Abstract
Isolation and analysis of cancer cells from body fluids have significant implications in diagnosis and therapeutic treatment of cancers. Circulating tumor cells (CTCs) are cancer cells circulating in the peripheral blood or spreading iatrogenically into blood vessels, which is an early step in the cascade of events leading to cancer metastasis. Therefore, CTCs can be used for diagnosing for therapeutic treatment, prognosing a given anticancer intervention, and estimating the risk of metastatic relapse. However, isolation of CTCs is a significant technological challenge due to their rarity and low recovery rate using traditional purification techniques. Recently microfluidic devices represent a promising platform for isolating cancer cells with high efficiency in processing complex cellular fluids, with simplicity, sensitivity, and throughput. This review summarizes recent methods of CTC isolation and analysis, as well as their applications in clinical studies.
1. INTRODUCTION
1.1 CTCs as Liquid Biopsy
Rare cell capture from body fluids has significant implications in diagnosis and therapeutic treatment of many diseases [1,2]. Circulating tumor cells (CTCs) were first discovered in 1869, when Ashworth described cells in the blood that appeared similar to those observed in the tumor at autopsy [3]. Since then, isolation and characterization of CTCs in patients suffering from a variety of cancers have been a topic of scientific investigations [4]. There were many claims that CTCs as determined by cytology were commonly seen in cancer patients. However, further studies indicated that hematopoietic cells were responsible for almost all of these results [5]. It had shown that the presence of cancer cells in peripheral blood of 17 patients was related to their clinical progress, and researchers have found cancer cells in venous blood draining the tumor as well as in peripheral blood [6,7]. CTCs have been shown to provide predictive and prognostic information in terms of disease relapse, overall survival, and tumor response to therapy in patients with metastatic colorectal [8–10], breast [11,12], prostate [13,14], lung [1], and ovarian cancers [15]. Research has showed that CTCs are cancer cells spontaneously circulating in the peripheral blood or spreading iatrogenic into blood vessels, which is an early step in the cascade of events leading to metastasis [16]. Since CTCs are cancer cells shed from the primary tumor into the peripheral blood circulatory system, they could spread to distant organs where they may reside and ultimately begin to form metastasis [17–20].
Biopsy is the current gold standard of cancer diagnosis for measuring the presence and extent of tumor. However, the process is invasive and prevents patients from being tested in an ongoing or repetitive basis. Besides, the spread of malignant cells can result from the tumor biopsy procedure due to the lack of cohesion of malignant cells and the ease of detachment of cancer cells from tumor. On the other hand, CTC examination in peripheral blood is much less invasive, and it can be regarded as “liquid biopsy” or “live biopsy” in effectively monitoring the progression of the disease and in determining the use of different treatments [20,21]. CTC monitoring enables noninvasive cancer diagnosis and rapid monitoring of therapeutic response with only 5–10 mL of patient blood needed. The number of CTCs present in patients can be a promising predictor in terms of their survival rates and the outcomes of their treatments. However, isolation of these rare cells is a significant technological challenge [2,22–25]. Technologies to exploit the biological characterization of CTCs have been challenged by the low abundance of CTCs (a few to hundreds per mL of whole blood) among a large number of erythrocytes (~109 per mL of whole blood) and leukocytes (~106 per mL of whole blood) [26–28] and the difficulty of separating CTCs among the background population of other hematopoietic components in the bloodstream. The fact that CTCs occur in low abundance has impeded the understanding of cancer evolution.
In more recent studies, CTCs isolated from blood samples potentially provide an accessible source for detection, characterization, and monitoring of cancer. Recently microfluidics-based devices offer a promising platform for capturing cancer cells from complex cellular fluids with high efficiency, sensitivity, and throughput.
1.2 Microfluidics Technology
Microfluidics, commonly associated with the term “lab-on-a-chip,” is a multidisciplinary field that studies the behaviors of fluids in the microscale and is derived from miniaturized total analysis systems (μTAS). The term μTAS, which refers to miniaturized devices that are integrated with all necessary components for analysis of a sample, was proposed and became popular in 1990s [29]. Developed from the field of miniaturization and microelectronics [30–32], μTAS was envisioned as a new concept for chemical sensing, characterized by its possibility to create a complete analytical microsystem by integrating different functional components to a single device [33,34]. μTAS has been known for its low consumption of reagents and samples [33], and it has been widely used in chemical and biomedical engineering [35,36], miniaturized polymerase chain reactions (PCRs) [37,38], and immunoassay [39–41].
The manipulation of fluids in channels with dimensions in tens of micrometers has emerged as a new field. Microfluidics, as an important part of microtechnology, is defined as the science and technology of systems that process or manipulate small (10–9 to 10−18 L) amounts of fluids, using channels with dimensions of tens to hundreds of micrometers; the technology has been used in different fields, especially for life science and chemistry [42]. Much of the original motivation for microfluidics calls for the ability to manipulate fluids on the cellular length scale, the desire to provide cheap and efficient diagnostic tools [29].
The first applications of microfluidic technologies have been in chemical analysis, for which they offer a number of useful capabilities: the ability to use very small quantities of samples and reagents, and to carry out separation and detection with high resolution and sensitivity; low cost; short time for analysis; and small footprints for the analytical devices [43]. Microfluidic devices are fabricated using techniques developed in the semiconductor industry and are often referred to as microfluidic chips. These microfluidic chips of the stamp size are often made of silicon, glass, or polymers, with the component of microsensors, micropumps, and microvalves. In addition to the materials mentioned above, polydimethylsiloxane (PDMS) was widely used in micro-fluidic device fabrication, and more complicated device could combine different microfluidic components. The well-developed PDMS devices were fabricated using soft lithography [44]. Microfluidic systems have now been improved to the state where they are commercially available for biomolecular separations and emerging as promising tools for high-throughput discovery and screening studies in chemistry and materials science [45,46].
Recently microfluidic devices immobilized with capture agents, including antibodies and nucleic acid aptamers, represent a promising approach to isolate cancer cells by processing complex cellular fluids with great simplicity, sensitivity, and throughput [47–55]. Microfluidic devices have many advantages over conventional bench-top systems to capture tumor cells. These advantages include reduced size of operating systems, flexibility in design, less reagent consumption, reduced production of wastes, decreased requirements for power, increased speed of analyses, and portability. The confined space and shorter diffusion distance in a microdevice result in high capture efficiency and cell purity. Microfluidic devices have been widely recognized as a powerful technology that will play an important role in future medical analysis to meet the large-scale and high-throughput requirements. These lead to the recent development of new methodologies for microfluidics-enabled CTC analysis.
2. METHODS OF ISOLATION AND ANALYSIS
2.1 Isolation Methods
Methods for CTC isolation have been developed to obtain greater efficiency using different principles (Table 1). The CTC isolation methods are based either on the biological properties of tumor cells or on their physical properties.
Table 1.
Isolation Methods for Circulating Tumor Cells (CTCs)
| Isolation methods based on physical properties | Size |
| Deformability | |
| Density | |
| Electric charge | |
| Other physical properties | |
| Isolation methods based on biological properties | Immunoseparation |
| EPISPOT assay | |
| Invasion assay | |
| Other biological properties |
The most widely used method is immunomagnetic separation. In this method, CTCs can be positively or negatively enriched based on the expression of surface proteins. This approach utilizes capture agent-labeled magnetic beads for either positive selection [56–58,11,59] of CTCs using cell-surface markers, or negative enrichment that depletes white blood cells (WBCs) using anti-CD45, which is widely expressed on WBCs [60,61]. Based on the positive selection mechanism, CellSearch assay [11,59] is the only platform, up to now, approved by the Food and Drug Administration (FDA) for CTC enumeration. This method enriches CTCs by using ferrofluid particles coated with antibodies against epithelial cell adhesion molecule (EpCAM), which is widely expressed on the surface of epithelial cells and epithelial-derived tumor cells [62]. In the past decades, along with novel CTC detection methods, magnetic-activated cell sorter [63] and Cel-lSearch [64] are commercially available for CTC detection in the clinical setting. But, CellSearch assay is challenged by its limited sensitivity [65].
The microfluidic devices have been applied for CTC detection since a “CTC-chip” was developed, which captured CTCs as blood flowed past EpCAM-coated microposts in 2007 [66]. More recently, several EpCAM-based microfluidic devices with improved features have been reported for highly efficient capture of CTCs [50–53,47,55,49,54]. Furthermore, devices were designed based on the difference in size and dielectric properties of CTCs, and they showed an ability to capture a significant percentage of rare cells. Microfluidics-based technologies have been developed for CTC isolation and detection [54,66–69], enabling efficient processing of complex cellular fluids, with minimal damage to sensitive cell populations and minimal blood volume; the low volume requirement could become important for some applications such as pediatric clinical care and small animal studies.
The advantages of microfluidics include its capacity for automatic programming, flexibility in performing a large number of samples, and the capacity for further molecular analysis. However, currently available technologies still suffer from low purity of the captured cells. Recent progress has been made in the development of various microfluidic devices to enrich CTCs, but some of them depend on the discovery and validation of new CTC markers.
Current CTC isolation technologies could be classified according to cells’ biology properties or physical properties, or the combination of properties, such as in the “CTC-iChip” reported in 2014 [61]. Other CTC detection methods have also been developed by incorporating multiple principles to achieve optimal cell isolation, such as processing whole blood by density gradient centrifugation and immunomagnetic isolation in parallel with the use of a microfluidic device to sort cells of interest [53,70–72].
2.1.1 Isolation Based on Physical Properties
CTCs can also be positively or negatively enriched on the basis of physical properties, including size, density, deformability, or electric charges. Those methods consist of centrifugation, membrane- or filtration-based systems, and dielectrophoresis (DEP). Each of these methods is briefly discussed as follows.
2.1.1.1 Gradient Centrifugation
Gradient centrifugation to isolate CTCs from other hematopoietic components is based on density differences. The mononuclear cells and CTCs have a density <1.077 g/mL, while the other blood cells and granulocytes have a density >1.077 g/mL [73]. This process generates a layered separation of different cell types based on their cellular density. Density gradient solutions, such as Ficoll (Amersham) and Lymphoprep (Nycomed), have been used for cell separation.
The advantage of this method is that it offers a quick and simple way to isolate CTCs. But the drawback of this technique is its poor sensitivity, due to the loss of some CTCs migrating to the plasma layer, or the formation of CTC aggregates settling to the bottom of the gradient.
The Ficoll solution separates whole blood into heavier particles (which includes erythrocytes and neutrophils) and lighter particles (which contains mononuclear cells, CTCs, and plasma) [74]. However, whole blood tends to mix with the density gradient, if the sample was not centrifuged immediately. This will result in high contamination with WBCs and potential loss of CTCs. To address the problem, OncoQuick developed layered separation by adding a porous barrier above the density gradient to prevent its mix with whole blood [75,76]. Consequently, density gradient centrifugation with OncoQuick can achieve higher CTC isolation efficiency than Ficoll density gradient centrifugation. The method would simplify the further immunocytochemical CTC detection, offering a method for the detection of CTCs in blood of cancer patients.
2.1.1.2 Microfluidic Inertial Focusing
Microfluidic inertial focusing achieves cell separation with a microfluidic device utilizing the centrifugal, Coriolis, and Euler force to transport and manipulate liquids through their interaction with microstructures. Micro-fluidic flows with dominant viscous drag forces (low Reynolds number, Re) are responsible for laminar flow profiles, entraining suspended particles and cells along streamlines. Separation of CTCs in spiral microfluidic channels has been developed to separate CTCs based on their size differences under the influence of Dean drag forces. The smaller blood cells, including red blood cells and leukocytes, migrate along the Dean vortices toward the inner wall and then back to the outer wall again, while the larger CTCs experience additional strong inertial lift forces and focus along the microchannel inner wall. The term, inertial focusing, refers to migration of cells across streamlines into equilibrium positions within the flow cross-section (after balancing all forces acting on them) as they travel downstream in a microchannel.
The advantage of the spiral microfluidic system is that it can have high-throughput, label-free isolation of CTCs, and unbiased detection of CTCs from all types of cancer, including both epithelial and mesenchymal cell types. However, the use of inertial focusing microfluidics for CTC separation in blood samples is greatly limited by the large number of RBCs and WBCs as well as heterogeneity in the size of CTCs. Also, cell–cell interactions can severely affect the cell-focusing behavior, decreasing the separation efficiency.
Warkiani et al. reported a spiral microfluidic device with a trapezoidal cross-section for isolation of CTCs from clinically relevant blood samples [77]. By using a trapezoidal cross-section, instead of a traditional rectangular cross-section, the position of the Dean vortex core was altered to achieve separation, resulting more than 80% cell capture efficiency of the tested cancer cells.
2.1.1.3 Microfabricated Filters
Because the majority of blood cells are smaller than CTCs, using filters to separate CTCs from whole blood can remove the majority of peripheral blood cells [78–82]. The parylene-based membrane microfilter device utilizes two parylene membrane layers and a photolithography-defined gap to minimize stress, yielding viable cells for further molecular analysis with high efficiency [79].
The advantage of this technology lies in its label-free isolation of CTCs, unbiased detection of CTCs from all types of cancer. However, the greatest limitation of this technology is its sensitivity to size. Questions have been raised regarding the loss of CTCs smaller than the filter’s pores, since the size of CTCs is extremely variable.
2.1.1.4 Dielectrophoresis
Dielectrophoresis (DEP) refers to the movement of a neutral but polarizable particle when it is subjected to a nonuniform electric field due to the interaction of the particle’s dipole and spatial gradient of the electric field. Since biological cells have diverse dielectric properties, DEP can be used to manipulate, transport, separate, and sort different types of cells.
DEP separation techniques can achieve a single-cell-level purification. However, the process could be slow (because low fluid velocity compatible with the DEP force is required), resulting in low sample throughput. The commercially available DEPArray™ technology combines synergistically the power of microelectronics with the precision of microfluidics in an automatic platform to identify and isolate individual cells with high accuracy and precision. The core of this technology is a disposable microsystem that integrates a microelectronic chip with microfluidic chambers and valves. With this method, the exposure of CTCs to strong electric fields and temperature gradients around the electrode can be avoided.
Fabbri et al. demonstrated the use of the DEPArray™ platform to isolate CTCs in patient blood with colon cancer in 2013 [83]. Further, Peeters et al. demonstrated successful molecular characterization of breast cancer tumor cells using the DEPArray™ system to perform predictive biomarker analysis and heterogeneity analysis [84].
2.1.1.5 Other Physical-Property-Based Methods
Various methods have been developed based on physical properties of cells. One of them is acoustophoresis, which has been used to separate tumor cells from healthy blood cells by using acoustic standing wave forces [85]. In this method, CTCs were isolated by subjecting them into free-flow acoustophoresis in a microfluidic device. CTCs were focused in the center of the microchannel and further collected in central outlet, while the non-targeted cells were randomly distributed along the channel.
2.1.2 Isolation Based on Biological Properties
CTCs can be distinguished from other blood components by utilizing their biological properties, such as surface antigens, cytoplasmic protein expression, and invasion capacity.
2.1.2.1 Immunomagnetic Separation
This approach utilizes capture agent-labeled magnetic beads for either positive selection of CTCs using cell-surface markers [11,56–59] or negative depletion of WBCs using anti-CD45 [61,86,87]. The leading example of the methods is the FDA-approved CellSearch™ assay. Through the clinical practices, however, oncologists have come to the conclusion that the use of CellSearch™ assay is challenged by its limited sensitivity [65]. As a result, several sophisticated systems, including VerIFAST [56], magnetic sifter [57], MagSweeper [88], and IsoFlux [89], have been developed to further improve the detection speed and efficiency.
2.1.2.2 Microfluidics-Enabled Immunoseparation
Microfluidics-based devices have been developed to advance CTC detection and isolation [66,68,67,69,90–93]. Microfluidics-based CTC devices using capturing agents, either antibodies [66,67,54] or nucleic acid aptamers [94,95], offer a promising platform to isolate cancer cells from complex cellular fluids with high efficiency, sensitivity, and throughput [48,50–53,47,54]. The advantages of this technology include its capacity for automatic programming, flexibility in performing a large number of samples, and possibility for further molecular analysis. The downside of this technology is that the most of the research efforts relied on the assessment of microscopy imaging for CTC confirmation after isolation.
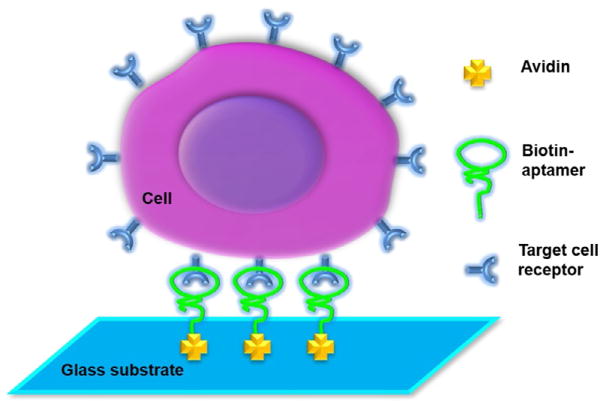
We have developed a microfluidic system that is capable of efficiently isolating cancer cells from whole blood with either aptamers or antibodies. Cell-affinity microstructures were built inside the channel, which was able to selectively capture suspended cancer cells from a heterogeneous cell solution through binding with immobilized high-affinity capture agents [69,94–98]. In our study, avidin was first immobilized onto the channel surfaces by physical adsorption. Aptamers or antibodies were then conjugated onto the device surfaces using biotin–avidin chemistry (Fig. 1).
Fig. 1.
Scheme of capturing cancer cells in a microfluidic device. Avidin is immobilized on the surface of microchannels via physical adsorption, followed by conjugation with biotinylated aptamers through biotin–avidin chemistry. Target cancer cells are then captured via the interaction between the aptamers and the receptors on cell surfaces. W. Sheng, et al., Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device. Anal. Chem. 84(9) (2012) 4199–4206. Reproduced with a permission of The American Chemical Society.
We have developed an aptamer-enabled method to isolate cancer cells [94,95,97]. Further, microfluidic devices consisted of arrays of micropillars inside microchannels, enhancing the surface areas and interactions between aptamers and target cancer cells [95]. The cell capture efficiency was about 95% with a cell purity of ~81% at a flow rate of 600 nL/s. The capture efficiency was defined as the ratio of the number of target cells captured to the number of target cells initially introduced. The capture purity was defined as the ratio of the number of target cells captured to the number of cells captured totally including the target cells and nontarget cells. We used the device for isolating colorectal tumor cells from unprocessed whole blood. Ten tumor cells were captured from 1 mL of whole blood in 28 min. We found that about 93% of the captured cells were viable, making them possible for subsequent molecular and cellular studies. We have also demonstrated that we can perform cell release and subsequent cell analysis after the cell capture. These results indicate that the microfluidic cell isolation system has many advantages including high efficiency, rapid analysis, no pre-treatment of blood samples, and low detection limit.
Further, we have developed a geometrically enhanced mixing chip for high-efficiency and high-purity tumor cell capture [69] (Fig. 2A). We have successfully demonstrated the isolation of CTCs from pancreatic cancer patients, as well as the release and culture of the captured tumor cells. The high performance of the device is based on its geometrically optimized micromixer structures, which enhance the transverse flow and flow folding, maximizing the interaction between cells and antibody-coated surfaces (Fig. 2B).
Fig. 2.
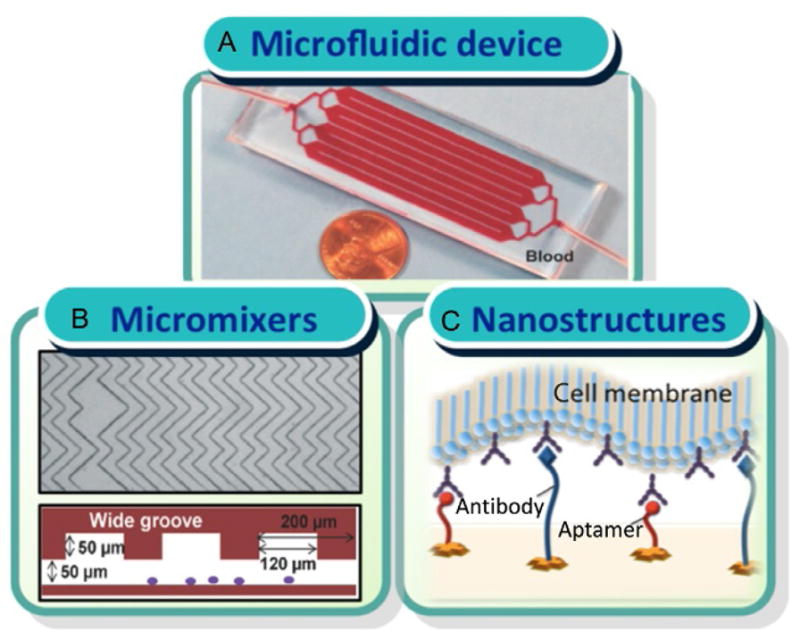
(A) Picture of the device. The size of the device is 1 in. ×3 in., the same size of a microscope slide. (B) Image of the geometrically optimized micromixer structures inside microchannels. (C) Scheme of using an ensemble of aptamers and antibodies as capture reagents inside microchannels. W. Sheng, et al., Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 14(1) (2014) 89–98; J. Zhang, W. Sheng, Z.H. Fan, An ensemble of aptamers and antibodies for multivalent capture of cancer cells. Chem. Commun. 50(51) (2014) 6722–6725. Reproduced with a permission of The Royal Society of Chemistry.
To improve cell capture efficiency at a higher flow rate, we have also developed a system that combines antibodies with aptamers to form a multivalent affinity surface for tumor cell isolation [96]. As shown in Fig. 2C, an ensemble of antibodies and aptamers could bind to cell-surface receptors in a cooperative manner due to their differences in the size. Due to the morphology of a cell and its surface structure with nanoscale microvilli and filopodia [99], the aptamer–antibody ensemble can increase the accessibility of receptors on cell surfaces and the frequency of interactions between receptors and ligands, permitting efficient cell capture. The advantages of enhanced binding avidity through the multivalent effect can generate enhanced local topographic interactions between the substrate and nano-scale cellular surface components [99,100], significantly improving the isolation of tumor cells. We have achieved >90% capture efficiency of human acute lymphoblastic leukemia cells (CCRF-CEM) at a flow rate of 2.0 μL/s.
Also, we have developed a system that utilized multivalent DNA nanospheres to isolate cancer cells in microfluidic devices [98]. In this study, gold nanoparticles immobilized with a number of aptamers were used as efficient capture reagents for high-efficiency cancer cell isolation. Under the optimized condition, the system showed cell capture efficiency of 92% of CCRF-CEM cells at a flow rate of 1.2 μL/s.
2.1.2.3 Other Biological-Property-Based Methods
Most of the current CTC isolation technologies are based on EpCAM expression. However, an epithelial-to-mesenchymal transition (EMT) may occur, in particular during tumor cell dissemination, some emerging technologies have focused on capturing EpCAM-negative CTCs. For example, Satelli et al. explored using cell-surface vimentin as a marker for detecting mesenchymal CTCs from sarcoma tumors [101].
The EPISPOT (EPithelial Immuno SPOT) assay and invasion assay can also achieve CTC enumeration. These assays are normally combined with other methods to partially enrich CTCs first, such as depletion of WBCs. Details of these two assays are discussed in Sections 2.2.2.2 and 2.2.2.3.
An in vivo CTC isolation method has been developed by GILUPI in Germany [102]. In the study, EpCAM-positive CTCs were successfully enriched from over 90% of patients with breast cancer or non-small cell lung cancer. The GILUPI method overcomes the limitation of a certain blood sample volume—which is encountered with most CTC isolation methods—by inserting their CellCollector™ directly into the peripheral blood stream of a patient. After 30 min of in vivo application in an arm vein, the GILUPI CellCollector™ have been in contact with a large volume of the patient’s blood, with a potential to collect rare CTCs.
2.2 CTC Analysis
Beside enumeration, further genetic and phenotypic studies of CTCs would provide important information to define the nature of individual tumor cells and study cancer metastasis. As a result, the number of studies that focus on the characterization of CTCs has increased during the past few years. Some of these technologies are listed in Table 2.
Table 2.
Analysis Approaches for CTCs
| Genetic analysis | qRT-PCR |
| FISH | |
| CGH | |
| Other genetic/genomic methods | |
| Protein analysis | Immunoseparation |
| EPISPOT assay | |
| Invasion assay | |
| Other proteomic methods |
Note: qRT-PCR, quantitative reverse transcription polymerase chain reaction; FISH, fluorescent in situ hybridization; CGH, comparative genomic hybridization; EPISPOT, epithelial immunospot.
2.2.1 Genetic Analysis of CTCs
2.2.1.1 Quantitative Real-Time Polymerase Chain Reaction
The methods of analyzing nucleic acids have been used to quantify and characterize CTCs. The methods utilize primers designed to target the specific gene of interest [103,104]. Researches have been conducted to investigate the transcriptional landscape of CTCs from different types of cancers using high-throughput quantitative reverse transcription polymerase chain reaction (qRT-PCR) and transcriptomic analysis [105–109]. The characterization of gene expression of CTCs has identified several important signatures of regulatory networks and biomarkers for CTCs derived from breast, pancreatic, prostate cancer, and melanoma. However, the limitations of such technologies include lack of stability and reproducibility due to sample contamination, leading to possible false analysis. Besides, they are usually unable to distinguish between viable and apoptotic cells [110].
Stott et al. used a microvortex-generating herringbone-chip to capture CTCs from patients with metastatic prostate cancer. Then, tumor-specific TMPRSS2-ERG translocation was identified by RT-PCR analysis [68]. A study on prostate cancer CTCs using microfluidic qRT-PCR showed that EMT-related genes are overexpressed in castration-resistant CTCs compared to castration-sensitive CTCs [106]. Studies also indicate that EMT and the formation of CTCs and metastasis are closely associated [111,112].
2.2.1.2 Fluorescent In Situ Hybridization
Fluorescence in situ hybridization (FISH) is a cytogenetic technique to detect the locus presence of specific DNA or RNA sequences on chromosomes in individual cells in a heterogeneous population [113]. Labeled DNA or RNA sequences are used as probes to specifically bind to the parts of the chromosome with high degree of sequence complementarity. Because of their greater safety, stability, and ease of detection, the technology allows studying more biomarkers of captured CTCs at the same time in a non-time-consuming way and providing a way to better understand CTCs.
FISH has been successfully applied to analyze CTCs isolated from cancer patients’ blood [114,115]. Specific genes were identified in captured cells to further conform that the cells isolated were CTCs. Recently, a quantifiable, dual-colorimetric RNA-in situ hybridization (ISH) assay was established to characterized EMT in CTCs from breast cancer patients [108]. They have used seven pooled epithelial transcripts (keratin 5, 7, 8, 18, and 19; EpCAM; and cadherin 1) and three mesenchymal transcripts (fibronectin 1, cadherin 2, and serpin peptidase inhibitor, clade E) to examine the expression in tumor cells. In their study, they have demonstrated the evidence of EMT in human breast cancer, showing an association of mesenchymal CTCs with disease progression. They also provided potential biomarkers of therapeutic resistance and potential drug targets of breast cancer.
2.2.1.3 Comparative Genomic Hybridization
Comparative genomic hybridization (CGH) allows the assessment of structural rearrangements in the cell, without the need for culturing cells. Briefly, differentially labeled tumor and genomic DNA are cohybridized to normal human metaphase chromosomes. Each DNA sample was labeled with different fluorescent molecules of different colors. Differences in the fluorescence ratios are used to evaluate the DNA copy number along the chromosome. Therefore, CGH can only detect unbalanced chromosomal changes.
In conjunction with the use of DNA array analysis, the more specific form ACGH (array comparative genomic hybridization) has been developed, allowing for increased resolution for gene analysis. By ACGH analysis, Heitzer et al. demonstrated the feasibility of copy number analysis and mutation screening in CTCs from patients diagnosed with metastatic colorectal cancer patients [116]. Their results showed copy number aberrations in CTCs. ACGH profiles also revealed different aberrations among single CTC.
2.2.2 Protein and Functional Assays of CTCs
2.2.2.1 Immunostaining
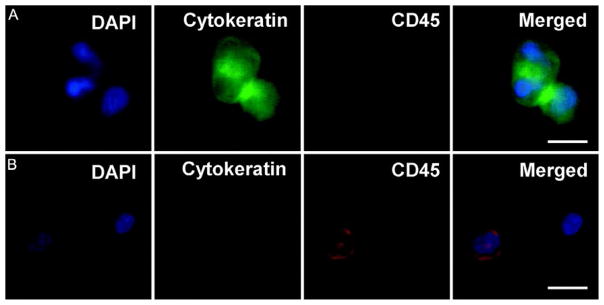
In the FDA-approved CellSearch method, captured CTCs are verified and characterized using immunostaining of cytokeratin (CK) 7/8 and CD45 in addition to 4′,6-diamidino-2-phenylindole (DAPI) for nucleus staining. In our study of pancreatic cancer patients, CTCs were also defined as DAPI positive, CK positive, and CD45 negative (Fig. 3) [69]. In addition to EpCAM and CK, more biomarkers have been used for identification of CTCs for specific type of cancer, such as prostate-specific antigen (PSA) for prostate cancer cells and human epidermal receptor 2 (HER2) for breast tumor cells. This immunostaining method is somewhat limited by the availability and specificity of antibodies.
Fig. 3.
Fluorescence microscope images of CTCs captured from patient bloods: (A) a representative image of CTCs, with DAPI+, cytokeratin+, and CD45−; (B) typical image of white blood cells (WBCs), with DAPI+, CK−, and CD45+. Scale bar=10 μm. W. Sheng, et al., Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 14(1) (2014) 89–98. Reproduced with a permission of The Royal Society of Chemistry.
2.2.2.2 EPISPOT Assay
The EPISPOT assay was developed to detect tumor-specific proteins released by CTCs. First, nitrocellulose membranes of the EPISPOT plates were coated with an antibody against a specific protein marker (eg, cytokeratin). Then, cells are seeded and cultured for 24–48 h. During this incubation step, the specific secreted proteins are directly captured on the antibody-coated membranes. Next, cells are washed off and the specific protein marker is detected by a secondary antibody conjugated with a fluorophore; the number of immunospots is counted. In this way, one immunospot corresponds to the fingerprint of one viable marker-secreting cell, thus the number of CTCs can be counted. EPISPOT can detect living CTCs from cancer patients since it is based on the detection of proteins secreted by these cells [117–119].
By using this assay, researchers showed that full-length CK-19 is released by viable epithelial tumor cells, and CK-19 secreting cells might constitute a biologically active subset of breast cancer cells with high metastatic properties [120]. Also, CTCs have been detected by EPISPOT assay in the peripheral and mesenteric blood of colorectal cancer patients, which are lower than in other cancer types [121]. In this study, CTCs were enriched first with an EpCAM-independent enrichment method. Then, the researchers performed CTC enumeration by EPISPOT assay that detected only viable CK19-releasing CTCs.
2.2.2.3 Invasion Assay
The invasion assay method is based on the ability of CTC to digest a fluorescently labeled cell adhesion matrix. This method can evaluate the invasion ability of CTCs.
Fan et al. examined the invasive growth of CTCs for the propagation of cancer metastasis [15]. They used invasion assay to evaluate the association of invasive CTCs with disease stage. They have found that invasive CTCs can be detected in a majority of epithelial ovarian cancer patients, and they proved that late-stage patients had more invasive CTCs.
2.2.3 Other Methods for CTC Analysis
A variety of other methods have been developed for analyzing CTCs. For instance, CTCs with stem cell properties can give rise to tumor growth in an immunodeficient mouse host [122]. This technology allows analyzing CTCs in vivo. The results of these experiments depend on the experimental conditions and mouse strains. Also, the lack of interaction of these circulating cells with a functional immune system may affect its results, and therefore cannot direct the study of human CTCs. Table 3 summarized the advantages and disadvantages of various methods for CTC analysis and characterization.
Table 3.
Comparison of the Methods for CTC Analysis and Characterization
| Analytical Methods | Advantages | Disadvantages |
|---|---|---|
| Immunocytochemistry | Allow CTC morphological analysis; labeling of specific ligands; quantification; and identification | Time-consuming; sometimes subjective evaluation due to expression variation |
| qRT-PCR | Able to detect specific biomarkers; high sensitivity | Possible false positive results due to WBC contamination |
| EPISPOT | Ability to detect viable CTCs; to identify specific secreted proteins | Time-consuming; proteins must be actively secreted or released by CTCs |
| Invasion assay | Examines the ability of CTCs to digest cell adhesion matrix | Time-consuming |
3. MODELING FOR CTC STUDIES
When a microfluidic device is developed for CTC isolation, simulation is often used to optimize the device design as well as to analyze the device properties. Since the Reynolds number in the microfluidic device is normally less than one, the fluid flow in microchannels is laminar flow. For flow field simulation, a common simplification is that blood cells or CTCs have negligible effect on the flow. A Newtonian flow is thus used in most cases, though whole blood flow is non-Newtonian.
Simulation of CTC isolation is usually based on fluid dynamic models, thermodynamic models, or biophysical models. Compared with experimental methods, simulation is usually less time consuming and of less cost. Although theoretical models still need to be improved (with better assumption and simplification), the currently available models can give a good description of the physics behind. CTC isolation-related simulations are mainly of two categories: device design simulation and CTC–device interaction simulation.
3.1 Device Design Simulation
To enhance the device performance, simulation may be used to optimize microfluidic device geometry, flow field, and other physical properties as discussed below.
Simulation is widely used for optimizing the microfluidic device geometry, such as microstructure arrays. Nagrath et al. optimized the hydrodynamic efficiency of a micropost-based CTC-Chip by comparing different arrangement of post arrays with computational analysis to optimize the device geometry [66]. Dickson et al. used computational fluid dynamic to optimize the alignment of microposts in the fluid channel, suggesting that randomization of different rows of microposts is more efficient than the staggering arrangement [123]. They established a formula to describe the CTC adhesion probability in different section of a microchannel. They considered not only the fluid parameters such as velocity field, shear force, and vorticity but also the encounter probability between CTCs and microposts. In addition, the “stickiness” of the microposts for CTCs was explored, indicating that it affected the capture efficiency of different sections in the micro-channel under different flow rates.
The flow field simulation is of great importance for microfluidic CTC analysis. Murlidhar et al. applied COMSOL to analyze the flow field in a radial flow microfluidic device, called OncoBean chip [124]. The velocity field and shear rate field were simulated. By using the Particle Tracing model, they were able to predict the CTC encounter probability with bean-shaped microposts in the device. Using the commercially available software, the velocity and shear limit of the device were tested, helping to optimize the design of the device. Shear forces added on CTCs not only affect the capture efficiency but also alter the captured cells’ viability. By using the Particle Tracing model, they predicted the encounter probability between CTCs and bean posts. However, the predicted result is a rough estimation because cell–post interaction due to biological properties is not considered, and only streamlines are illustrated.
Another important parameter in flow field simulation is flow rates. Shim et al. optimized the slot design for dielectrophoretic field-flow fractionation (DEP-FFF) using COMSOL MUTIPHYSICS models [125]. In the DEP-FFF device, CTCs were first separated from other blood cells in the dielectrophoretic field along the main channel, then withdrew from the main channel and entered a slot. Through controlling the slot dimension in the simulation, the fractional output flow rate was revised and the CTC isolation efficiency as well as purity was optimized.
Hydrodynamic effect is also critical for device design. Hyun et al. evaluated the velocity field fluctuations in the branch channels to optimize the design of a parallel multi-orifice flow fractionation (p-MOFF) device [126]. In their simulation, fluidic resistance was adjusted to achieve similar flow rates in different channels. Besides of fluidic parameters, other physical properties, such as acoustic properties and magnetic properties, are also of interests. For example, Kang et al. simulated magnetic density around permanent magnets using finite element method and calculated magnetic field-induced velocity of magnetic-bead-bound CTCs [127].
3.2 CTC–Device Interaction Models
As discussed in Section 3.1, parameters such as flow field, magnetic field, electrical field, can play an important role in CTC isolation using micro-fluidic system. However, modeling of those parameters themselves are not enough. The interactions between microfluidic devices and cells inside are of prominent consideration.
For affinity-based CTC isolation, enhancing the interactions between CTCs and the microfluidic device is critical for improving cell capture efficiency. Different modeling methods have been proposed to achieve this goal. Smith et al. applied a theoretical model to describe the cell capture mechanism in microfluidic devices [128]. They showed the collision frequency variation between CTCs and obstacles under different arrangements of obstacles, based on which the device pattern could be optimized.
CTCs are often modeled as rigid spheres with receptors distributed on the surface, while device substrate was modeled as a two-dimensional surface containing ligands (ie, capture agents). The model was discussed in detail by Decuzzi et al. [129]. By combining the shear force, receptor density, ligands density, and affinity constant, they gave an expression of probability of cell adhesion. The combined formula offered a simple mathematical expression of adhesion probability for the particle–substrate interaction. In this modeling, they simplified the problem by ignoring the convection and thermal diffusion of particles. Also, receptors and ligands were considered as the properties of the particle and the substrate, respectively. In this study, the single receptor–ligand bond was not modeled.
A more detailed model that described CTC adhesion to a device substrate was reported by Zheng et al. [130]. In this study, CTCs were introduced into a flat microchannel. When a CTC flow went over a flat surface, Langevin equation was applied to depict the motion of the cells [131,132]. The receptors were modeled as strings randomly distributed on cell surfaces as shown in Fig. 4. There was a capture zone for each receptor, within which a ligand–receptor bond can be formed. The velocity of the cell was altered by shear force as well as bond force caused by ligand–receptor bonds.
Fig. 4.
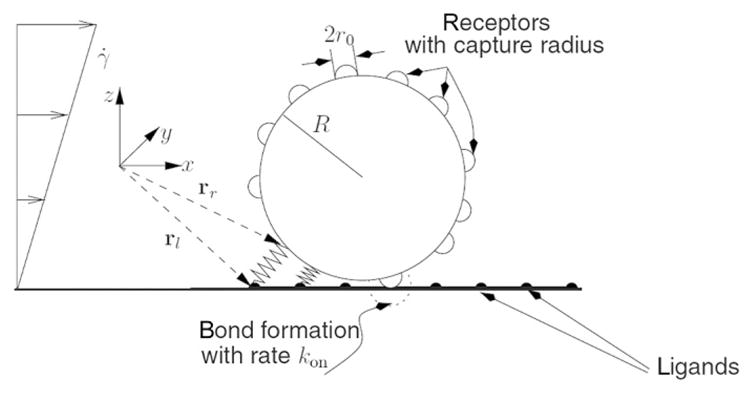
CTC–substrate interaction model. The CTC is modeled as a rigid sphere. Receptors are modeled as strings randomly distributed on the surface of the CTC. Each receptor has a length of r0, forming a capture zone with a radius of r0. When ligands, which are modeled as an array of points on the device surface, are located within the capture zone, it is possible that a ligand–receptor is formulated. The ligand–receptor bond is considered as a spring. C.B., Korn, U.S. Schwarz, Dynamic states of cells adhering in shear flow: from slipping to rolling. Phys. Rev. E 77(4) (2008) 041904. Reproduced with a permission of American Physical Society.
To determine formation of every single receptor–ligand bond, adhesive dynamics was introduced [133]. The ligand–receptor bonds were considered as springs, which made the bond force calculation much easier. This model not only showed how a CTC moved in a microchannel but also illustrated the reactions between the receptors on a CTC and the ligands on the device surface. The theoretical model actually zoomed in the CTC–substrate interaction and showed more physical insights for the CTC capture process. However, a more complicated model could require a lot of computation power and time and would be difficult to be applied for CTC isolation.
4. CTCs IN CLINICAL APPLICATIONS
The detection of CTC in peripheral blood has been demonstrated to provide useful information for clinical study, such as cancer prognostication, treatment monitoring, and drug development. Since CTC is continuously released by primary or metastatic tumor, it has a great potential to be an independent biomarker for clinical applications.
4.1 CTCs as Biomarkers
CTCs have been used as biomarkers in a number of clinical trials. During the past decades, CTCs have received enormous attention as new biomarkers and are one of the hot subjects of basic cancer research. However, their clinical utility is still under investigation.
4.1.1 Definition of Biomarkers
A biomarker is defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or biologic responses to a therapeutic intervention” [134]. Bio-markers are categorized based on clinically utility as diagnostic, prognostic, predictive, pharmacodynamic, or as a surrogate for clinical endpoints. The use of biomarkers has clearly helped to improve cancer survival and the overall morbidity, but there are significant limitations to the application of current biomarkers in clinical practice. Most of blood biomarkers are organ specific rather than disease specific. Therefore, they may not be completely representative of the status of cancer disease. Additionally, biopsy has been the current gold standard for cancer diagnosis [135], which is invasive, preventing patients from being tested in an ongoing or repetitive basis. CTC has received enormous attention as a new cancer bio-marker, because its detection is much less invasive. CTCs can be regarded as “liquid biopsy” and considered to be more effective in monitoring the progression of the disease and choosing different treatments, which allows for real-time monitoring [20,21]. Besides, CTC, originated from the primary tumor, has a potential to guild the development of personalized treatment to optimize the selection of targeted therapies and to monitor the responses.
4.1.2 Clinical Studies
In 2004, Cristofanilli et al. have showed CTCs presented as an independent prognosticator for metastatic breast cancer. They have showed that patients with a cut-off of 5 CTCs/7.5 mL in blood had highly predictive differences of treatment therapy [11]. In a comprehensive meta-analysis of the reported work on the prognostic relevance of CTCs in patients with breast cancer, 6825 patients were included [136]. This analysis has showed CTC can be an accurate prognostic factor in patients with early stage and metastatic breast cancer.
In 2008, Bono et al. have showed that the number of CTCs can accurately and independently predict the overall survival (OS) for castration-resistant prostate cancer [137]. Patients were grouped into Favorable (<5 CTC/7.5 mL) or Unfavorable (≥5 CTC/7.5 mL) according to CTC number. For Favorable group, OS was improved. For Unfavorable group, shorter OS was observed both before treatment and after treatment. Additionally, the transition from Unfavorable group before treatment to Favorable group after treatment has associated with improved OS. On the other hand, the transition from Favorable group to Unfavorable group post treatment has associated with poorer survival. Besides, the CTC number has been shown to be a better prognosticator than PSA decrement in prostate cancer [137–139].
Also, the studies of using CTCs as a diagnostic factor in various cancers were performed [140–143]. However, as CTCs were undergoing EMT, the current isolation methods are not able to identify this subtype of CTCs. Further developments in new technologies for the detection and characterization of CTCs are needed to verify CTCs as new biomarkers, such as CTC subpopulation analysis. Yu et al. monitored the changes of epithelial and mesenchymal markers in CTCs from breast cancer patients. The mesenchymal cells have been showed to associate with disease progression [108].
4.2 CTC Detection for Treatment Monitoring
Current biomarkers and imaging assessments for treatment monitoring are not able to optimally manage individual patient due to deficient specificity to clinical outcomes. For neoadjuvant chemotherapy, systemic responses to the treatment seem to be irrelevant to clinic-pathological features. Therefore, the CTC levels during treatments can better define the effect of therapy [144]. Besides, molecular analyses of CTCs have showed a potential to realize real-time monitoring disease aggressiveness and treatment response [145,146].
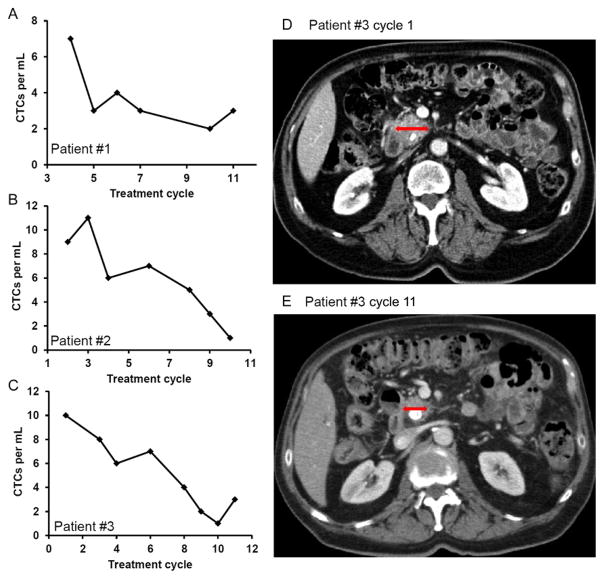
In one of our recent studies, we tested the potential utility of capture and enumeration of CTCs with a microfluidic device for monitoring the response to anticancer drug treatment in pancreatic cancer patients [69]. We have evaluated the correlation between the CTC number and tumor size in patients undergoing chemotherapy. As shown in Fig. 5, three patients with stage IV metastatic pancreatic cancer were included in the study. In general, the CTC number decreased with continuation of treatment, and the result modeled the computed tomography (CT) results. Fig. 5D and e showed that the tumor size decreased as treatment progressed for patient #3, which was in agreement with the trend of CTC number in Fig. 5C. The results indicate that CTC enumeration has a potential to become a tool for monitoring early response or failure to cancer treatment.
Fig. 5.
(A–C) The number of CTCs per mL of blood from pancreatic cancer patients at different treatment cycles for three patients: (A) patient #1; (B) patient #2; (C) patient #3. (D and E) CT scan image of patient #3 at (D) the beginning of the treatment (cycle 1); (E) the latter stage of treatment (cycle 11); the red (dark gray in the print version) arrows indicate the regression of the primary pancreatic cancer. Each treatment cycle is 14 days. W. Sheng, et al., Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 14(1) (2014) 89–98. Reproduced with a permission of The Royal Society of Chemistry.
4.3 Potential Applications in Drug Discovery
The cost of cancer drug development can reach billions and the process typically takes 8–10 years from discovery to registration [147]. The study of CTC under treatment responses in real time can be applied to the clinic drug development. The patients’ CTC can predict treatment failure faster and more accurately than current modalities. Thus, it can indicate drug’s curative effect and predict early success or failure for anticancer drugs [148]. The biological studies on CTC can lead to new drug targets and anticancer therapies. In addition, the clinical trials were established to use CTCs to determine maximal tolerate dose [149]. Studies have also utilized CTC analysis to guild the selection of optimal dose for anticancer drugs [150].
5. CONCLUSION
Since the first identification of CTCs in the blood stream of a patient with metastatic cancer in 1869, significant advancement has been made in the isolation and analysis of CTCs. With these advancement and development, CTCs have been used to predict cancer metastasis and disease progress, to monitor clinical treatment in patients undergoing therapy, and to track disease progression along with treatment efficacy and outcome prediction. This allows individualized therapy (personalized medicine) to identify drug targets and better drug dose, and study the nature of cancer metastasis.
However, the current technologies have been challenged by specificity and sensitivity, compromising cell viability for further CTC studies. Progresses have been made in several directions. The technologies reviewed in this paper represent the potential advances of CTC studies in cancer research. With the development of micro- and nanotechnology, CTC isolation and follow-up analysis can be realized to achieve high capture efficiency, with accuracy and sensitivity.
Acknowledgments
We acknowledge the financial support from National Institute of Health (R21GM103535 and K25CA149080), National Science Foundation (OISE-0968313 and DBI-1353423), University of Florida (UF) Health Cancer Center for Pilot Project Award, and UF Division of Sponsored Research for Preparatory Grant and Opportunity Fund.
References
- 1.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8(5):329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 3.Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:146–147. [Google Scholar]
- 4.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 5.Christopherson WM. Cancer cells in the peripheral blood: a second look. Acta Cytol. 1965;9:169–174. [PubMed] [Google Scholar]
- 6.Drye JC, Rumage WT, Jr, Anderson D. Prognostic import of circulating cancer cells after curative surgery: a long time follow up study. Ann Surg. 1962;155:733–740. doi: 10.1097/00000658-196205000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engell HC. Cancer cells in the circulating blood; a clinical study on the occurrence of cancer cells in the peripheral blood and in venous blood draining the tumour area at operation. Acta Chir Scand Suppl. 1955;201:1–70. [PubMed] [Google Scholar]
- 8.Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 9.Wong SCC, et al. Clinical significance of cytokeratin 20-positive circulating tumor cells detected by a refined immunomagnetic enrichment assay in colorectal cancer patients. Clin Cancer Res. 2009;15(3):1005–1012. doi: 10.1158/1078-0432.CCR-08-1515. [DOI] [PubMed] [Google Scholar]
- 10.Uen YH, et al. Persistent presence of postoperative circulating tumor cells is a poor prognostic factor for patients with stage I-III colorectal cancer after curative resection. Ann Surg Oncol. 2008;15(8):2120–2128. doi: 10.1245/s10434-008-9961-7. [DOI] [PubMed] [Google Scholar]
- 11.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in met-astatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 12.Pachmann K, et al. Monitoring the response of circulating epithelial tumor cells to adjuvant chemotherapy in breast cancer allows detection of patients at risk of early relapse. J Clin Oncol. 2008;26(8):1208–1215. doi: 10.1200/JCO.2007.13.6523. [DOI] [PubMed] [Google Scholar]
- 13.Danila DC, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(23):7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 14.Morgan TM, et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clin Cancer Res. 2009;15(2):677–683. doi: 10.1158/1078-0432.CCR-08-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan T, et al. Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol. 2009;112(1):185–191. doi: 10.1016/j.ygyno.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 17.Bernards R, Weinberg RA. Metastasis genes: a progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 18.Kling J. Beyond counting tumor cells. Nat Biotechnol. 2012;30(7):578–580. doi: 10.1038/nbt.2295. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser J. Cancer’s circulation problem. Science. 2010;327(5969):1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 20.van de Stolpe A, et al. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. 2011;71(18):5955–5960. doi: 10.1158/0008-5472.CAN-11-1254. [DOI] [PubMed] [Google Scholar]
- 21.Crowley E, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 22.Helo P, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with Cel-lSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55(4):765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J, et al. Detecting carcinoma cells in peripheral blood of patients with hepato-cellular carcinoma by immunomagnetic beads and rt-PCR. J Clin Gastroenterol. 2007;41(8):783–788. doi: 10.1097/01.mcg.0000247996.19710.f2. [DOI] [PubMed] [Google Scholar]
- 24.Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the Cell Search system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 25.Dharmasiri U, et al. Microsystems for the capture of low-abundance cells. Annu Rev Anal Chem. 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 26.Krivacic RT, et al. A rare-cell detector for cancer. Proc Natl Acad Sci USA. 2004;101(29):10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42(2):155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 28.Racila E, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA. 1998;95(8):4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manz A, Graber N, Widmer HM. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sensors Actuators B1. 1990;1:244–248. [Google Scholar]
- 30.Feynman RP. There’s plenty of room at the bottom. J Micromech Syst. 1959;1(1):60–66. [Google Scholar]
- 31.Whitesides GM. The ‘right’ size in nanobiotechnology. Nat Biotechnol. 2003;21(10):1161–1165. doi: 10.1038/nbt872. [DOI] [PubMed] [Google Scholar]
- 32.Moore GE. Cramming more components onto integrated circuits. Electronics. 1965;38:114–117. [Google Scholar]
- 33.Reyes DR, et al. Micro total analysis systems. 1. Introduction, theory, and technology. Anal Chem. 2002;74(12):2623–2636. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- 34.Lee SJ, Lee SY. Micro total analysis system (μ-TAS) in biotechnology. Appl Microbiol Biotechnol. 2004;64(3):289–299. doi: 10.1007/s00253-003-1515-0. [DOI] [PubMed] [Google Scholar]
- 35.Woolley AT, Mathies RA. Ultra-high-speed DNA sequencing using capillary electrophoresis chips. Anal Chem. 1995;67(20):3676–3680. doi: 10.1021/ac00116a010. [DOI] [PubMed] [Google Scholar]
- 36.Woolley AT, Mathies RA. Ultra-high-speed DNA fragment separations using microfabricated capillary array electrophoresis chips. Proc Natl Acad Sci USA. 1994;91(24):11348–11352. doi: 10.1073/pnas.91.24.11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp MU, de Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280(5366):1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 38.Krishnan M, Ugaz VM, Burns MA. PCR in a Rayleigh-Bénard convection cell. Science. 2002;298(5594):793. doi: 10.1126/science.298.5594.793. [DOI] [PubMed] [Google Scholar]
- 39.Sanders GHW, Manz A. Chip-based microsystems for genomic and proteomic analysis. Trends Anal Chem. 2000;19(6):364–378. [Google Scholar]
- 40.Hadd AG, Jacobson SC, Ramsey JM. Microfluidic assays of acetylcholinesterase inhibitors. Anal Chem. 1999;71(22):5206–5212. [Google Scholar]
- 41.Hatch A, et al. A rapid diffusion immunoassay in a T-sensor. Nat Biotechnol. 2001;19(5):461–465. doi: 10.1038/88135. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen N-T, Wereley Steven T. Fundamentals and Applications of Microfluidics. Artech House; Boston/London: 2002. [Google Scholar]
- 43.Manz A, et al. Planar chips technology for miniaturization and integration of separation techniques into monitoring systems—capillary electrophoresis on a chip. J Chromatogr. 1992;593(1–2):253–258. [Google Scholar]
- 44.Sia SK, Whitesides GM. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis. 2003;24(21):3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 45.Hansen CL, et al. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci USA. 2002;99(26):16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi J, et al. A microfluidic device for conducting gas-liquid-solid hydrogenation reactions. Science. 2004;304(5675):1305–1308. doi: 10.1126/science.1096956. [DOI] [PubMed] [Google Scholar]
- 47.Wang ST, et al. Highly efficient capture of circulating tumor cells by using nano-structured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed. 2011;50(13):3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dharmasiri U, et al. High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal Chem. 2011;83(6):2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng X, et al. A high-performance microsystem for isolating circulating tumor cells. Lab Chip. 2011;11(19):3269–3276. doi: 10.1039/c1lc20331b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Owens GE, Tseng HR. Nano “fly paper” technology for the capture of circulating tumor cells. Methods Mol Biol. 2011;726:141–150. doi: 10.1007/978-1-61779-052-2_10. [DOI] [PubMed] [Google Scholar]
- 51.Saliba AE, et al. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci USA. 2010;107(33):14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikolajczyk SD, et al. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol. 2011;2011:252361. doi: 10.1155/2011/252361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoshino K, et al. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip. 2011;11(20):3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams AA, et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130(27):8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed. 2009;48(47):8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casavant BP, et al. The VerIFAST: an integrated method for cell isolation and extra-cellular/intracellular staining. Lab Chip. 2013;13(3):391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 57.Earhart CM, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14(1):78–88. doi: 10.1039/c3lc50580d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ozkumur E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5(179):179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaffer DR, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(7):2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 60.Tong X, et al. Application of immunomagnetic cell enrichment in combination with RT-PCR for the detection of rare circulating head and neck tumor cells in human peripheral blood. Cytometry B Clin Cytom. 2007;72(5):310–323. doi: 10.1002/cyto.b.20177. [DOI] [PubMed] [Google Scholar]
- 61.Karabacak NM, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9(3):694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moldenhauer G, et al. Epithelium-specific surface glycoprotein of Mr 34,000 is a widely distributed human carcinoma marker. Br J Cancer. 1987;56(6):714–721. doi: 10.1038/bjc.1987.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNiece I, et al. Large-scale isolation of CD34+ cells using the Amgen cell selection device results in high levels of purity and recovery. J Hematother. 1997;6(1):5–11. doi: 10.1089/scd.1.1997.6.5. [DOI] [PubMed] [Google Scholar]
- 64.Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the cell search system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Attard G, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69(7):2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 66.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by micro-chip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gleghorn JP, et al. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10(1):27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107(43):18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheng W, et al. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip. 2014;14(1):89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang YY, et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed Microdevices. 2013;15(4):673–681. doi: 10.1007/s10544-012-9718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kojima T, et al. A simple biological imaging system for detecting viable human circulating tumor cells. J Clin Invest. 2009;119(10):3172–3181. doi: 10.1172/JCI38609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shigeyasu K, et al. Fluorescence virus-guided capturing system of human colorectal circulating tumour cells for non-invasive companion diagnostics. Gut. 2014;64:627–635. doi: 10.1136/gutjnl-2014-306957. [DOI] [PubMed] [Google Scholar]
- 73.Morgan TM, Lange PH, Vessella RL. Detection and characterization of circulating and disseminated prostate cancer cells. Front Biosci. 2007;12:3000–3009. doi: 10.2741/2290. [DOI] [PubMed] [Google Scholar]
- 74.Leise EM, et al. Lymphocyte and polymorphonuclear enzymes in stress. 1. Method of separation of leukocytes from small amounts of blood into lymphocyte and poly-morphonuclear leukocytes by density-gradient centrifugation. Biochem Med. 1970;4(3–4):327–335. doi: 10.1016/0006-2944(70)90058-x. [DOI] [PubMed] [Google Scholar]
- 75.Rosenberg R, et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49(4):150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 76.Gertler R, et al. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149–155. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 77.Warkiani ME, et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14(1):128–137. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 78.Vona G, et al. Isolation by size of epithelial tumor cells—a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156(1):57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng S, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162(2):154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 80.Tan SJ, et al. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11(4):883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 81.Tan SJ, et al. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens Bioelectron. 2010;26(4):1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 82.Lecharpentier A, et al. Detection of circulating tumour cells with a hybrid (epithelial/ mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105(9):1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fabbri F, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett. 2013;335(1):225–231. doi: 10.1016/j.canlet.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 84.Peeters DJE, et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from Cell Search enriched blood samples using dielectrophoretic cell sorting. Br J Cancer. 2013;108(6):1358–1367. doi: 10.1038/bjc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li P, et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci USA. 2015;112(16):4970–4975. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lara O, et al. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004;32(10):891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 87.Tong XD, et al. Application of immunomagnetic cell enrichment in combination with RT-PCR for the detection of rare circulating head and neck tumor cells in human peripheral blood. Cytometry B Clin Cytom. 2007;72B(5):310–323. doi: 10.1002/cyto.b.20177. [DOI] [PubMed] [Google Scholar]
- 88.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA. 2009;106(10):3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harb W, et al. Mutational analysis of circulating tumor cells using a novel micro-fluidic collection device and qPCR assay. Transl Oncol. 2013;6(5):528–538. doi: 10.1593/tlo.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong Y, et al. Microfluidics and circulating tumor cells. J Mol Diagn. 2013;15(2):149–157. doi: 10.1016/j.jmoldx.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 91.Huang SB, et al. High-purity and label-free isolation of circulating tumor cells (CTCs) in a microfluidic platform by using optically-induced-dielectrophoretic (ODEP) force. Lab Chip. 2013;13(7):1371–1383. doi: 10.1039/c3lc41256c. [DOI] [PubMed] [Google Scholar]
- 92.Ohnaga T, et al. Polymeric microfluidic devices exhibiting sufficient capture of cancer cell line for isolation of circulating tumor cells. Biomed Microdevices. 2013;15(4):611–616. doi: 10.1007/s10544-013-9775-7. [DOI] [PubMed] [Google Scholar]
- 93.Casavant BP, et al. A negative selection methodology using a microfluidic platform for the isolation and enumeration of circulating tumor cells. Methods. 2013;64(2):137–143. doi: 10.1016/j.ymeth.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu Y, et al. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal Chem. 2009;81(17):7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheng W, et al. Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device. Anal Chem. 2012;84(9):4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang J, Sheng W, Fan ZH. An ensemble of aptamers and antibodies for multivalent capture of cancer cells. Chem Commun. 2014;50(51):6722–6725. doi: 10.1039/c4cc02002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Phillips JA, et al. Enrichment of cancer cells using aptamers immobilized on a micro-fluidic channel. Anal Chem. 2009;81(3):1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sheng WA, et al. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano. 2013;7(8):7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang S, et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed Engl. 2009;48(47):8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen W, et al. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano. 2013;7(1):566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Satelli A, et al. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin Chem. 2015;61(1):259–266. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saucedo-Zeni N, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol. 2012;41(4):1241–1250. doi: 10.3892/ijo.2012.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.von Knebel Doeberitz M, Lacroix J. Nucleic acid based techniques for the detection of rare cancer cells in clinical samples. Cancer Metastasis Rev. 1999;18(1):43–64. doi: 10.1023/a:1006208303075. [DOI] [PubMed] [Google Scholar]
- 104.Keilholz U, et al. Reliability of reverse transcription-polymerase chain reaction (RT-PCR)-based assays for the detection of circulating tumour cells: a quality-assurance initiative of the EORTC melanoma cooperative group. Eur J Cancer. 1998;34(5):750–753. doi: 10.1016/s0959-8049(97)10105-8. [DOI] [PubMed] [Google Scholar]
- 105.Welty CJ, et al. Single cell transcriptomic analysis of prostate cancer cells. BMC Mol Biol. 2013;14:6. doi: 10.1186/1471-2199-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen CL, et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate. 2013;73(8):813–826. doi: 10.1002/pros.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cann GM, et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS One. 2012;7(11):e49144. doi: 10.1371/journal.pone.0049144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Powell AA, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7(5):e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zippelius A, et al. Limitations of reverse-transcriptase polymerase chain reaction analyses for detection of micrometastatic epithelial cancer cells in bone marrow. J Clin Oncol. 1997;15(7):2701–2708. doi: 10.1200/JCO.1997.15.7.2701. [DOI] [PubMed] [Google Scholar]
- 111.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119(6):1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6(10):782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 114.Engel H, et al. Detection of circulating tumour cells in patients with breast or ovarian cancer by molecular cytogenetics. Br J Cancer. 1999;81(7):1165–1173. doi: 10.1038/sj.bjc.6690825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Swennenhuis JF, et al. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry A. 2009;75(6):520–527. doi: 10.1002/cyto.a.20718. [DOI] [PubMed] [Google Scholar]
- 116.Heitzer E, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73(10):2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 117.Jin A, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. 2009;15(9):1088–1092. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- 118.Alix-Panabieres C, et al. Detection of circulating prostate-specific antigen-secreting cells in prostate cancer patients. Clin Chem. 2005;51(8):1538–1541. doi: 10.1373/clinchem.2005.049445. [DOI] [PubMed] [Google Scholar]
- 119.Alix-Panabieres C, et al. Detection and characterization of putative metastatic precursor cells in cancer patients. Clin Chem. 2007;53(3):537–539. doi: 10.1373/clinchem.2006.079509. [DOI] [PubMed] [Google Scholar]
- 120.Alix-Panabieres C, et al. Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. 2009;11(3):R39. doi: 10.1186/bcr2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deneve E, et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem. 2013;59(9):1384–1392. doi: 10.1373/clinchem.2013.202846. [DOI] [PubMed] [Google Scholar]
- 122.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dickson MN, et al. Efficient capture of circulating tumor cells with a novel immu-nocytochemical microfluidic device. Biomicrofluidics. 2011;5(3):034119. doi: 10.1063/1.3623748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murlidhar V, et al. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small. 2014;10(23):4895–4904. doi: 10.1002/smll.201400719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shim S, et al. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics. 2013;7(1):011807. doi: 10.1063/1.4774304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hyun KA, et al. Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients. Biosens Bioelectron. 2013;40(1):206–212. doi: 10.1016/j.bios.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 127.Kang JH, et al. A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab Chip. 2012;12(12):2175–2181. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]
- 128.Smith JP, et al. Parametric control of collision rates and capture rates in geometrically enhanced differential immunocapture (GEDI) microfluidic devices for rare cell capture. Biomed Microdevices. 2014;16(1):143–151. doi: 10.1007/s10544-013-9814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Decuzzi P, Ferrari M. The adhesive strength of non-spherical particles mediated by specific interactions. Biomaterials. 2006;27(30):5307–5314. doi: 10.1016/j.biomaterials.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 130.Zheng X, et al. Cell receptor and surface ligand density effects on dynamic states of adhering circulating tumor cells. Lab Chip. 2011;11(20):3431–3439. doi: 10.1039/c1lc20455f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Korn C, Schwarz U. Mean first passage times for bond formation for a Brownian particle in linear shear flow above a wall. J Chem Phys. 2007;126(9):095103. doi: 10.1063/1.2464080. [DOI] [PubMed] [Google Scholar]
- 132.Korn CB, Schwarz US. Dynamic states of cells adhering in shear flow: from slipping to rolling. Phys Rev E. 2008;77(4):041904. doi: 10.1103/PhysRevE.77.041904. [DOI] [PubMed] [Google Scholar]
- 133.Hammer DA, Apte SM. Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion. Biophys J. 1992;63(1):35–57. doi: 10.1016/S0006-3495(92)81577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lesko LJ, Atkinson AJ. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu Rev Pharmacol Toxicol. 2001;41:347–366. doi: 10.1146/annurev.pharmtox.41.1.347. [DOI] [PubMed] [Google Scholar]
- 135.King JD, Casavant BP, Lang JM. Rapid translation of circulating tumor cell bio-markers into clinical practice: technology development, clinical needs and regulatory requirements. Lab Chip. 2014;14(1):24–31. doi: 10.1039/c3lc50741f. [DOI] [PubMed] [Google Scholar]
- 136.Zhang LL, et al. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18(20):5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 137.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 138.Scher HI, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10(3):233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Halabi S, et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol. 2003;21(7):1232–1237. doi: 10.1200/JCO.2003.06.100. [DOI] [PubMed] [Google Scholar]
- 140.Lim E, et al. Clinical results of microfluidic antibody-independent peripheral blood circulating tumor cell capture for the diagnosis of lung cancer. J Thorac Cardiovasc Surg. 2014;147(6):1936–1938. doi: 10.1016/j.jtcvs.2013.09.052. [DOI] [PubMed] [Google Scholar]
- 141.Qi FM, et al. Quantitation of rare circulating tumor cells by folate receptor alpha ligand-targeted PCR in bladder transitional cell carcinoma and its potential diagnostic significance. Tumor Biol. 2014;35(7):7217–7223. doi: 10.1007/s13277-014-1894-0. [DOI] [PubMed] [Google Scholar]
- 142.Jin T, Peng H, Wu H. Clinical value of circulating liver cancer cells for the diagnosis of hepatocellular carcinoma: a meta-analysis. Biomed Rep. 2013;1(5):731–736. doi: 10.3892/br.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khoja L, et al. Biomarker utility of circulating tumor cells in metastatic cutaneous melanoma. J Investig Dermatol. 2013;133(6):1582–1590. doi: 10.1038/jid.2012.468. [DOI] [PubMed] [Google Scholar]
- 144.Serrano MJ, et al. Dynamics of circulating tumor cells in early breast cancer under neoadjuvant therapy. Exp Ther Med. 2012;4(1):43–48. doi: 10.3892/etm.2012.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nakagawa T, et al. Detection of circulating tumor cells in early-stage breast cancer metastasis to axillary lymph nodes. Clin Cancer Res. 2007;13(14):4105–4110. doi: 10.1158/1078-0432.CCR-07-0419. [DOI] [PubMed] [Google Scholar]
- 146.Amir E, et al. Tissue confirmation of disease recurrence in breast cancer patients: pooled analysis of multi-centre, multi-disciplinary prospective studies. Cancer Treat Rev. 2012;38(6):708–714. doi: 10.1016/j.ctrv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 147.DiMasi JA, Grabowski HG. Economics of new oncology drug development. J Clin Oncol. 2007;25(2):209–216. doi: 10.1200/JCO.2006.09.0803. [DOI] [PubMed] [Google Scholar]
- 148.Tan DSW, et al. Biomarker-driven early clinical trials in oncology a paradigm shift in drug development. Cancer J. 2009;15(5):406–420. doi: 10.1097/PPO.0b013e3181bd0445. [DOI] [PubMed] [Google Scholar]
- 149.Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res. 2007;96:213–268. doi: 10.1016/S0065-230X(06)96008-4. [DOI] [PubMed] [Google Scholar]
- 150.Devriese LA, et al. Circulating tumor cells as pharmacodynamic biomarker in early clinical oncological trials. Cancer Treat Rev. 2011;37(8):579–589. doi: 10.1016/j.ctrv.2011.04.006. [DOI] [PubMed] [Google Scholar]