Abstract
The MDM4 protein plays an important part in the negative regulation of the tumor suppressor p53 through its interaction with MDM2. In line with this, MDM4 amplification has been observed in several tumor forms. A polymorphism (rs4245739 A>C; SNP34091) in the MDM4 3′ untranslated region has been reported to create a target site for hsa‐miR‐191, resulting in decreased MDM4 mRNA levels. In this population‐based case–control study, we examined the potential association between MDM4 SNP34091, alone and in combination with the MDM2 SNP309T>G (rs2279744), and the risk of breast‐, colon‐, lung‐, and prostate cancer in Norway. SNP34091 was genotyped in 7,079 cancer patients as well as in 3,747 gender‐ and age‐matched healthy controls. MDM4 SNP34091C was not associated with risk for any of the tumor forms examined, except for a marginally significant association with reduced risk for breast cancer in a recessive model (OR = 0.77: 95% CI = 0.59–0.99). Stratifying according to MDM2 SNP309 status, we observed a reduced risk for breast cancer related to MDM4 SNP34091CC among individuals harboring the MDM2 SNP309GG genotype (OR = 0.41; 95% CI = 0.21–0.82). We conclude, MDM4 SNP34091 status to be associated with reduced risk of breast cancer, in particular in individuals carrying the MDM2 SNP309GG genotype, but not to be associated with either lung‐, colon‐ or prostate cancer.
Keywords: Cancer risk, MDM4, population based, SNP309, SNP34091
Introduction
The tumor suppressor p53 plays a pivotal role in many physiological processes, including metabolism and maintenance of genomic stability. In order to allow normal cell proliferation and to maintain cell viability during absence of stress signals, the activity of p53 is kept under strict control, predominantly by the protein product of the murine double minute 2 gene, MDM2, and its homolog MDM4, acting in concert 1. It is well established that MDM2 and p53 are linked in an autoregulatory negative feedback loop, where p53 transcriptionally induces MDM2 and MDM2 downregulates p53 2, mainly by direct inhibition and/or proteolytic degradation 3, 4, 5. Although MDM4 alone is unable to target p53 for ubiquitin‐proteasome‐dependent degradation 6, the MDM2/MDM4 heterodimer has been shown more potent degrading the p53 protein as compared to the MDM2 homodimer 7, 8. Additionally, using Mdm2/Mdm4/p53 triple knockout MEFs, Yuan and colleagues showed that an Mdm2/Mdm4 heterodimer is required for the E3 ligase activity of Mdm2 9. These data suggest that elevated levels of MDM4 may contribute to reduced p53 activity and tumor development. In line with this, the MDM4 gene has been found amplified in malignant gliomas with no TP53 mutations or MDM2 amplifications 10, 11 as well as in breast cancer 12, and acute lymphoblastic leukemia 13. Furthermore, studies in transgenic mice show that overexpression of Mdm4 induced spontaneous tumor formation and accelerated tumorigenesis 14.
Single‐nucleotide polymorphisms (SNP) affecting the levels of both MDM2 and MDM4 have been reported 15, 16, 17, 18. While MDM2 SNP309T>G (rs2279744) and SNP285G>C (rs117039649) both affect MDM2 transcription, MDM4 SNP34091A>C (rs4245739) has been found to affect MDM4 mRNA stability and protein levels 17, 18. SNP34091 is located in the 3′ untranslated region of MDM4, and was found to create a functional target site for hsa‐miR‐191 and hsa‐miR‐887. Both miRs bind to the MDM4 SNP34091 C‐allele with higher affinity than to the MDM4 SNP34091 A‐allele, leading to miR‐mediated decrease in MDM4 protein levels in cells carrying the MDM4 SNP34091C variant 17, 18. Genotype AA was recorded to be more frequent in patients with high‐grade than low‐grade ovarian carcinoma 18. Furthermore, previous studies have indicated the SNP34091C allele to be associated with a reduced risk for non‐Hodgkin lymphoma 19, breast cancer 20, esophageal squamous cell carcinoma 21, and prostate cancer 22.
Contrasting these results, genome wide association studies (GWAS) reported the C allele to be associated with an increased risk for estrogen receptor negative, and in particular, triple negative breast cancer 23, 24, 25.
In this study, we assessed the impact of MDM4 SNP34091 status on the risk of cancer of the breast, lung, prostate, and colon in a large population‐based cohort of Caucasian descent.
Materials and Methods
Study population
From the population‐based Cohort of Norway (CONOR) study 26, we genotyped 7079 incident cancer cases and 3747 healthy controls, described in detail previously 27. Thus, we examined the potential effect of MDM4 SNP34091A>C by analyzing the four major cancer forms; breast (n = 1,717), lung (n = 1,331), colon (n = 1,531), and prostate (n = 2,500). On the basis of previously published allele frequencies in healthy controls and breast cancer cases 23, we found our study design to provide adequate statistical power (β‐values ranging from 0.83 to 0.95 for the four cancer sample sets, given an α‐value of 0.05).
MDM4 SNP34091 genotyping
All samples were genotyped for MDM4 SNP34091 status using a custom LightSNiP assay (TIB MOLBIOL Syntheselabor GmbH, Berlin, Germany) on a LightCycler 480 II instrument (Roche, Basel, Switzerland). The reactions were performed in a final reaction volume of 10 μL, containing 1 μL LightCycler®FastStart DNA Master HybProbe mix (Roche Diagnostics), 0.5 μL LightSNiP mix (TIB MOLBIOL), 3 mmol/L MgCl2 and 10–50 ng DNA. The thermocycling and melting curve conditions were as follows: 10 min initial denaturation/activation at 95°C, followed by 45 cycles of denaturation at 95°C for 10 sec, annealing for 10 sec at 60°C and elongation at 72°C for 15 sec. Subsequent to the thermocycling amplification the high‐resolution melting (HRM) step was initiated with a denaturation step at 95°C for 30 sec, followed by melting from 40°C to 75°C with a ramp rate of 0.19°C/sec and finally a cooling step at 40°C for 30 sec. The HRM curve profiles were analyzed by the Melt Curve Genotyping software (version 1.5) on the LightCycler® 480 II instrument (Roche Diagnostics).
Statistical analysis
Potential deviations from Hardy–Weinberg equilibrium were assessed by calculating the expected genotype distribution based on the observed allele frequencies and comparing the output with the observed genotype distribution using Chi‐square tests.
Potential associations between MDM4 SNP34091 and the risk of any of the cancer types tested as well as cancer risk within different subgroups were estimated by calculating Odds Ratios (OR) with 95% confidence intervals (CI) and logistic regression adjusting for sex and age. In addition, for colon‐ and lung cancer, overall calculations were performed including both genders using the Mantel–Haenszel test (sex adjusted).
All statistical analyses were performed using the IBM SPSS 22 software package (IBM Corp, Armonk, NY, USA) and Stata 13.0 for Windows (Stata Corp, College Station, TX, USA). All P‐values are given as two‐sided.
Results
Distribution of MDM4 SNP34091
In this study, 7,079 cancer cases and 3,747 healthy controls were analyzed for MDM4 SNP34091 status. Among the healthy individuals, the percentages harboring the three different genotypes (MDM4 SNP34091AA, AC, and CC) were recorded to be 54.5%, 38.4%, and 7.1%, respectively. The genotype frequencies were found to be in Hardy–Weinberg equilibrium (P > 0.9). A comprehensive overview of the MDM4 SNP34091 distribution in the healthy controls as well as the four cancer types analyzed is given in Table 1. Among the healthy controls, no substantial gender difference with respect to genotype distribution was observed (P = 0.193).
Table 1.
MDM4 SNP34091 distribution and cancer risk
| Cases/controls | Genotype | OR (95% CI) | P‐value | OR (95% CI) | P‐value | ||
|---|---|---|---|---|---|---|---|
| SNP34091 n (%) | SNP34091 | SNP34091 | |||||
| AA | AC | CC | CC versus AA+AC | CC+AC versus AA | |||
| Controls | 2042 (54.5) | 1439 (38.4) | 266 (7.1) | 1.00 | – | 1.00 | – |
| Women | 1021 (54.6) | 703 (37.6) | 146 (7.8) | 1.00 | – | 1.00 | – |
| Men | 1021 (54.4) | 736 (39.2) | 120 (6.4) | 1.00 | – | 1.00 | – |
| Colon cancera | 823 (53.8) | 600 (39.2) | 108 (7.1) | 1.04 (0.82–1.32) | 0.737 | 1.04 (0.93–1.18) | 0.484 |
| Womenb | 429 (55.1) | 293 (37.7) | 56 (7.2) | 1.02 (0.73–1.41) | 0.919 | 1.01 (0.85–1.20) | 0.941 |
| Menc | 394 (52.3) | 307 (40.8) | 52 (6.9) | 1.09 (0.78–1.54) | 0.601 | 1.10 (0.92–1.30) | 0.295 |
| Lung cancera | 715 (53.7) | 515 (38.7) | 101 (7.6) | 1.11 (0.87–1.41) | 0.396 | 1.03 (0.91–1.17) | 0.662 |
| Womenb | 264 (53.1) | 194 (39.0) | 39 (7.9) | 1.05 (0.73–1.53) | 0.781 | 1.07 (0.87–1.30) | 0.535 |
| Menc | 451 (54.1) | 321 (38.5) | 62 (7.4) | 1.19 (0.86–1.64) | 0.288 | 1.01 (0.86–1.19) | 0.912 |
| Prostate cancerc | 1412 (56.5) | 927 (37.1) | 161 (6.4) | 1.01 (0.79–1.29) | 0.946 | 0.92 (0.82–1.04) | 0.182 |
| Breast cancerb | 966 (56.3) | 643 (37.5) | 108 (6.3) | 0.77 (0.59–0.99) | 0.045 | 0.93 (0.82–1.07) | 0.317 |
Sex and age adjusted (logistic regression).
Calculations with female controls only, age adjusted.
Calculations with male controls only, age adjusted.
MDM4 SNP34091 status and cancer risk in four major cancer forms
In order to assess the potential impact of MDM4 SNP34091 status on cancer risk, we compared the frequency of the MDM4 SNP34091 genotypes among breast‐ (n = 1,717), lung‐ (n = 1,331), colon‐ (n = 1,531), and prostate cancer (n = 2,500) patients to healthy controls (n = 3,747). We observed no significant correlation between MDM4 SNP34091 status and the risk of either cancer in the colon, lung, or prostate, either in a dominant or a recessive model (SNP34091 CC+AC vs. AA, or CC vs. AA+AC, respectively). Furthermore, analyzing tumors of the right or the left side of the colon separately, revealed no significant effect of MDM4 SNP34091 status and cancer risk in either of the two groups (Table S1). We observed, however, a marginally significant association with reduced risk for breast cancer among individuals harboring the SNP34091CC genotype (recessive model; OR = 0.77; 95% CI = 0.59–0.99; Table 1, Fig. 1A).
Figure 1.
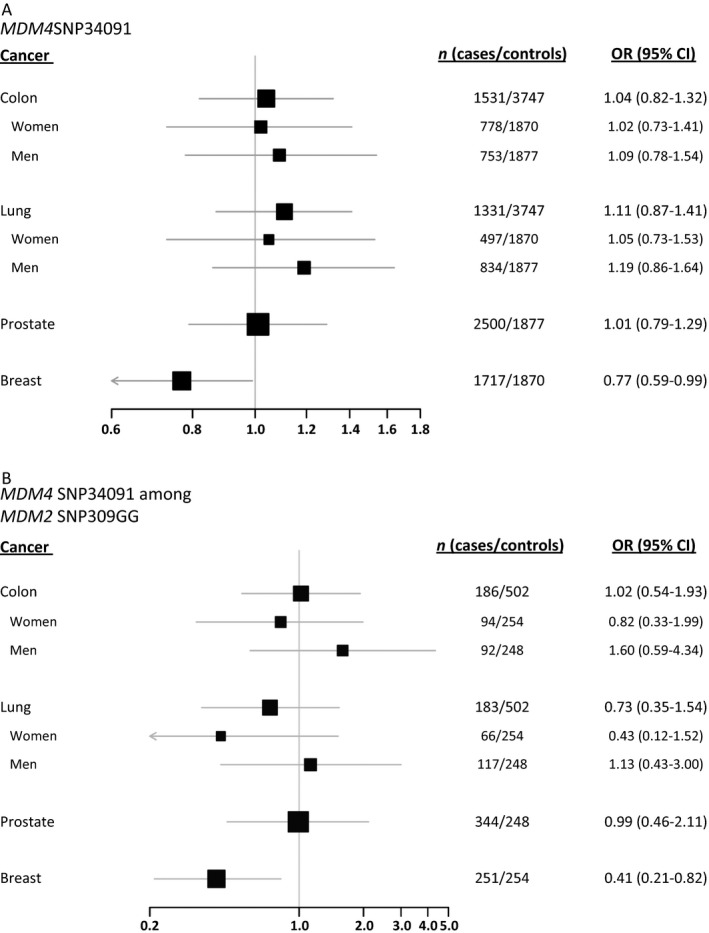
Impact of MDM4 SNP34091 on cancer risk. Forest plots showing the effect of SNP34091 on cancer of the colon, lung, prostate, and breast, as compared to healthy controls, among the total study population (A) and among individuals harboring the MDM2 SNP309GG genotype (B).
Since 92.6% of the lung cancer patients (from whom we had data) were smokers, excluding nonsmokers from the analysis had no impact on the estimates (Table S2).
Potential interactions between MDM4 SNP34091 status and MDM2 promoter SNPs
Previously, we assessed SNP status of the MDM4 partner MDM2 across the same population of cancer patients and healthy controls 27. While the MDM2 SNP309GG genotype has been associated with a nonsignificantly increased risk for breast cancer, breast cancer patients carrying the SNP309GG genotype have been found particularly sensitive to cancer risk reduction by a second MDM2 SNP (SNP285G>C) 27. This observation has also been recorded in another separate sample set of breast cancer patients 16.
Since MDM4 forms a heterodimer with MDM2 and promotes MDM2‐mediated polyubiquitination and subsequent degradation of p53, we investigated potential interactions/synergistic effects between MDM4 SNP34091 and MDM2 SNPs with respect to cancer risk. Stratifying according to MDM2 SNP309 status (SNP309TT, SNP309TG, and SNP309GG) we found the MDM4 SNP34091CC genotype (recessive model) to be significantly associated with reduced risk of breast cancer among patients carrying the SNP309GG genotype (OR = 0.41; 95% CI = 0.21–0.82; Table 2, Fig. 1B). Notably, when refining the OR estimates by removing individuals harboring the less frequent MDM2 SNP285C allele, which antagonizes SNP309G‐induced transcriptional enhancement 16, this negative association became slightly stronger (gender adjusted OR = 0.40; 95% CI = 0.19–0.85; Table S3).
Table 2.
MDM4 SNP34091 among MDM2 SNP309GG
| Cases/controls | Genotype | OR (95% CI) | P‐value | OR (95% CI) | P‐value | ||
|---|---|---|---|---|---|---|---|
| SNP34091 n (%) | SNP34091 | SNP34091 | |||||
| AA | AC | CC | CC versus AA+AC | CC+AC versus AA | |||
| Controls | 294 (58.6) | 167 (33.3) | 41 (8.2) | 1.00 | – | 1.00 | – |
| Women | 149 (58.7) | 77 (30.3) | 28 (11.0) | 1.00 | – | 1.00 | – |
| Men | 145 (58.5) | 90 (36.3) | 13 (5.2) | 1.00 | – | 1.00 | – |
| Colon cancera | 111 (59.7) | 60 (32.3) | 15 (8.1) | 1.02 (0.54–1.93) | 0.947 | 0.98 (0.69–1.39) | 0.903 |
| Womenb | 56 (59.6) | 30 (31.9) | 8 (8.5) | 0.82 (0.33–1.99) | 0.653 | 1.09 (0.65–1.83) | 0.745 |
| Menc | 55 (59.8) | 30 (32.6) | 7 (7.6) | 1.60 (0.59–4.34) | 0.356 | 1.03 (0.63–1.70) | 0.898 |
| Lung cancera | 100 (54.6) | 73 (39.9) | 10 (5.5) | 0.73 (0.35–1.54) | 0.413 | 1.18 (0.83–1.68) | 0.357 |
| Womenb | 38 (57.6) | 25 (37.9) | 3 (4.6) | 0.43 (0.12–1.52) | 0.190 | 1.17 (0.66–2.10) | 0.588 |
| Menc | 62 (53.0) | 48 (41.0) | 7 (6.0) | 1.13 (0.43–3.00) | 0.799 | 1.30 (0.82–2.05) | 0.264 |
| Prostate cancerc | 184 (53.5) | 143 (41.6) | 17 (4.9) | 0.99 (0.46–2.11) | 0.974 | 1.26 (0.90–1.77) | 0.176 |
| Breast cancerb | 160 (63.8) | 77 (30.7) | 14 (5.6) | 0.41 (0.21–0.82) | 0.012 | 0.75 (0.52–1.10) | 0.139 |
Sex and age adjusted (logistic regression).
Calculations with female controls only, age adjusted.
Calculations with male controls only, age adjusted.
In addition to assessing the effect of MDM4 SNP34091 within subgroups of MDM2 SNP309 genotypes, we also explored differences between all possible combinations of MDM4 SNP34091/MDM2 SNP309 genotypes. By doing so, we confirmed the MDM4 SNP34091CC/MDM2 SNP309 GG genotype to associated with reduced risk of breast cancer (OR = 0.47; 95% CI = 0.24–0.92, when compared with the highest risk genotype (MDM4 SNP34091AA/MDM2 SNP309GG), data not shown).
No effect on the risk of any of the other cancer forms with respect to MDM4 SNP34091 status within the different MDM2 genotypes, either when stratifying cancer of the colon according to tumors of the right or left side, or when excluding the nonsmokers in lung cancer, was recorded.
Discussion
In this study, we observed no association between MDM4 SNP34091 status and the risk for colon‐, prostate‐, or lung cancer, while a marginally significant association with reduced risk of breast cancer was observed. Our observation in breast cancer is similar to, but weaker than the observations of Liu and colleagues, who found the SNP34091 AC and CC genotypes to be significantly associated with reduced breast cancer risk compared with the AA genotype in two different Chinese populations 20. In contrast, GWAS have found an elevated OR for ER negative breast cancer related to the SNP34091C allele in Caucasians but not among Asians 23, 24, 25. Regrettably, information on receptor status was not available for the breast cancer patients examined in this study; thus, a potential effect of SNP43091 status in the minor group of patients harboring ER negative tumors may have been overlooked. Regarding prostate cancer risk, we observed a weak, non‐significant association between MDM4 SNP34091C and reduced risk in the dominant model. This is in line with data from the majority of individual datasets from European populations, included in a recent large GWAS 22, and mirrors our previous findings related to MDM2 polymorphisms [27, 28, 29].
This study is, to our knowledge, the first population‐based case–control study assessing the impact of MDM4 SNP34091 on cancer risk in lung‐ and colon cancer. Previous case–control studies assessing this variant in other cancer forms (esophageal squamous cell carcinoma and non‐Hodgkin lymphoma), including breast cancer, have found the SNP34091C allele to be associated with reduced risk, but have all been performed in Chinese populations 19, 20, 21.
Regarding the variations in the results between studies of different ethnic groups, notably, there is a large difference in the distribution of MDM4 SNP34091 between Europeans and Asians with a MAF of 0.26 and 0.05, respectively [30], possibly affecting the power of studies in Asian populations even though the numbers of patients included are large. Also, a possible explanation for the discrepancy may be yet unknown functional SNP(s) that are in linkage disequilibrium (LD) with SNP34091: There are examples of functional SNPs in LD where the SNPs have different geographical distributions and thus confer diverging risk estimates between Europeans and Asians, for example, the two MDM2 SNPs; SNP309 and SNP285 [30, 31, 32, 33]. On the other hand, the possibility of publication bias, where case–control studies reporting positive results are favored cannot be excluded.
After stratifying according to MDM2 SNP309 status, we found a reduced risk for breast cancer among individuals harboring the MDM4 SNP34091CC/MDM2 SNP309GG genotype, and this association was stronger after removing individuals harboring the MDM2 SNP285C allele, previously shown to antagonize SNP309G‐induced transcription elevation 16. Interestingly, the MDM4 SNP34091C allele, similar to SNP285G>C seems to execute their effects on breast cancer risk among individuals carrying the SNP309GG genotype only 16, 27.
In conclusion, we found no association between the MDM4 SNP34091 status and risk for lung‐, prostate‐, or colon cancer, and a weak association with breast cancer, applying the candidate gene approach. The latter finding was substantiated by the observation of a seemingly synergistic effect between the MDM2 SNP309GG and MDM4 SNP34091AA genotypes on increased risk for breast cancer.
Conflict of Interest
None declared.
Supporting information
Table S1. MDM4 SNP34091 distribution and left versus right colon cancer risk.
Table S2. MDM4 SNP34091 distribution and lung cancer risk in smokers.
Table S3. MDM4 SNP34091 among MDM2 SNP309GG without MDM2 SNP285C.
Acknowledgments
The authors thank Beryl Leirvaag for technical assistance. Most of this work was performed in the Mohn Cancer Research Laboratory. This study was supported by grants from the Bergen Research Foundation, the Norwegian Cancer Society's Pink Ribbon Campaign, the Norwegian Health Region West and the Norwegian Research Council.
Cancer Medicine 2015; 4(12): 1901–1907
References
- 1. Junttila, M. R. , and Evan G. I.. 2009. p53 ‐ a Jack of all trades but master of none. Nat. Rev. Cancer 9:821–829. [DOI] [PubMed] [Google Scholar]
- 2. Wu, X. , Bayle J. H., Olson D., and Levine A. J.. 1993. The p53‐mdm‐2 autoregulatory feedback loop. Genes Dev. 7:1126–1132. [DOI] [PubMed] [Google Scholar]
- 3. Haupt, Y. , Maya R., Kazaz A., and Oren M.. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. [DOI] [PubMed] [Google Scholar]
- 4. Honda, R. , Tanaka H., and Yasuda H.. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27. [DOI] [PubMed] [Google Scholar]
- 5. Kubbutat, M. H. G. , Jones S. N., and Vousden K. H.. 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303. [DOI] [PubMed] [Google Scholar]
- 6. Marine, J.‐C. , and Jochemsen A. G.. 2005. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun. 331:750–760. [DOI] [PubMed] [Google Scholar]
- 7. Linares, L. K. , Hengstermann A., Ciechanover A., Muller S., and Scheffner M.. 2003. HdmX stimulates Hdm2‐mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA 100:12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang, X. , Wang J., and Jiang X.. 2011. MdmX protein is essential for Mdm2 protein‐mediated p53 polyubiquitination. J. Biol. Chem. 286:23725–23734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawai, H. , Lopez‐Pajares V., Kim M. M., Wiederschain D., and Yuan Z. M.. 2007. RING domain‐mediated interaction is a requirement for MDM2′s E3 ligase activity. Cancer Res. 67:6026–6030. [DOI] [PubMed] [Google Scholar]
- 10. Riemenschneider, M. J. , Buschges R., Wolter M., Reifenberger J., Bostrom J., Kraus J. A., et al. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 59:6091–6096. [PubMed] [Google Scholar]
- 11. Riemenschneider, M. J. , Knobbe C. B., and Reifenberger G.. 2003. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int. J. Cancer 104:752–757. [DOI] [PubMed] [Google Scholar]
- 12. Danovi, D. , Meulmeester E., Pasini D., Migliorini D., Capra M., Frenk R., et al. 2004. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol. 24:5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han, X. , Garcia‐Manero G., McDonnell T. J., Lozano G., Medeiros L. J., Xiao L., et al. 2007. HDM4 (HDMX) is widely expressed in adult pre‐B acute lymphoblastic leukemia and is a potential therapeutic target. Mod. Pathol. 20:54–62. [DOI] [PubMed] [Google Scholar]
- 14. Xiong, S. , Pant V., Suh Y. A., Van Pelt C. S., Wang Y., Valentin‐Vega Y. A., et al. 2010. Spontaneous tumorigenesis in mice overexpressing the p53‐negative regulator Mdm4. Cancer Res. 70:7148–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bond, G. L. , Hu W., Bond E. E., Robins H., S. G. Lutzker , Arva N. C., et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119:591–602. [DOI] [PubMed] [Google Scholar]
- 16. Knappskog, S. , Bjornslett M., Myklebust L. M., Huijts P. E., Vreeswijk M. P., Edvardsen H., et al. 2011. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell 19:273–282. [DOI] [PubMed] [Google Scholar]
- 17. Stegeman, S. , Moya L., Selth L. A., Spurdle A. B., J. A. Clements , and Batra J.. 2015. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr. Relat. Cancer 22:265–276. [DOI] [PubMed] [Google Scholar]
- 18. Wynendaele, J. , Bohnke A., Leucci E., Nielsen S. J., Lambertz I., Hammer S., et al. 2010. An illegitimate microRNA target site within the 3 ‘UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 70:9641–9649. [DOI] [PubMed] [Google Scholar]
- 19. Fan, C. , Wei J., Yuan C., Wang X., Jiang C., Zhou C., et al. 2014. The functional TP53 rs1042522 and MDM4 rs4245739 genetic variants contribute to non‐Hodgkin lymphoma risk. PLoS ONE 9:e107047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu, J. , Tang X., Li M., Lu C., Shi J., Zhou L., et al. 2013. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res. Treat. 140:151–157. [DOI] [PubMed] [Google Scholar]
- 21. Zhou, L. , Zhang X., Li Z., Zhou C., Li M., Tang X., et al. 2013. Association of a genetic variation in a miR‐191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS ONE 8:e64331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eeles, R. A. , Olama A. A., Benlloch S., Saunders E. J., Leongamornlert D. A., Tymrakiewicz M., et al. 2013. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 45:385–391, 91e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia‐Closas, M. , Couch F. J., Lindstrom S., Michailidou K., Schmidt M. K., Brook M. N., et al. 2013. Genome‐wide association studies identify four ER negative‐specific breast cancer risk loci. Nat. Genet. 45:392–398, 8e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Purrington, K. S. , Slager S., Eccles D., Yannoukakos D., Fasching P. A., Miron P., et al. 2014. Genome‐wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple‐negative breast cancer. Carcinogenesis 35:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stevens, K. N. , Vachon C. M., and Couch F. J.. 2013. Genetic susceptibility to triple‐negative breast cancer. Cancer Res. 73:2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Naess, O. , Sogaard A. J., Arnesen E., Beckstrom A. C., Bjertness E., Engeland A., et al. 2008. Cohort profile: cohort of Norway (CONOR). Int. J. Epidemiol. 37:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gansmo, L. B. , Knappskog S., Romundstad P., Hveem K., Vatten L., and Lonning P. E.. 2015. Influence of MDM2 SNP309 and SNP285 status on the risk of cancer in the breast, prostate, lung and colon. Int. J. Cancer 137:96–103. [DOI] [PubMed] [Google Scholar]
- 28. Knappskog, S. , Trovik J., Marcickiewicz J., Tingulstad S., Staff A. C., et al. 2012. MoMaTEC study group, SNP285C modulates oestrogen receptor/Sp1 binding to the MDM2 promoter reduces risk of endometrial but not prostatic cancer. Eur. J. Cancer 48:1988–1996. [DOI] [PubMed] [Google Scholar]
- 29. Knappskog, S. , Gansmo L. B., Romundstad P., Bjørnslett M., Trovik J., Huijts P., et al. 2012. MDM2 promoter SNP344T>A (rs1196333) status does not affect cancer risk. Plos One 7:e36263;1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Genomes Project C , Abecasis, G. R. , Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., and McVean G. A.. 2012. An integrated map of genetic variation from 1092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Economopoulos, K. P. , and Sergentanis T. N.. 2010. Differential effects of MDM2 SNP309 polymorphism on breast cancer risk along with race: a meta‐analysis. Breast Cancer Res. Treat. 120:211–216. [DOI] [PubMed] [Google Scholar]
- 32. Hu, Z. , Jin G., Wang L., Chen F., Wang X., and Shen H.. 2007. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: evidence from 21 case‐control studies. Cancer Epidemiol. Biomark. Prev. 16:2717–2723. [DOI] [PubMed] [Google Scholar]
- 33. Knappskog, S. , Gansmo L. B., Dibirova K., Metspalu A., Cybulski C., Peterlongo P., et al. 2014. Population distribution and ancestry of the cancer protective MDM2 SNP285 (rs117039649). Oncotarget 5:8223–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. MDM4 SNP34091 distribution and left versus right colon cancer risk.
Table S2. MDM4 SNP34091 distribution and lung cancer risk in smokers.
Table S3. MDM4 SNP34091 among MDM2 SNP309GG without MDM2 SNP285C.


