failure patients with dyssynchronous contraction respond well to pacemaker therapy for cardiac resynchronisation, but approximately one third do not. Patient-specific information to predict response to cardiac resynchronisation therapy, including where to place the pacing device leads and how to select interventricular pacing delays, would be highly beneficial. In sub-clinical disease, interactions between environmental and genetic factors together with adverse events lead to adaptations in cardiac shape and motion. With recent advances in medical imaging, large-scale cohort studies and medical image analysis, it is now possible to address these problems from two perspectives: first by examining how the heart changes its shape and function in response to disease and exposure to risk factors; and second by identifying biophysical parameters that characterise physiological and biomechanical behaviours in health and disease.
Cardiac shape and function continuously adapt (remodel) in response to pre-clinical and symptomatic disease, as well as vascular events. Better quantification of this remodelling could provide more predictive information on the status of heart health and the progression of disease, since these adaptations reflect initially compensatory mechanisms leading eventually to decompensated remodelling and heart failure. For example, concentric remodelling (relative thickening of the heart walls), increased left ventricular (LV) end-systolic volume, and increased LV sphericity have all been associated with decreased survival in patients with myocardial infarction (Sutton and Sharpe, 2000). Although remodelling has typically been characterised as changes in morphology due to vascular events, such as myocardial ischaemia or infarction, hypertensive and idiopathic cardiomyopathies (i.e. those of unknown origin) also give rise to remodelling features, which are important clinical markers of disease progression. Pre-clinical remodelling can occur in asymptomatic individuals, prior to the establishment of clinical manifestations of disease, in response to exposure to risk factors and genetic interactions. This type of remodelling has also been associated with adverse outcomes (Bluemke et al., 2008).
Changes in physiological parameters, such as contractility and muscle stiffness, are also indicative of disease processes. For example, resting myocardial stiffness is a major determinant of ventricular function, with large changes in stiffness associated with heart failure and myocardial infarction. Increased muscle stiffness is detrimental to the filling function of the heart, which in turn increases blood pressure and the amount of contraction force required by the muscle. Some forms of heart failure may be associated with increased myocardial stiffness, but it is difficult to characterise patients effectively to determine if the clinically observed symptoms are due to increased passive tissue stiffness, impaired relaxation, impaired contractility or some combination of these.
Mathematical modelling of cardiac shape, motion and physiology is a rapidly developing field with the potential for providing detailed information on the mechanisms of disease processes and cardiac dysfunction. Models of cardiac function can incorporate geometry, motion, microstructure, nonlinear and anisotropic constitutive behaviour, loading conditions, and kinematic constraints. Activation models comprise initiation and propagation of action potentials, calcium transients and cross-bridge activation and de-activation, active force generation and relaxation. Patient-specific biophysical parameters governing myocardial stiffness and contractility can be estimated by optimally matching the behaviour of these models to data from medical imaging. In this way, medical imaging examinations can be augmented with model-based interpretation and thereby provide new information on mechanisms of compensatory and decompensated adaptations.
Medical imaging now enables precise quantification of structural and functional information on cardiac status and performance, but each modality has particular strengths and weaknesses. Multi-detector computed tomography (CT) is very rapid and provides detailed 3D images at approximately 0.5 mm isotropic resolution. However, exposure to ionising X-ray radiation prevents this method from being widely used in routine assessments or evaluation of children with congenital heart disease. Echocardiography provides lower-cost rapid evaluation of function, with more than 50 3D frames per second possible with modern 3D transducers. However, signal dropout due to poor acoustic windows, particularly in the right heart, limits this method in many patients. Transesophageal echocardiography can provide better delineation of the right heart, but it is semi-invasive and the patient may need sedation during acquisition.R1.1 Cardiac magnetic resonance imaging (MRI) provides a range of contrast mechanisms from motion to T1 mapping and perfusion quantification, but typically cannot be used in patients with implanted devices. Analysis methods that exploit medical imaging must therefore be compatible with a range of modalities, and must integrate information from a variety of sources.
This review will examine applications of model-based analysis of cardiac images, with emphasis on the breadth available in large population-based imaging studies, and depth available in patient specific physiological modelling. We also provide a potential roadmap for the future, which will require closer links between algorithm development and clinical applications.
2. Remodelling in Pre-Clinical and Clinical Disease
Much of what is known about multivariable risk factors of cardiac disease has been derived from the large cohort Framingham Heart Study, which does not include cardiac geometry and function. Recently, several cohort studies have used medical imaging as part of a suite of investigations into the effects of risk factors and disease events on heart function. The Multi-Ethnic Study of Atherosclerosis (MESA) was the first major population study to employ cardiac MRI as part of a large-scale epidemiological study to examine the progression of disease from pre-clinical manifestations to clinical symptoms, and apply modern imaging methods to develop new biomarkers and risk factors to augment those identified by Framingham and other population-based studies (Bluemke et al., 2008). Similarly, the UK Biobank is an extensive population-based study that recruited 500, 000 people aged between 40 and 69 years in 2006–2010 from across the UK, with over 6000 participants already imaged using cardiac MRI, abdominal MRI, brain MRI, carotid ultrasound, and dual-energy X-ray absorptiometry (Petersen et al., 2013). This is now being extended to image 100,000 participants over the next 6 years. Several other more localised cardiac imaging studies are currently being performed in order to identify novel cardiac disease risk factors based on cardiac shapes and function.
Atlas-based shape analysis is a powerful tool to quantify shape changes in pre-clinical and clinicial disease (Fonseca et al., 2011). Preliminary results in the MESA cohort have shown that atlas-based shape measures are more sensitive than traditional remodelling indices for describing associations with common risk factors (Medrano-Gracia et al., 2014).
Principal component analysis (PCA) has been used to quantify the major determinants of shape variation in MESA participants (Fonseca et al., 2011). In 1,991 MESA participants, after correction for height, the major principal modes of shape variation were associated with known clinical indices of adverse remodelling, including heart size, sphericity and concentricity (Medrano-Gracia et al., 2014). Geometric variations can also be associated with traditional risk factors and demographic data. For example, significant differences were found between PCA shape modes in sub-cohorts grouped by traditional risk factors including sex, ethnicity, smoking and alcohol. Males and African Americans tended to have larger hearts and females and Chinese tended to have smaller hearts for their height. Heart size increased with history of smoking or alcohol. Female hearts were more spherical than males, and Chinese were less spherical than Whites. Differences in sphericity were also found due to alcohol use (more spherical with current consumption), and presence of diabetes (more spherical with untreated diabetes) at end-systole. Shape indices derived from multidimensional atlas-based analysis were more powerfully associated with known risk factors such as sex, ethnicity, smoking, hypertension and diabetes than traditional imaging markers such as ejection fraction, volume, and LV mass (Medrano-Gracia et al., 2014).
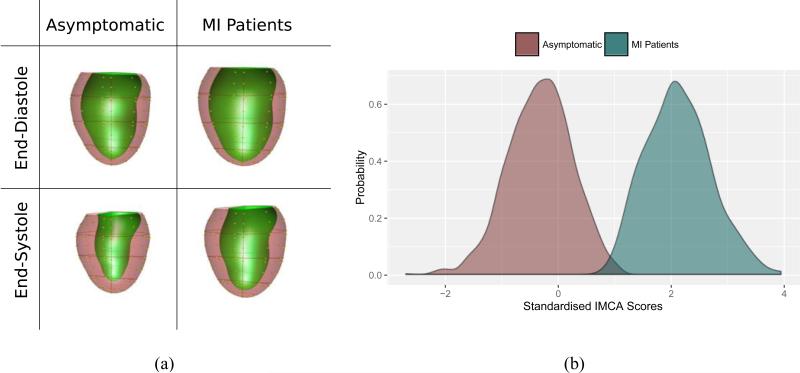
Combinations of shape components may enable calculation of remodelling indices that are specifically associated with traditional risk factors, presence of disease, or adverse outcomes. For example, linear discriminant analysis was used to determine that atlas-based components are more sensitive to traditional risk factors than standard imaging indices such as mass and volume (Zhang et al., 2014). Supervised dimension reduction methods can be used to define a single integrated remodelling component that best describes the remodelling process in relation to a specific disease process (Zhang et al., 2015). As shown in Figure 1, a single shape mode with the strongest association with myocardial infarction was able to discriminate patients from asymptomatic subjects with 95% accuracy (Zhang et al., 2015).
Figure 1.
Information maximising component analysis (IMCA) of shape differences between asymptomatic subjects and patients with myocardial infarction (MI). Left: visualisation of the remodelling index. Right: discrimination of remodelling score (Zhang et al., 2015).
Future applications include characterization of the developing heart. Foetal MR imaging is provides a promising avenue for early diagnosis of disease and improves pregnancy management. Foetal cardiac MRI is currently hampered by the difficulty to measure foetal ECG signals, but when method such as metric-optimized gating (Roy et al., 2013) becomes clinically applicable, this will open a new exciting area of cardiac image analysis for early assessment of congenital heart disease (Wielandner et al., 2013). R1.2 Another important application is the quantification of change in longitudinal studies. For example, remission of remodelling could identify the benefits of treatment effects. Also, in patients at risk of heart failure, longitudinal changes could identify when interventions are required. It may be possible that the longitudinal changes in shape which are most important for these applications are different from the cross-sectional indices discussed above. R1.4
An important future application of atlas-based analysis is for use in investigating congenital heart disease (CHD), which is the most common birth defect with a prevalence of approximately 75 in every 1000 births. Due to an improvement in interventions, survival rates are increasing, and 90% of infants born with CHD now survive to adulthood. However, heart failure is a significant problem for adults with some CHDs, such as hypoplastic left heart syndrome and tricuspid atresia, who have a single functioning ventricle and increased risk of heart failure remodelling. As a result, CHD patients undergo regular imaging investigations in order to identify adverse remodelling in time to intervene. Although there has been substantial work on improving analysis of MRI data, analysis of CHD cases remains a significant problem, particularly because right ventricular function and interdependence between the ventricles is of particular importance, and right and left biventricular geometry is complex and variable. A recent analysis method, which incorporates representations of all four valves, shows promise for the rapid customisation of biventricular models (Gilbert et al., 2014). A regularisation method which penalises deviation from affine (D-affine) deformations, together with a polar prediction step to improve preconditioning, enabled real-time updates within an interactive segmentation framework (Gilbert et al., 2014).
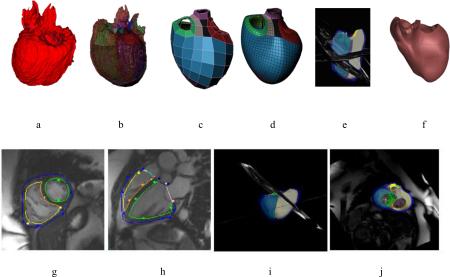
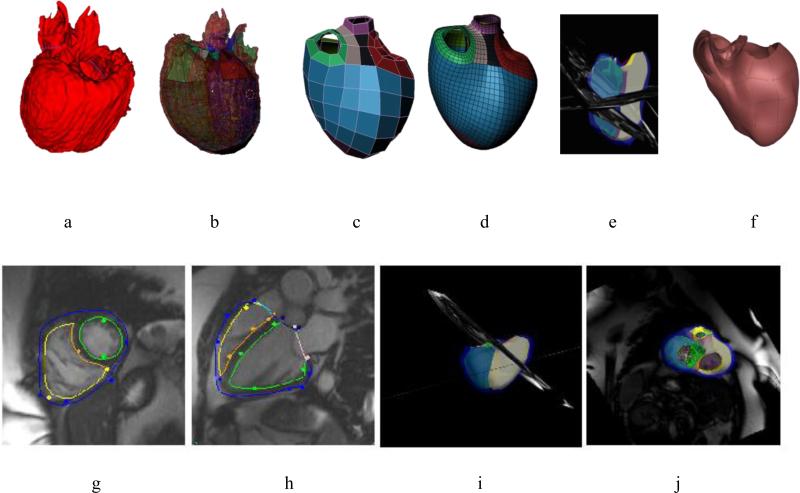
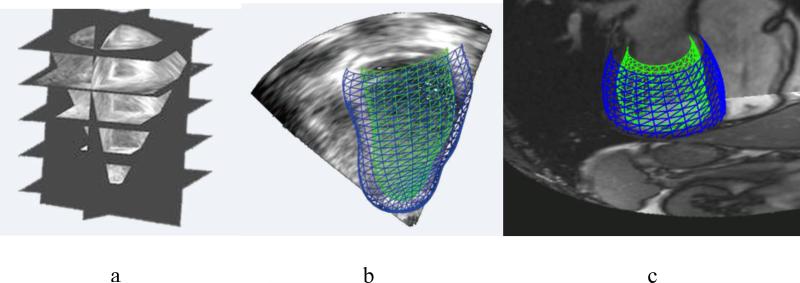
With the variety of shape changes due to different CHD lesions, it is likely that a variety of shape templates will be required to analyse patient specific geometry and function. A pipeline for creating such templates is possible using image analysis and shape modelling methods (Gonzales et al., 2013; Zhang et al., 2012) (Figure 2). Hexahedral bilinear elements were used to define the surface mesh topology, allowing for extraordinary nodes (i.e. nodes with valence not equal to 4). The mesh was registered to data from CT or MRI, and subdivided using a Li-Kobbelt algorithm, which results in a C1 surface (approximately C1 at extraordinary nodes). This mesh was converted to a Bezier cubic mesh and input as a template in the interactive customisation process described above (Gilbert et al., 2014). Thus, templates describing characteristic lesions, such as congenitally corrected transposition of the great arteries, can be interactively customised to MRI data from a range of patients with this condition.
Figure 2.
Top: Pipeline for generating templates for CHD lesions: a) 3D image from MRI or CT; b) registration with initial model; c) coarse shape customisation; d) refinement through subdivision surfaces; e) customisation to different patients; f) patient specific model. Bottom: Patient specific model at end-diastole for a 42 year old female with repaired tetralogy of Fallot: g) short axis view; h) long axis view; i) model with images; j) valves.
3. Physiology and Biomechanics
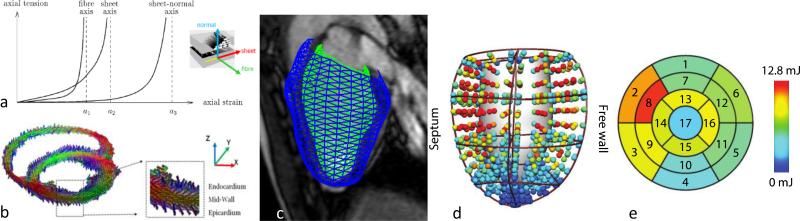
Although the above geometric analyses precisely quantify cardiac function and remodelling during disease, extracting information on the underlying biophysical parameters is now possible using computational physiological modelling of cardiac activation and contraction (Marchesseau et al., 2013). This approach offers a platform with which clinical information from a variety of sources can be integrated in a manner that is consistent with the laws of physics to predict the complex mechanical function of the heart, and estimate local stresses and contractile forces in cardiac muscle, which cannot be measured directly. This approach enables estimation of tissue-specific parameters, which are theoretically independent of chamber geometry and loading conditions, unlike traditional indices such as ejection fraction and stroke volume. One major issue is that it is difficult to non-invasively estimate myocardial stiffness in vivo. In the heart, passive myocardial stiffness increases non-linearly with increasing load (Figure 3a). In addition to its non-linear behaviour, myocardial stiffness is anisotropic owing to its fibrous and layered architecture (Figure 3b). Typically, stiffness is greatest in the fibre direction, intermediate transverse to the fibres in the plane of the layer, and least in the direction orthogonal to the layers.
Figure 3.
a) Anisotropic mechanical behaviour. b) Diffusion tensor MRI showing muscle fibre orientation. c) Left ventricle model customised to patient images. d) Stress derived from a biomechanical model. e) Local myocardial work estimated for each of the 17 AHA segments.
Finite element analysis of cardiac mechanics can incorporate realistic non-linear, anisotropic material properties, active force generation, resulting in realistic simulations of cardiac mechanics (Chabiniok et al., 2016; Wang et al., 2015). The cardiac cycle is typically simulated in five phases: 1) passive diastolic filling, 2) isovolumic contraction, 3) ejection, 4) isovolumic relaxation and 5) early filling, though by coupling the biventricular model to a closed-loop model of the pulmonary and systemic circulations and atria, it is possible for the model itself to generate the entire cycle (Kerckhoffs et al., 2007). Physiological parameters governing passive myocardial stiffness and the active contraction force can be “reverse engineered” from a knowledge of the shape and motion of the heart obtained from MRI (Figure 3c), combined with knowledge of the pressure boundary conditions and muscle fibre architecture (Wang et al., 2009). Contractility can be quantified by parameters related to calcium mediated force production in the muscle sarcomeres, and relaxation can be quantified by de-activation of the sarcomeres. In addition to the fibre tension, there is also a significant cross-fibre force due to the cross-bridge binding angle, which affects the anisotropic dynamic stiffness (Tangney et al., 2013). Patient specific models can be customised to image and clinical information (Tangney et al., 2013; Wang et al., 2016). Multi-scale models show promise in CHD and predicting outcomes of surgical procedures (Meoli et al., 2015).
Although these methods have been applied to data from CT and MRI, echocardiography is by far the most common modality for cardiac imaging exams. With recent advances in 3D transducers, it is now possible to obtain 50 3D frames per second. In the near future, it should be possible to translate methods between modalities, and integrate the best information available from each modality (Figure 4).
Figure 4.
a) 3D echo dataset showing short and long axis reformatted slices. b) 3D model customised to a 3D echo dataset from a healthy volunteer. c) 3D model customised to MRI dataset in the same healthy volunteer.
Another promising area for future research is the integration of finite element heart models with elastography, for example magnetic resonance elastography (MRE), in order to obtain information about the dynamic stiffness properties of the heart. MRE is a non-invasive imaging technique that quantifies harmonic small (i.e. less than 100 microns) perturbations in displacement at a frequency of around 80 Hz. It is possible to apply finite element analysis to MRE data to recover anisotropic material properties of heart tissue (Miller et al., 2015). This method may provide complimentary information to the large deformation finite elasticity analysis methods described above. The advantage of MRE is that it does not require invasive pressure recordings to estimate myocardial stiffness parameters.
4. Benchmarking and Validation in Cardiac Image Analysis
Research fields can advance more rapidly when researchers are able to reproduce and verify results from other studies. Open data sharing, algorithm benchmarking, and validation through unit tests are mechanisms to ensure reproducible research. Recently there has been significant work on validation of computational models of the heart (Land et al., 2015; Niederer et al., 2011). These studies provide important tests for cardiac mechanics modelling, even though closed-form (“ground truth”) solutions do not exist. By comparing results between code implementations and different methodologies, modelling issues and problems can be identified. For example, a cardiac electrophysiology benchmark highlighted differences in convergence and solution accuracy between different numerical schemes (Niederer et al., 2011).
Open benchmark challenges also provide excellent test beds for comparing algorithms. A recent challenge in LV segmentation and calculation of ejection fraction1 resulted in a large number of machine learning implementations. Other open challenges in cardiac image analysis2, have attracted stimulating research activities from diverse groups to benchmark their new developments. One promising avenue of future research is in the training of machine learning algorithms using simulated images. Since the availability of ground truth data is very limited, and variable between analysts, images can be simulated from parametric models of heart shape and motion, with statistical variation derived from population studies (Prakosa et al., 2013; Tobon-Gomez et al., 2011) and added noise and image artefacts, so that the underlying ground truth is known exactly.
5. Future Directions
Models of cardiac function and physiology provide a tool to analyse large numbers of patient studies, as well as patient specific estimation of physiological parameters. In the future, medical imaging data on tissue characteristics can be incorporated into these models, such as in vivo diffusion tensor imaging myocardial T1 and extracellular volume maps, and tissue fingerprinting. The amount of data that are available for cardiovascular practice and research is growing at an unprecedented pace. Big data analytics, which lies at the intersection of cardiac imaging, biomechanical modelling, data mining and machine learning, will lead to improved cardiovascular patient care (Suinesiaputra et al., 2015). There is a strong potential to discover novel, specific risk factors in addition to traditional cardiovascular assessment. Data from administrative, clinical registry, electronic health records, and imaging devices can be merged with biometrics, genomics, proteomics, simulation data, and other sources, including experimental studies and social media. The breadth of population studies and depth of physiological and biomechanical analysis from the myocyte level to organ structures will position cardiac image modelling at the heart of quantitative cardiovascular medicine.
The key to successfully combine population-based cardiac image modelling with biophysical parameters identification is the generation of personalised scores that can be mapped within population norms. The ability to customise biomechanical and physiological parameters into individual patients could significantly transform clinical care. Although population databases will be less accurate than could be obtained with targeted studies using invasive procedures for parameter characterization, clinicians are well used to making the most from limited resources.R2.3 A future scenario could be rapid imaging sessions with echocardiography at clinics for monitoring current status of cardiac remodelling, where physicians can investigate 3D biophysical customization of a probabilistic computational heart model with visualisation of differential adaptations of cardiac shape and function associated with possible adverse effects. With a reference to atlas-based asymptomatic cardiac remodelling pathways using studies with long-term outcome measures, the combined image-based cardiac model with personalised biomechanical and physiological parameters can be extrapolated to predict future remodelling and events. For example, the power of large scale physiological modelling may be in nested case-control studies where biophysical parameters prove particularly useful in particular patient groups. For example, myocardial stiffness may be important in particular types of heart failure (those with preserved rather than reduced ejection fraction). Even though the estimate of stiffness may not be as accurate as a targeted study with invasive measurements, evaluation on a larger number of cases with limited data may be useful in highlighting where targeted detailed research studies may be most useful.R2.2-R2.6
Highlights.
Population-based large cohort studies give an unprecedented breadth to the quantification of population variation and disease development in cardiac performance
Biophysical properties of myocardial tissue in health and disease can be obtained by interpreting imaging data using computational modelling
Open benchmarks for algorithm comparison and validation, open sharing of data and algorithms, and demonstration of clinical efficacy in patient management and care are needed for future developments in this field
Acknowledgements
The authors acknowledge the support of NHLBI R01HL121754 and The Health Research Council of New Zealand.
Footnotes
References
- 1.Bluemke DA, Kronmal RA, Lima JA, Liu K, Olson J, Burke GL, Folsom AR. The relationship of left ventricular mass and geometry to incident cardiovascular events: the MESA (Multi-Ethnic Study of Atherosclerosis) study. J Am Coll Cardiol. 2008;52:2148–2155. doi: 10.1016/j.jacc.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chabiniok R, Wang VY, Hadjicharalambous M, Asner L, Lee J, Sermesant M, Kuhl E, Young AA, Moireau P, Nash MP, Chapelle D, Nordsletten DA. Multiphysics and multiscale modelling, data–model fusion and integration of organ physiology in the clinic: ventricular cardiac mechanics. Interface Focus. 2016;6:20150083. doi: 10.1098/rsfs.2015.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonseca CG, Backhaus M, Bluemke DA, Britten RD, Chung JD, Cowan BR, Dinov ID, Finn JP, Hunter PJ, Kadish AH, Lee DC, Lima JA, Medrano-Gracia P, Shivkumar K, Suinesiaputra A, Tao W, Young AA. The Cardiac Atlas Project--an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27:2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert K, Cowan BR, Suinesiaputra A, Occleshaw C, Young AA. Medical Image Computing and Computer-Assisted Intervention. Springer; Boston: 2014. Rapid D-Affine Biventricular Cardiac Function with Polar Prediction; pp. 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzales MJ, Sturgeon G, Krishnamurthy A, Hake J, Jonas R, Stark P, Rappel WJ, Narayan SM, Zhang Y, Segars WP, McCulloch AD. A three-dimensional finite element model of human atrial anatomy: new methods for cubic Hermite meshes with extraordinary vertices. Med Image Anal. 2013;17:525–537. doi: 10.1016/j.media.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerckhoffs RCP, Neal M, Gu Q, Bassingthwaighte JB, Omens JH, McCulloch AD. Coupling of a 3D finite element model of cardiac ventricular mechanics to lumped systems models of the systemic and pulmonic circulation. Annals of Biomedical Engineering. 2007;35:1–18. doi: 10.1007/s10439-006-9212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Land S, Gurev V, Arens S, Augustin CM, Baron L, Blake R, Bradley C, Castro S, Crozier A, Favino M, Fastl TE, Fritz T, Gao H, Gizzi A, Griffith BE, Hurtado DE, Krause R, Luo X, Nash MP, Pezzuto S, Plank G, Rossi S, Ruprecht D, Seemann G, Smith NP, Sundnes J, Rice JJ, Trayanova N, Wang D, Jenny Wang Z, Niederer SA. Verification of cardiac mechanics software: benchmark problems and solutions for testing active and passive material behaviour. Proceedings. Mathematical, physical, and engineering sciences / the Royal Society. 2015;471:20150641. doi: 10.1098/rspa.2015.0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchesseau S, Delingette H, Sermesant M, Cabrera-Lozoya R, Tobon-Gomez C, Moireau P, Figueras i Ventura RM, Lekadir K, Hernandez A, Garreau M, Donal E, Leclercq C, Duckett SG, Rhode K, Rinaldi CA, Frangi AF, Razavi R, Chapelle D, Ayache N. Personalization of a cardiac electromechanical model using reduced order unscented Kalman filtering from regional volumes. Med Image Anal. 2013;17:816–829. doi: 10.1016/j.media.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Medrano-Gracia P, Cowan BR, Ambale-Venkatesh B, Bluemke DA, Eng J, Finn JP, Fonseca CG, Lima JAC, Suinesiaputra A, Young AA. Left ventricular shape variation in asymptomatic populations: The Multi-Ethnic Study of Atherosclerosis. Journal of Cardiovascular Magnetic Resonance. 2014;16:56. doi: 10.1186/s12968-014-0056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meoli A, Cutri E, Krishnamurthy A, Dubini G, Migliavacca F, Hsia TY, Pennati G, Modeling of Congenital Hearts Alliance, G. Taylor A, Giardini A, Khambadkone S, Schievano S, de Leval M, Hsia TY, Bove E, Dorfman A, Baker GH, Hlavacek A, Migliavacca F, Pennati G, Dubini G, Marsden A, Feinstein J, Vignon-Clementel I, Figliola R, McGregor J. A multiscale model for the study of cardiac biomechanics in single-ventricle surgeries: a clinical case. Interface Focus. 2015;5:20140079. doi: 10.1098/rsfs.2014.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller R, Jiang HD, Mazumder R, Cowan BR, Nash MP, Kolipaka A, Young AA. Determining Anisotropic Myocardial Stiffness from Magnetic Resonance Elastography: A Simulation Study. Lect Notes Comput Sc. 2015;9126:346–354. [Google Scholar]
- 12.Niederer SA, Kerfoot E, Benson AP, Bernabeu MO, Bernus O, Bradley C, Cherry EM, Clayton R, Fenton FH, Garny A, Heidenreich E, Land S, Maleckar M, Pathmanathan P, Plank G, Rodriguez JF, Roy I, Sachse FB, Seemann G, Skavhaug O, Smith NP. Verification of cardiac tissue electrophysiology simulators using an N-version benchmark. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences. 2011;369:4331–4351. doi: 10.1098/rsta.2011.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen SE, Matthews PM, Bamberg F, Bluemke DA, Francis JM, Friedrich MG, Leeson P, Nagel E, Plein S, Rademakers FE, Young AA, Garratt S, Peakman T, Sellors J, Collins R, Neubauer S. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J Cardiovasc Magn Reson. 2013;15:46. doi: 10.1186/1532-429X-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakosa A, Sermesant M, Delingette H, Marchesseau S, Saloux E, Allain P, Villain N, Ayache N. Generation of synthetic but visually realistic time series of cardiac images combining a biophysical model and clinical images. IEEE transactions on medical imaging. 2013;32:99–109. doi: 10.1109/TMI.2012.2220375. [DOI] [PubMed] [Google Scholar]
- 15.Roy CW, Seed M, van Amerom JF, Al Nafisi B, Grosse-Wortmann L, Yoo SJ, Macgowan CK. Dynamic imaging of the fetal heart using metric optimized gating. Magnetic resonance in medicine. 2013;70:1598–1607. doi: 10.1002/mrm.24614. [DOI] [PubMed] [Google Scholar]
- 16.Suinesiaputra A, Medrano-Gracia P, Cowan BR, Young AA. Big heart data: advancing health informatics through data sharing in cardiovascular imaging. IEEE journal of biomedical and health informatics. 2015;19:1283–1290. doi: 10.1109/JBHI.2014.2370952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 18.Tangney JR, Chuang JS, Janssen MS, Krishnamurthy A, Liao P, Hoshijima M, Wu X, Meininger GA, Muthuchamy M, Zemljic-Harpf A, Ross RS, Frank LR, McCulloch AD, Omens JH. Novel role for vinculin in ventricular myocyte mechanics and dysfunction. Biophysical journal. 2013;104:1623–1633. doi: 10.1016/j.bpj.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobon-Gomez C, Sukno FM, Bijnens BH, Huguet M, Frangi AF. Realistic simulation of cardiac magnetic resonance studies modeling anatomical variability, trabeculae, and papillary muscles. Magnetic resonance in medicine. 2011;65:280–288. doi: 10.1002/mrm.22621. [DOI] [PubMed] [Google Scholar]
- 20.Wang VY, Lam HI, Ennis DB, Cowan BR, Young AA, Nash MP. Modelling passive diastolic mechanics with quantitative MRI of cardiac structure and function. Med Image Anal. 2009;13:773–784. doi: 10.1016/j.media.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang VY, Nielsen PM, Nash MP. Image-Based Predictive Modeling of Heart Mechanics. Annual review of biomedical engineering. 2015;17:351–383. doi: 10.1146/annurev-bioeng-071114-040609. [DOI] [PubMed] [Google Scholar]
- 22.Wang ZJ, Wang VY, Bradley CP, Nash MP, Young AA, Cao JJ. Quantifying passive myocardial stiffness and wall stress in heart failure patients using personalized ventricular mechanics. Journal of Cardiovascular Magnetic Resonance. 2016;18(Suppl 1):O17. [Google Scholar]
- 23.Wielandner A, Mlczoch E, Prayer D, Berger-Kulemann V. Potential of magnetic resonance for imaging the fetal heart. Semin Fetal Neonatal Med. 2013;18:286–297. doi: 10.1016/j.siny.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Ambale-Venkatesh B, Bluemcke DA, Cowan BR, Finn JP, Fonseca CG, Kadish AH, Lee DC, Lima JAC, Hundley WG, Suinesiaputra A, Young AA, Medrano-Gracia P. Information maximizing component analysis of left ventricular remodeling due to myocardial infarction. Journal of Translational Medicine. 2015;13 doi: 10.1186/s12967-015-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Cowan BR, Bluemcke DA, Finn JP, Fonseca CG, Kadish AH, Lee DC, Lima JAC, Suinesiaputra A, Young AA, Medrano-Gracia P. Atlas-based quantification of cardiac remodeling due to myocardial infarction. PLoS One. 2014;9:e110243. doi: 10.1371/journal.pone.0110243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Liang X, Ma J, Jing Y, Gonzales MJ, Villongco C, Krishnamurthy A, Frank LR, Nigam V, Stark P, Narayan SM, McCulloch AD. An atlas-based geometry pipeline for cardiac Hermite model construction and diffusion tensor reorientation. Med Image Anal. 2012;16:1130–1141. doi: 10.1016/j.media.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]