Abstract
Quantitative characterization of membrane defects (pores) is important for elucidating molecular basis of many membrane active peptides. We study kinetic defects induced by melittin in vesicular and planar lipid bilayers. Fluorescence spectroscopy measurements indicate that melittin induces time-dependent calcein leakage. Solution atomic force microscopy (AFM) is used to visualize melittin induced membrane defects. After initial equilibration, the most probable defect radius is ~3.8 nm in 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) bilayers. Unexpectedly, defects become larger with longer incubation, accompanied by substantial shape transformation. The initial defect radius is ~4.7 nm in 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) bilayers. Addition of 30mol% cholesterol to DOPC bilayers suppresses defect kinetics, although the inhibitory impact is negated by longer incubation. Overall, the kinetic rate of defect development follows DLPC > DOPC > DOPC/cholesterol. Kinetic defects are also observed when anionic lipids are present. Based on the observation that defects can occupy as large as 40% of the bilayer surface, we propose a kinetic defect growth model. We also study the effect of melittin on the phase behavior of DOPC/egg-sphingomyelin/cholesterol bilayers. We find that melittin initially suppresses or eliminates liquid-ordered (Lo) domains; Lo domains gradually emerge and become the dominant species with longer incubation. Defects in phase coexisting bilayers have a most probable radius of ~5 nm, and are exclusively localized in the liquid-disordered (Ld) phase. Our experimental data highlight that melittin induced membrane defects are not static; conversely, spontaneous defect growth is intrinsically associated with membrane permeabilization exerted by melittin.
Graphical abstract
Introduction
It has been known for almost a half century1 that the 26-residue amphipathic peptide melittin has an extraordinary high affinity for lipid membranes2. Sequence analysis shows that four charged and two polar residues are sequestered at the C-terminal (residues 21–26), while the N-terminal is primarily composed of hydrophobic residues. Atomistic crystal structure3 reveals that melittin monomer contains two helices that are joined by a short Gly-Leu-Pro loop (Fig. S1, Supporting Information). The inside of the wedge-shaped monomer contains a stretch of hydrophobic residues (except the C-terminal), which are proposed to preferentially interact with lipid hydrocarbon chains. The wedge-shaped monomer can lie parallel to membrane surface by partially inserting into lipid backbone region, thus disrupting proximal lipid chain packing and allowing solutes to permeate through the membrane barrier (the “wedge” effect). Alternatively, melittin monomers can line up at the edge of lipid membranes and form transmembrane (TM) pores (the “edge” effect). The two mechanisms are not necessarily exclusive, but can take effect in different regimes depending on melittin concentration.
The large interest in melittin induced membrane disruption lies in the emerging health concern of infectious bacterial pathogens. By nonspecifically targeting and damaging bacterial membranes, small cationic antimicrobial peptides (AMPs) including melittin are thought as promising substitutes for conventional antibiotics. More recently, melittin has been shown to exhibit anti-inflammation and anticancer activities.4–5 As a well-studied prototype, melittin has been instrumental in elucidating the action mode of the AMP family. Consistent with the wedge and edge effects, melittin is generally thought to bind to membrane surface at low peptide concentration. Monomers with rapid in-plane motion6 can transiently translocate between opposing leaflets to reach symmetrical distribution, as well as to form transient pores. TM configuration becomes the preferential state when melittin concentration becomes large. Stable pores are formed by grouping a few TM monomers together. Based on ion conductivity7–8 and neutron in-plane scattering9 measurements, it was proposed that melittin pores are lined by a mixture of monomers and lipids, the so-called toroidal pore model. The transition from surface state to TM state was formulated into a two-phase model.10 The model predicts that above a peptide/lipid threshold, P/L*, TM pores are spontaneously formed. One limitation of the two-phase model is that it only addresses the behavior of static pores. Vesicle based studies have suggested that melittin pores might be more dynamic than previously thought.11–12 To explore the kinetic aspect of melittin pores, it is important to study pore structure as a function of time. In addition, the existence of nanometer-sized pores was primarily based on indirect measurements, such as neutron in-plane scattering and fluorescence spectroscopy. It will be advantageous to directly see these pores in a membrane environment. Currently, there is only one study that has directly observed pore-like structures in lipid monolayers induced by melittin.13 Since atomic force microscopy (AFM) in air was used in that study,13 the reported structures need to be verified or refuted by measurements performed in solution environment.
Cell membranes are crowded with proteins, including those forming ion channels and pores. It has been proposed that the interplay between proteins and lipids is important in governing membrane lateral organization.14 Since melittin is a prototype of TM pores, it will be interesting to study whether and how melittin modulates lipid membrane lateral structure. Indeed, AMPs have been shown to impart a profound effect on lipid membrane phase behavior.15–17 For example, by using lipid bilayers exhibiting liquid-ordered (Lo) and liquid-disordered (Ld) phase coexistence, it was found that the morphology and size of Lo domains could be modified by AMPs or alike peptides. In this paper we are interested in exploring (i) whether and how phase coexistence inhibits melittin pore formation, and (ii) the role of melittin in governing the heterogeneous organization of phase coexisting bilayers.
We use vesicular and planar lipid bilayers to study the kinetic behavior of melittin induced pores. We first use calcein leakage to study the disruptive effect of melittin on unilamellar vesicles (ULVs). Time-course measurements are used to reveal membrane permeabilization as a function of incubation time and melittin concentration. We next use solution AFM to visualize melittin induced defects (pores) in mica supported lipid bilayers. Note that we will use "defects" instead of "pores" to describe features observed in our solution AFM study. It has been shown that mica is a suitable substrate for studying membrane-associated phenomena. There is a ~2-nm-thick water layer separating the supported bilayer from the mica substrate.18–19 Water layer mediated decoupling gives rise to reasonable diffusion coefficients of lipids (~1 µm2/s) in mica supported bilayers.20 Mobility of peptides and lipids is important for our kinetic study. In particular, we visualize nanometer-sized defects in several types of lipid bilayers with different melittin concentrations. Unexpectedly, membrane defects are found to evolve as a function of incubation time. To explain the kinetic behavior of membrane defects, we propose a kinetic defect growth model, which extends the existing two-phase model by introducing a lipid removal process. In addition to single-phase bilayers, we also explore the kinetic effect of melittin on Lo+Ld phase coexisting bilayers.
Materials and Methods
1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol), egg-sphingomyelin (eSM), and cholesterol (Chol) are purchased as lyophilized powders from Avanti Polar Lipids (Alabaster, AL). Melittin purified from bee venom is purchased from Fisher Scientific (Waltham, MA).
Fluorescence spectroscopy
DOPC dry film is hydrated using 20 mM NaCl, 30 mM calcein, and 20 mM HEPES at pH 7.0 (buffer A). Freeze-thaw cycles between −80 and 50°C are carried out to ensure uniform distribution of calcein. Lipid dispersion is extruded using an Avanti mini-extruder and a filter with 100-nm-diameter pores. External calcein is removed by a gel filtration column (Superdex 200 10/300 GL). The elution buffer contains 108 mM NaCl and 20 mM HEPES at pH 7.0 (buffer B). The equality of the osmotic pressure between buffer A and buffer B is confirmed using a Wescore 5500 vapor pressure osmometer (Logan, Utah).
For kinetic leakage study, an aliquot of melittin stock solution (in buffer B) is diluted with buffer B to make a final volume of 980 µL. The solution is transferred to a 1-mL quartz cuvette placed in the sample holder of a Jasco FP-8300 fluorometer (Easton, MD). A small magnetic rod is used to stir the solution (300 rpm). Right before each measurement, 20 µL of calcein-enclosed DOPC ULVs is pipetted into the cuvette. Time-course fluorescence intensity is collected every 1 sec. The excitation and emission wavelength is 494 and 514 nm, respectively. Based on the time-course spectrum, the initial mixing time for ULV and melittin solution in the cuvette is ~10 s. The mixing time becomes much longer without magnetic stirring. To obtain the maximum fluorescence intensity (Imax) when complete leakage is reached, 100 µL of 100 mM Triton X-100 (in buffer B) is added to the reacted ULV/melittin mixture (dilution is taken into account). The percentage of fluorescence enhancement (FE) is defined as: FE=(I(t)−I0)/(Imax−I0) × 100%, where I0 is the fluorescence intensity of the ULV solution without melittin.
Solution AFM
Unless noted otherwise, all concentrations, ratios, fractions, or percentages are molar based. Lipid mixtures are prepared by mixing appropriate ratios of stock solutions in glass test tubes. Organic solvents are removed by a gentle stream of argon gas using a 12-position N-EVAP evaprator (Organomation Associates, Inc., Berlin, MA), and then pumped under vacuum for > 2 h. Lipid dry films are hydrated by 10 mM HEPES at pH 7.0 and appropriate volume of melittin stock solution (in the same HEPES buffer). Small unilamellar vesicles (SUVs) are produced by ultrasonicating lipid/peptide suspension using a Sonic Dismembrator operated at ~40 W with total duration of 10 min. The obtained SUVs are centrifuged before planar bilayer preparation.
Solution AFM measurements (at room temperature) are performed using a liquid-compatible Multimode 8 AFM and a Nanoscope V controller (Bruker, Santa Barbara, CA). Experimental procedure has been described elsewhere.21–23 Briefly, mica supported planar bilayers are formed by injecting SUVs into the AFM liquid cell using a syringe pump. Once a complete bilayer is formed (~20 min), 10 mM HEPES at pH 7.0 is injected to remove excess SUVs. To test whether and how melittin disrupts preformed bilayers, a modified procedure is carried out. Specifically, pure lipid SUVs (without melittin) are injected into the AFM liquid cell to form a bilayer. After flushing with HEPES buffer (to remove excess SUVs), melittin dissolved in HEPES buffer (e.g., 1 µM) is injected and remains in the liquid cell for the rest of the AFM measurement. For both procedures, kinetic bilayer disruption is monitored by in situ AFM scans.
PeakForce quantitative nanomechanic (QNM) mode coupled with a silicon nitride probe (Bruker model: ScanAsyst-Fluid+) is used for bilayer scanning. The peak force is set to ~300 pN. Square images are acquired at a scan rate of 1.0 Hz. The typical scanning time for one image is ~5 min. For kinetic study, images are acquired at different time points after initial SUV injection. To eliminate potential influence of repetitive scanning on bilayer structure, AFM tip is withdrawn and moved to a different location after one or a few scans with different magnifications. AFM images are leveled by subtracting linear backgrounds and analyzed using Image Processing Toolbox of Matlab (The MathWorks, Inc., Natick, Massachusetts).
Results
Calcein leakage induced by melittin
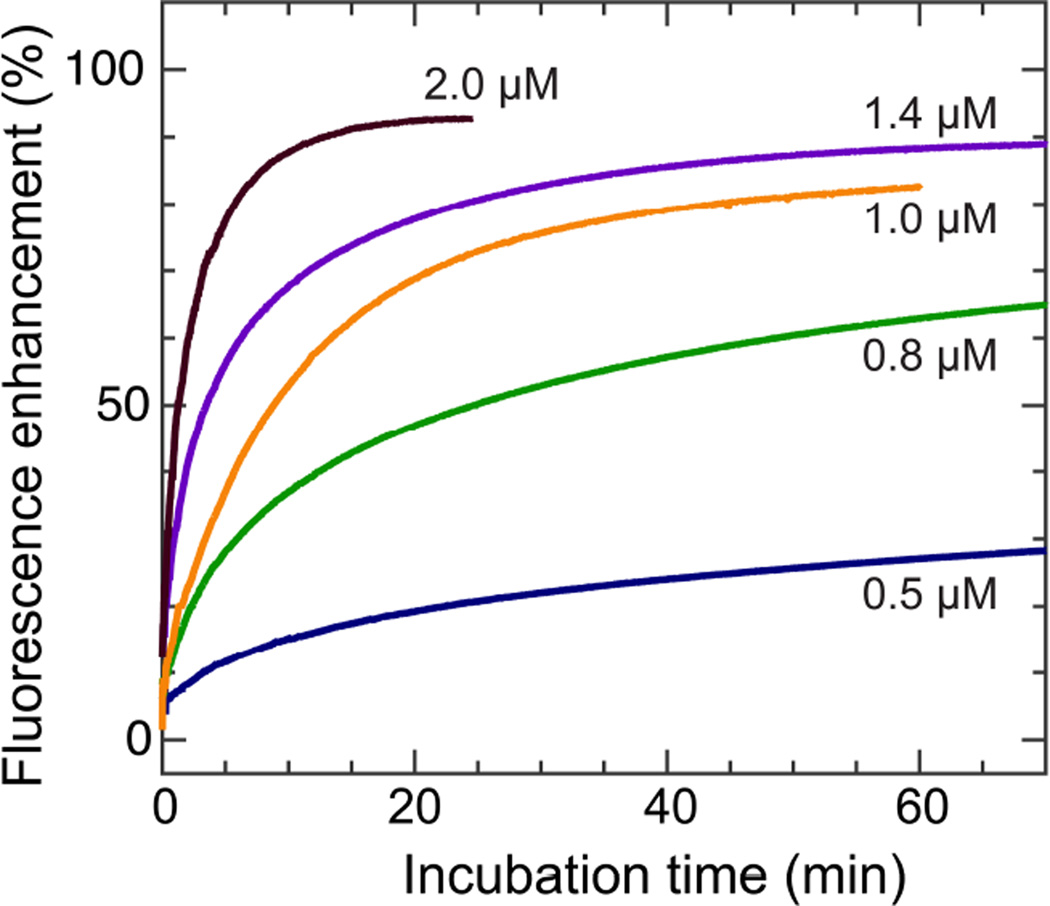
Formation of membrane defects (pores) can be monitored by influx or efflux of fluorescent markers that are initially compartmentalized at the exterior or interior of membrane vesicles. We use time-dependent calcein leakage from DOPC ULVs to study the membrane disruptive effect of melittin. Calcein-encapsulated ULVs are exposed to different concentrations of melittin. Note that DOPC lipid concentration is ~0.1 mM for all samples based on phosphorus assay of ULV stock solution.18 By monitoring fluorescence intensity as a function of incubation time, the kinetic leakage behavior is obtained (Fig. 1). We use an exponential expression to fit the fluorescence enhancement data, FE=FEmax(1−exp(−(t+t0)/τ)), where FEmax is the maximum fluorescence enhancement, t0 is a time shift due to mixing, and τ is the exponential time constant. The resulting time constant is 23, 18, 12, 9, and 3 min for melittin concentration of 0.5, 0.8, 1.0, 1.4, and 2.0 µM, respectively (Fig. S2, Supporting Information). The inverse relationship between the time constant and the peptide concentration indicates that the efficiency of membrane disruption increases with melittin concentration.
Figure 1.
Time-course measurements of calcein leakage induced by melittin at different concentrations.
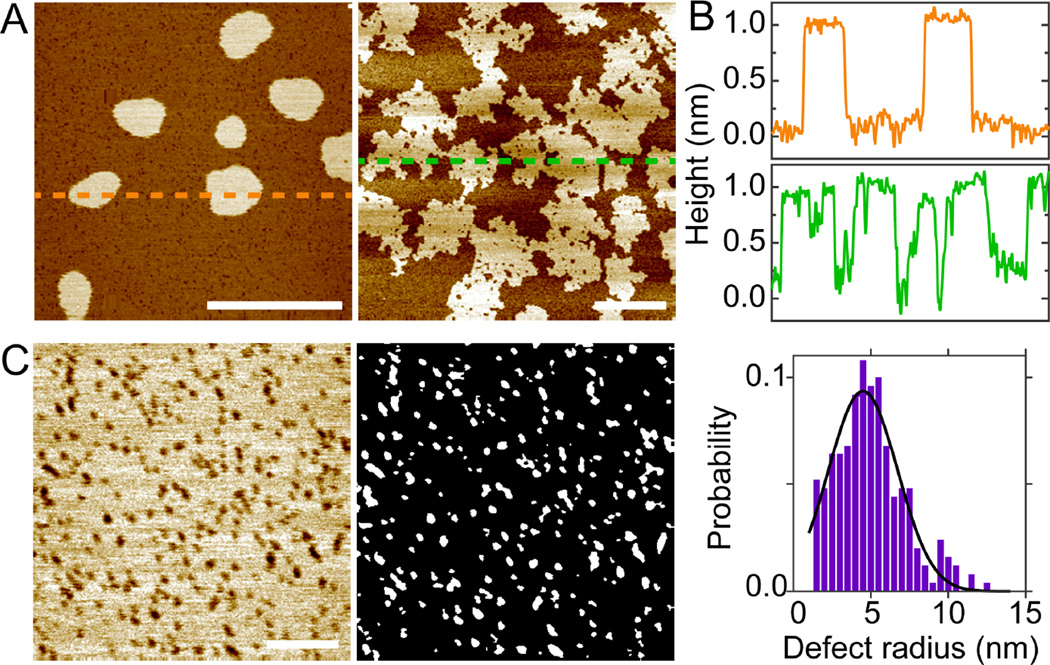
Melittin induced defects in DLPC bilayers
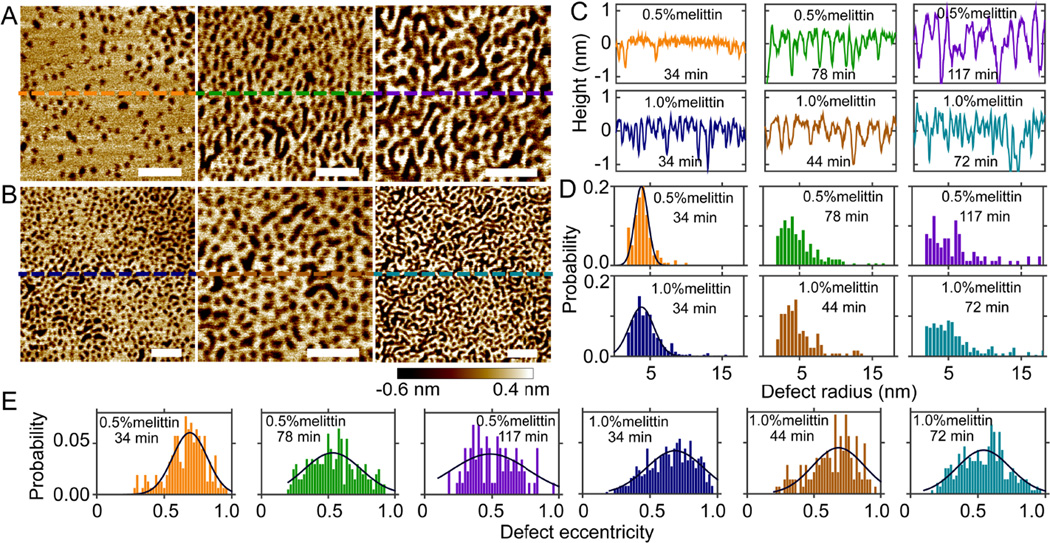
We perform solution AFM measurements to visualize melittin induced defects in mica supported planar bilayers. (We use "defects" instead of "pores" to describe the features observed in our solution AFM study.) Height images of DLPC bilayers doped with 0.5 and 1.0mol% melittin are shown in Fig. 2. A time stamp is used in each image to notate the incubation time of the bilayer in the AFM liquid cell (from SUV injection to the time when the image is acquired). At both peptide concentrations, round defects are seen after initial incubation of 34 min (Fig. 2A and 2B). Longer incubation results in curvilinear defects. To examine the depth of defects, height profiles along horizontal lines crossing the center of each bilayer are displayed (Fig. 2A and 2B). The apparent defect depth is ~1 nm after initial equilibration; the apparent depth increases to ~2 nm at 117 min for DLPC+0.5mol% melittin bilayer. Compared to the known thickness of DLPC bilayers (~3 nm),24 melittin induced defects do not seem to penetrate the entire bilayer. This is not consistent with the contemporary notion that melittin forms TM pores (defects). The discrepancy is likely due to the finite size of AFM tips. When defect width is comparable to the size of the AFM tip, the tip cannot go all the way down to the bottom of the defect. As the defect width becomes larger, the apparent depth becomes more accurate compared to the actual depth. This trend is illustrated by comparing DLPC+0.5mol% melittin bilayer at 34 and 117 min, i.e., the apparent depth increases from ~1 to ~2 nm as the defect width increases (Fig. 2C).
Figure 2.
Solution AFM height images of DLPC bilayers doped with 0.5mol% (A) and 1.0mol% (B) melittin. Scale bars = 50 nm. From left to right, the incubation time is 34, 78, and 117 min in (A), and 34, 44, and 72 min in (B). (C) Height profiles along dashed lines indicated in (A) and (B). (D) Normalized probabilities of defect radius. (E) Normalized probabilities of defect eccentricity. Solid lines (D and E) are Gaussian fits.
We next perform image analysis to quantitatively characterize defect size and shape. Proper height thresholds are used to distinguish defects from the rest of the bilayer. (Pixels with heights smaller than the threshold are considered as defects.) To account for pixels with occasional small heights (i.e., noise), a cutoff is used to remove artificial defects (e.g., regions with areas smaller than 7 nm2 are not considered as defects). The resulting defects (i.e., bright regions in the binary image) are shown in Fig. S3 (Supporting Information). Defect area is determined by the total pixels (scaled by pixel size) occupied by each defect. The area fraction covered by defects is 0.15, 0.34, and 0.39 for DLPC+0.5mol% melittin bilayer at 34, 78, and 117 min, respectively; and 0.32, 0.36, and 0.40 for DLPC+1.0mol% melittin bilayer at 34, 44, and 72 min, respectively. These values indicate that larger area of DLPC/melittin bilayer is perforated with longer incubation. Since defect (pore) radius is more commonly used in the literature, we convert defect area into defect radius by assuming that defects are circularly shaped. To account for non-circular shapes, we use a parameter referred to as defect eccentricity. This is done by identifying an equivalent ellipse that has the same second-moments as the defect (Matlab function: regionprops). Defect eccentricity is defined by the ratio of the minor and major axis of the ellipse. Qualitatively, defect eccentricity of one corresponds to a circular defect, while defect eccentricity of zero corresponds to a line.
Normalized probabilities of defect radius and defect eccentricity are shown in Fig. 2D and 2E. The most probably defect radius (based on Gaussian curve fitting) is 3.8 nm at both peptide concentrations after 34-min incubation. The probability of observing ~4 nm sized defects decreases with longer incubation. This is paralleled by enhanced probabilities of larger defect radii. For shape transformation, the most probable defect eccentricity is ~0.7 at 34 min; the most probable defect eccentricity shifts to ~0.5 at later time points. Based on the definition of defect eccentricity, it is clear that defects become elongated with longer incubation. We note that defect eccentricity is not linearly related to the actual shape. This explains the moderate decrease of defect eccentricity while defect shape seems to change remarkably (Fig. 2A and 2B).
Melittin induced defects in DOPC bilayers
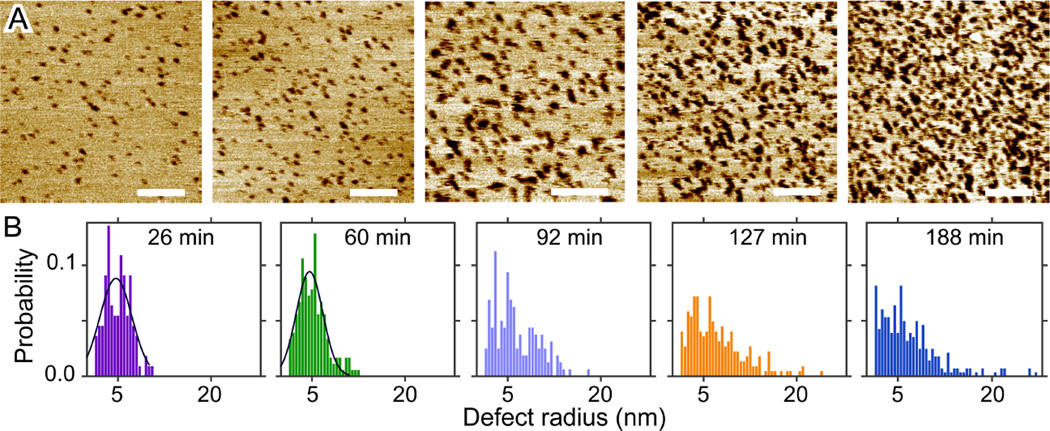
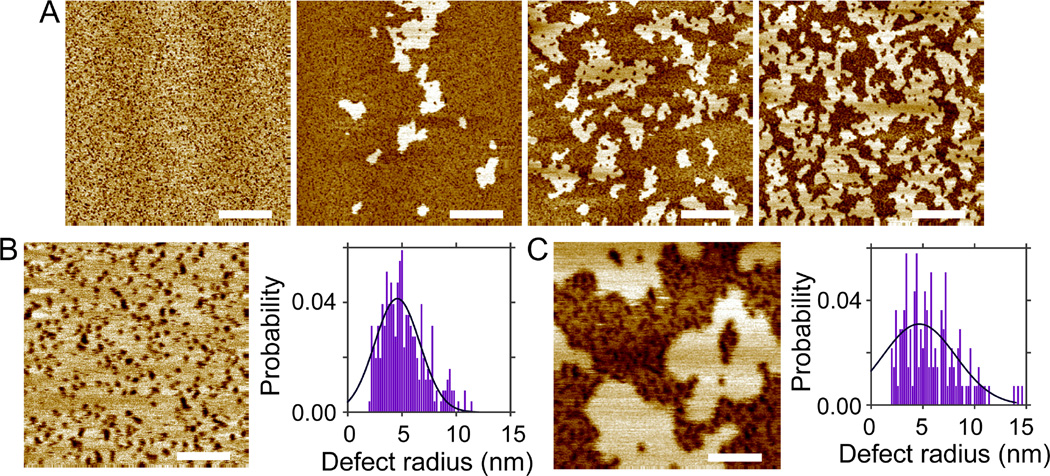
DOPC bilayers were reported to exhibit a peptide/lipid threshold of ~1/100, above which melittin induced defects were detected.25 The existence of a threshold is recapitulated by our solution AFM measurements. At 1.0mol% melittin, only a few defects are observed with incubation time up to 141 min (Fig. S4, Supporting Information). Increasing melittin concentration to 2.0mol% results in many defects immediately after initial incubation; defects become more densely packed with longer incubation (Fig. 3). Height profiles crossing bilayer surfaces (Fig. S5, Supporting Information) show similar apparent defect depths as in DLPC bilayers. The same image analysis procedure is used to extract defects from the rest of the bilayer. The total area fraction of defects is found to increase from 0.04 to 0.25 as the incubation time increases from 26 to 188 min (Fig. S5, Supporting Information). The most probable defect radius is ~4.7 nm for incubation time ≤ 60 min; the probability of observing larger defects increases with incubation time (Fig. 3). Interestingly, no curvilinear defects are observed even after ~3 h incubation. This is supported by little change of the eccentricity distribution as a function of incubation time (Fig. S5, Supporting Information).
Figure 3.
(A) Solution AFM height images of the DOPC+2.0mol% melittin bilayer. From left to right, the incubation time is 26, 60, 92, 127, and 188 min. Scale bars = 100 nm. (B) Normalized probabilities of defect radius. Solid lines are Gaussian fits. The most probable defect radius is 4.7 and 4.6 nm for incubation time of 26 and 60 min, respectively.
For both DLPC and DOPC bilayers, defects are observed by mixing melittin with lipids before bilayer formation. Since AFM based studies have reported that (i) pore-like structures were not detected16 or (ii) bilayers underwent large-scale dissolution16, 26–27 when melittin was added to preformed bilayers, it is interesting to explore whether lipid-melittin premixing is a prerequisite for defect formation. We test this hypothesis by injecting 1.0 µM melittin into the AFM liquid cell with a preformed DOPC bilayer. The process is very similar to calcein leakage experiment where melittin is introduced to the exterior of lipid vesicles. In accord with calcein leakage data, 1.0 µM melittin is able to induce defects in mica supported DOPC bilayers as a function of incubation time (Fig. S6, Supporting Information). Image analysis reveals that the most probable defect radius is 4.8 nm after 79 min incubation; the number of defects, as well as the defect size, increases with incubation time. We also tested melittin concentrations ≥ 5.0 µM. Consistent with previous reports16, 26–27, the majority of bilayers are immediately dissolved after a few minutes of incubation. Altogether, our data indicate that kinetic defects can be obtained by incubating low concentrations of melittin (e.g., a few µM) with preformed lipid bilayers, while larger concentrations of melittin result in fast bilayer dissolution.
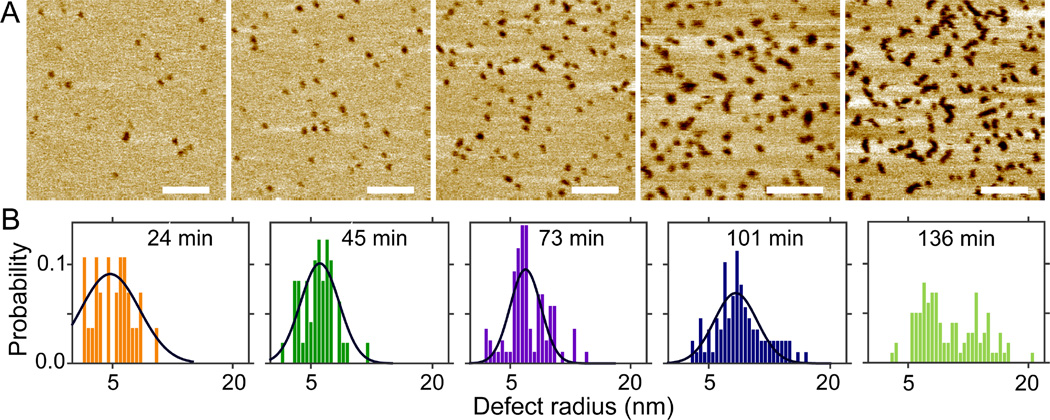
Melittion induced defects in DOPC/cholesterol bilayers
By modulating lipid membrane structural and mechanical properties, cholesterol was reported to change membrane response to melittin disruption.28–29 We study cholesterol effect by adding 30mol% cholesterol to DOPC+2.0mol% melittin bilayer. Solution AFM height images are shown in Fig. 4. Sporadic defects are observed after 24 min incubation; the number of defects and defect size increase with incubation time. Height profiles reveal similar apparent defect depths compared to those when no cholesterol is added (Fig. S7, Supporting Information). The total area fraction of defects is determined to be 0.01, 0.03, 0.07, 0.16, and 0.17 for incubation time of 24, 45, 73, 101, and 136 min, respectively. Defect growth is illustrated by the probability distribution of defect radius (Fig. 4B). Gaussian curve fitting indicates that the most probable radius is 4.7, 6.1, 6.9, and 8.3 nm for incubation time of 24, 45, 73, and 101 min, respectively. Defect shape transition is reflected by the change of the eccentricity distribution (Fig. S7, Supporting Information). Collectively, melittin induced defects with similar sizes are observed in DOPC and DOPC/cholesterol bilayers. One noticeable difference is the number of defects as a function of incubation time. For example, after incubation of ~25 min, the number (and the area fraction) of defects is larger in DOPC bilayer than in DOPC/cholesterol bilayer (Figs. 3 and 4). Similar difference is observed after incubation of ~2 h. It seems that cholesterol acts by suppressing the kinetics of defect development, although the inhibitory effect is negated by longer incubation.
Figure 4.
(A) Solution AFM height images of the DOPC+2.0mol% melittin bilayer doped with 30mol% Chol. From left to right, the incubation time is 24, 45, 73, 101, and 136 min. Scale bars = 100 nm. (B) Normalized probabilities of defect radius. Solid lines are Gaussian fits.
Melittin induced defects in phase coexisting bilayers
In addition to single-phase bilayers, we are interested in elucidating the interplay of melittin induced defects and the lateral organization of phase coexisting bilayers. We choose the well-studied DOPC/eSM/Chol as the model system.21, 30–31 The heterogeneous structure of DOPC/eSM/Chol 0.48/0.32/0.20 is shown in Fig. S8 (Supporting Information). Round domains (average radius of ~230 nm) corresponding to the Lo phase are surrounded by the Ld bulk phase. The height contrast between the coexisting phases is ~1 nm, and the total area fraction of the Lo phase is ~0.27. Addition of 1.2mol% melittin to the heterogeneous bilayer yields smaller Lo domains after 38 min incubation (Fig. 5). Specifically, the average radius of Lo domains decreases to ~120 nm, and the area fraction of the Lo phase decreases to 0.11. A marked change is observed after incubating the bilayer for 142 min. The total area fraction of the Lo phase increases to 0.47 (Fig. 5A). Moreover, Lo domains have irregular boundaries and the interiors of Lo domains are perforated by Ld domains. Despite these changes, the height contrast between the coexisting phases remains at ~1 nm. Close examination reveals that melittin induced defects are exclusively localized in the Ld phase. We choose a bilayer patch in the Ld phase (incubation of 66 min) to analyze defect structure (Fig. 5C). Image analysis indicates that the most probable defect radius is 4.5 nm. This value is similar to those obtained from DOPC-containing single-phase bilayers (Figs. 3 and 4).
Figure 5.
(A) Solution AFM height images of the DOPC/eSM/Chol 0.48/0.32/0.20 bilayer doped with 1.2mol% melittin. From left to right, the incubation time is 38 and 142 min. Scale bars = 800 nm. (B) Height profiles along dashed lines highlighted in (A). (C) Defects in the Ld-phase bilayer (incubation of 66 min). Scale bar = 100 nm. Solid line is Gaussian fit.
Increasing melittin to 2.0mol% reveals a more dramatic transition (Fig. 6). After incubation of 47 min, no Lo domains are observed; the bilayer only contains Ld phase with uniformly dispersed defects. Lo domains gradually emerge with longer incubation; the area fraction of Lo phase increases to ~0.38 at 140 min. Similar to the ternary bilayer with 1.2mol% melittin (Fig. 5), defects are sequestered in the Ld phase; the newly formed Lo domains have irregular boundaries and are perforated by Ld domains; the height contrast between the coexisting phases is ~1 nm (Fig. S9, Supporting Information). Using a bilayer patch in the Ld phase (incubation of 56 min), we find that the most probable defect radius is 4.5 nm (Fig. 6B). Image analysis of defects in the Lo+Ld phase coexisting bilayer is problematic due to the height contrast between the coexisting phases. To circumvent the difficulty, we select several regions corresponding to the Ld phase (Fig. S9, Supporting Information). Defects in these regions are then determined using the same image analysis method. Compared to the bilayer at 56 min, defects become slightly larger at 129 min (Fig. 6B and 6C). Defect elongation is also seen by comparing the eccentricity distribution (Fig. S9, Supporting Information).
Figure 6.
(A) Solution AFM height images of the DOPC/eSM/Chol 0.48/0.32/0.20 bilayer doped with 2.0mol% melittin. From left to right, the incubation time is 47, 95, 117, and 140 min. Scale bars = 400 nm. (B) Defects in the Ld-phase bilayer (incubation of 56 min). Scale bar = 100 nm. (C) Defects in the Ld-phase region of the Ld+Lo phase coexisting bilayer (incubation of 129 min). Scale bar = 100 nm. Solid lines (B and C) are Gaussian fits.
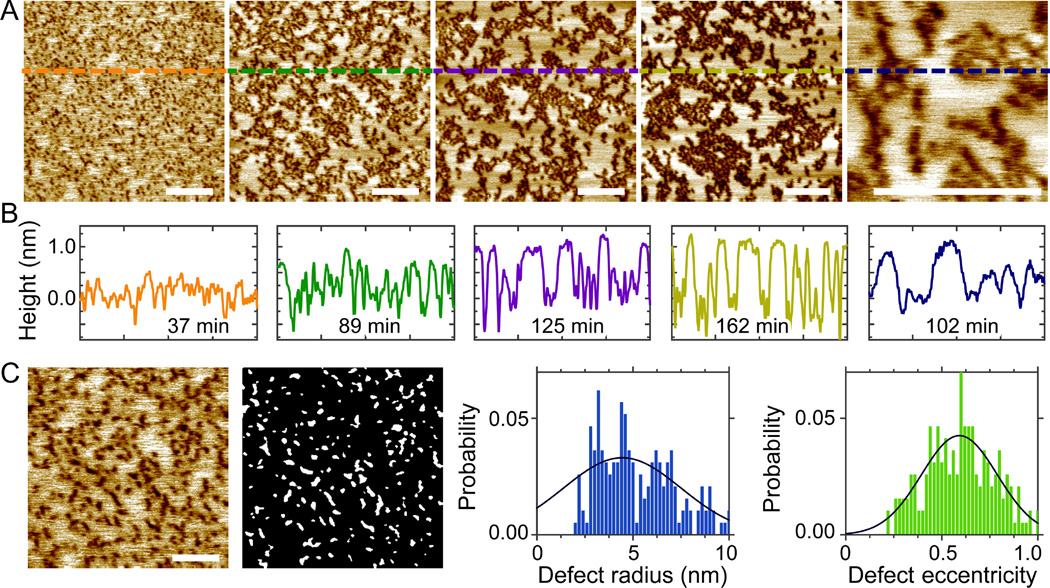
To explore whether the retardation effect of cholesterol on defect kinetics inferred from single-phase bilayers also play a role in phase coexisting bilayers, we fix the ratio of DOPC/eSM to 3:2 while increasing cholesterol concentration to 32mol% (DOPC/eSM/Chol 0.41/0.27/0.32). The topographic image is shown in Fig. S10 (Supporting Information). Height profiles reveal that the height contrast between coexisting phases is ~0.5 nm. The small height contrast reduces line tension at domain edges,32 thus yielding non-circularly shaped Lo domains. Such a transition has been reported before.21, 30 In addition to domain shape and height contrast, the area fraction of the Lo phase becomes 0.37 at 32mol% Chol.
Addition of 2.0mol% melittin results in time-dependent phase remodeling (Fig. 7). Small height contrast (~0.3 nm) between coexisting phases is observed after 37 min incubation. Defects are dispersed in regions with smaller height (i.e., the Ld phase). The height contrast between the coexisting phases gradually increases to ~0.5, ~0.9, and ~1.0 nm at 89, 125, and 162 min, respectively (Fig. 7B). The progressively increasing height contrast indicates that melittin is able to induce lipid redistribution between coexisting phases. The largest increase of the Lo phase content occurs between 37 and 89 min; the area fraction of the Lo phase remains more or less constant (~0.5) after 89 min. Defect structure is determined for the bilayer at 51 min (Fig. 7C). Gaussian curve fitting reveals that the most probable radius and eccentricity are 4.5 nm and 0.59, respectively. For the bilayer with longer incubation, the peculiar heterogeneity renders quantitative defect analysis difficult. Nevertheless, close inspection indicates that defects preferentially partition into the Ld phase; defect size does not increase significantly with incubation time. Together, increasing cholesterol content from 20 to 32mol% in the ternary bilayer system results in retarded response of height contrast between coexisting phases, whereas the overall impact of melittin on phase restructuring remains similar, including irregular Lo domain boundaries and spontaneous increase of the Lo phase content.
Figure 7.
(A) Solution AFM height images of the DOPC/eSM/Chol 0.41/0.27/0.32 bilayer doped with 2.0mol% melittin. From left to right, the incubation time is 37, 89, 125, 162, and 102 min. Scale bars = 200 nm. (B) Height profiles along dashed lines highlighted in (A). (C) Defects in the ternary bilayer incubated for 51 min. Scale bar = 100 nm. The gray-scale image shows defects determined by image analysis. Solid lines are Gaussian fits.
Melittin induced defects in anionic lipid-containing bilayers
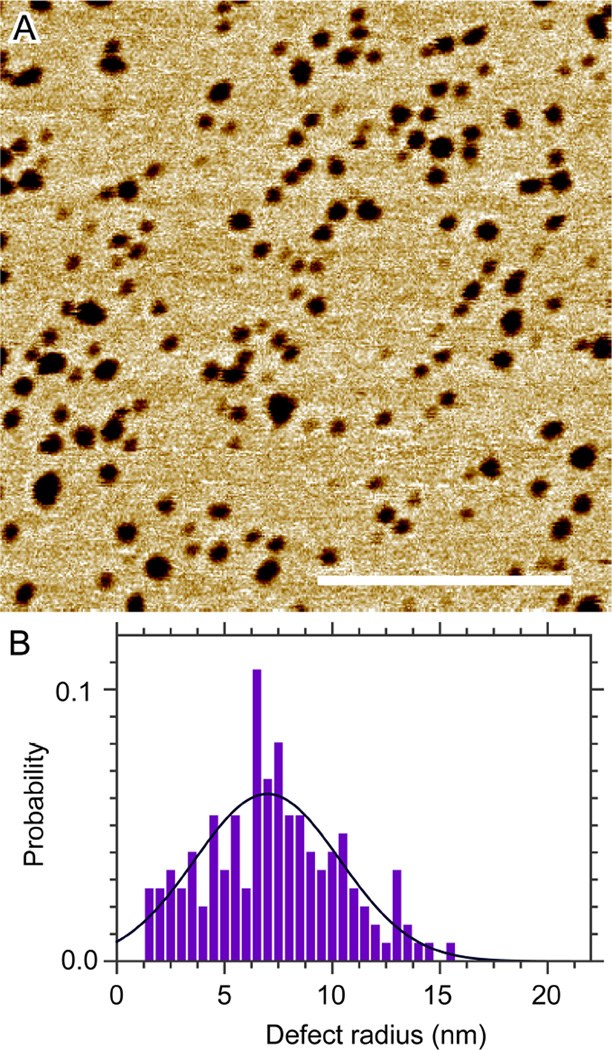
Anionic lipids were proposed as the driving force of membrane selectivity for many cationic AMPs against eukaryotic and bacterial membranes.33–35 In addition, the degree of membrane disruption induced by AMPs was found to be dependent on the charge state of lipids.36–38 Compared to zwitterionic lipids, it has been reported that cationic lipids not only reduced vesicle permeabilization caused by melittin, but also exhibited no apparent selectivity against vesicle-encapsulated fluorescence markers with different sizes.39 We use a binary lipid mixture, POPC+10mol% POPG, to explore the effect of anionic lipid POPG on melittin induced defects. After bilayer formation and removal of excess SUVs, 0.7 µM melittin is injected into the AFM liquid cell. Bilayer defects are detected after initial equilibration. An example is shown in Fig. 8. Image analysis indicates that the most probable defect radius is 7.0 nm after equilibrating for ~2 h. This value is similar to the defect size of DOPC bilayers after being exposed to 1.0 µM melittin for similar time (Fig. S6, Supporting Information). It seems that for peptide concentrations used in this study, melittin is able to bind to lipid bilayers and induce TM defects, regardless of the bilayers' charge state. This could be due to the high affinity of melittin to lipid membrane environment.
Figure 8.
(A) Solution AFM height image of the POPC+10mol% POPG bilayer after being exposed to 0.7 µM melittin for ~2 h. Scale bar = 200 nm. (B) Defect radius distribution. Gaussian curve fitting (solid line) indicates that the most probable defect radius is 7.0 nm.
Discussion
Calcein leakage
We use calcein leakage to monitor membrane perturbation imparted by melittin. Note that efflux of calcein cannot distinguish the wedge or edge effect, but rather provides a collective assessment of membrane disruption as a function peptide concentration and/or incubation time. Another fact that is often neglected is that fluorescence intensity is not linearly related to calcein concentration. Therefore, the obtained fluorescence enhancement data cannot be directly translated into the actual amount of leaked calcein (or the number of membrane defects). Nevertheless, leakage of fluorescent markers has been broadly used to explore membrane permeabilization induced by melittin.12, 28, 40–45 In addition to vesicle leakage, aspiration of micron-sized giant unilamellar vesicles has also been used to study the response of a single vesicle after being exposed to melittin.46–48 Our time-course measurements indicate that the exponential time constant derived from fluorescence enhancement data is inversely related to melittin concentration. Membrane permeabilization takes place over a time scale of minutes to hours depending on melittin concentration. This result ensures the feasibility of AFM based kinetic study since it normally requires a few minutes to acquire one AFM image.
Defect size
Nanometer-sized defects in mica supported planar bilayers are visualized using solution AFM. Similar defects have been observed before.49 Since defect size governs the largest molecule that can permeate through the membrane, it is interesting to quantitatively compare our results to literature values. Neutron in-plane scattering measurement suggested an inside radius of 2.2 nm in DLPC and POPC bilayers.9 By co-encapsulating fluorescent markers, an inside defect radius of 1.3–1.5 nm was inferred.50 Using solution AFM, we find that after ~0.5 h incubation, the most probable defect radius is 3.8 nm in DLPC bilayers, and 4.7 nm in DOPC bilayers. Our values are larger than previous estimations. Several sources could contribute to the discrepancy. For example, neutron scattering relies on the assumption of a uniform water column residing at defect lumen, whereas vesicle leakage is based on size estimation of fluorescent markers. Defect radius obtained from our AFM study corresponds to defect opening at the distal leaflet (relative to the substrate) of the supported bilayer. Considering the wedge-like structure of melittin monomers, it is possible that defects might become narrower near the bilayer center. In addition, our size estimation is affected by the shape of AFM tips and the step size used during AFM scan (i.e., pixel size). Most of our analyses are performed using pixel sizes of 0.8–2 nm. This gives rise to an estimated upper boundary of 2 nm uncertainty for the reported defect radius. Finally, based on our observation that defects can continuously grow, size estimation is complicated by when measurements are performed relative to bilayer formation. Other factors such as peptide concentration and water content may also play a role.51
Defect kinetics
The most remarkable result from our solution AFM study is the observation that melittin induced defects can continuously grow – through defect enlargement, merging, or generation of new defects. Note that although we do not monitor the growth of individual defects, the broadening of defect radius distributions clearly shows that at least some defects become larger with longer incubation. Such a conclusion is also supported by directly examining AFM height images. Defect growth requires more materials to line defect edges. Since the amount of melittin monomers do not increase, defect growth can only be achieved by recruiting more lipids to defect edges. The picture that defects are lined by TM peptides and lipids fits the proposed toroidal pore model.9 In addition to defect size, Table S1 (Supporting Information) shows that the total area fraction covered by defects (empty space) also increases with incubation time. Take the DLPC+0.5mol% melittin bilayer for example, the empty space occupies 40% of the total bilayer surface after ~2 h incubation. If no material is lost, 40% reduction of DLPC lipid area is required (0.5mol% melittin contributes little to the total bilayer area). This obviously is not physical, considering the minor impact of melittin on lipid area.52 In fact, studies have suggested a disordering effect of melittin on lipid chain packing.53 Then the only reasonable explanation that can account for the significantly increased defect area fraction is the removal of lipids from the bilayer environment, probably through micellization process (some peptides might also be removed from the bilayer). Our hypothesis of melittin facilitated lipid removal is supported by rapid dissolution of solid supported bilayers after being exposed to higher concentrations of melittin.27
Kinetic defect growth model
We propose a kinetic defect growth model to describe our data. The model encompasses following processes: (i) depending on lipid bilayer properties and peptide concentration, surface-binding melittin monomers can reorient and take TM configuration; (ii) defects are nucleated by lining a few TM monomers and lipid headgroups at defect edges (i.e., toroidal pore model); (iii) compared to lipids in the bulk phase, defect lining lipids have lower free energy barrier ΔG of being transferred to the aqueous phase. Removal of metastable defect lining lipids prompts in-plane movement of melittin monomers (away from defect edges); (iv) water exposure of hydrophobic lipid chains in the vicinity of defect edges results in reorientation of nearby lipid headgroups to line the enlarged defect. Steps (iii) and (iv) repeat as defect grows.
Using X-ray diffraction and oriented circular dichroism measurements, Huang and coworkers reported that melittin (and several other AMPs) forms stable defects only when the peptide/lipid ratio is above a threshold.10, 54 Depending on lipid composition, the threshold can be infinitesimally small or as large as 1/30.25, 55 To account for the disparate thresholds, a two-phase model – originally referred to as the two-state model – was proposed that defects spontaneously form when the chemical potential per monomer is the same between the surface-binding and the TM phase. Different thresholds were attributed to lipid dependent parameters describing the chemical potential in each phase (or state).10 One assumption made by the two-phase model is that defect size does not vary once formed. This obviously is not compatible with the kinetics observed in our study. Our proposed kinetic defect growth model extends the two-phase model by introducing a third process that defect lining lipids can be removed from the bilayer. Note that the first two steps in our model follow similar principle as the two-phase model. Therefore, experimental observations such as the existence of peptide/lipid threshold25, 55 and membrane dependent defect formation28 are compatible with our model.
Our model predicts that defect growth is accomplished by lipid removal from the membrane. This implies that defect growth rate will be influenced by the energy barrier ΔG, which is required to transfer defect-lining lipids to the aqueous phase. Our AFM study of single-phase lipid bilayers indicate that defect growth rate is in the order of DLPC > DOPC > DOPC/cholesterol (Figs. 2–4). Such a trend can be reconciled by considering that (i) short chain lipid DLPC has the largest tendency of dissociating from defect edges, and (ii) addition of cholesterol modifies lipid properties (e.g., hydrocarbon chain order), thus augmenting the energy barrier of removing DOPC lipids from defect edges.
Lipid removal from the bilayer environment by melittin resembles the action mode of the nonionic detergent Triton X-100.56 Figure S11 (Supporting Information) shows an example of lipid dissolution for a DOPC bilayer after incubating with 0.5 mM Triton X-100 for ~50 min. TM defects with irregular edges are observed. Compared to the defects induced by melittin, the defects induced by Triton X-100 exhibit larger heterogeneity in terms of size and shape. The difference could be due to different degrees of cooperativity when extracting lipids from defect edges. Nevertheless, the similarity between the two types of defects supports our proposition that melittin induces kinetic defects by extracting lipids from defect edges to the aqueous phase.
Membrane phase behavior modulated by melittin induced defects
Lateral heterogeneity is important for various membrane-associated activities. Despite large interest, the molecular basis governing membrane lateral organization is unclear. For a lipid bilayer exhibiting Lo+Ld phase coexistence, the content of Lo (or Ld) phase is determined by the overall lipid composition (i.e., phase content is determined by the coordinate in a triangular Gibbs phase diagram). Our kinetic defect growth model suggests that (i) defects are formed by recruiting lipids to defect edges, and (ii) defect growth is accomplished by lipid removal. It is conceivable that depending on lipid chemical composition, certain lipids will have a larger tendency of being attracted to defect edges and/or being removed from the bilayer. Lipid redistribution will alter the overall lipid composition in defect-free region, thus yielding modulated phase content. Indeed, by using a three-component lipid bilayer system (DOPC/eSM/Chol), we find that Lo domains can be suppressed or eliminated by changing melittin concentration. Longer incubation results in large increase of the Lo phase content. Based on the reported phase diagrams,31, 57–58 the Lo phase is enriched with eSM and cholesterol, and the Ld phase is enriched with DOPC. To explain the enhanced Lo phase content, we propose that compared to eSM and cholesterol, DOPC lipid is preferentially attracted to defect edges and later removed from the bilayer. This hypothesis is in line with the observation that defects are exclusively localized in the DOP-Cenriched Ld phase. Similar phase remodeling induced by AMPs and other peptides has been reported.16–17 The authors attributed their findings to altered line tension at domain edges. Since line tension is dependent on height contrast between coexisting phases,32 our observation of time-dependent height contrast (Fig. 7) supports the proposition of line tension modulation.
Conclusions
We use lipid vesicles and mica supported planar bilayers to study melittin induced defects in single-phase and Lo+Ld phase coexisting bilayers. Time-course fluorescence spectroscopy measurements reveal that melittin induces defects that allow calcein to permeate through; leakage efficiency is proportionally correlated with peptide concentration. Solution AFM is used to directly visualize membrane defects. In particular, we find that defects are readily observed in DLPC bilayers with 0.5 and 1.0mol% melittin. Image analysis indicates that the most probable defect radius is 3.8 nm after initial equilibration. In contrast to the concept of static defects (pores), both defect size and shape evolve as a function of incubation time. The area fraction of membrane defects can be as large as 40% of the total bilayer surface. This value cannot be simply explained by the reduction of area per lipid. For DOPC bilayers, the most probable defect radius is 4.7 nm (incubation ≤ 60 min); defects become larger with longer incubation; area fraction covered by defects increases up to 25% after ~3 h incubation. Interestingly, addition of 30mol% cholesterol to DOPC+2.0mol% melittin bilayer delays defect development. Overall, the kinetic rate of melittin induced defect formation follows DLPC > DOPC > DOPC/cholesterol. Similar TM defects are observed when exposing POPC+10mol% POPG bilayers to melittin solution. Therefore, for the peptide concentrations used in this study, addition of anionic lipids does not seem to modify bilayer disruption imparted by melittin. To explain the observed kinetics, we propose a kinetic defect growth model, which extends the established two-phase model by introducing a third process that defect lining lipids can be transferred from defect edges to the aqueous phase.
We also study the effect of melittin induced defects on the phase behavior of DOPC/eSM/Chol bilayers. We find that (i) defects are exclusively localized in the Ld phase; (ii) depending on its concentration, melittin can either suppress or eliminate Lo phase formation; (iii) Lo domains with irregular boundaries and perforated interiors gradually emerge and become the dominant species as the incubation time increases; (iv) the retardation effect of cholesterol is evidenced by the gradually increasing height contrast between liquid coexisting phases; (v) for all the phase coexisting bilayers studied, the most probable defect radius is ~5 nm after initial equilibration; and (vi) longer incubation leads to defect enlargement (at least for some defects), a similar trend observed in single-phase bilayers. The observed phase remodeling behavior can be accounted for by our proposed kinetic defect growth model, which predicts that lipid redistribution in defect-free region can be induced by kinetic defects that selectively attract and remove DOPC lipids.
Supplementary Material
Acknowledgments
The research is supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R15GM117531 (J.P.).
Footnotes
Supporting Information Available: Figure S1, melittin crystal structure; Figure S2, calcein leakage data fitting; Figure S3, defect determination for DLPC/melittin bilayers; Figure S4, AFM images of DOPC + 1.0mol% melittin bilayers; Figure S5, AFM images and defect analysis of DOPC + 2.0mol% melittin bilayer; Figure S6, AFM images and defect analysis of DOPC + 1.0 µM melittin bilayer; Figure S7, AFM images and defect analysis of DOPC/cholesterol/melittin bilayer; Figure S8, AFM images of DOPC/eSM/cholesterol bilayer; Figure S9, AFM images and defect analysis of DOPC/eSm/cholesterol + 2mol% melittin bilayer; Figure S10, AFM images of DOPC/eSM/cholesterol bilayer; Figure S11, AFM images of DOPC + 0.5 mM Triton X-100 bilayer; Table S1, area fraction of melittin induced defects in different lipid bilayers. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sessa G, Freer JH, Colacicc G, Weissman G. Interaction of a Lytic Polypeptide, Melittin, with Lipid Membrane Systems. J Biol Chem. 1969;244:3575–3582. [PubMed] [Google Scholar]
- 2.Dempsey CE. The Actions of Melittin on Membranes. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 3.Terwilliger TC, Weissman L, Eisenberg D. The Structure of Melittin in the Form-I Crystals and Its Implication for Melittins Lytic and Surface-Activities. Biophys J. 1982;37:353–361. doi: 10.1016/S0006-3495(82)84683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer A, Fries A, Cornelsen I, Speck N, Koch-Nolte F, Gimpl G, Andra J, Bhakdi S, Reiss K. Melittin Modulates Keratinocyte Function through P2 Receptor-dependent ADAM Activation. J Biol Chem. 2012;287:23678–23689. doi: 10.1074/jbc.M112.362756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajski G, Garaj-Vrhovac V. Melittin: A Lytic Peptide with Anticancer Properties. Environ Toxicol Phar. 2013;36:697–705. doi: 10.1016/j.etap.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Toraya S, Nishimura K, Naito A. Dynamic Structure of Vesicle-Bound Melittin in a Variety of Lipid Chain Lengths by Solid-State NMR. Biophys J. 2004;87:3323–3335. doi: 10.1529/biophysj.104.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanke W, Methfessel C, Wilmsen HU, Katz E, Jung G, Boheim G. Melittin and a Chemically Modified Trichotoxin Form Alamethicin-Type Multistate Pores. Biochim Biophys Acta. 1983;727:108–114. doi: 10.1016/0005-2736(83)90374-7. [DOI] [PubMed] [Google Scholar]
- 8.Tosteson MT, Tosteson DC. The Sting - Melittin Forms Channels in Lipid Bilayers. Biophys J. 1981;36:109–116. doi: 10.1016/S0006-3495(81)84719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. Barrel-Stave Model or Ttoroidal Model? A Case Study on Melittin Pores. Biophys J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang HW. Free Energies of Molecular Bound States in Lipid Bilayers: Lethal Concentrations of Antimicrobial Peptides. Biophys J. 2009;96:3263–3272. doi: 10.1016/j.bpj.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokot G, Mally M, Svetina S. The Dynamics of Melittin-Induced Membrane Permeability. Eur Biophys J Biophy. 2012;41:461–474. doi: 10.1007/s00249-012-0800-1. [DOI] [PubMed] [Google Scholar]
- 12.Wiedman G, Herman K, Searson P, Wimley WC, Hristova K. The Electrical Response of Bilayers to the Bee Venom Toxin Melittin: Evidence for Transient Bilayer Permeabilization. Bba-Biomembranes. 2013;1828:1357–1364. doi: 10.1016/j.bbamem.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimenez D, Sanchez-Munoz OL, Salgado J. Direct Observation of Nanometer-Scale Pores of Melittin in Supported Lipid Mono layers. Langmuir. 2015;31:3146–3158. doi: 10.1021/la504293q. [DOI] [PubMed] [Google Scholar]
- 14.Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Saez AJ, Chiantia S, Salgado J, Schwille P. Pore Formation by a Bax-Derived Peptide: Effect on the Line Tension of the Membrane Probed by AFM. Biophys J. 2007;93:103–112. doi: 10.1529/biophysj.106.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw JE, Epand RF, Hsu JCY, Mo GCH, Epand RM, Yip CM. Cationic Peptide-Induced Remodelling of Model Membranes: Direct Visualization by in situ Atomic Force Microscopy. J Struct Biol. 2008;162:121–138. doi: 10.1016/j.jsb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Won A, Ruscito A, Ianoul A. Imaging the Membrane Lytic Activity of Bioactive Peptide Latarcin 2A. Bba-Biomembranes. 2012;1818:3072–3080. doi: 10.1016/j.bbamem.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Khadka NK, Cheng XL, Ho CS, Katsaras J, Pan JJ. Interactions of the Anticancer Drug Tamoxifen with Lipid Membranes. Biophys J. 2015;108:2492–2501. doi: 10.1016/j.bpj.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das C, Sheikh KH, Olmsted PD, Connell SD. Nanoscale Mechanical Probing of Supported Lipid Bilayers with Atomic Force Microscopy. Phys Rev E. 2010;82:041920. doi: 10.1103/PhysRevE.82.041920. [DOI] [PubMed] [Google Scholar]
- 20.Honigmann A, Mueller V, Hell SW, Eggeling C. STED Microscopy Detects and Quantifies Liquid Phase Separation in Lipid Membranes Using a New Far-Red Emitting Fluorescent Phosphoglycerolipid Analogue. Faraday Discuss. 2013;161:77–89. doi: 10.1039/c2fd20107k. [DOI] [PubMed] [Google Scholar]
- 21.Khadka NK, Ho CS, Pan JJ. Macroscopic and Nanoscopic Heterogeneous Structures in a Three-Component Lipid Bilayer Mixtures Determined by Atomic Force Microscopy. Langmuir. 2015;31:12417–12425. doi: 10.1021/acs.langmuir.5b02863. [DOI] [PubMed] [Google Scholar]
- 22.Ho CS, Khadka NK, Pan J. Sub-Ten-Nanometer Heterogeneity of Solid Supported Lipid Membranes Determined by Solution Atomic Force Microscopy. Bba-Biomembranes. 2015;1858:181–188. doi: 10.1016/j.bbamem.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Ho CS, Khadka NK, She F, Cai J, Pan J. Polyglutamine Aggregates Impair Lipid Membrane Integrity and Enhance Lipid Membrane Rigidity. Bba-Biomembranes. 2016;1858:661–670. doi: 10.1016/j.bbamem.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kucerka N, Nieh MP, Katsaras J. Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature. Bba-Biomembranes. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Lee MT, Chen FY, Huang HW. Energetics of Pore Formation Induced by Membrane Active Peptides. Biochemistry-Us. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 26.Steinem C, Galla HJ, Janshoff A. Interaction of Melittin with Solid Supported Membranes. Phys Chem Chem Phys. 2000;2:4580–4585. [Google Scholar]
- 27.Juhaniewicz J, Sek S. Atomic Force Microscopy and Electrochemical Studies of Melittin Action on Lipid Bilayers Supported on Gold Electrodes. Electrochim Acta. 2015;162:53–61. [Google Scholar]
- 28.Allende D, Simon SA, McIntosh TJ. Melittin-Induced Bilayer Leakage Depends on Lipid Material Properties: Evidence for Toroidal Pores. Biophys J. 2005;88:1828–1837. doi: 10.1529/biophysj.104.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin JJ, Cheng CW, Lin JJ, Chen WY. Exploring the Effect of Cholesterol in Lipid Bilayer Membrane on the Melittin Penetration Mechanism. Anal Biochem. 2007;367:49–55. doi: 10.1016/j.ab.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 30.Connell SD, Heath G, Olmsted PD, Kisil A. Critical Point Fluctuations in Supported Lipid Membranes. Faraday Discuss. 2013;161:91–111. doi: 10.1039/c2fd20119d. [DOI] [PubMed] [Google Scholar]
- 31.Bezlyepkina N, Gracia RS, Shchelokovskyy P, Lipowsky R, Dimova R. Phase Diagram and Tie-Line Determination for the Ternary Mixture DOPC/eSM/Cholesterol. Biophys J. 2013;104:1456–1464. doi: 10.1016/j.bpj.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzmin PI, Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Line Tension and Interaction Energies of Membrane Rafts Calculated from Lipid Splay and Tilt. Biophys J. 2005;88:1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epand RM, Vogel HJ. Diversity of Antimicrobial Peptides and Their Mechanisms of Action. Bba-Biomembranes. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 34.Hancock REW, Sahl HG. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 35.Zasloff M. Antimicrobial Peptides of Multicellular Organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 36.Ladokhin AS, Selsted ME, White SH. Bilayer Interactions of Indolicidin, a Small Antimicrobial Peptide Rich in Tryptophan, Proline, and Basic Amino Acids. Biophys J. 1997;72:794–805. doi: 10.1016/s0006-3495(97)78713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallock KJ, Lee DK, Omnaas J, Mosberg HI, Ramamoorthy A. Membrane Composition Determines Pardaxin's Mechanism of Lipid Bilayer Disruption. Biophys J. 2002;83:1004–1013. doi: 10.1016/S0006-3495(02)75226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takano T, Konoki K, Matsumori N, Murata M. Amphotericin B-Induced Ion Flux is Markedly Attenuated in Phosphatidylglycerol Membrane as Evidenced by a Newly Devised Fluorometric Method. Bioorgan Med Chem. 2009;17:6301–6304. doi: 10.1016/j.bmc.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 39.Ladokhin AS, White SH. 'Detergent-Like' Permeabilization of Anionic Lipid Vesicles by Melittin. Bba-Biomembranes. 2001;1514:253–260. doi: 10.1016/s0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- 40.Portlock SH, Clague MJ, Cherry RJ. Leakage of Internal Markers from Erythrocytes and Lipid Vesicles Induced by Melittin, Gramicidin-S and Alamethicin - a Comparative-Study. Biochim Biophys Acta. 1990;1030:1–10. doi: 10.1016/0005-2736(90)90231-c. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz G, Zong RT, Popescu T. Kinetics of Melittin Induced Pore Formation in the Membrane of Lipid Vesicles. Biochim Biophys Acta. 1992;1110:97–104. doi: 10.1016/0005-2736(92)90299-2. [DOI] [PubMed] [Google Scholar]
- 42.Benachir T, Lafleur M. Study of Vesicle Leakage Induced by Melittin. Bba-Biomembranes. 1995;1235:452–460. doi: 10.1016/0005-2736(95)80035-e. [DOI] [PubMed] [Google Scholar]
- 43.Hincha DK, Crowe JH. The Lytic Activity Of The Bee Venom Peptide Melittin Is Strongly Reduced By The Presence Of Negatively Charged Phospholipids Or Chloroplast Galactolipids In The Membranes Of Phosphatidylcholine Large Unilamellar Vesicles. Bba-Biomembranes. 1996;1284:162–170. doi: 10.1016/s0005-2736(96)00122-8. [DOI] [PubMed] [Google Scholar]
- 44.Matsuzaki K, Yoneyama S, Miyajima K. Pore Formation and Translocation of Melittin. Biophys J. 1997;73:831–838. doi: 10.1016/S0006-3495(97)78115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rex S, Schwarz G. Quantitative Studies on the Melittin-Induced Leakage Mechanism of Lipid Vesicles. Biochemistry-Us. 1998;37:2336–2345. doi: 10.1021/bi971009p. [DOI] [PubMed] [Google Scholar]
- 46.Longo ML, Waring AJ, Gordon LM, Hammer DA. Area Expansion and Permeation of Phospholipid Membrane Bilayers by Influenza Fusion Peptides and Melittin. Langmuir. 1998;14:2385–2395. [Google Scholar]
- 47.Lee MT, Hung WC, Chen FY, Huang HW. Mechanism and Kinetics of Pore Formation in Membranes by Water-Soluble Amphipathic Peptides. P Natl Acad Sci USA. 2008;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MT, Sun TL, Hung WC, Huang HW. Process of Inducing Pores in Membranes by Melittin. P Natl Acad Sci USA. 2013;110:14243–14248. doi: 10.1073/pnas.1307010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliynyk V, Kaatze U, Heimburg T. Defect Formation of Lytic Peptides in Lipid Membranes and Their Influence on the Thermodynamic Properties of the Pore Environment. Bba-Biomembranes. 2007;1768:236–245. doi: 10.1016/j.bbamem.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Ladokhin AS, Selsted ME, White SH. Sizing Membrane Pores In Lipid Vesicles By Leakage Of Co-Encapsulated Markers: Pore Formation By Melittin. Biophys J. 1997;72:1762–1766. doi: 10.1016/S0006-3495(97)78822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun DL, Forsman J, Woodward CE. Multistep Molecular Dynamics Simulations Identify the Highly Cooperative Activity of Melittin in Recognizing and Stabilizing Membrane Pores. Langmuir. 2015;31:9388–9401. doi: 10.1021/acs.langmuir.5b01995. [DOI] [PubMed] [Google Scholar]
- 52.Hristova K, Dempsey CE, White SH. Structure, Location, And Lipid Perturbations Of Melittin At The Membrane Interface. Biophys J. 2001;80:801–811. doi: 10.1016/S0006-3495(01)76059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dufourc EJ, Smith ICP, Dufourcq J. Molecular Details of Melittin-Induced Lysis of Phospholipid-Membranes as Revealed by Deuterium and Phosphorus Nmr. Biochemistry-Us. 1986;25:6448–6455. doi: 10.1021/bi00369a016. [DOI] [PubMed] [Google Scholar]
- 54.Huang HW. Action of Antimicrobial Peptides: Two-State Model. Biochemistry-Us. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 55.Lee MT, Hung WC, Chen FY, Huang HW. Many-Body Effect Of Antimicrobial Peptides: On The Correlation Between Lipid's Spontaneous Curvature And Pore Formation. Biophys J. 2005;89:4006–4016. doi: 10.1529/biophysj.105.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morandat S, El Kirat K. Membrane Resistance To Triton X-100 Explored By Real-Time Atomic Force Microscopy. Langmuir. 2006;22:5786–5791. doi: 10.1021/la0604228. [DOI] [PubMed] [Google Scholar]
- 57.Veatch SL, Keller SL. Seeing Spots: Complex Phase Behavior in Simple Membranes. Bba-Mol Cell Res. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 58.Feigenson GW. Phase Diagrams and Lipid Domains in Multicomponent Lipid Bilayer Mixtures. Bba-Biomembranes. 2009;1788:47–52. doi: 10.1016/j.bbamem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.