Abstract
BACKGROUND
Pre-exposure prophylaxis (PrEP) using antiretroviral (ARV) agents has been shown to effectively prevent human immunodeficiency virus type 1 (HIV-1) acquisition in high-risk populations. However, the efficacy of these regimens is highly variable, which is thought to be largely due to the varying degrees of adherence to a daily intervention in the populations. Passive immunization using broadly neutralizing antibodies (bNAbs) against HIV-1, with their relatively long half-life and favorable safety profile, could provide an alternative to daily PrEP. However, most bNAbs have a limited breadth, only neutralizing 70% to 90% of all HIV-1 strains.
METHODS
To overcome the problem of limited antiviral breadth, we proposed that targeting human CD4 and HIV-1 envelope proteins simultaneously may improve virus-neutralization breadth and potency. Therefore, we constructed bispecific Abs (biAbs) using scFvs of anti-gp120 bNAbs fused to ibalizumab (iMab), a humanized monoclonal antibody that binds human CD4, the primary receptor for HIV-1.
RESULTS
Some of our biAbs neutralized 100% of HIV-1 strains tested in vitro at clinically achievable concentrations. Distinct neutralization patterns were observed in this panel of biAbs. Those biAbs with specificity for the CD4-binding site on gp120 demonstrated 100% breadth, as well as slightly improved potency compared to iMab. In contrast, biAbs with specificity for the V1-V2 apex epitope or the V3-glycan epitope on gp120 demonstrated dramatically improved potency; some showed limited gain in neutralization breadth while others (e.g., PGT128-LM52 and 123-iMab) improved to 100% breadth.
CONCLUSION
Our data suggest that this panel of iMab-based biAbs could be used to probe the parameters for potent HIV-1 neutralization. Moreover, a few of these biAbs warrant further studies and possibly clinical development.
Keywords: Anti-HIV-1, bispecific antibody, CD4, passive immunization
INTRODUCTION
Despite 30 years of intense effort, an effective vaccine against HIV-1 has remained elusive. Alternative methods of HIV-1 prevention, such as the use of small molecule antiretroviral drugs (ARVs) as pre-exposure prophylaxis (PrEP), have demonstrated a degree of success, especially when there is good adherence to the daily ARV regimens (1-6). When compared with most small molecules, monoclonal antibodies generally have longer half-lives, requiring less frequent administration. The recent discovery of potent broadly neutralizing antibodies (bNAb) such as VRC01 (7), PG9 (8), 3BNC117 and 3BNC60 (9), PGT antibodies (10, 11), NIH45-46G54W (12), and 10E8 (13) has given momentum to the approach of passive administration of bNAbs for HIV-1 prevention. Compared to the first-generation HIV-1-neutralizing monoclonal antibodies such as 2G12 (14), 4E10 (15), 2F5 (16), and IgG1b12 (17), much lower concentrations of one such next-generation antibody, PGT121, or bNAb combinations protected monkeys from virus challenges (10) and led to therapeutic effects (18, 19). Also, AAV-based expression of another bNAb, VRC01, conferred protection against HIV-1 infection in a humanized mouse model (20). Nevertheless, with the exception of 10E8, these next-generation bNAbs could only neutralize around 70% to 90% of circulating HIV-1 strains in vitro, even at concentrations as high as 50 μg/ml.
Passive immunization strategies may also include mAbs specific for the HIV-1 receptors, CD4 (21-24) and CCR5 (25), and these mAbs also show potent and broad neutralizing activity against HIV-1. One such example is ibalizumab (iMab), a humanized IgG4 mAb that blocks entry of HIV-1 isolates from multiple subtypes (26) by highly-specific binding to human CD4 (22-24, 27, 28). The iMab epitope is located at the interface between domains 1 and 2 of CD4 (29, 30), and positioned on the opposite side from the site of CD4 that engages major histocompatibility complex class II or HIV-1 gp120. Consistently, iMab does not inhibit binding of CD4 to monomeric gp120 (21) and thus is thought to inhibit a post CD4-binding step required for virus entry. In Phase 1 and 2 clinical trials in HIV-1 positive patients, iMab treatment resulted in an average of 1 log decrease in viral load and a corresponding increase in CD4+ T-cell numbers without any serious drug-related adverse events (22, 24).
However, when iMab was tested in the TZM-bl/pseudotype assay against a panel of 118 diverse viral strains, eight percent of the viruses in this panel were resistant to this mAb (26). In a phase 1b clinical trial, resistant virus emerged in some patients in the presence of continuous ibalizumab therapy (22). In both studies, the principal cause for HIV-1 resistance to iMab is the loss of a glycan in the N-terminal region of the V5 loop of gp120. Here, we sought to improve the anti-viral potency and breadth of iMab by the construction of bispecific antibodies (biAbs). A panel of biAbs was constructed using iMab as the backbone onto which the antigen-binding domains of select human anti-gp120 bNAbs were fused. Among these iMab-based biAbs, distinct anti-HIV-1 neutralizing activity profiles were observed, including some with exceptional potency and breadth.
RESULTS
Construction, expression, and binding kinetics of iMab-based biAbs
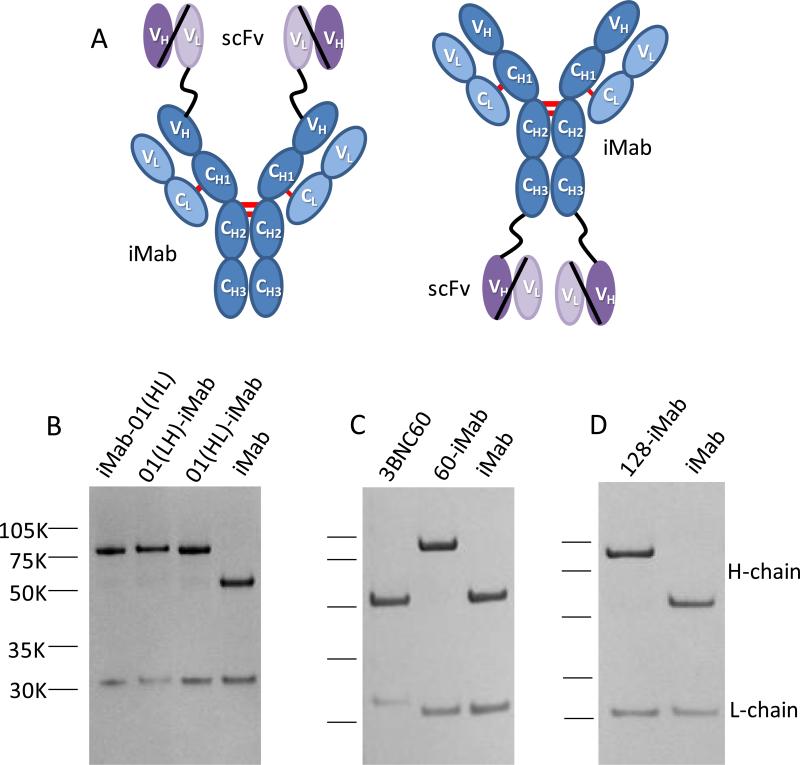
We sought to improve the anti-HIV-1 neutralization breadth and potency of iMab by generating a panel of biAbs utilizing iMab as the backbone. Initially, the single-chain Fv (scFv) of VRC01 was tethered to the N-terminus or C-terminus of the heavy- (H-) chain of iMab via a glycine-serine linker of 15-20 amino acids in length (Figure 1A). VRC01 is a well-characterized bNAb directed to the CD4-binding site on the envelope trimer (7). To investigate which orientation would be better for the design of this panel of iMab-based biAbs, two scFv forms of VRC01 (VH-VL and VL-VH) were fused to the N-terminus of iMab H-chain to create VRC01(VH-VL)-iMab (01(HL)-iMab) and VRC01(VL-VH)-iMab (01(LH)-iMab), and the VH-VL form of VRC01 scFv was fused to the C-terminus of iMab H-chain to create iMab-VRC01(VH-VL) (iMab-01(HL)) (Figure 1A). The molecular weight of each of the VRC01-iMab biAbs H-chain is ~80kD, compared to ~50 kD for the H-chain of iMab alone (Figure 1B). By ELISA, 01(HL)-iMab bound HIV-1 BaL gp120 comparably to VRC01 IgG; however, iMab-01(HL) bound less well (Figure S1). The binding kinetics for BaL gp120 were further evaluated using surface plasmon resonance and were similar for 01(HL)-iMab, 01(LH)-iMab, and the parental VRC01 (Table S1). The affinity of 01(LH)-iMab to soluble CD4 was comparable to that of the iMab (Table S1). These data suggest that the binding of iMab to CD4 was not greatly altered by placing a scFv at the N-terminus of the iMab H-chain. Therefore, the scFvs of 3BNC60 (Figure 1C), NIH45-46G54W, PGT123, or PGT128 (Figure 1D) were each fused in the VH-VL format to the N-terminus of iMab H-chain to create 3BNC60-iMab (60-iMab), NIH45-46G54W-iMab (45-46-iMab), PGT123-iMab (123-iMab), or PGT128-iMab (128-iMab). The molecular weight of each of these biAbs in this panel is ~200kD, compared to ~150 kD for each of the parental mAbs alone. Notably, the binding kinetics of 60-iMab, 45-46-iMab, or 128-iMab for BaL or HXBc2 gp120 were also comparable to those of the parental gp120-specific bNAbs (Table S1).
Figure 1.
Ibalizumab (iMab)-based biAb construction and confirmation. (A) Schematic depicting the structure of an iMab-based biAb. The variable fragments (VH and VL) of VRC01, 3BNC60, NIH45-46G54W, PGT123, PGT128 or 10E8 were tethered by a flexible glycine-serine linker to create a single-chain Fv (scFv), which was fused to the N-terminus or C-terminus of the iMab heavy chain. (B) SDS-PAGE analysis of purified VRC01-ibalizumab (01-iMab) bispecific Abs: iMab-VRC01(VH-VL) (iMab-01(HL)), VRC01(VL-VH)-iMab (01(LH)-iMab), and VRC01(VH-VL)-iMab (01(HL)-iMab). (C) 3BNC60-iMab (60-iMab) biAb. (D) PGT128-iMab (128-iMab) biAb.
iMab-based biAbs show improved anti-HIV-1 activity compared to the parental mAbs
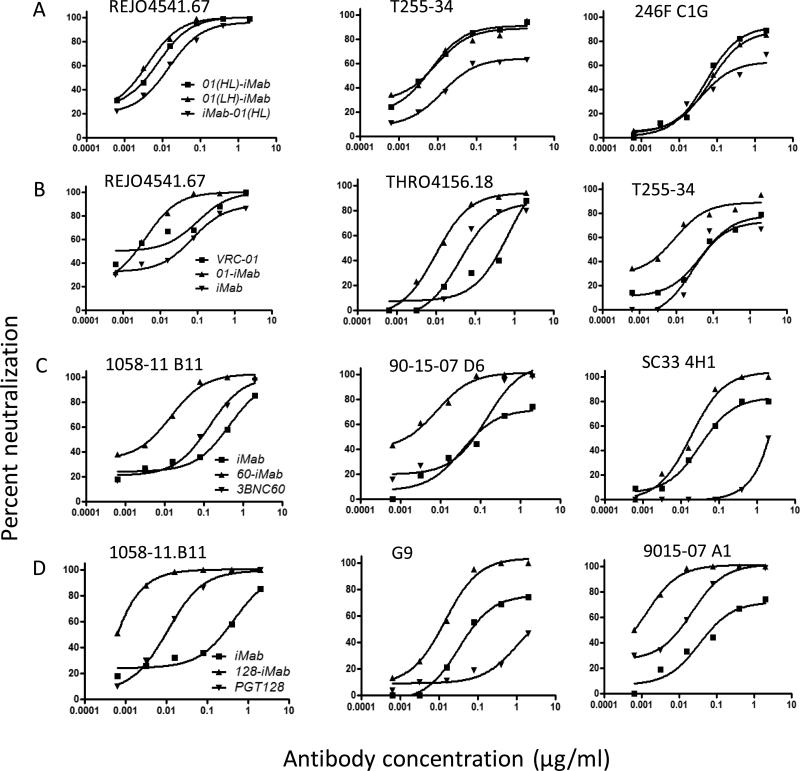
We next sought to evaluate the neutralization profiles of this panel of iMab-based biAbs against pseudotyped HIV-1 in TZM-bl cells. Among the three forms of VRC01/iMab biAbs, 01(HL)-iMab and 01(LH)-iMab showed comparable neutralization activities (Figure 2A) against all three viruses tested, while the activity of iMab-01(HL) was reduced. This correlated with the reduced binding to gp120 of iMab-01(HL) (Figure S1). Therefore, 01(LH)-iMab was selected to represent 01-iMab biAbs henceforth. Improved neutralization activities were observed with 01-iMab against three viruses tested compared to the two parental mAbs VRC01 and iMab (Figure 2B). Similarly, 60-iMab and 128-iMab also demonstrated marked enhancement over their two parental Abs against three viruses tested (Figure 2C and 2D).
Figure 2.
Neutralization data for iMab-based biAbs against HIV-1 env pseudoviruses in the TZM-bl assay. (A) Comparison of three forms of 01-iMab biAbs: iMab-01(HL), 01(HL)-iMab, and 01(LH)-iMab. (B) Enhanced neutralization activity of 01-iMab comparing with its two parental mAbs. (C) Enhanced neutralization activity of 60-iMab compared to iMab and 3BNC60. (D) The neutralization activity of 128-iMab compared to iMab and PGT128. (E) The neutralization activity of 128-LM52 in comparison with 128-iMab.
iMab-based biAbs lead to enhanced anti-HIV-1 activity compared to the co-administration of the two parental mAbs
When the neutralization profiles of 60-iMab or 128-iMab were compared with the combination of their respective parenteral antibodies at 1:1 ratio, the neutralization activity of the biAbs was consistently better (Figure S2). These data suggest that the linkage of anti-HIV-1 mAbs through iMab to human CD4 is necessary for the markedly improved potency of this panel of iMab-based biAbs. These findings are consistent with our previous observations with PG9-iMab (31).
Distinct neutralization profiles were observed with iMab-based biAbs against a large HIV-1 panel
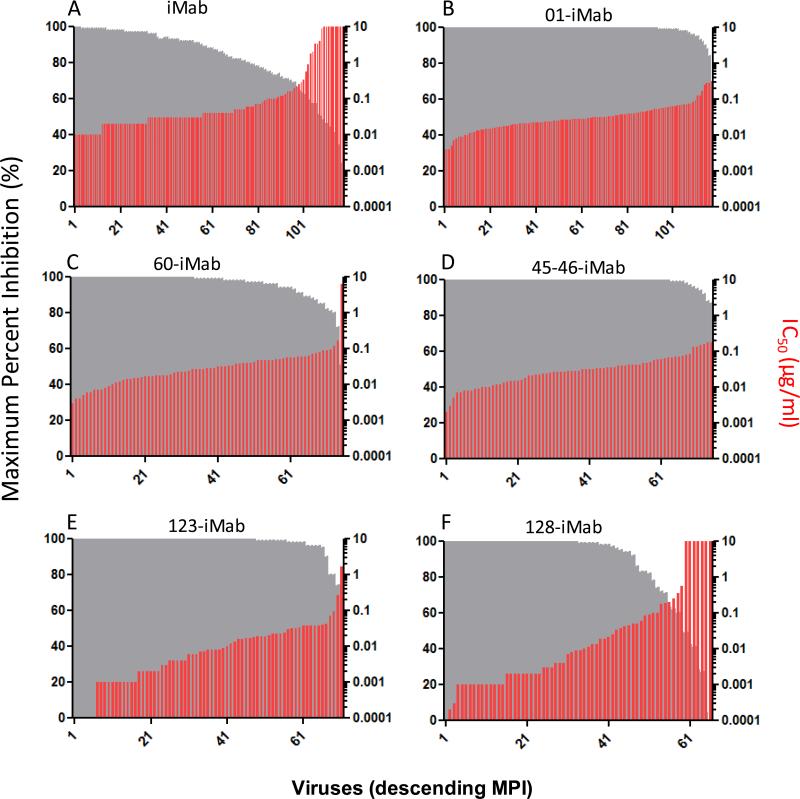
To further assess the anti-HIV-1 neutralization profiles of these iMab-based biAbs, we tested them in the TZM-bl/pseudotype assay against a panel of 118 diverse viral strains (for 01-iMab, and 128-LM52) in the Antibody Core Lab of the Collaboration for AIDS Vaccine Discovery (32). 60-iMab, 45-46-iMab, and 128-iMab were tested in a subset of the viruses in this panel including all the viruses that are known to be resistant to iMab. 123-iMab was tested in all the subtype B, C, A, and BC viruses in this panel (71 viruses total). Compared to the parental iMab (Figure 3A), significantly improved anti-HIV-1 breadth was observed with 01-iMab (Figure 3B), 60-iMab (Figure 3C), 45-46-iMab (Figure 3D), and 123-iMab (Figure 3E). These four biAbs, tested at a maximum concentration of 10 μg/ml, neutralized 100% of viruses as defined by ≥50% inhibition. For comparison, iMab inhibited 92% of the same panel of viruses (26). Although 60-iMab and 45-46-iMab were not tested against the entire panel of viruses, all of the iMab-resistant viruses were included in order to assess the improvement in breadth of these biAbs relative to their parental counterparts. In contrast, seven of the ten iMab-resistant viruses were also resistant to 128-iMab (Figure 3F).
Figure 3.
Neutralization data for iMab and iMab-based biAbs against a panel (or a part of the panel) of 118 tier-2 HIV-1 Env pseudoviruses in the TZM-bl assay. For each virus, grey bars indicate maximum percent inhibition (MPI, %) when tested up to 10 μg/ml and red bars indicate the antibody concentrations that confer 50% neutralization (IC50) values. Viruses are ordered by descending MPI. iMab and 01-iMab were tested against the entire panel of viruses, while 128-iMab, 123-iMab, 60-iMab, and 45-46-iMab were tested against 65, 71, 75, and 75 viruses in the same virus panel, respectively. The data for iMab (26) were obtained from the literature.
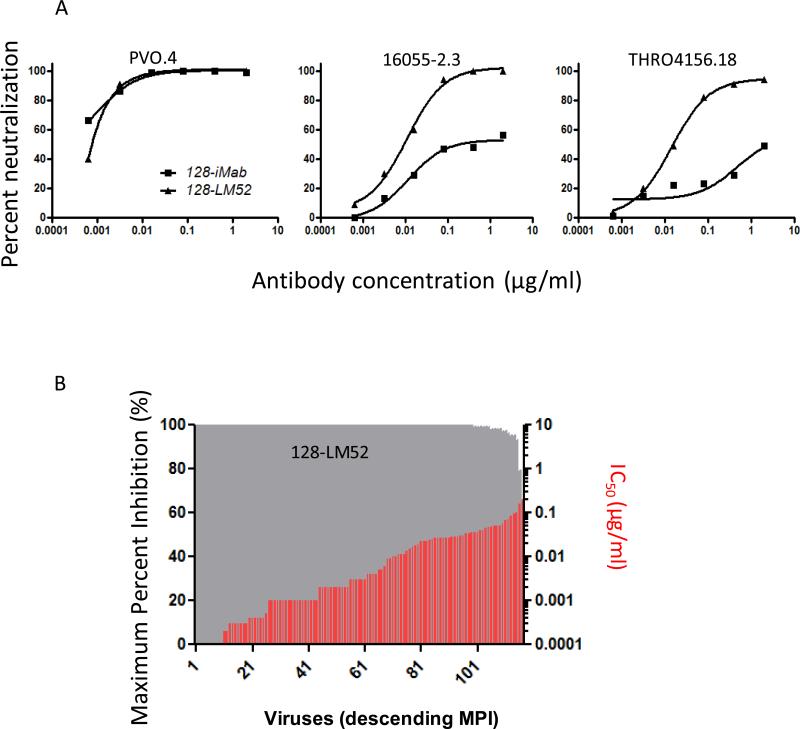
To further improve the breadth of 128-iMab, 128-LM52 was created by pairing of 128-iMab H-chain with LM52 L-chain that contains an extra N-glycan at AA52 in the variable region (33), since we have previously shown that LM52 could significantly improve the anti-HIV-1 breadth of iMab. Indeed, 128-LM52 could neutralize two viruses (16055-2.3 and THRO4156.18) that were resistant to 128-iMab (Figure 4A). When tested against the 118 virus panel, 128-LM52 could neutralize all of the viruses, including those that were resistant to 128-iMab (Figure 4B).
Figure 4.
Neutralization data for 128-LM52 against HIV-1 env pseudoviruses in the TZM-bl assay. (A) Comparison of 128-LM52 to 128-iMab. (B) Neutralization activity of 128-LM52 against a panel of 118 tier-2 HIV-1 Env pseudoviruses in the TZM-bl assay. For each virus, grey bars indicate maximum percent inhibition (MPI, %) when tested up to 10 μg/ml and red bars indicate the antibody concentrations that confer 50% neutralization (IC50) values. Viruses are ordered by descending MPI.
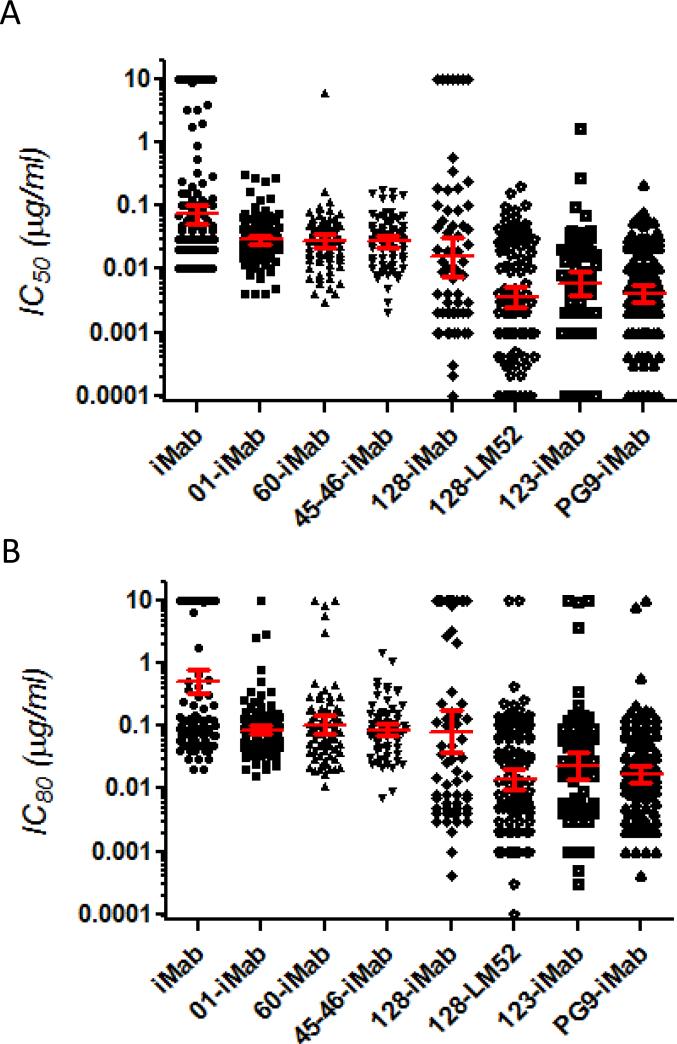
The IC50 values of our biAbs are plotted differently in Figure 5A. Although 100% breadth was achieved with 01-iMab, 60-iMab, or 45-46-iMab, their geometric mean IC50 values against this panel of viruses ranged from 25 ng/ml to 31 ng/ml, only approximately 3-fold lower than iMab (74 ng/ml). Among all the viruses tested with these iMab-based biAbs with a scFv targeting the CD4-binding site, no IC50 value was below 3 ng/ml (only 1 value was at 3 ng/ml for 45-46-iMab). On the other hand, although 128-iMab did not neutralize 7 of the 65 viruses tested, the geometric mean IC50 against this panel of viruses was only 16 ng/ml and 24 viruses were neutralized at an IC50 of < 3 ng/ml. Interestingly, 123-iMab neutralized 100% of the 71 viruses tested in the same panel with a geometric mean IC50 of 6 ng/ml, and neutralized 25 viruses at IC50 values < 3 ng/ml. In addition, the geometric mean IC50 value for 128-LM52 in this panel of viruses was only 4 ng/ml, almost 20 times lower compared to that of iMab (74 ng/ml). Furthermore, this biAb could neutralize 50 out of the 118 viruses at IC50 values below or at 3 ng/ml.
Figure 5.
Neutralization data for iMab and iMab-based biAbs against a panel (or panel subset) of 118 tier-2 HIV-1 Env pseudoviruses in the TZM-bl assay. The corresponding (A) IC50 (μg/ml) and (B) IC80 (μg/ml) against each virus are showed. Red lines indicate the geometric means with 95% confidence intervals. IMab, 01-iMab, and 128-LM52 were tested against the entire panel of viruses, while 128-iMab, 123-iMab, 60-iMab, and 45-46-iMab were tested against 65, 71, 75, and 75 viruses from the same virus panel, respectively. The data for iMab were obtained from the literature (26).
When evaluating neutralization activity based on 80% inhibitory concentrations, 01-iMab, 60-iMab, 45-46-iMab, 123-iMab, or 128-LM52 neutralize 99, 97, 100, 97, and 99% viruses tested (Figure 5B), respectively, at concentrations up to 10 μg/ml, compared to only 66% for iMab. Also, the geometric mean of IC80 values for 01-iMab, 60-iMab, or 45-46-iMab ranged from 76 ng/ml to 92 ng/ml compared to 510 ng/ml for iMab (Figure 5B). In comparison, the geometric mean IC80 values for 128-iMab, 123-iMab, and 128-LM52 were 82, 24, and 14 ng/ml (36 times better than iMab), respectively.
These data indicate that the addition of a scFv of any of the three anti-CD4 binding site mAbs (VRC01, 3BNC60, or NIH45-46G54W) to the N-terminus of iMab resulted in ~ 100% breadth but with only marginally improved potency, while the addition of PGT128 or PGT123 scFv to the N-terminus of iMab led to markedly improved potency in at least a subset of viruses. The best neutralization profiles were observed for 128-LM52 (Figure 4B) and 123-iMab (Figure 3E), both of which could inhibit 100% of the viruses tested in this panel with geometric mean IC50 values below 6 ng/ml.
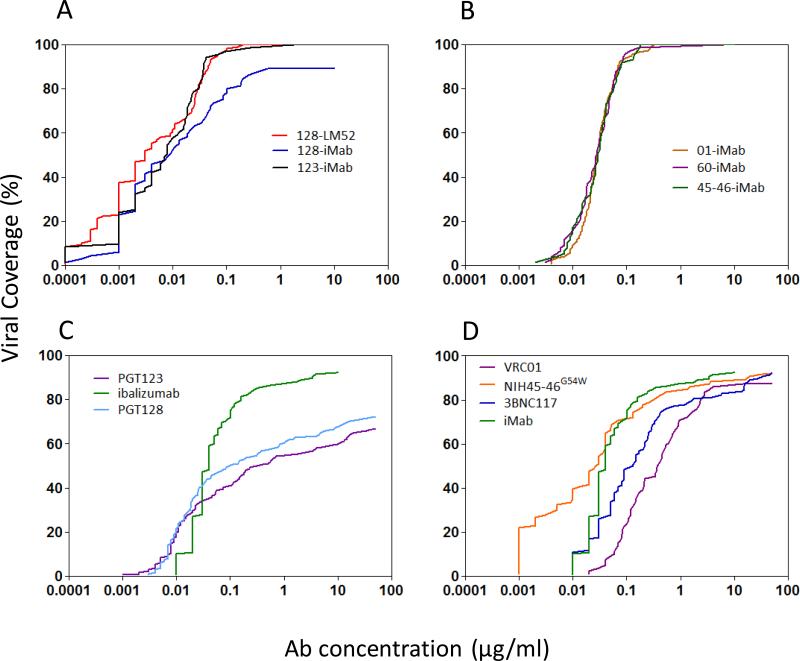
The distinct neutralization activities of these iMab-based biAbs were most evident in plots of their anti-HIV-1 coverage with increasing antibody concentrations (Figure 6). In general, better anti-HIV-1 coverage was observed with this panel of biAbs (Upper panels) compared to their parental mAbs (Lower panels). The viral coverage curves for 01-iMab, 60-iMab, and 45-46-iMab were nearly superimposable even though their parental anti-gp120 antibodies have marked varying neutralization activities (3BNC117 was shown instead of 3BNC60 since these two bNAbs had comparable neutralization activities (9) while 3BNC60 was not tested in a large virus panel, Figure 6, right panels). On the other hand, 128-iMab, 123-iMab, and 128-LM52 each gained substantial breadth and potency when compared to the parental antibodies (Figure 6, left panels).
Figure 6.
Percent viral coverage achieved by iMab-based biAbs versus those observed for mAbs. (A) Viral coverage curves achieved with iMab-based 123-iMab, 128-iMab, and 128-LM52. (B) Viral coverage curves achieved with 01-iMab, 60-iMab, and 45-46-iMab. (C) Viral coverage curves achieved with selected parental mAbs, including iMab, PGT128, and PGT123. (D) Viral coverage curves achieved with VRC01, 3BNC117, iMab, and NIH45-46G54W. 3BNC117 was showed instead of 3BNC60 because it demonstrated comparable anti-viral potency compared to 3BNC60 (9). IMab-based biAbs and iMab were tested up to 10 μg/ml, while the other mAbs were tested up to 50 μg/ml.
A twenty amino-acid linker is optimal for the potency of iMab-based biAbs
Our data suggest that the HIV-1 epitope targeted by iMab-based biAbs influences whether the biAb exhibits enhanced potency, as observed for 128-iMab and 123-iMab, or whether the biAb exhibits improvement mostly in breadth, as observed for biAbs targeting the CD4 binding site of gp120 (01-iMab, 60-iMab, and 45-46-iMab). Because the PGT128 and PGT123 epitopes appear to be more readily accessible than the CD4-binding site on the virus, we assessed whether increasing the length of the linker used to tether the anti-HIV-1 scFvs to iMab would improve the potency of iMab-based biAbs directed to the CD4 binding site (Figure S4). Lengthening the linker from 20 to 25, or even to 47, amino acids did not enhance the activity of 60-iMab or 01-iMab. On the other hand, shortening of the linker to 10 amino acids resulted in some loss of activity. These findings suggest that the lack of markedly improved potency of biAbs targeting the CD4-binding site is not due to constraints imposed by a defined linker length used to tether the scFvs to iMab.
DISCUSSION
The epitope on human CD4 targeted by iMab is invariant (29, 30), and by definition the bNAb epitopes are relatively conserved across all HIV-1 isolates. Therefore, we postulate that simultaneously targeting these two sites may provide enhanced potency and breadth. To this end, we generated a panel of iMab-based biAbs with dramatically improved antiviral properties in vitro. Placing the scFvs of VRC01, 3BNC60, NIH45-46G54W, PGT123, or PGT128 at the N-terminus of iMab H-chain maintained comparable binding affinities to HIV-1 gp120 (Table S1) as their parental counterparts. However, when the scFv of VRC01 was placed at the C-terminus of iMab H-chain, decreased binding affinity to gp120 (Figure S1) and inferior virus-neutralizing activity against HIV-1 (Figure 2) were observed. In general, our iMab-based biAbs showed enhanced potency and breadth compared to the parental mAbs, and even to their combinations (Figures 2 and 3). The only exception was iMab-01(HL), consistent with previous findings that the placement of PG9 scFv on the C-terminus of iMab was not favorable (31). This suggests that there is a preferred orientation for the anti-gp120 scFv when linked to the H-chain of iMab.
The neutralization activities of 60-iMab or 128-iMab were consistently better than the combination of their parenteral antibodies (Figure S2), indicating that the linkage of the two binding elements is important. Anchoring the scFv directed to gp120 on CD4 could well have increased the “local concentration” of the antiviral component at the site of virus entry. Furthermore, distinct neutralization patterns were observed in this panel of novel biAbs. 01-iMab, 60-iMab, and 45-46-iMab inhibited 100% breadth and slightly improved potency (Figures 5 and S3). On the other hand, 128-iMab demonstrated markedly improved potency but only slightly improved breadth. 123-iMab showed dramatically improved potency and 100% breadth. When an N glycan was introduced to the iMab L-chain of 128-iMab to create 128-LM52 (33), both 100% breadth and exceptional potency were observed (Figures 4 and 5).
The work described herein demonstrates that the anti-HIV-1 activity of iMab could be substantially enhanced in either breadth or potency, or both, by the strategic placement of an Env-specific scFv in the H-chain. Indeed, our data indicate that ~100% antiviral breadth can be reached by a number of iMab-based biAbs: 01-iMab, 60-iMab, 45-46-iMab, 123-iMab, and 128-LM52. However, distinct anti-HIV-1 neutralization patterns were observed with this panel of biAbs. The degree of potency enhancement for biAbs with specificity for the V3-glycan epitope 128-LM52, 123-iMab, and 128-iMab is striking, compared to biAbs with specificity for the CD4-binding site 01-iMab, 60-iMab, and 45-46-iMab. Interestingly, the latter three biAbs show superimposable anti-HIV-1 activities although the affinities of the three parental anti-CD4-binding-site mAbs (VRC01, 3BNC60, and NIH45-46G54W) are very different (Figure 6).
We lack a proper explanation for this observed phenomenon. The best neutralization profiles came from 128-LM52 and 123-iMab, both of which could neutralize all of the viruses tested in this panel with excellent potencies (geometric mean IC50 values of 4 and 6 ng/ml, respectively, Figures 3-5). It is worth noting that 128-LM52 reached 100% coverage at 0.2 μg/ml. When these anti-viral coverage curves were also compared to those of PG9-iMab (31), and other mAbs in clinical development, including VRC01, 10-1074 (34), 3BNC117, iMab (26), LM52 (33) , and eCD4-Ig (35) (Figure S3), the best coverage was achieved with 128-LM52, 123-iMab and PG9-iMab (31). The potency of 128-LM52 was even slightly better than our previously published PG9-iMab biAb (31) against the same panel of viruses. What accounts for the remarkable gain in anti-HIV-1 potency of 128-iMab, 123-iMab, and PG9-iMab compared to 01-iMab, 60-iMab, and 45-46-iMab? We could only speculate, since the afore mentioned “local concentration” effect should apply to all of these bispecific antibodies. We suspect the answer lies with the Env epitopes being targeted by the scFv arm. The scFvs of PGT128 and PGT123 target the V3-N332-glycan epitope region, while that of PG9 targets the V2-apex epitope region. Both of these spitope clusters are at the tip of the trimer spike (Figure S5A), rendering them to be more readily accessible. Binding of these epitopes by the biAbs (e.g., 128-iMab or PG9-iMab) is likely to be reached earlier during the process of virus entry. On the other hand, the CD4-binding site is located lower down on the trimer spike (Figure S5B) and situated in a recessed pocket. Binding of the CD4-binding site by the biAbs (e.g., 01-iMab) may occur at a later time in viral entry, and with less favorable kinetics, especially when the movement of the scFv may be restricted due to its tethering to iMab and CD4. This limitation in the freedom of movement may offset any affinity differences in the scFvs of the CD4-binding mAbs, thereby accounting for the overlapping antiviral profiles observed in Figure 6B. More work will be required to confirm or refuse such speculations.
BiAbs continue to be an area of great interest in the pursuit of next-generation antibodies. Dual-targeting iMab-based biAbs, such as our previously described PG9-iMab and PG16-iMab (31), iMabm36 (38), and now 01-iMab, 60-iMab, 45-46-iMab, 123-iMab, 128-iMab, and 128-LM52, exhibit markedly improved activity in vitro. In particular, 123-iMab and 128-LM52 have antiviral profiles superior to many mAbs in clinical development (Figure S3). However, these biAbs must be properly evaluated for their developability, including characteristics such as stability, aggregation propensities, pharmacokinetic profiles, and ease of manufacturing. Nonetheless, their favorable breadth and potency suggest their inclusion as candidates for clinical development as preventive agents against HIV-1 transmission.
MATERIALS AND METHODS
Cell Lines, reagents, and pseudotyped viruses
TZM-bl cells (cat#8129) and CD4-IgG2 (cat#11780, Progenics Pharmaceuticals) were obtained through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH. TZM-bl cells are a genetically engineered HeLa cell line that express CD4, CXCR4, and CCR5 and contain Tat-responsive reporter genes for luciferase and β-galactosidase under the control of an HIV-1 long terminal repeat. The Standard Reference Panels of Subtype B HIV-1 Env clones from acute and early infections and Env-deficient backbone plasmid (SG3ΔEnv, cat#11051) were also obtained through the NIH ARRRP. HIV-1 env pseudotyped viruses were prepared by co-transfection of 293A cells (Invitrogen) with an Env-expression plasmid and SG3ΔEnv. Recombinant sCD4 comprising the full-length extracellular domain of human CD4 was obtained from Progenics Pharmaceuticals, Inc. (Tarrytown, NY). Ibalizumab (iMab) protein was provided by TaiMed Biologics (Irvine, CA). Plasmids pMV1 and pLC, which encode for iMab H-chain and L-chain, respectively, were amplified from cDNA and cloned into pCDNA3.1 (+) (Invitrogen).
Construction of iMab-based BiAb H-chain
The single-chain variable fragment (scFv) genes of VRC01, 3BNC60, NIH45-46G54W, PGT123, PGT128, or 10E8 were designed as VH-VL or VL-VH orientation, with a (Gly4Ser)4 linker in between. The genes were chemically synthesized by Genscript (Piscataway, NJ) and tethered to the N-terminal or C-terminus of iMab H-chain with a (Gly4Ser)3 linker for VRC01-iMab, and 3BNC60-iMab; or (Gly4Ser)4 linker for NIH45-46G54W-iMab, PGT123-iMab, and PGT128-iMab to create the H-chain of the BiAb by overlapping PCR. The H-chains were then cloned into pCDNA3.1(+), sequenced and transiently transfected into HEK293A cells using a 1:1 ratio of H- and L-chain plasmids with 25-kDa linear polyethylenimine (PEI)-DNA complex. Supernatants were harvested on day 5 post transfection and BiAbs were purified with a protein-A agarose (Thermo Scientific, Rockford, IL) column and analyzed by SDS-PAGE.
Virus neutralization assay using TZM-bl cells
Neutralization assay was performed based on the method of Wei et al.(39) with modification (32). Briefly, 10,000 cells per well were seeded in a 96-well plate in 100 μl/well of DMEM supplemented with 10% fetal bovine serum (D10) and incubated overnight. The next day, serial diluted iMab or BiAbs were added to the cells and incubated for 1 h. Then, 200 times of 50%-tissue-culture-infective-doses (TCID50) of pseudotyped HIV-1 were prepared in D10 containing DEAE-Dextran (Sigma, St. Louis, MO) and added to the cells. The cells were incubated for 48 h and the β-galactosidase activity was measured using the Galacto-Star System (Applied Biosystems, Cedarville, OH). The percentage of inhibition of viral infectivity was calculated as 1 minus the ratio of antibody-treated wells versus untreated-infected wells multiplied by 100. The IC50 and IC80 values (the antibody concentrations that confer 50% and 80% neutralization, respectively) were calculated by a nonlinear regression analysis.
Surface Plasmon Resonance
Binding affinity analyses were performed with a Biacore T3000 optical biosensor (GE Healthcare, Piscataway, NJ). Immobilization of iMab, anti-Env mAbs, and the BiAbs were performed following the standard amine coupling procedure. Briefly, carboxyl groups on the sensor chip surface were activated by injection of 35 μl of a solution containing 0.2 M N-(3-dimethylaminopropyl)-N-ethylcarbodiimide and 0.05 M N-hydroxysuccinimide at a flow rate of 5 μl/minute. Next, the antibody, at a concentration of 2 μg/ml in 10 mM sodium-acetate buffer, pH 4.5, was allowed to flow over the chip surface at a rate of 10 μl/minute until the desired level of response units of reacted protein (150-200 RU) was achieved. After unreacted protein was washed out, excess active ester groups on the sensor surface were capped by the injection of 35 μl of 1 M ethanolamine, pH 8.0, at a flow rate of 5 μl/minute. To correct for instrument and buffer artifacts, a reference was generated under the same conditions with omission of the protein ligand. Binding experiments were performed at 25°C in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% vol/vol surfactant P20 (GE Healthcare). Binding kinetics were measured by passing various concentrations of analyte (human sCD4 or gp120 protein) over the chip surface at a flow rate of 30 μl/minute for 3 min. The dissociation of bound analytes was monitored while the surface was washed for 10 min. Remaining analytes were removed at a flow rate of 50 μl/minute with two 30 sec injections of 10 mM glycine-HCl, pH 2.0. The kinetic parameters were determined by collectively fitting the overlaid sensograms locally using the BIAevaluation 4.1 software to the 1:1 Langmuir binding model.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Tsuji, X. Wu, D. Franco, M.-W. Chen, A. Kao, D. Oren, and Y. Huang for helpful input. D.D.H. was supported by the Bill and Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery (Grants OPP50714 and OPP1040732) and by the National Institutes of Health (Grant DP1DA033263). M.S.S. was supported by the Bill and Melinda Gates Foundation's Comprehensive Ab Vaccine Immune Monitoring Consortium (Grant 1032144).
Abbreviations used
- iMab
ibalizumab
- HIV
human immunodeficiency virus
- bNAbs
broadly neutralizing Abs
- PrEP
pre-exposure prophylaxis
- biAb
bispecific antibody
- scFv
single-chain Fv
- IC50
the concentration of Ab to inhibit 50% of virus
- IC80
the concentration of Ab to inhibit 50% of virus
Footnotes
AUTHOR CONTRIBUTIONS
R.S. and D.D.H. conceived the study and designed the experiments. R.S., C.P., M.S.S., Q.F., M.S., C.A., A.W., and N.P. performed the experiments. R.S. and D.D.H. analyzed the data and wrote the manuscript.
REFERENCE
- 1.Baeten JM, Donnell D, Ndase P, et al. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. The New England Journal of Medicine. 2012 Aug 2;367(5):399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thigpen MC, Kebaabetswe PM, Paxton LA, et al. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. The New England Journal of Medicine. 2013 Aug 2;367(5):423–34. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- 3.Van Damme L, Corneli A, Ahmed K, et al. Preexposure prophylaxis for HIV infection among African women. The New England Journal of Medicine. 2012 Aug 2;367(5):411–22. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant RM, Lama JR, Anderson PL, et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. The New England Journal of Medicine. 2010 Dec 30;363(27):2587–99. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdool Karim Q, Abdool Karim SS, Frohlich JA, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science (New York, NY. 2010 Sep 3;329(5996):1168–74. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choopanya K, Martin M, Suntharasamai P, et al. Antiretroviral prophylaxis for HIV infection in injecting drug users in Bangkok, Thailand (the Bangkok Tenofovir Study): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2013 Jun 15;381(9883):2083–90. doi: 10.1016/S0140-6736(13)61127-7. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, NY. 2010 Aug 13;329(5993):856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science (New York, NY. 2009 Oct 9;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science (New York, NY. 2011 Sep 16;333(6049):1633–7. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldt B, Rakasz EG, Schultz N, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18921–5. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011 Sep 22;477(7365):466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diskin R, Scheid JF, Marcovecchio PM, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science (New York, NY. 2011 Dec 2;334(6060):1289–93. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012 Nov 15;491(7424):406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trkola A, Purtscher M, Muster T, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of Virology. 1996 Feb;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwick MB, Labrijn AF, Wang M, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. Journal of Virology. 2001 Nov;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muster T, Steindl F, Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of Virology. 1993 Nov;67(11):6642–7. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton DR, Pyati J, Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science (New York, NY. 1994 Nov 11;266(5187):1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 18.Barouch DH, Whitney JB, Moldt B, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013 Nov 14;503(7475):224–8. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shingai M, Nishimura Y, Klein F, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013 Nov 14;503(7475):277–80. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011 Jan 5;481(7379):81–4. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkly LC, Olson D, Shapiro R, et al. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal antibody. Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. J Immunol. 1992 Sep 1;149(5):1779–87. [PubMed] [Google Scholar]
- 22.Jacobson JM, Kuritzkes DR, Godofsky E, et al. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrobial Agents and Chemotherapy. 2009 Feb;53(2):450–7. doi: 10.1128/AAC.00942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitrov A. Ibalizumab, a CD4-specific mAb to inhibit HIV-1 infection. Curr Opin Investig Drugs. 2007 Aug;8(8):653–61. [PubMed] [Google Scholar]
- 24.Kuritzkes DR, Jacobson J, Powderly WG, et al. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. The Journal of Infectious Diseases. 2004 Jan 15;189(2):286–91. doi: 10.1086/380802. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson JM, Saag MS, Thompson MA, et al. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. The Journal of Infectious Diseases. 2008 Nov 1;198(9):1345–52. doi: 10.1086/592169. [DOI] [PubMed] [Google Scholar]
- 26.Pace CS, Fordyce MW, Franco D, Kao CY, Seaman MS, Ho DD. Anti-CD4 Monoclonal Antibody Ibalizumab Exhibits Breadth and Potency Against HIV-1, With Natural Resistance Mediated by the Loss of a V5 Glycan in Envelope. J Acquir Immune Defic Syndr. 2013 Jan 1;62(1):1–9. doi: 10.1097/QAI.0b013e3182732746. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XQ, Sorensen M, Fung M, Schooley RT. Synergistic in vitro antiretroviral activity of a humanized monoclonal anti-CD4 antibody (TNX-355) and enfuvirtide (T-20). Antimicrobial Agents and Chemotherapy. 2006 Jun;50(6):2231–3. doi: 10.1128/AAC.00761-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boon L, Holland B, Gordon W, et al. Development of anti-CD4 MAb hu5A8 for treatment of HIV-1 infection: preclinical assessment in non-human primates. Toxicology. 2002 Apr 2;172(3):191–203. doi: 10.1016/s0300-483x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 29.Song R, Franco D, Kao CY, Yu F, Huang Y, Ho DD. Epitope mapping of ibalizumab, a humanized anti-CD4 monoclonal antibody with anti-HIV-1 activity in infected patients. Journal of Virology. 2010 Jul;84(14):6935–42. doi: 10.1128/JVI.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman MM, Seaman MS, Rits-Volloch S, et al. Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody. Structure. 2010 Dec 8;18(12):1632–41. doi: 10.1016/j.str.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace CS, Song R, Ochsenbauer C, et al. Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13540–5. doi: 10.1073/pnas.1304985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seaman MS, Leblanc DF, Grandpre LE, et al. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology. 2007 Oct 10;367(1):175–86. doi: 10.1016/j.virol.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song R, Oren DA, Franco D, Seaman MS, Ho DD. Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat Biotechnol. 2013 Nov;31(11):1047–52. doi: 10.1038/nbt.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouquet H, Scharf L, Euler Z, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012 Nov 20;109(47):E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardner MR, Kattenhorn LM, Kondur HR, et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015 Feb 18; doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galimidi RP, Klein JS, Politzer MS, et al. Intra-Spike Crosslinking Overcomes Antibody Evasion by HIV-1. Cell. 2015 Jan 29;160(3):433–46. doi: 10.1016/j.cell.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Myszka DG, Tendian SW, et al. Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding. Proc Natl Acad Sci U S A. 1996 Dec 24;93(26):15030–5. doi: 10.1073/pnas.93.26.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun M, Pace CS, Yao X, et al. Rational design and characterization of the novel, broad and potent bispecific HIV-1 neutralizing antibody iMabm36. J Acquir Immune Defic Syndr. 2014 Aug 15;66(5):473–83. doi: 10.1097/QAI.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003 Mar 20;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.