Summary
A class of cis-regulatory elements called enhancers plays a central role in orchestrating spatio-temporally precise gene expression programs during development. Consequently, divergence in enhancer sequence and activity is thought to be an important mediator of inter- and intra-species phenotypic variation. Here, we give an overview of emerging principles of enhancer function, current models of enhancer architecture, genomic substrates from which enhancers emerge during evolution and the influence of three-dimensional genome organization on long-range gene regulation. We discuss intricate relationships between distinct elements within complex regulatory landscapes and consider their potential impact on specificity and robustness of transcriptional regulation.
Introduction
The accurate, precise and robust regulation of gene expression during development is a cornerstone for complex biological life. Much of this information is encoded by cis-regulatory elements called enhancers, canonically defined as short (~100-1000bp) noncoding DNA sequences that act to drive transcription independent of their relative distance, location or orientation to their cognate promoter (for a historical perspective on the discovery of enhancers, see Schaffner, 2015). Although enhancers share many features with other classes of cis-regulatory elements, especially promoters (reviewed in (Kim and Shiekhattar, 2015)), it is their ability to activate transcription over long genomic distances that sets them apart. This feature allows a gene to be regulated by multiple distal enhancers with different spatiotemporal activities, facilitating enormous combinatorial complexity of gene expression repertoires (and, in turn, a vast array of cellular states) using a relatively limited set of genes. In addition to their central role in developmental gene regulation, enhancers are fertile targets for evolutionary change as they are both cell type-specific (allowing for modulation of target gene expression in specific tissue context without affecting other pleiotropic gene functions) and commonly exist in groups of redundant elements (facilitating accumulation of genetic variation by buffering the risk of lethality) (Levine, 2010; Wittkopp and Kalay, 2011).
For decades most of the key insights into enhancer function and divergence have come from Drosophila genetics. However, recent years have seen a renewed interest in long-range gene regulation in many species including mammals, fueled by the increasing availability of genome sequences, technological progress in genomics, genome editing and imaging, as well as the realization that disease- and trait-associated genetic variants frequently map to enhancers. Insights into genome folding have revealed that most metazoan chromosomes are partitioned into self-associating topological domains that restrict enhancer search space for target promoters. Furthermore, comparative epigenomics from both closely related and more distant species have afforded us a better glimpse into cis-regulatory evolution and constraint. Here, we discuss these new developments and give an overview of our current understanding of the general principles of enhancer function and evolution.
I. General principles of enhancer function
Enhancers as clusters of TF binding sites
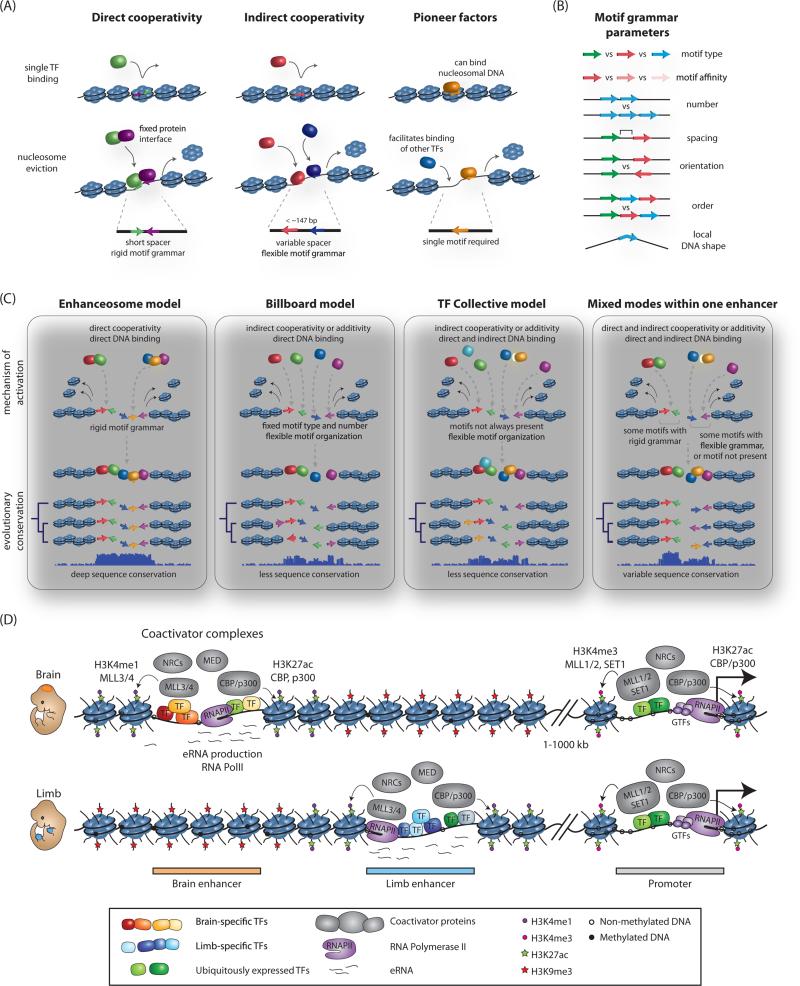
Functional enhancers are composed of concentrated clusters of transcription factor (TF) recognition motifs. Generally, enhancer activation requires the binding of multiple TFs, often including both lineage-specific factors and sequence-dependent effectors of signal transduction pathways, thereby ensuring integration of intrinsic and extrinsic signaling cues at these elements (reviewed in Buecker and Wysocka, 2012) (Figure 1). One reason why the action of multiple TFs is essential is that regulatory element sequences have high intrinsic affinity for histone octamers (Tillo et al., 2010), creating a strong barrier for access of TFs to the underlying DNA. Cooperative binding of multiple factors in close proximity (i.e. less than one nucleosome length) is thought to play a major role in overcoming the energetic barrier of nucleosome eviction, thus facilitating downstream effector binding and enhancer activation (Spitz and Furlong, 2012). Such cooperativity can rely on the direct physical association between TFs before or concurrent with DNA binding, called ‘direct cooperativity’ (Figure 1A, left panel). Importantly, cooperative binding of TFs to DNA can also occur in the absence of direct protein-protein interactions through a process referred to as ‘indirect cooperativity’ or ‘collaborative competition’, in which a cohort of TFs collectively competes with the same histone octamer for access to underlying DNA as demonstrated experimentally (Miller and Widom, 2003) and predicted by in silico modelling (Mirny, 2010) (Figure 1A, middle panel). Thus, as a general rule, increased numbers of TF motifs at an enhancer should be positively correlated with increased nucleosomal eviction, DNA binding and consequently, elevated gene expression output. Numerous studies looking both at endogenous enhancers (Burz et al., 1998) and using synthetic reporters (Erceg et al., 2014; Smith et al., 2013a) support this view, but also highlight an often complex interplay between different regulatory sequences, as discussed in more detail below. In addition, at some developmental enhancers there is evidence for step-wise licensing by lineage-determining TFs, also known as master regulators, or pioneer factors which directly bind nucleosomal DNA to prime enhancers for activation (Figure 1A, right panel). These factors can recruit chromatin remodeling activities, which then facilitate removal and post-translational modification of histones meaning that subsequent TF and coactivator binding is less strictly dependent on direct competition between TFs and nucleosomes (Zaret and Carroll, 2011).
Figure 1. Nucleosome eviction, enhancer grammar and models of enhancer architecture.
(A) TF mechanisms for overcoming energetic barrier to nucleosomal eviction and underlying motif requirements. (B) Parameters of motif grammar at enhancers. (C) Models of enhancer architecture and their primary mechanism of TF binding, flexibility of motif organization, and selective constraint across evolution. (D) Coactivator binding and chromatin signatures at active enhancers. Presence of cell-type specific and broadly-expressed TFs, RNA Polymerase II (RNAPII) and associated enhancer RNAs (eRNAs), coactivator complexes such as Mediator (Med), nucleosome remodeling complexes (NRCs), histone acetyltransferases (CBP/p300) and methyltransferases (MLL3/4) at tissue-specific enhancer elements is schematically depicted for a brain (top) or limb (bottom) specific enhancer. Select modifications of neighboring nucleosomes associated with active enhancer states are highlighted. An overlapping set of protein complexes and modifications is also present at promoters, with the distinction that enhancers have high ratio of H3K4me1 to H3K4me3, and the reverse is true at promoters. Both active enhancers and promoters are characterized by DNA hypomethylation, with the methylation status of enhancers being more dynamic and tracking with their cell-type specificity.
Motif organization, enhancer grammar and models of enhancer architecture
Because of this complex interplay and hierarchical logic between DNA-binding regulators at enhancers, the underlying organization of consensus motifs can have dramatic effects on enhancer activity and robustness. The universal principles of this organization – referred to as enhancer “grammar” - are quite complex and still an area of active research. The enhancer lexicon incorporates the number, type, order, spacing, orientation, local DNA shape and binding affinity of TF motifs, all of which can affect the functional output of any given enhancer, and are subject to varying degrees of selective constraint during evolution (Figure 1B). From these principles, two main distinct models for describing enhancer architecture have emerged, with the first ‘enhanceosome’ model requiring rigid motif organization and spacing for function, while the alternative ‘billboard’ model allows greater flexibility of binding, with the presence of a specifc set of TFs being more important than the defined order or orientation of their underlying motifs (Figure 1C).
Several examples of enhancers falling at both ends of this architectural spectrum have been described. The archetypal model of an enhanceosome is the mammalian viral-inducible interferon-β (IFNβ) enhancer (Thanos and Maniatis, 1995), which requires the cooperative binding of eight TFs to create a composite surface for DNA binding. While documented examples of enhanceosomes are rare, simulations of TF binding have suggested that this mode of enhancer regulation may be in fact relatively common in mammalian genomes (Guturu et al., 2013). On the other end of the spectrum, so-called ‘billboard’ enhancers preserve function by maintaining TFBS content, but with significant flexibility as to the order or spacing of those sites within the larger enhancer, likely by relying on indirect TF cooperativity for activation and also allowing for independent contribution of multiple sub-elements to gene expression (Arnosti and Kulkarni, 2005). Interestingly, these sorts of enhancers often contain suboptimal binding sites that not only tolerate rapid motif turnover across species, but that also may serve an important role in promoting enhancer specificity (Crocker et al., 2015; Farley et al., 2015). For example, the neural plate-specific Otx-a enhancer in Ciona, contains GATA and ETS motifs that are imperfect matches to the consensus (Farley et al., 2015). Strikingly, optimization of motif sequence or spacing causes stronger and ectopic enhancer activity patterns, as presumably the enhancer is now responsive to lower levels of GATA or ETS. Thus, combinatorial deployment of multiple suboptimal enhancers may help to promote specificity of regulation without sacrificing signal strength. In addition, often a number of TFs are localized to enhancers without underlying consensus binding sites, and are instead recruited through protein-protein interactions, as proposed for the ‘transcription factor collective’ model of enhancer organization (Figure 1C), (Junion et al., 2012).
Both large-scale comparisons of enhancer conservation across species (Taher et al., 2011) as well as synthetic enhancer reporter studies (Smith et al., 2013a) have broadly supported the more flexible organizational models whereby a collection of TF binding events can drive reporter activity in different orders or orientations. However, reexamination of the prevalence of more rigid motif grammar may be warranted, as recent work has challenged the widely held assumption that direct cooperativity of TFs should be evident on the DNA level by the presence of a composite motif (i.e., complete motifs of both TFs with a defined spacing). In fact, SELEX analysis using consecutive affinity-purification to interrogate pairwise TF binding specificities (CAP-SELEX, (Jolma et al., 2015)) has demonstrated that the majority of TF-TF interactions have a novel consensus motif, bound only weakly by each TF in isolation. Importantly, these novel long motifs detected by SELEX analysis are enriched in mammalian ChIP-seq TF clusters, suggesting that this sort of cooperative binding is commonplace throughout the genome. Therefore, it was likely the inability to recognize sites of direct cooperativity between TFs rather than their true scarcity, which contributed to the notion that most mammalian enhancers are devoid of defined TF motif spacing. Indeed, evidence is beginning to emerge that spacing and orientation constraints on pair-wise TF binding are more prevalent than initially anticipated (Ng et al., 2014). Thus, in reality, the architecture of most enhancers likely falls into a spectrum between the rigid motif organization of the enhanceosome and flexible organization of the billboard and TF collective, with defined spacing and orientation being required for some motifs within the enhancer but not others (Figure 1C).
Implications of enhancer architecture on regulatory landscape evolution
It is interesting to consider what these different models of enhancer architecture could mean during regulatory landscape evolution. At enhanceosomes, any binding site mutation will preclude binding of the entire complex, demanding deep conservation at the DNA sequence level to maintain function. Such strictly conserved enhancer sequences do indeed exist, and have been studied for quite some time (reviewed in (Visel et al., 2007)). The question remains, however, whether there is anything unique about these deeply conserved enhancers. One hypothesis is that enhanceosomes may have potential roles in mediating switch-like transcriptional activation of genes requiring strict regulatory inputs from multiple signaling pathways (Spitz and Furlong, 2012). Consistent with this theory, deeply-conserved enhancers tend to fall as clusters near genes encoding developmentally-important transcription factors (see (Boffelli et al., 2004) and references within). Because of their developmental importance and strict structural requirements, mutations at conserved regulatory sites are also associated with human disease, such as seen for the deeply conserved Shh enhancer that drives preaxial polydactyly upon disruption (Lettice et al., 2003).
Despite their association with developmental TFs and human disease, deeply conserved enhancers are an exception rather than the rule. Instead, de novo gains and losses of enhancers appear to be relatively common across evolution (discussed in more detail below), and furthermore, even when an ancestral function is conserved, these enhancers often retain no recognizable sequence similarity or have only very weak conservation signatures. For example, only ~24% of murine human heart enhancers with p300 occupancy showed any detectable conservation with other vertebrates, yet these poorly-conserved enhancers still demonstrated heart-specific activity in transgenic mouse embryo reporter assays regardless of their level of sequence conservation (Blow et al., 2010; May et al., 2012). Similarly, poised developmental enhancers identified in human embryonic stem cells drove cell type- and stage-specific expression when introduced to zebrafish embryos, even in the absence of detectable sequence conservation between the two species (Rada-Iglesias et al., 2011). One mechanism that could explain this conservation of ancestral function but not of sequence is compensatory TFBS turnover, which is tolerated at enhancers following more flexible organizational principles. A classic example is the even-skipped stripe 2 enhancer (S2E) in the Drosophila genus, which has undergone dramatic reshuffling of TFBSs across different species, yet directs the same expression pattern in transgenic reporter assays (Ludwig et al., 2000)(Arnosti et al., 1996). Chimeric enhancer assays provided strong evidence for stabilizing selection at this site, as swapping the 3’ and 5’ halves of the native S2E enhancers from different species did not recapitulate the endogenous activity patterns (Ludwig et al., 2000). A more systematic enhancer discovery strategy called STARR-seq confirms that compensatory motif turnover in functionally-conserved enhancers is common even between closely-related species (Arnold et al., 2014). More generally, TFBS loss or gain without concomitant alteration of enhancer function appears to be a common event at enhancers that employ a more flexible mode of motif grammar over evolutionary timescales.
Impact of sequences that affect DNA shape
In addition to the core TF motif recognition, emerging evidence supports the role of local DNA shape in influencing binding affinity, site accessibility and choice, known as ‘shape readout’ (Slattery et al., 2014). The DNA sequence at sites flanking a core TF binding sites can thus profoundly influence binding by TFs that are sensitive to DNA shape (Gordân et al., 2013; Levo et al., 2015). Similarly, changes in protein structure that affect shape readout can dramatically affect binding preference. For example, altering amino acids involved in DNA shape recognition by the Scr protein abrogated preferential binding to a narrowed minor groove DNA structure, while introduction of these shape-recognizing residues into another Hox factor, Antp, caused a switch in its binding specificity in vitro, and transcriptional targets in vivo to that of Scr (Abe et al., 2015). Thus, at least in some defined cases, local DNA shape readout influences the binding preference of TF family members. Consequently, mutations falling outside core TFBS may affect local DNA shape and contribute to modulation or loss of enhancer activity in both an evolutionary and disease context.
Role of coactivators in enhancer activity
The ability of TFs to activate transcription on chromatin templates is dependent on the recruitment of coactivator proteins (reviewed in (Roeder, 2005)(Weake and Workman, 2010)). Coactivators are defined as factors that are ‘required for the function of DNA-binding activators, but not for basal transcription per se, and do not show site-specific binding by themselves’ (Malik and Roeder, 2010). Coactivators typically act through modifying and remodeling the chromatin context of enhancers, and include histone acetyltransferases (such as p300/CBP, SAGA complex, MOF, TIP60 and others), histone methyltransferases (e.g. MLL3/4, CARM1), chromatin remodeling factors (e.g. Brg1, CHD7) and factors that promote crosstalk with the basal transcriptional machinery at promoters (e.g. Mediator complex) (Krasnov et al., 2016; Malik and Roeder, 2010; Taatjes et al., 2004). Coactivator complexes are often broadly and highly expressed, and can be recruited to chromatin by a diverse range of TFs. Consequently, coactivators tend to be associated with most active enhancers in a given cell type, regardless of the examined tissue (see Figure 1D). This property has been widely utilized for annotations of putative enhancers in a myriad of cellular contexts, where occupancy of coactivators (e.g. p300, Mediator, BRG1) or their enzymatic products (e.g. H3K27ac, catalyzed by p300/CBP or H3K4me1, catalyzed by MLL3/4) can be mapped using ChIP-seq (for reviews on enhancer chromatin signatures and epigenomic annotation strategies see (Calo and Wysocka, 2013; Sakabe and Nobrega, 2013; Whitaker et al., 2015)).
Consistent with the idea that coactivators are the workhorses behind enhancer-mediated transcriptional activation, RNA-guided recruitment of the acetyltransferase domain of p300 fused to dCas9 is sufficient to bypass the requirement for TFs during enhancer activation in an endogenous chromosomal context (Hilton et al., 2015). Conversely, RNA-guided recruitment of the H3K4me1/2 demethylase LSD1 is sufficient to repress enhancer activity and gene transcription (Kearns et al., 2015). A comprehensive study in Drosophila used GAL4 DNA-binding domain (DBD) fusions of 338 coactivators and corepressors in enhancer complementation assays to test their capacity to bypass the requirement for key TFs by replacing their motifs with GAL4 recognition sites. Interestingly, most (80%) of the examined coactivators and corepressors activated or repressed, respectively, transcription in at least one enhancer context, whereas some coactivators including p300, Mediator subunits MED15 and MED25, and Lpt1 (a fly homolog of the N-terminal part of MLL3/4) were sufficient to activate transcription in all examined contexts (Stampfel et al., 2015). While not without caveats, taken together these results suggest that the main role of TFs is to provide a specific genomic address for coactivator complexes, which in turn enable activation of transcription by influencing the activity of RNA polymerase and facilitating establishment of a transcriptionally permissive chromatin environment.
Nonetheless, from an evolutionary perspective, alterations of regulatory sequences themselves are the major driver of change in enhancer activity, as enhancer elements are subject to high rates of turnover (Arnold et al., 2014; Villar et al., 2015) while both coactivator-TF interfaces and TF DNA binding specificities appear to be well conserved over evolutionary timescales (Minezaki et al., 2006; Nitta et al., 2015). It is therefore important to consider the genomic substrates from which enhancers emerge over evolutionary time and how the availability of such substrates may either act to drive or limit the rates of enhancer genesis.
II. Genomic substrates for evolving new enhancers
The birth of new enhancers can be mediated by a number of mechanisms, which will be discussed here in turn.
Regulatory innovation following genomic duplication
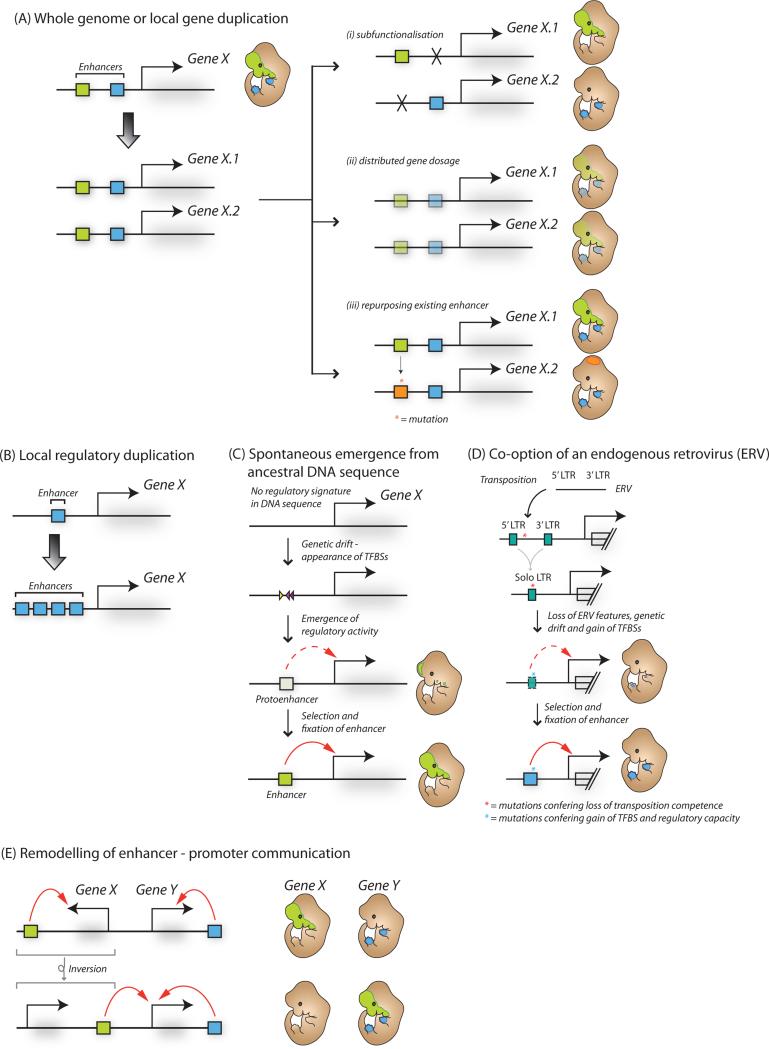
Genomic duplication is an important event driving evolutionary change as it generates new gene loci with the potential to evolve divergent functions and drives the formation of new regulatory programs (Taylor and Raes, 2004). Following a duplication event, often one copy of a pair of duplicated genes is simply lost due to lack of selective pressure to maintain both paralogs. However, subfunctionalization of enhancer activity (the loss of activity of a subset of enhancers leading to different expression domains of gene paralogs), distributed gene dosage (Lan and Pritchard, 2015) or rapid divergence in the expression domain of one or both of the gene paralogs by repurposing existing enhancer(s) for novel regulatory functions (Goode et al., 2011) can result in retention of both duplicated genes (Figure 2Ai-iii). For example, teleost fish experienced a relatively recent whole genome duplication 350 million years ago, with many paralogous duplicated genes retaining an equivalent protein function but showing divergent expression domains that arose through changes in their local cis-regulatory landscape (for example, the duplicated pax6a/b in zebrafish (Kleinjan et al., 2008)). Smaller duplications of non-coding DNA can also generate copies of enhancer elements in cis to the original enhancer and promoter pair (Figure 2B). While in many cases these duplicated enhancers are lost due to negative selection or genetic drift, in some instances locally duplicated enhancers are retained, for example enhancers at the human apoE/C-I/C-I′/C-IV/C-II gene cluster (Allan et al., 1995). Finally, genomic duplication of previously non-regulatory sequences can also provide new raw material from which primordial regulatory sequences can emerge through neutral drift, as discussed below.
Figure 2. The birth and death of enhancers during evolution.
(A) Following whole genome or local duplication events, enhancers and associated genes can become duplicated (left), and subsequently undergo: (i) loss of enhancer function and sub-functionalization of associated gene activity, (ii) enhancer repurposing with novel tissue or developmental stage expression for the duplicated gene or (iii) reduction in enhancer activity and gene dosage sharing between alleles. (B) Local duplication events can lead to increased enhancer copy number. (C) Novel enhancer activity can emerge from ancestral DNA through genetic drift and spontaneous appearance of transcription factor binding sites (TFBSs) and protoenhancer activity. (D) Enhancer elements can be exapted from transposable elements. For example, following endogenization and unequal homologous recombination, the long terminal repeats (LTRs) of endogenous retroviral (ERV) elements can gain tissue-specific regulatory activity through accumulation of mutations and emergence of TFBSs. (E) Enhancer activity can be transferred to a new gene target for example through a genomic inversion event.
Repurposing of ancestral regulatory or coding sequences
With or without genomic duplication events, novel regulatory function can arise from functional ancestral DNA sequences through the repurposing of older enhancer elements (Rebeiz et al., 2011). The evolutionary barrier to the formation of a novel enhancer is potentially lower at these sites because they already contain motifs for DNA binding factors, which can act cooperatively with newly emerging sites. Recent genome-wide mapping of DNase hypersensitive sites (DHSs) (Vierstra et al., 2014) revealed that DHSs at orthologous loci between human and mouse were functionally active in distinct tissues, revealing frequent regulatory element repurposing across 90 million years of evolution. Importantly, modulation of existing enhancers can drive regulatory innovation and evolution of morphological change, such as morphogenesis of trichomes in Drosophila larvae (Frankel et al., 2011) and limb length in bats (Cretekos et al., 2008).
Interestingly, enhancer function can also arise within protein-coding sequences, for example in mouse liver, 6% of epigenomically-defined enhancers overlap exons (enhancer exons, or eExons), a number of which have been implicated in regulation of nearby liver-specific genes (Birnbaum et al., 2012). A further extensive investigation of this phenomenon in 81 cell types found that ~15% of coding exons have DNase hypersensitivity footprints, suggestive that they are dual-use codons with both protein-coding potential and transcription factor binding capacity, or “duons” (Stergachis et al., 2013). However, whether duons are subject to selective constraint beyond that associated with their protein-coding potential remains controversial (Agoglia and Fraser, 2016; Xing and He, 2015).
Spontaneous emergence of enhancer sequences from non-regulatory DNA
Attempts to reverse engineer enhancers (Smith et al., 2013b) have revealed that short (i.e, 15-mer) random sequences are often sufficient to drive tissue-specific expression, suggesting that enhancer activity can emerge spontaneously from neutral DNA sequences purely by random genetic drift. Modeling theoretical rates of enhancer genesis in Drosophila concluded that it should take 0.5 – 10 million years to evolve novel anterior-posterior (A-P) patterning enhancer elements from neutral DNA sequence (Duque and Sinha, 2015). These predictions match well with observations from across the Drosophila genus whereby hundreds of species-specific enhancers have arisen over similar timescales (Arnold et al., 2014), and provide an initial framework for understanding emergence of new enhancers from the genetic void. Recent studies emphasize that indeed most species-specific enhancers evolve de novo from previously non-regulatory non-coding DNA (Figure 2C). For example, using comparative epigenomic analysis of enhancers from livers of 20 diverse mammalian species Villar et al demonstrated that the majority (52-77%) of species-specific enhancers arose through exaptation of ancestral DNA sequences (i.e. present in the genomes of the ancestral species for at least 100 million years), with the remainder being derived primarily from transposable elements (Villar et al., 2015).
Co-option of transposable elements
A large proportion of eukaryotic genomes, including almost half the human genome, are composed of repetitive transposable elements (TEs) (Lander et al., 2001). TEs are a rich source of regulatory innovation, as they are disseminated throughout the genome via TE expansion, carrying with them cis-regulatory sequences that exploit the host trans environment for their transcription (Feschotte, 2008), and have the potential to acquire mutations that allow for their tissue-specific exaptation for host gene regulation (Bourque et al., 2008). Indeed, evidence is beginning to emerge that TEs play a central role in rewiring gene regulatory networks, and can facilitate rapid evolution of ecologically relevant traits, such as sweet perception in primates (Ting et al., 1992) or the diversification of tissues at the boundary between mother and fetus in eutherian mammals (Lynch et al., 2011).
Indeed, according to some estimations, the majority of primate-specific regulatory sequences are derived from TEs (Jacques et al., 2013). For example, Alu elements, which comprise ~10% of the human genome, contain suboptimal TFBSs and can acquire enhancer-like features with few mutations (Su et al., 2014), while ERV1 elements are enriched at sites of cis-regulatory divergence in primate neural crest cells (Prescott et al., 2015), suggesting that many enhancers that changed their activity since the separation of humans and chimpanzees did so through exaptation of pre-existing TEs. Importantly, direct evidence for LTR-mediated gene regulation has been demonstrated for human innate immunity genes (Chuong et al., 2016). This is consistent with the emerging idea that motif-rich LTRs of endogenous retroviruses are particularly fertile substrate for evolving new enhancers, with many LTRs acquiring inducible or cell-type specific enhancer function during evolution (Figure 2D). As endogenous retroviruses are remnants of ancient retroviral infections, one can speculate that the ancestral evolutionary history of viral infections and endogenization can and will continue to influence the future cis-regulatory evolution of a species.
DNA methylation and biased gene conversion in regulatory evolution
The frequency with which TFBSs emerge by chance from ancestral DNA sequences and TEs is greatly influenced by local mutational rate. Certain processes can act to increase or to suppress the basal mutagenesis rate, and have been implicated in rapid evolution of sequences and emergence of novel regulatory function. For example, the majority of vertebrate genomes are methylated at cytosine residues in the context of a CpG dinucleotide (Jones, 2012). Mutation of methylated cytosines to thymine due to spontaneous deamination is subject to error prone repair (Duncan and Miller, 1980). Thus C to T mutations are the most common mutation among all base substitutions (Sved and Bird, 1990), favoring de novo genesis of TFBSs that contain TpG dinucleotides. For example, there is evidence that spontaneous deamination has helped to generate thousands of p53 binding sites genome-wide (Zemojtel et al., 2009).Therefore, in addition to its role in genome silencing, DNA methylation may play a role in regulatory evolution. A second mechanism causing rapid change of local nucleotide composition is known as GC-biased gene conversion (gBGC), in which mismatch repair processes following meiotic recombination favor G or C alleles over A or T alleles, thus leading to increase in GC content at these regions (Galtier and Duret, 2007). gBGC might have played an important role in recent human evolution, as around 20% of sequences which are conserved across vertebrates but have undergone rapid change in the human lineage (e.g. human accelerated regions or HARs) can be explained by gBGC alone (Kostka et al., 2012).
Remodeling of the chromosomal context
Novel enhancer function can arise not only through change of the enhancer sequence itself, but also through the rearrangement of the neighboring chromosomal context (Figure 2E). For example, in the beetle Tribolium castaneum the expression of the ladybird gene is absent from the dorsal mesoderm as compared to the honeybee Apis mellifera and fruit fly Drosophila melanogaster and replaced by expression of C15, a neighboring gene. This switch in expression from ladybird to C15 appears to have arisen from a genomic inversion redirecting a conserved ladybird 3' enhancer to regulate C15 (Cande et al., 2009). Thus, despite minimal alteration to the enhancer itself, change in its genomic location can rapidly introduce a new gene or enhancer into a pre-existing gene regulatory network in one step. Recently, much progress has been made towards understanding how three-dimensional genome organization influences enhancer function, poising the field to uncover the frequency with which structural genomic variation can drive evolutionary rewiring of enhancer targets.
III. Topological constraints on enhancer function and relationships between enhancers within complex regulatory landscapes
Topological Associated Domains (TADs) as structural elements of chromatin organization
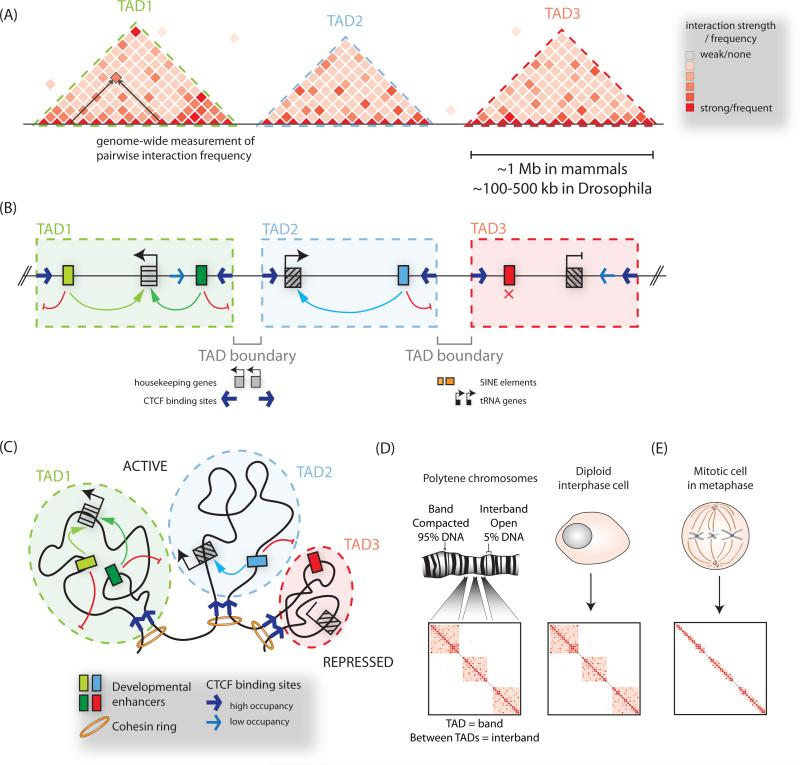
A defining feature of enhancers is their ability to activate transcription at a distance. Indeed, in mammalian genomes such distances can be over a megabase long, as exemplified by the Shh limb enhancer (Lettice et al., 2003), raising the question as to what constrains enhancer search space such that high regulatory precision is preserved. A key insight into this question came from studies of chromosome folding using techniques such as Hi-C and 5C, which are chromosome conformation capture (3C)-based assays, and rely on the principle that digestion and re-ligation of chromatin can capture the three dimensional proximity of DNA sequences in the nucleus (extensively reviewed elsewhere, see (de Wit and de Laat, 2012)). These studies have revealed that the chromosomes of many eukaryotic genomes are segmented into self-interacting domains reported as physical domains in Drosophila (Hou et al., 2012; Sexton et al., 2012), and topological associated domains (TADs) in mammalian genomes (Dixon et al., 2012; Nora et al., 2012) (Figure 3A-B). High resolution mapping has estimated TAD median length at 185 kb in mammals, although importantly a subset of domains are over a megabase in size (Rao et al., 2014). Importantly, compartmentalization of the genome in this manner partitions the chromosome into ‘regulatory neighborhoods’ by limiting the activity of cis-regulatory elements to genes which fall within the same TAD and by preventing spreading of heterochromatic marks (Dixon et al., 2012; Nora et al., 2012) (Figure 3B-C, also discussed in more detail below).
Figure 3. Organization of chromatin into topologically associated domains.
(A) Hi-C or 5C heatmaps visualize three-dimensional interactions, or compartmentalization of chromosomes into topologically associated domains (TADs), visible as triangular blocks of increased interaction frequencies. (B) TAD boundaries restrict the influence of regulatory elements to genes within a given TAD, and limit the spread of chromatin modifications. The boundary regions between TADs have been observed to be associated with CTCF binding sites, housekeeping genes, SINE elements and tRNA genes. (C) A model of chromosome folding corresponding to (B), with convergent CTCF sites and cohesin depicted at loop anchors. (D) Correspondence between TADs and cytological bands on polytene chromosomes, and TAD boundaries with decondensed interbands. (E) During mitosis TAD structure is lost.
Strikingly, TAD boundaries appear to be largely invariant between cell-types, and are conserved across even distantly related species, suggesting that TADs represent a fixed structural unit of chromatin organization within which tissue-specific regulatory interactions can occur and evolve (Dixon et al., 2012)(Vietri Rudan et al., 2015a)(Harmston et al., 2016). Such an organizational role for TADs is supported by the demonstration of a one-to-one correspondence between TADs and bands on polytene chromosomes from Drosophila larval salivary glands, with interbands corresponding to highly-transcribed TAD boundary regions (Eagen et al., 2015) (Figure 3D). Endoreplicated polytene chromosomes have long served as a cytological model for understanding relationship between chromatin structure and function, with highly stereotyped banding patterns observed across cells within a given cell type, and only minor variations seen between tissues (Bridges, 1935)(Mavragani-Tsipidou et al., 1990). This recent observation further suggests that TADs and their boundaries represent fundamental features of chromatin organization. Notably, the invariance of TADs across cell states is not due to a passive maintenance from one cell division to the next, as TAD structures are lost during mitosis to accommodate an alternative conformational state of metaphase chromosomes (Naumova et al., 2013) (Figure 3E). This implies that with each cell division TADs must reproducibly refold, but the mechanisms that drive their reformation and maintenance are still under investigation.
Conservation of topological domain structures across the eukaryotic tree of life
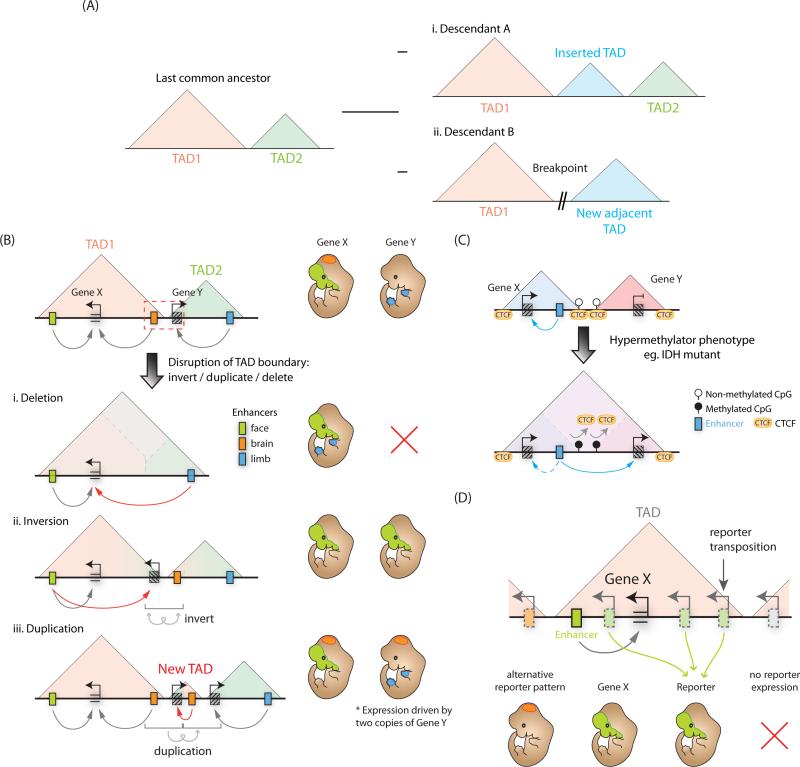
Given the importance of TAD boundaries in demarcating the limits of gene regulatory domains, changes to domain architecture would represent a means for saltatory (or non-gradual) change in gene regulation over evolutionary time, as shifting of domain boundaries would expose multiple genes to a novel regulatory environment. However, this sort of restructuring of TADs appears to be rare during evolution, as TADs tend to overlap with highly syntenic genomic blocks of conserved enhancers and non-coding elements (Harmston et al., 2016). Interestingly, structural rearrangements between mammalian genomes appear to instead occur between the boundaries of adjacent domains (Farré et al., 2015)(Vietri Rudan et al., 2015b) (Figure 4Ai-ii).
Figure 4. Topologically associated domains define discrete units of gene regulation.
(A) TAD boundaries are highly conserved across mammalian evolution, therefore structural changes tend to occur at TAD boundaries, for example (i) insertion of a new TAD or (ii) chromosomal breaks. (B) Disruption of a TAD boundary (red, dashed box) can impact gene expression. (i) Boundary deletion fuses two adjacent TADs facilitating de novo regulation of genes from one TAD by the enhancers in another. (ii) Boundary inversion can translocate genes or enhancers into an adjacent TAD where they are then incorporated into the regulatory environment of the new TAD. (iii) TAD boundary duplication can create a new TAD and potentially expose any duplicated genes to a novel regulatory environment. (C) CTCF binding sites can be DNA methylation sensitive, therefore aberrant DNA methylation can abrogate CTCF binding, causing TAD boundary defects and misregulation of gene expression. (D) Reporter constructs transposed into different regions of a TAD containing a developmental enhancer, but not into genomic regions outside the TAD, can recapitulate gene expression pattern of the endogenous gene (Gene X) controlled by this enhancer.
In the context of high conservation of TAD boundaries in vertebrates and their ostensible role in organizing regulatory landscapes from Drosophila to man, it is surprising that some multicellular organisms appear to lack conventional TAD organization. For example, in C. elegans, TAD-like structures seem to predominantly occur on the X chromosomes of XX hermaphrodites, and are driven by the dosage compensation complex (DCC), whereas autosomes are largely devoid of long-range chromosomal interactions (Crane et al., 2015). This could be perhaps rationalized by the fact that C. elegans has a compact genome with most of the cis-regulatory information usually contained within 10 kb from the TSS, thus alleviating the need for long-range gene regulatory domains. In plants, which appear to have long-range cell-type specific enhancers (Zhu et al., 2015), TAD structures are also not detectable by Hi-C, albeit both boundary-like elements and self-interacting heterochromatic regions have been reported (Feng et al., 2014; Grob et al., 2014). Interestingly, both C. elegans and A. thaliana do not encode a CTCF homolog (Heger et al., 2012), suggesting that these species exploit alternative mechanisms of genome organization and functional segmentation (the role of CTCF in mammalian TAD formation is discussed below). Interestingly, the inactive mammalian X chromosome also mostly lacks TADs and is instead divided into two megadomains (Giorgetti et al., 2016). Therefore in addition to being absent in a number of eukaryotic species, TAD-like structures are not a required feature of mammalian interphase chromosome folding.
It is unclear when and how TAD-like structures first appeared during evolution. However, recent studies from both bacteria and yeast suggest that self-interacting domains may be an ancient feature of chromosome organization. Hi-C experiments in Caulobacter cells found evidence of independent spatial domains called “chromosomally interacting domains” (CIDs), ranging in length from 30 to 420 kb which are proposed to correspond to supercoiled DNA loops, or plectonemes, arranged in a bottle brush–like fiber and flanked by highly-transcribed genes at CID boundaries (Le et al., 2013). In the fission yeast S. pombe, the cohesin complex has been shown to be important for the formation of globule structures at the 50-100 kb scale (Mizuguchi et al., 2014). Further explorations in S. cerevisiae also found CIDs which are much shorter self-interacting regions than those detected in other organisms, on the order of around 2-10 kb in size, and bounded once again by highly transcribed genes (Hsieh et al., 2015). Interestingly, these self-associating topological domains in S. cerevisiae encompass ~1-5 genes, approximately the same order of gene number as mammalian TADs, suggesting that the size of genes and intergenic spacing may influence the size and formation of topological domains. Further interrogation of the topological landscapes at high resolution across the eukaryotic tree of life will be necessary to understand the evolution and functional role of topological domain architecture.
Formation of TAD boundaries and CTCF-mediated loops
Since TAD boundaries appear to restrict both enhancer function (Dowen et al., 2014)(Flavahan et al., 2015)(Guo et al., 2015)(Lupiáñez et al., 2015) and spreading of chromatin marks (Narendra et al., 2015), they fulfill a canonical definition of insulator elements. Indeed, TAD boundaries are enriched for insulator-binding proteins such as CP190, CTCF, and BEAF-32 in Drosophila (Van Bortle et al., 2014)(Sexton et al., 2012)(Phillips-Cremins et al., 2013) and by CTCF and cohesin in mammals (Dowen et al., 2014; Sofueva et al., 2013). In fact, high resolution Hi-C maps in mammalian cell lines revealed that 86% of roughly 10,000 long-range contact peaks, interpreted as anchors for chromatin loops, are associated with CTCF (Rao et al., 2014). Moreover, CTCF sites engaged in these contacts are characterized by a unique motif orientation, with convergent (inward facing) CTCF motifs found in >90% of loop anchors (Figure 3B) (Rao et al., 2014)(Vietri Rudan et al., 2015b). Subsequent work has led to an as of yet experimentally untested ‘loop extrusion model’ of TAD formation whereby a chromosomal loop is randomly initiated then continuously fed through an extrusion complex containing a cohesin ring until convergent CTCF binding sites are encountered, at which point the loop structure is stabilized, likely via CTCF dimerization (Fudenberg et al., 2015; Sanborn et al., 2015). Although this hypothetical model is attractive, at present it remains unclear to what extent convergent CTCF-cohesin sites are the major driver of genome segmentation into TAD domains. Interestingly, neighboring topological domains do not completely collapse or merge upon deletion of domain boundary regions nor when cohesin is depleted in non-cycling cells (Seitan et al., 2013; Sofueva et al., 2013; Zuin et al., 2014). This can be rationalized by inherent interactions within a TAD mediating TAD structure organization by preventing interactions with a neighboring TAD and thus contributing to the presence of a boundary between them (Giorgetti et al., 2014)(Ulianov et al., 2016). Taken together, current results suggest that multiple parallel mechanisms may contribute to separating the genome into highly reproducible TAD structures.
Consequences of TAD organization on enhancer function and gene regulation
Regardless of the specific mechanisms, genome segmentation into TADs has profound implications on how cis-regulatory landscapes are organized and evolve by limiting the genomic search space within which enhancers can act. As such, perturbations of TAD boundary elements either through structural variation (Figure 4B) or aberrant DNA methylation resulting in loss of CTCF binding (Figure 4C), can lead to misregulation of genes by exposing them to the regulatory influence of enhancers in the adjacent TAD, in some cases resulting in congenital malformations or cancer (Nora et al., 2012)(Gómez-Marín et al., 2015; Guo et al., 2015)(Flavahan et al., 2015)(Lupiáñez et al., 2015).
Importantly, genes lying within the same topological domain are often co-regulated (Nora et al., 2012)(Gómez-Marín et al., 2015) suggesting that enhancers may sample multiple, if not all promoters confined within a TAD. This idea is supported by mouse reporter assays, in which transposed LacZ reporter sensors maintain a similar expression readout across a range of distances from a known enhancer, until they cross a TAD boundary (Symmons et al., 2014) (Figure 4D). However, given that in many cases genes in the same TAD in fact exhibit distinct expression patterns, this apparent capacity of enhancers to activate transcription across their designated topological domain raises an issue of enhancer-promoter specificity. More quantitative analysis of genomic space sampling by enhancers is warranted, as some genomic positions within a topological domain may be favored targets due to preferential intra-domain chromatin folds. Moreover, additional mechanisms such as promoter-specific repression, complex synergistic or competitive relationships between simultaneously active enhancers within the same TAD or biochemical compatibility between enhancers and promoters may all contribute to achieving greater regulatory specificity of enhancer-promoter interactions within a topological domain.
What is also becoming clear is that while there are certainly well documented examples of enhancer-promoter loops, typical enhancer-promoter contacts are likely less stable and/or less frequent than structural loops mediated by CTCF that are readily detectable by Hi-C methods. For example, most enhancers and promoters active in a given cell type are not detected as peak points (a.k.a. loop anchors) on high-resolution Hi-C maps (Rao et al., 2014). Regrettably, the authors chose to de-emphasize this point by focusing on a subset of promoters and enhancers that coincide with stable loop anchor sites. However, the elements in this subset are distinct from most enhancers and promoters as they are bound by CTCF, and therefore may only represent the minority of enhancer-promoter pairs which serve dual roles as both structural and regulatory elements.
Intriguingly, recent live-imaging studies in Drosophila uncovered the capacity of a single enhancer to drive synchronized transcriptional bursting from two equidistant promoters (Fukaya et al., 2016), raising the question of whether or how chromatin topology regulates this behavior. Another recent study supports a highly dynamic view of enhancer-promoter contacts in mammalian cells and shows that enforcing promoter-enhancer looping can increase transcriptional burst frequency, but doesn't effect burst size (Bartman et al., 2016). One should also consider that alternative mechanisms of communication may be at play such as the previously proposed tracking model (Zhu et al., 2007), lateral propagation of torsional strain, indirect association in transcriptional hot spots or ‘factories’ (Kolovos et al., 2012), enhancer-mediated trapping of activator complexes in a ‘reaction vessel’, or other yet-to-be-conceived mechanisms. With carefully measured parameters of transcriptional bursting in relation to changes in nuclear positions of relevant regulatory elements and intervening sequences in living cells, it may soon become possible to gain insights into the role of chromatin topology in long-range gene regulation by enhancers.
Regulatory relationships among simultaneously active enhancers
With our greater appreciation of the complexity of genomic organization, it is becoming apparent that cis-regulatory elements do not function in isolation. Many (if not most) developmental genes are regulated by multiple enhancers with both overlapping and distinct spatiotemporal activities. Indeed, key lineage genes in a given cell type tend to be associated with dense cluster(s) of highly active enhancers, often referred to as super-enhancers (Hnisz et al., 2013; Whyte et al., 2013). These observations beg several questions: What are the regulatory relationships among simultaneously active enhancers? What are the consequences of multiple enhancers regulating one gene?
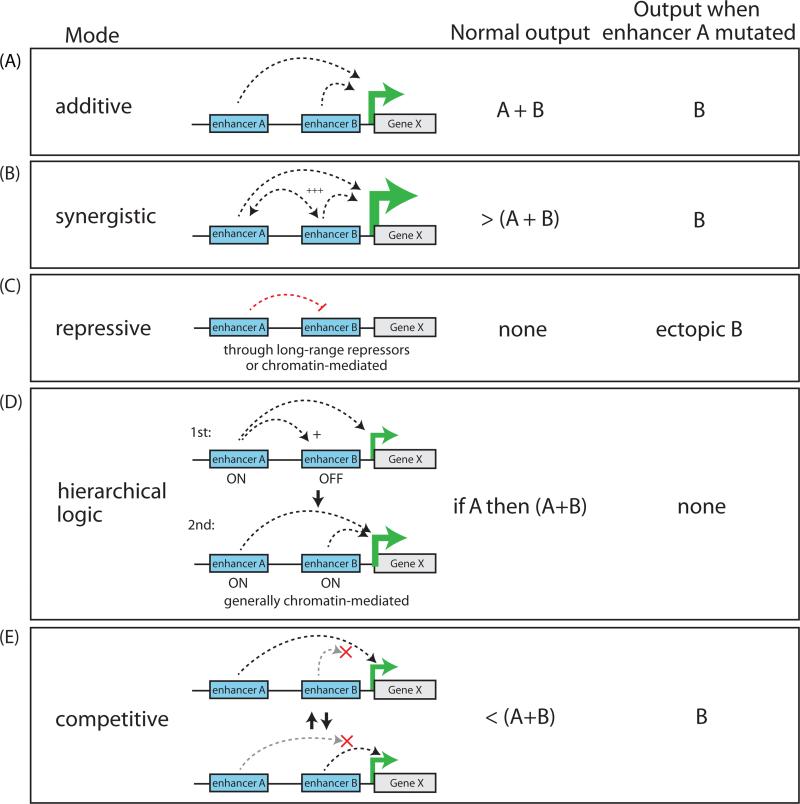
The classic mode of enhancer behavior is described as autonomous, modular, and additive - a feature which has been suggested to confer evolvability to enhancers over other functional parts of the genome (Carroll, 2008) (Figure 5A). Many demonstrations of the modular and additive behavior of enhancers exist, both for elements controlling the same locus across different tissues and for simultaneously active individual elements within a super-enhancer (for example (Hay et al., 2016; Maeda and Karch, 2011; Visel et al., 2009)). This relative modularity is thought to confer highly tissue-specific phenotypes of enhancer deletions near pleiotropically-expressed genes.
Figure 5. Regulatory crosstalk between enhancers operating within the same cis-regulatory landscape.
(A-E) Potential modes of enhancer relationships and their effect on transcriptional output under wildtype or mutant conditions.
In other cases, however, the overall expression pattern of a given gene is different from the sum of the individual activities of each enhancer element. While the pervasiveness of such non-additive interactions between enhancers acting within regulatory landscapes is still unclear, some studies suggest that such interactions may be in fact extremely common. For example, comparison of individual enhancer activity with endogenous gene expression patterns estimate that the activity of up to ~37% of enhancers may be modulated by their extended endogenous loci in Drosophila (Kvon et al., 2014).
The first appreciation of this regulatory complexity came from detailed analysis of the endo16 regulatory region in sea urchins. Using a quantitative reporter and in situ hybridizations, these studies discovered that some elements within the broader regulatory region were required for the activity of other elements, or could linearly amplify the output of another element by a factor of ~4 (Yuh and Davidson, 1996; Yuh et al., 1998). Since this early proof of principle, multiple studies in other species, including mammals, have documented similar multiplicative or greater-than-multiplicative effects of enhancers acting together to boost expression (for example (Maekawa et al., 1989; Stine et al., 2011)) (Figure 5B).
Importantly, enhancers acting together can drive not only quantitative but also qualitative changes in spatiotemporal activity patterns in vivo. A number of reports using transgenes in Drosophila and mice have demonstrated that the spatial expression patterns of multiple enhancers tested together can be distinct from the sum of each enhancer tested individually, and that this interplay is required to recapitulate the endogenous expression pattern of their target genes (examples include (Dib et al., 2011; Dunipace et al., 2011; Prazak et al., 2010)). Interestingly, one recurrent observation from such studies is that often the effect of multiple enhancers acting together is not to generate novel activity domains but to prevent ectopic expression outside of its proper context. Several careful examples of this in Drosophila have been described, one being the restriction of gap gene expression patterns to proper contexts by specific enhancers near hunchback and knirps (Perry et al., 2011). Similarly, synergistic enhancers restrict aberrant activity domains at the murine Fgf8 locus (Marinić et al., 2013). One suggested mechanism for this behavior is through the recruitment of long-range repressors which suppress the activity of the neighboring enhancer in ectopic regions (Dunipace et al., 2011; Perry et al., 2011) (Figure 5C). Thus, it appears that fine-tuning of gene activation patterns in vivo is in part mediated by repressive influences between neighboring regulatory elements.
In some cases, cis-regulatory landscapes appear to follow a more complex hierarchical logic. A classic example of this is seen at the Drosophila bithorax complex (BX-C), responsible for the restricted expression of Ubx, abd-A or Abd-B along the fly's A-P axis. A shared ~300kb regulatory region for the BX-C is divisible into nine parasegment-specific chromosomal domains, with each domain shown to control the activation of one of the three BX-C homeotic genes in a pattern appropriate for that segment (reviewed in (Maeda and Karch, 2011)). Interestingly, while each domain contains several regulatory elements, generally only one or two of these enhancers is limited in activity along the A-P axis to their correct parasegment while the rest drive activity in tissues along the entire axis when tested alone. These elements with limited activity were termed ‘initiator’ elements, as they were proposed to read the appropriate parasegmental address and determine if the surrounding regulatory domain is to be altogether active or silenced (Iampietro et al., 2010; Mihaly et al., 2006). Importantly, when initiator elements are switched between domains it results in a homeotic transformation (Iampietro et al., 2010) demonstrating that the initiator is sufficient to coordinate the various enhancers within the domain. While such a well-characterized domain-restricted system hasn't been discovered in mammals, some examples of hierarchical logic at enhancers have been described, such as a conditional relationship between two enhancers near the PU.1 locus in mouse myeloid cells where an upstream regulatory element (URE) directly initiates the activity of a nearby enhancer (the “−12 kb enhancer”), possibly through a chromatin-mediated mechanism (Leddin et al., 2011) (Figure 5D).
Enhancer redundancy and competition
Another mode of non-additive behavior in cis-regulatory regions is enhancer redundancy, with functionally redundant enhancers referred to as “shadow enhancers” (reviewed in (Barolo, 2012)), as coined by Mike Levine and colleagues to describe “remote secondary enhancers mapping far from the target gene and mediating activities overlapping the primary enhancer” (Hong et al., 2008). Importantly, under this definition shadow enhancers are redundant only in that they have overlapping activity patterns, and are not necessarily functionally identical. For example, despite having overlapping activity, shadow enhancers may have different temporal dynamics, or may serve to fine-tune temporal or spatial gene expression boundaries (see below). Interestingly, shadow enhancers may also confer robustness against environmental or genetic variability (also known as canalization), since mutation of a shadow enhancer can remain cryptic under normal conditions but be revealed under stress conditions (Frankel et al., 2010).
A potential mechanism for enhancer redundancy would be a competition model, where two enhancers compete for association with a shared promoter, buffering individual enhancer activity to facilitate a constant transcriptional output (Figure 5E). In this model, strongly activated enhancers would function sub-additively, while weaker enhancers would be expected to function in a more additive manner as a consequence of lower rates of promoter competition. Live monitoring of transcription at the hunchback and snai loci in Drosophila suggests that enhancer pairs operate in exactly this sub-additive manner when they are strongly activated due to competition for the target promoter (Figure 5E) (Bothma et al., 2015). This model could explain phenotypic robustness (or canalization) in a population, as loss-of-function polymorphisms in one enhancer would be compensated for by increased frequency of promoter contacts of the remaining enhancer, resulting in maintenance of normal expression levels of the target gene. In addition, this enhancer competition model could also create sharp boundaries of gene expression during development through the recruitment of long-range repressors. For example, interrogation of the primary and shadow enhancers at the knirps and Kruppel loci suggests that they act additively at the center of an expression domain, but can become dominantly repressive in the posterior regions, facilitating establishment of a sharp expression boundary (El-Sherif and Levine, 2016). Therefore, enhancer competition can buffer against fluctuations in gene expression levels in the ‘on’ state, yet impart sharp on/off boundaries at the edges of expression domains when combined with tissue-specific recruitment of repressors.
The ability of enhancers to cooperate or compete adds an additional layer of complexity to understanding how regulatory landscapes evolve. Redundant ‘shadow’ enhancers for example may facilitate accumulation of neutral mutations, which may then be unmasked during times of environmental stress, conferring “evolvability” to the species by increasing the phenotypic diversity to allow more rapid selection to a changing fitness landscape. In contrast, cooperative enhancers may experience weak negative selection to co-evolve across evolution, with potential positional constraints to remain within the same topological domain, or to retain enough spacer to minimize short-range cross-repressive interactions (Dunipace et al., 2011). Consequently, changes in synergistic relationships may be a source of evolutionary innovation that cannot be easily captured by conventional sequence comparisons, and will require more complex functional analyses to detect systematically, such as genetic perturbation screens to manipulate function of enhancers in their native chromosomal context.
Concluding remarks and future perspective
As ever, progress in our mechanistic understanding of gene regulation has thrown into relief the remaining gaps in our knowledge, and opened up many new questions regarding enhancer function. Firstly, despite identification of a huge number of putative enhancer sequences, our understanding of the importance of orientation, spacing, and copy number of TFBSs for enhancer function remains rudimentary. Efforts to engineer synthetic enhancers have made strides towards unpicking the enhancer lexicon, however, thus far they had limited combinatorial power, were often performed in an episomal context, and designed without much appreciation for the distinct sequence preferences of TFs in cooperative versus individual binding contexts. Given that: (i) the sampling size for potential enhancer sequences is so large, (ii) multiple distinct sequences may confer equivalent activities, and (iii) the local regulatory context may alter enhancer output, uncovering a universally predictive set of rules for enhancer grammar, activity and specificity in different tissues will be a major challenge for future research.
Chromosomal conformation studies (3C and derivatives) have recently provided key insights into long-range regulation by showing that most metazoan genomes are partitioned into TADs, which delimit boundaries of self-interacting chromatin and thus organize regulatory landscapes. However, 3C-type approaches also have significant limitations, as they involve chromatin crosslinking, are typically performed on the population level, and it is difficult to normalize the data to reflect realistic background models. As a result, not only may the extent to which enhancers and promoters form ‘loops’ have been overestimated, but also the kinetic information underlying enhancer activation at a single cell level has been lost. Orthogonal approaches, including high resolution imaging of fixed cells has already provided some insight into the dynamics of enhancer-promoter interactions (Giorgetti et al., 2014)(Fabre et al., 2015; Williamson et al., 2016). We anticipate that future live-cell imaging approaches will further help to address how the information from enhancers is dynamically transduced to promoters to allow for precise activation of transcription.
It recently became clear that the majority of genetic variants associated with human complex disease or normal range trait variation maps to the non-coding parts of the genome (reviewed in (Tak and Farnham, 2015)). A substantial fraction of such variants is thought to modulate cis-regulatory element function, and a number of examples of disease-associated non-coding mutations that ablate or change enhancer function or impact the folding of topological domains have been identified (reviewed in (Scacheri and Scacheri, 2015)). Indeed, genetic variation within the human population coupled with quantitative epigenomic approaches can be leveraged to link sequence changes to chromatin state divergence, both locally and distally within interacting chromosomal regions, providing an avenue for an interpretation of GWAS studies and future investigations of mechanisms underlying disease traits (Grubert et al., 2015; Waszak et al., 2015). Similarly, evidence has accumulated that enhancer sequence changes mediate morphological divergence between species, and ‘cellular anthropology’ approaches utilizing pluripotent stem cells from great apes have enabled investigation of recent hominid cis-regulatory landscape evolution in developmentally and evolutionary relevant cell types (Prescott et al., 2015)(Gallego Romero et al., 2015). Further understanding of the contribution of enhancer sequence variation to human disease susceptibility, normal range variation and evolutionary innovation will likely soon come from human genetics and follow-up functional studies in cells and model organisms. A key challenge will be to understand how combinations of such regulatory variants make us uniquely human and uniquely individual.
Acknowledgements
We thank Wysocka lab members and anonymous reviewers for insightful comments on the manuscript. We apologize to colleagues whose important primary studies we were unable to cite due to space constraints. This work was supported by Howard Hughes Medical Institute, NIH R01 GM112720-01 (J.W.) and Wellcome Trust, Sir Henry Wellcome Postdoctoral Fellowship 106051/Z/14/Z (H.K.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe N, Dror I, Yang L, Slattery M, Zhou T, Bussemaker HJ, Rohs R, Mann RS. Deconvolving the Recognition of DNA Shape from Sequence. Cell. 2015;161:307–318. doi: 10.1016/j.cell.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoglia RM, Fraser HB. Disentangling sources of selection on exonic transcriptional enhancers. Mol. Biol. Evol. 2016;33:585–590. doi: 10.1093/molbev/msv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan CM, Walker D, Taylor JM. Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. Identification of a second region that confers high level and liver-specific expression of the human apolipoprotein E gene in transgenic mice. J. Biol. Chem. 1995;270:26278–26281. doi: 10.1074/jbc.270.44.26278. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 2014;46:685–692. doi: 10.1038/ng.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays. 2012;34:135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CCS, Raj A, Blobel GA. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol. Cell. 2016;62:237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger a. M., Nguyen L, et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22:1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama J. a, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong C-T, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife. 2015;4:1–14. doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew J, Ruan Y, Wei C, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. 2008:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. Salivary chromosome maps with a key to the banding of the chromosomes of Drosophila melanogaster. J. Hered. 1935;26:60–64. [Google Scholar]
- Buecker C, Wysocka J. Enhancers as information integration hubs in development: Lessons from genomics. Trends Genet. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burz DS, Rivera-Pomar R, Jäckle H, Hanes SD. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of Enhancer Chromatin: What, How, and Why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande JD, Chopra VS, Levine M. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development. 2009;136:3153–3160. doi: 10.1242/dev.038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science (80−. ) 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015 doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretekos CJ, Wang Y, Green ED, Martin JF, Rasweiler IV JJ, Behringer RR. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–151. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. Low Affinity Binding Site Clusters Confer Hox Specificity and Regulatory Robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib S, Denarier E, Dionne N, Beaudoin M, Friedman HH, Peterson AC. Regulatory modules function in a non-autonomous manner to control transcription of the mbp gene. Nucleic Acids Res. 2011;39:2548–2558. doi: 10.1093/nar/gkq1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque T, Sinha S. What Does It Take to Evolve an Enhancer? A Simulation-Based Study of Factors Influencing the Emergence of Combinatorial Regulation. Genome Biol. Evol. 2015;7:1415–1431. doi: 10.1093/gbe/evv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagen KP, Hartl TA, Kornberg RD. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell. 2015;163:934–946. doi: 10.1016/j.cell.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E, Levine M. Shadow Enhancers Mediate Dynamic Shifts of Gap Gene Expression in the Drosophila Embryo. Curr. Biol. 2016;26:1164–1169. doi: 10.1016/j.cub.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erceg J, Saunders TE, Girardot C, Devos DP, Hufnagel L, Furlong EEM. Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PJ, Benke A, Joye E, Nguyen Huynh TH, Manley S, Duboule D. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc. Natl. Acad. Sci. 2015:17972. doi: 10.1073/pnas.1517972112. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science. 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Robinson TJ, Ruiz-Herrera A. An Integrative Breakage Model of genome architecture, reshuffling and evolution. BioEssays. 2015;37:479–488. doi: 10.1002/bies.201400174. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. Genome-wide Hi-C Analyses in Wild-Type and Mutants Reveal High-Resolution Chromatin Interactions in Arabidopsis. Mol. Cell. 2014;55:694–707. doi: 10.1016/j.molcel.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. bioRxiv. 2015:024620. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:1–11. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, et al. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. 2015:1–29. doi: 10.7554/eLife.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. Structural organization of the inactive X chromosome in the mouse. Nature in press. 2016:1–5. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci U S A. 2015;112:201505463. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode DK, Callaway H. a, Cerda G. a, Lewis KE, Elgar G. Minor change, major difference: divergent functions of highly conserved cis-regulatory elements subsequent to whole genome duplication events. Development. 2011;138:879–884. doi: 10.1242/dev.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Grossniklaus U. Hi-C Analysis in Arabidopsis Identifies the KNOT, a Structure with Similarities to the flamenco Locus of Drosophila. Mol. Cell. 2014;55:678–693. doi: 10.1016/j.molcel.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guturu H, Doxey AC, Wenger AM, Bejerano G. Structure-aided prediction of mammalian transcription factor complexes in conserved non-coding elements. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20130029. doi: 10.1098/rstb.2013.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston N, Ing-simmons E, Tan G, Perry M, Merkenschlager M, Lenhard B. Topologically associated domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. 2016 doi: 10.1038/s41467-017-00524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen LLP, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 2016:1–12. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova A. a, Hoke H. a, Young R. a. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS. Shadow Enhancers as a Source of Evolutionary Novelty. Science (80−. ) 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Initiator Elements Function to Determine the Activity State of BX-C Enhancers. PLoS Genet. 2010;6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G. The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements. PLoS Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. A Transcription Factor Collective Defines Cardiac Cell Fate and Reflects Lineage History. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kearns N. a, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, et al. Subfunctionalization of Duplicated Zebrafish pax6 Genes by cis-Regulatory Divergence. PLoS Genet. 2008;4:e29. doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolovos P, Knoch T. a, Grosveld FG, Cook PR, Papantonis A. Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin. 2012;5:1. doi: 10.1186/1756-8935-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Hubisz MJ, Siepel A, Pollard KS. The role of GC-Biased gene conversion in shaping the fastest evolving regions of the human genome. Mol. Biol. Evol. 2012;29:1047–1057. doi: 10.1093/molbev/msr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov AN, Mazina MY, Nikolenko JV, Vorobyeva NE. On the way of revealing coactivator complexes cross-talk during transcriptional activation. Cell Biosci. 2016;6:15. doi: 10.1186/s13578-016-0081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- Lan X, Pritchard JK. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. bioRxiv. 2015:019166. doi: 10.1126/science.aad8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leddin M, Perrod C, Hoogenkamp M, Ghani S, Assi S, Heinz S, Wilson NK, Follows G, Schönheit J, Vockentanz L, et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117:2827–2838. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L. a., Heaney SJH, Purdie L. a., Li L, de Beer P, Oostra B. a., Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. gr. 2015:185033.114. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell. 2015:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Gene expression in time and space: additive vs hierarchical organization of cis-regulatory regions. Curr. Opin. Genet. Dev. 2011;21:187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Imamoto F, Merlino GT, Pastan I, Ishii S. Cooperative function of two separate enhancers of the human epidermal growth factor receptor proto-oncogene. J. Biol. Chem. 1989;264:5488–5494. [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinić M, Aktas T, Ruf S, Spitz F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell. 2013;24:530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Mavragani-Tsipidou P, Scouras ZG, Kastritsis CD. Comparison of the polytene chromosomes of the salivary gland, the fat body and the midgut nuclei of Drosophila auraria. Genetica. 1990;81:99–108. doi: 10.1007/BF00226448. [DOI] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama J. a, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 2012;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Barges S, Sipos L, Maeda R, Cléard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- Miller J. a, Widom J. Collaborative Competition Mechanism for Gene Activation In Vivo Collaborative Competition Mechanism for Gene Activation In Vivo. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human Transcription Factors Contain a High Fraction of Intrinsically Disordered Regions Essential for Transcriptional Regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science (80−. ) 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny L. a, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FSL, Schütte J, Ruau D, Diamanti E, Hannah R, Kinston SJ, Göttgens B. Constrained transcription factor spacing is prevalent and important for transcriptional control of mouse blood cells. Nucleic Acids Res. 2014;42:13513–13524. doi: 10.1093/nar/gku1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EEM, et al. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife. 2015;4:1–20. doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong CT, Hookway T. a., Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazak L, Fujioka M, Gergen JP. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev. Biol. 2010;344:1048–1059. doi: 10.1016/j.ydbio.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J. Enhancer Divergence and cis-Regulatory Evolution in the Human and Chimp Neural Crest. Cell. 2015:1–16. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann S. a, Flynn R. a, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl. Acad. Sci. 2011;108:10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Sakabe NJ, Nobrega MA. Beyond the ENCODE project: using genomics and epigenomics strategies to study enhancer evolution. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20130022. doi: 10.1098/rstb.2013.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. 2015;112:201518552. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri CA, Scacheri PC. Mutations in the noncoding genome. Curr. Opin. Pediatr. 2015;27:659–664. doi: 10.1097/MOP.0000000000000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W. Enhancers, enhancers - From their discovery to today's universe of transcription enhancers. Biol. Chem. 2015;396:311–327. doi: 10.1515/hsz-2014-0303. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. Cohesin-Based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat. Genet. 2013a;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Riesenfeld SJ, Holloway AK, Li Q, Murphy KK, Feliciano NM, Orecchia L, Oksenberg N, Pollard KS, Ahituv N. A compact, in vivo screen of all 6-mers reveals drivers of tissue-specific expression and guides synthetic regulatory element design. Genome Biol. 2013b;14:R72. doi: 10.1186/gb-2013-14-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, Stark A. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015:1–18. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, Reynolds A, Raubitschek A, Ziegler S, LeProust EM, Akey JM, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine ZE, McGaughey DM, Bessling SL, Li S, McCallion AS. Steroid hormone modulation of RET through two estrogen responsive enhancers in breast cancer. Hum. Mol. Genet. 2011;20:3746–3756. doi: 10.1093/hmg/ddr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Han D, Boyd-Kirkup J, Yu X, Han JDJ. Evolution of Alu Elements toward Enhancers. Cell Rep. 2014;7:376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Sved J, Bird a. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, Ovcharenko I. Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res. 2011;21:1139–1149. doi: 10.1101/gr.119016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and Divergence: The Evolution of New Genes and Old Ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:1–5. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- Ulianov SV, Khrameeva EE, Gavrilov AA, Flyamer IM, Kos P, Mikhaleva EA, Penin AA, Logacheva MD, Imakaev MV, Chertovich A, et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016;26:70–84. doi: 10.1101/gr.196006.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-sun S, Sabo PJ, Byron R, Humbert R, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. 2014;346:1007–1013. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. Cell Rep. 2015a;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. Cell Rep. 2015b;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. Enhancer Evolution across 20 Mammalian Species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 2007;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. Functional autonomy of distant-acting human enhancers. Genomics. 2009;93:509–513. doi: 10.1016/j.ygeno.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–1050. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Whitaker JW, Nguyen TT, Zhu Y, Wildberg A, Wang W. Computational schemes for the prediction and annotation of enhancers from epigenomic assays. Methods. 2015;72:86–94. doi: 10.1016/j.ymeth.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson WI, Lettice LA, Hill R, Bickmore WA. Shh and ZRS enhancer co localisation is specific to the zone of polarizing activity. bioRxiv. 2016:050849. doi: 10.1242/dev.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Xing K, He X. Reassessing the “Duon” hypothesis of protein evolution. Mol. Biol. Evol. 2015;32:1056–1062. doi: 10.1093/molbev/msu409. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Davidson EH. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996;122:1069–1082. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Kielbasa SM, Arndt PF, Chung H-R, Vingron M. Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet. 2009;25:63–66. doi: 10.1016/j.tig.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhang W, Zhang T, Liu B, Jiang J. Genome-Wide Prediction and Validation of Intergenic Enhancers in Arabidopsis Using Open Chromatin Signatures. Plant Cell. 2015;27:2415–2426. doi: 10.1105/tpc.15.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden M.I.J. a, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch T. a, van IJcken WFJ, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]