Abstract
The Arabidopsis ROS1/DEMETER family of 5mC DNA glycosylases are the first genetically characterized DNA demethylases in eukaryotes. However, the features of ROS1 targeted genomic loci are not well-understood. In this study, we characterized ROS1 target loci in Arabidopsis Col-0 and C24 ecotypes. We found that ROS1 preferentially targets transposable elements (TEs) and intergenic regions. Compared to most TEs, ROS1-targeted TEs are closer to protein coding genes, suggesting that ROS1 may prevent DNA methylation spreading from TEs to nearby genes. ROS1-targeted TEs are specifically enriched for H3K18Ac and H3K27me3, and depleted of H3K27me and H3K9me2. Importantly, we identified thousands of previously unknown RNA-directed DNA methylation (RdDM) targets upon depletion of ROS1, suggesting that ROS1 strongly antagonizes RdDM at these loci. In addition, we show that ROS1 also antagonizes RdDM-independent DNA methylation at some loci. Our results provide important insights into the genome-wide targets of ROS1 and the crosstalk between DNA methylation and ROS1-mediated active DNA demethylation.
Introduction
5-methylcytosine (5mC) is an important epigenetic mark present in many eukaryotes, and is involved in many crucial biological processes, such as gene imprinting, regulation of gene expression, and genome stability 1–3. In plants, DNA methylation frequently occurs in three sequence contexts, including CG, CHG, and CHH (H represents either A, C or T). In Arabidopsis, DNA methylation is established and maintained by different pathways. DNA methylation (de novo methylation) is established by domains rearranged methyltransferase2 (DRM2), through the RNA-directed DNA methylation (RdDM) pathway, in which RNA polymerase IV (Pol IV)-dependent 24-nt small interfering RNAs (siRNAs) function to guide DRM2 to target loci. Recently, it was found that Pol IV-dependent 25–35nt precursors of the 24-nt siRNAs can trigger DNA methylation independently of the 24-nt siRNAs 4–7. Four different enzymes maintain DNA methylation after DNA replication, depending on the sequence context: mCG is maintained by DNA methyltransferase 1 (MET1), mCHG is maintained by chromomethylase3 (CMT3), and mCHH is maintained by CMT2 and DRM2.
DNA methylation levels are dynamically regulated. DNA methylation can be passively lost due to lack of maintenance methylation, or can be actively removed by DNA demethylases 3. In plants, active DNA demethylation is initiated by the ROS1/Demeter family of proteins. ROS1 is the first genetically characterized DNA demethylase (the first enzyme in the active DNA demethylation pathway) in eukaryotes8. ROS1 can remove the 5mC base and nick the DNA backbone, leaving a single nucleotide gap that is filled with an unmethylated cytosine through a base excision repair pathway 9–13. An anti-silencing protein complex containing methyl-DNA binding protein 7 (MBD7), increased DNA methylation 1 (IDM1), IDM2, and IDM3, was recently discovered to regulate ROS1 targeting, and in turn DNA demethylation 14–16. In Arabidopsis, some TEs show lower expression levels in ros1 mutants due to increased DNA methylation 15,17. Some genes are silenced in ros1 mutants due to DNA hypermethylation of nearby TEs 14,17–19, suggesting that ROS1-mediated demethylation of TEs is important for regulation of gene expression by preventing nearby genes from being silenced. The methylomes of an Arabidopsis ros1/dml2/dml3 (rdd) triple mutant 25 and of a ros1 single mutant 14 have been analyzed, revealing thousands of genomic regions subjected to active DNA demethylation by ROS114. However, the features of ROS1 targets are not known.
Previous studies suggested interactions between ROS1-mediated DNA demethylation and RdDM 20–22. Recently, studies revealed that the expression of ROS1 is finely tuned by RdDM and DNA demethylation pathways23,24, although the genome-wide interaction between ROS1-mediated DNA demethylation and RdDM has not been investigated. In this study, we generated and analyzed the DNA methylomes of ros1 mutants in both Col-0 and C24 ecotypes of Arabidopsis, and characterized ROS1 target loci in these two genetic backgrounds. Our analyses identified and characterized thousands of genomic loci that are regulated by both ROS1 and RdDM. Interestingly, we discovered thousands of previously unidentified RdDM targets by analyzing the DNA methylome of ros1/nrpd1 double mutant plants that are defective in both active DNA demethylation and RdDM. In addition, we show that besides antagonizing RdDM, ROS1 can also antagonize RdDM-independent DNA methylation at over a thousand genomic loci. These results provide important insights into the genome-wide effect of ROS1-mediated active DNA demethylation and the interaction between DNA demethylation and methylation in plants.
Results
Characterization of ros1 mutant methylomes in Col-0 and C24 ecotypes
ros1-4 is an Arabidopsis mutant of Col-0 ecotype with T-DNA insertion in the ROS1 gene, causing complete loss of function of ROS1 26. ros1-1 is a loss-of-function mutant of ROS1 in C24 ecotype and has a single nucleotide substitution in ROS1 leading to a premature stop codon, and is likely a null allele 8. In this study, we generated single-base resolution maps of DNA methylomes of two-week-old seedlings of ros1-4 and ros1-1 mutants. Methylomes of Col-0 and C24 wild types at the same developmental stage were sequenced and served as controls.
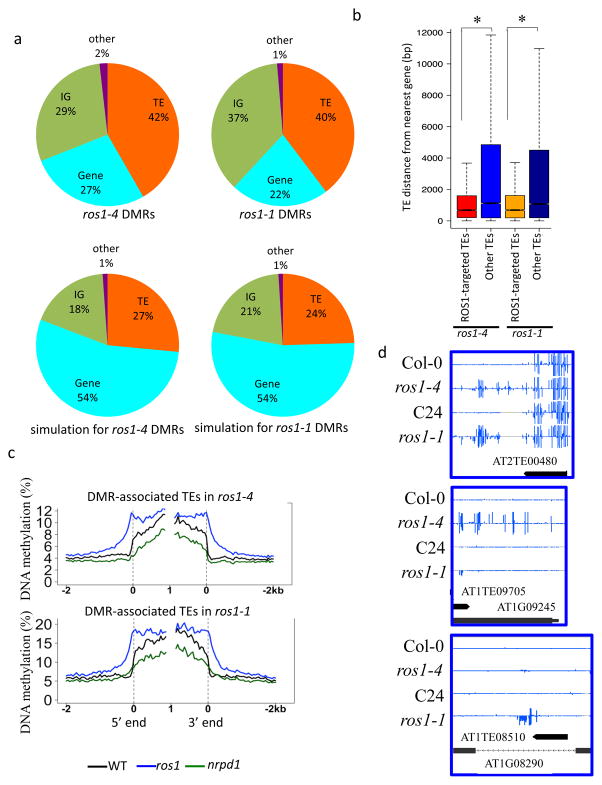
To identify potential genomic targets of ROS1 and compare the targets in different ecotypes, we identified differentially methylated regions (DMRs) in ros1-4 and ros1-1 mutants relative to their respective wild type plants. ros1-4 has 6902 hypermethylated DMRs (hyper DMRs) with an average length of 495 bp, and 1469 hypomethylated DMRs (hypo DMRs) with an average length of 193 bp, while 5011 hyper DMRs and 332 hypo DMRs were identified in ros1-1. The overwhelmingly higher numbers of hyper DMRs compared to hypo DMRs of ros1 mutants in both Col-0 and C24 are consistent with the ROS1 function in DNA demethylation. In ros1-4, 1887 (27%) hyper DMRs are in genic regions, 2878 (42%) in TE regions, 2010 (29%) in intergenic (IG) regions, and 127 (2%) in the category of others (Fig. 1a). Compared to the composition of randomly selected control regions that are composed of 27% TEs, 54% genes, and 18% IG regions (Fig. 1a), ros1-4 hyper DMRs have a decreased percentage in genes and increased percentages in TEs and IG regions, which is also observed in ros1-1 hyper DMRs (Fig. 1a). This indicates that ROS1 preferentially targets TEs and IG regions. ros1-1 and ros1-4 hyper DMRs are distributed throughout the five chromosomes of Arabidopsis (Supplementary Fig. 1a). For both ros1-1 and ros1-4 hyper DMRs, DNA hypermethylation was detected in all three contexts (CG, CHG and CHH) (Supplementary Fig. 1b). The length distribution of ROS1 targeted TEs is similar to that of all TEs (Supplementary Table 1), suggesting that ROS1 has no preference for short or long TEs. Interestingly, by analyzing the distance between TEs and its nearest genes, we found that ROS1-targeted TEs in both ros1-1 and ros1-4 tend to be located closer to genes relative to TEs that are not targeted by ROS1 (Fig. 1b).
Figure 1. Characterization of the DNA methylomes of ros1 mutants in Col-0 and C24 ecotypes.
a. Composition of the hyper DMRs in ros1-4, ros1-1 and of the corresponding simulated genomic regions.
b. Box plot showing the distances between ROS1-targeted or non-targeted TEs and their nearest protein coding genes (*p-value < 2.2e-16, one-tailed Wilcoxon rank sum test).
c. DNA methylation levels of ros1 hyper DMR-associated TEs in wild type, ros1 and nrpd1 mutants. TEs were aligned at the 5′ end or the 3′ end, and average methylation for all cytosines within each 50 bp interval was plotted.
d. Methylation levels at shared or non-shared hyper DMRs between ros1-1 and ros1-4. Integrated Genome Browser (IGB) display of whole-genome bisulfite sequencing data is shown in the screenshots. DNA methylation levels of cytosines were indicated with the heights of vertical bars on each track.
In both ros1-1 and ros1-4, DNA methylation is substantially increased around the boarders of TEs targeted by ROS1 (Fig. 1c). As expected, these TEs display decreased methylation in nrpd1 mutants, which are dysfunctional for RdDM due to disruption of NRPD1, the largest subunit of RNA polymerase IV (Fig. 1c). Interestingly, we noticed that, the hypermethylation in ros1 mutants extends from the TE borders to neighboring sequences before tempering off (Fig. 1c). These patterns support our previous hypothesis that ROS1 may counteract RdDM to prevent the spreading of methylation from highly methylated regions, such as TEs, to nearby genes 3.
To investigate the influence of different genetic backgrounds on ROS1 targeting, we compared DMRs of ros1-4 and ros1-1, which are mutants in Col-0 and C24 ecotypes respectively. We found that only 27% hyper DMRs in ros1-4 are also hyper DMRs in ros1-1, suggesting that ROS1 targeting is greatly influenced by genetic backgrounds. Interestingly, 35% of TE-type and 28% of intergenic type hyper DMRs are shared between ros1-4 and ros1-1, but only 15% of genic type hyper DMRs are shared between the two mutants. Thus, ROS1 targeting seems relatively conserved in TE regions in Col-0 and C24 ecotypes. Since TEs and genes typically display similar levels of genetic variation, these findings suggest that chromatin features important for active DNA demethylation might be more conserved at TEs than genes between the two ecotypes. Several examples of shared hyper DMRs and non-shared hyper DMRs are displayed in Fig. 1d and Supplementary Fig. 1c.
In summary, we identified, characterized, and compared targets of ROS1 in Col-0 and C24 genetic backgrounds. ROS1 targets in both Col-0 and C24 display a preference for TEs and intergenic regions, and the targeted TEs are located near genes. However, the specific genomic regions targeted by ROS1 are mostly distinct in Col-0 and C24 backgrounds.
Chromatin features associated with ROS1 targets
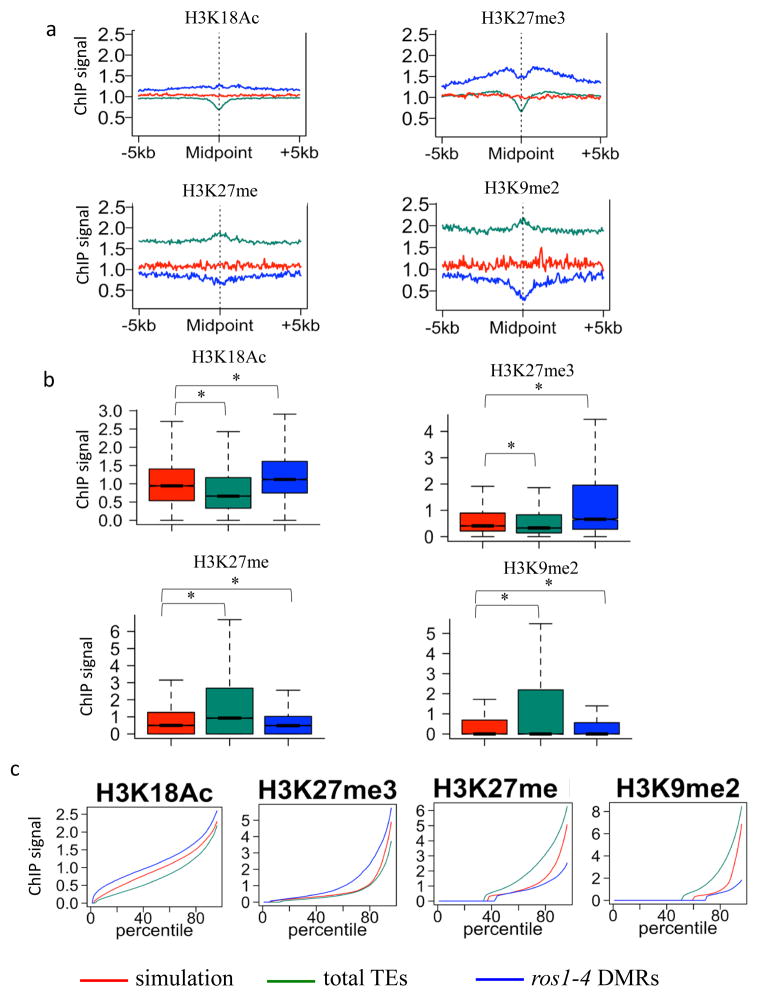
Histone modifications, such as histone methylation and acetylation, are known to interact with DNA methylation, therefore we determined which histone marks are associated with ROS1 targets. Compared to simulated regions, which are randomly selected genomic regions with the same length distribution as the DMRs, both total TEs and ROS1 hyper DMRs show a slight decrease in the level of H3 (Supplementary Fig. 2a–c), indicating a lower nucleosome density in TEs and ROS1 targets. ROS1 targets are negatively associated with most active histone marks compared to control regions, such as H3K36 di-/tri-methylation (H3K36me2/3), H3K4me2/3 and H3K9 acetylation (H3K9Ac) (Supplementary Fig. 2a–c), which was expected since a large proportion of ROS1 targets are within TEs (Fig. 1a). However, in contrast with most TEs, ROS1 targets are positively associated with the active histone mark H3K18Ac compared to control regions (Fig. 2a–c). Because only 42% of ros1-4 hyper DMRs are within TE regions (Fig. 1a), it is possible that the remaining 58% of ros1 targets that are not within TEs account for the enrichment of H3K18Ac. To investigate this possibility, we compared TE-, intergenic, and genic types of ros1 DMRs with simulated TEs, intergenic regions and genic regions, respectively. Consistently, we found that H3K18Ac is enriched in all types of ros1 DMRs (Supplementary Fig. 2d), suggesting that ros1 targets are indeed generally characterized by enrichment of H3K18Ac. The association with H3K18Ac is fully consistent with our previous finding that IDM1, an H3K18/23 acetyltransferase, is required for the demethylation of a subset of ROS1 targets 14.
Figure 2. Chromatin features associated with ROS1 targets.
a. Association of different histone modifications surrounding ros1-4 hyper DMRs. Association of histone modifications at total TEs and simulated regions served as controls. In contrast to total TEs, ros1-4 hyper DMRs are positively associated with H3K18Ac, H3K27me3, and negatively associated with H3K27me and H3K9me2.
b. Box plots display of the results in Fig. 2a (*p-value < 1e-15, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Fig. 2b. For each histone mark, simulated regions, TEs and ros1-4 DMRs were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.
We identified additional histone marks that distinguish ROS1 target regions. As shown in Fig. 2, TEs in general are negatively associated with H3K27me3, and are positively associated with H3K27me and H3K9me2. In contrast, ros1 DMRs have the opposite features, in that they are associated with enrichment of H3K27me3 and depletion of H3K27me and H3K9me2 (Fig. 2a–c). Similarly, we compared these chromatin features of ros1 targets and corresponding simulation for each type of regions (TE, intergenic and genic regions). All types of ros1 targets are enriched of H3K27me3 compared to their respective simulated regions (Supplementary Fig. 2d). TE-type ros1 targets have decreased H3K27me and H3K9me2 ChIP signals compared to the corresponding simulated regions (Supplementary Fig. 2d). We did not observe decreased H3K27me and H3K9me2 signals for genic and intergenic ros1 targets, since the levels of these histone marks are already very low in simulated genic and intergenic regions (Supplementary Fig. 2d). These results support that ros1 targets are associated with enrichment of H3K27me3 and depletion of H3K27me and H3K9me2.
A new class of RdDM targets
De novo DNA methylation, especially in the CHH context, is established through the RdDM pathway, which requires DNA dependent RNA polymerase IV for small RNA production 27. In previous studies, RdDM targets have been identified through the identification of hypo DMRs in RdDM mutants compared to wild type plants. In this study, we identified 4580 hypo DMRs and 2348 hyper DMRs in nrpd1 (Pol IV largest subunit) mutant compared to wild type plants; the large number of hypo DMRs in the Pol IV mutant is consistent with the role of Pol IV in DNA methylation. The more than 2000 hyper DMRs in Pol IV may be related to reduced expression level of ROS1 in nrpd1 mutant 23,24 (see below).
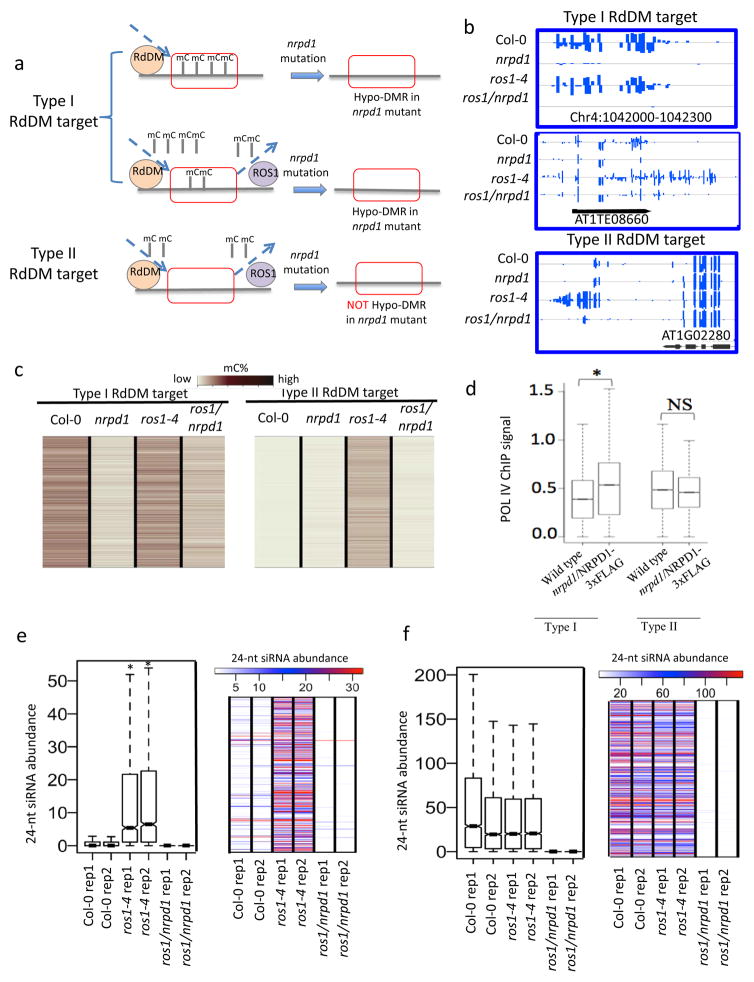
Homeostasis of DNA methylation is regulated by DNA methylation and active DNA demethylation processes 3,28. As diagrammed in Fig. 3a, regions identified as hypo DMRs (“type I”) in Pol IV mutants must be methylated in the wild type. The presence of methylation in the wild type implies that RdDM dominates over active DNA demethylation at these loci or that active DNA demethylation does not occur at these loci. We refer to the 4580 hypo DMRs in Pol IV mutant as type I RdDM targets.
Figure 3. Identification and characterization of type II RdDM targets.
a. Schematic hypothesis that different RdDM targets may be regulated differently by ROS1. Some type I RdDM targets are not regulated by ROS1 (Upper panel) whereas other type I RdDM targets are regulated by ROS1, although RdDM is more dominant at these loci (Middle panel). Type II RdDM targets are always regulated by ROS1, and ROS1 is more dominant at these loci (Lower panel).
b. Methylation levels of type I and type II RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1. The three panels are representative regions as diagramed in Fig. 3a respectively.
c. Heat maps showing DNA methylation levels of all type I and type II RdDM target loci in different genotypes.
d. Box plot showing Pol IV enrichment at type I and type II RdDM targets. Pol IV signal in wild type plants served as control. (*p-value<2.2e-16, one-tailed Wilcoxon rank sum test; NS, not significant).
e. Box plot and heat map showing 24-nt siRNA abundance of type II RdDM target loci in different genotypes (*p-value<2.2e-16, paired two-sample t-test).
f. Box plot and heat map showing 24-nt siRNA abundance of type I RdDM target loci in different genotypes.
We hypothesized that DNA demethylation may be dominant over RdDM at some genomic loci. These RdDM targets (“type II”) would not be methylated in wild type plants due to the dominance of active DNA demethylation (Fig. 3a). To uncover type II RdDM targets, we introduced the nrpd1 mutation into the ros1-4 mutant, and compared the methylome of the ros1/nrpd1 double mutant with that of the ros1-4 mutant (Fig. 3a,b). The type II regions would be predicted to gain cytosine methylation in ros1-4 mutants due to loss of ROS1 function, however, this gained cytosine methylation would be lost in ros1/nrpd1 mutant due to the dysfunction in RdDM. In total, we identified 6069 hypo DMRs in ros1/nrpd1 compared to ros1. Out of the 6069 hypo DMRs, 3750 display DNA methylation in wild type (mC% >= 2%), and about 60% of these 3750 regions overlap with the 4580 type I RdDM targets. Importantly, and consistent with our hypothesis, there are 2319 hypo DMRs that do not display DNA methylation in the wild type (mC% < 2%). These represent the type II RdDM targets, which have not been identified previously. Similarly, by using published methylome data of ros1-1/nrpd1 in C24 background, we found 4966 hypo DMRs in ros1-1/nrpd1 double mutant compared to ros1-1, and 1656 of them are type II RdDM targets, demonstrating that type II RdDM targets exist in both Col-0 and C24 ecotypes. As shown in Fig. 3b, c and Supplementary Fig. 3a, b, methylation of type I loci is decreased in nrpd1 relative to wild type. In contrast, type II loci do not display a change in methylation level in nrpd1 (Fig. 3b, c and Supplementary Fig. 3c, d). However, introducing the nrpd1 mutation into the ros1-4 mutant revealed the role of RdDM in DNA methylation at the type II loci (Fig. 3a–c and Supplementary Fig. 3c, d).
We evaluated 24-nt siRNA enrichment for type II loci and found that 39% of type II loci had 24-nt siRNAs (N>0 in either WT replicates), whereas 61% of type II loci did not display 24-nt siRNA reads (N=0 in both WT replicates). We cannot exclude the possibility that siRNA levels at these loci were too low to be detected by siRNA-seq. Similar with type I targets, the type II targets also have a decreased siRNA level in nrpd1 mutant relative to wild type (Supplementary Fig. 3e). This result further supports that type II loci are targets of RdDM. However, the siRNA level in type II loci is much lower than that in type I loci (Supplementary Fig. 3e), indicating weaker RdDM at type II loci.
We examined Pol IV occupancy at type I and type II loci using previously published Pol IV ChIP-seq data29. Pol IV is enriched in type I loci (Fig. 3d). However, we did not observe a significant enrichment of Pol IV in type II loci (Fig. 3d). The low siRNA level and low Pol IV enrichment are consistent with weak RdDM effects at these type II loci. However, type II loci display Pol IV-dependent increased DNA methylation in ros1 mutant (Fig. 3c), suggesting enhanced RdDM at these loci in ros1 mutant.
We performed small RNA-Seq in ros1-4 mutant and ros1-4/nrpd1 double mutant plants. We found that type II RdDM targets have a significantly elevated 24-nt siRNA level in ros1 relative to WT plants, and this increase in siRNAs can be suppressed by nrpd1 mutation (Fig. 3e). In contrast, type I RdDM targets do not display increased 24-nt siRNA levels in ros1 mutant compared to WT (Fig. 3f). The results suggest that RdDM becomes stronger at type II loci when ROS1 is removed.
DRD1 is a component of the RdDM pathway, and a previous study showed that DRD1-mediated CHH methylation was positively correlated with the histone marks H3K27me3, H3K4me2, H3K4me3, and H3K36me3, and was negatively correlated with H3, H3K9me2 and H3K27me 30. Consistent with this previous study, type I and type II RdDM targets display a slight decrease in H3 enrichment (Supplementary Fig. 4a–c), suggesting a reduced nucleosome density in RdDM target loci than in control regions. Our results show that both type I and type II targets are associated with a depletion of euchromatic histone marks, including H3K4me2/3, H3K36me2/3 and H3K9meAc (Supplementary Fig. 4a–c).
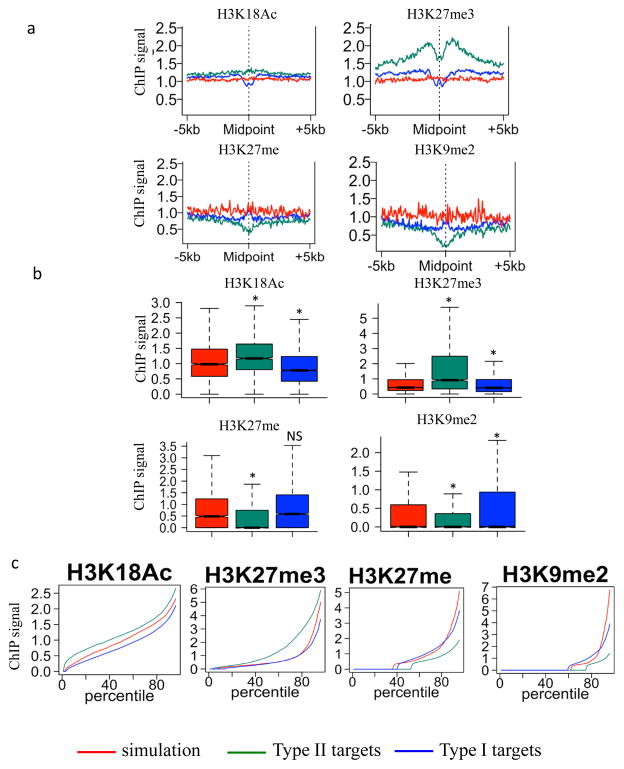
Type I and type II targets also display distinct chromatin features as shown in Fig. 4a–c. Chromatin features of type II RdDM targets are similar to ROS1 targets, including enrichment of H3K18Ac and H3K27me3 (Fig. 2 and 4), as expected. In contrast, type I targets display decreased H3K18Ac and slightly decreased H3K27me3 (Fig. 4). These distinct chromatin features are supported by examination of type I and type II targets for different categories of regions (TE, intergenic and genic regions) (Supplementary Fig. 4d). Consistently, we found enrichment of H3K18Ac and H3K27me3 in all categories of type II targets compared to the corresponding categories of type I targets. Also unlike type I targets, type II targets are depleted of H3K9me2 and H3K27me. The depletion of H3K9me2 and H3K27me was found only in TE and intergenic regions of type II targets compared to type I targets, but not in genic regions of type II targets (Supplementary Fig. 4d).
Figure 4. Chromatin features associated with type I and type II RdDM targets.
a. Association of different histone modifications at regions surrounding the mid-points of type I and type II targets. Simulation regions served as control regions.
b. Box plots showing the same results as in Fig. 4a. (*p-value<0.005, one-tailed Wilcoxon rank sum test; NS, not significant compared to simulation).
c. Percentile plots of the same data as in Fig. 4b. For each histone mark, simulated regions, type I and type II RdDM targets were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis.
In summary, type I RdDM targets show DNA methylation in the wild type, and they may or may not be regulated by ROS1. In contrast, the newly discovered type II RdDM targets are all regulated by ROS1 and are essentially depleted of DNA methylation in the wild type due to ROS1 activity. The two types of RdDM targets are also characterized by distinct small RNA profiles and histone modification marks.
Relationship between ROS1-mediated DNA demethylation and RdDM pathway
ROS1-mediated active DNA demethylation counteracts the RdDM pathway to prevent DNA hypermethylation at some specific loci 8,31,32. However, the genome-wide crosstalk between these two pathways has not been studied. To identify genomic regions targeted by both ROS1 and RdDM, we compared two groups of DMRs: hyper DMRs in ros1 mutant and hypo DMRs in nrpd1 mutants. We found that there are 1136 shared DMRs between ros1 hyper DMRs and nrpd1 hypo DMRs, suggesting that 16.5% (1136/6902) of ROS1 targets are antagonized by RdDM in wild type plants, however, this ratio increased to 60.1 % (4146/6902) by using hypo DMRs identified in ros1/nrpd1 mutant relative to ros1 (Supplementary Fig. 4e). These results suggest that the antagonistic effects between ROS1-mediated active DNA demethylation and RdDM have been underestimated, since type II RdDM targets were previously unappreciated.
It has been reported recently that there is a regulatory link between RdDM and ROS1-mediated active DNA demethylation. It was found that ROS1 expression is dramatically reduced in RdDM mutants, including nrpd1, due to the change of DNA methylation at the promoter region of ROS1 gene 23,24. Thus, we speculated that the hyper DMRs in nrpd1 might be caused by reduced ROS1 expression. We found that 1026 of the 2348 hyper DMRs in nrpd1 overlapped with hyper DMRs in ros1, suggesting that nearly half of the hyper methylated loci in nrpd1 might be caused by reduction of ROS1 expression in nrpd1. ROS1 expression is reduced in not only nrpd1, but also other RdDM mutants, such as nrpe123. As shown in Fig. 5a,b and Supplementary Fig. 5a,b, hyper DMRs of different RdDM mutants, including nrpd1 and nrpe1, also have increased DNA methylation in ros1 mutants, suggesting that the decreased ROS1 expression level contributes to the hyper methylation in the two examined RdDM mutants. We then determined whether the methylome of the nrpd1 single mutant, which has a dramatically reduced ROS1 expression level, is similar to the methylome of ros1/nrpd1 double mutant (where ROS1 was knocked out). After comparing ros1/nrpd1 with nrpd1, we identified 3411 hyper DMRs in ros1/nrpd1 relative to nrpd1. Interestingly, only 35% of the 3411 hyper DMRs are included in the 6902 hyper DMRs in ros1-4, suggesting that ROS1 may have new target loci in nrpd1 mutant compared to those in wild type. This finding suggests that the remaining ROS1 expression in nrpd1 mutants still functions at thousands of loci, although other DMLs may also contribute to these hyper DMRs.
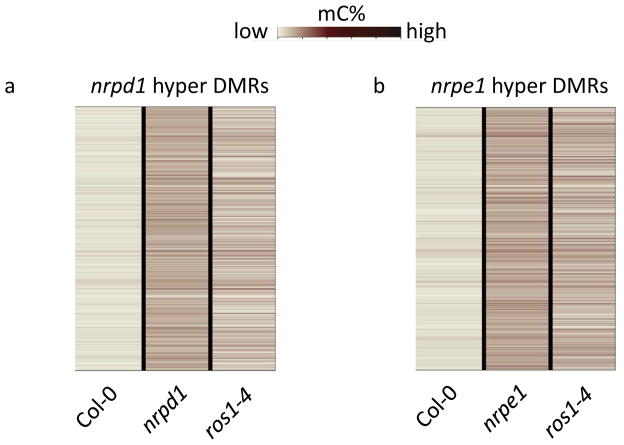
Figure 5. Reduced ROS1 expression contributes to DNA hypermethylation in RdDM mutants.
a. Heat map showing total C methylation levels of nrpd1 hyper DMRs in Col-0, nrpd1 and ros1-4.
b. Heat map showing total C methylation levels of nrpe1 hyper DMRs in Col-0, nrpe1 and ros1-4.
ROS1 antagonizes RdDM-independent DNA methylation
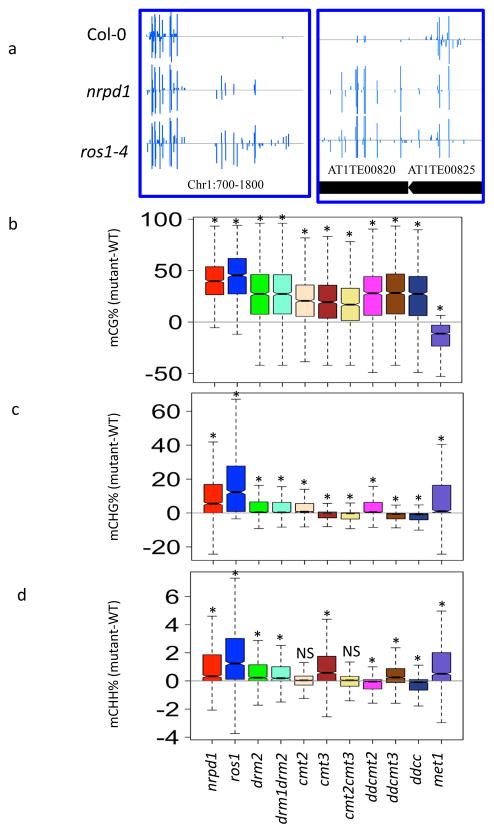
We identified 1026 shared hypermethylated loci in nrpd1 and ros1 (Fig. 6a), which are distributed across five chromosomes (Supplementary Fig. 6a). Although these loci have similar chromatin features to ROS1 targets, such as H3K18Ac, H3K4me2, and H3K4me3, they display slightly increased levels of H3 compared to general ROS1 targets (Supplementary Fig. 6b). At these loci, ROS1 prevents hypermethylation, and the methylation must be independent of RdDM since the methylation can occur in nrpd1 mutants. This indicates that there are RdDM-independent pathways responsible for the methylation and are antagonistic to ROS1 at these loci. Using previously published methylome data (Supplementary Table 2), we examined methylation levels of these 1026 loci in wild type, nrpd1, ros1, drm2, drm1drm2, cmt2, cmt3, cmt2cmt3, drm1/drm2/cmt2 (ddcmt2), drm1/drm2/cmt3 (ddcmt3), drm1/drm2/cmt2/cmt3 (ddcc), and met1 mutants. The nrpd1, ros1, and drm2 mutants display increased mCG, mCHG and mCHH levels at these loci (Fig. 6b–d), suggesting that MET1, DRM1, CMT2 and CMT3 may all contribute to the methylation at these loci. Indeed, we found that the mCG level of these loci is significantly reduced in met1 (Fig. 6b), whereas the mCHG levels are significantly reduced in cmt3 (Fig. 6c). Although CMT2 and CMT3 have been shown to function redundantly in mCHG methylation 33, it seems that the mCHG methylation at these loci mainly depends on CMT3 (Fig. 6c). For mCHH methylation level, there are no significant changes in cmt2 and cmt2cmt3 double mutants. However, mCHH is significantly reduced in ddcmt2 and ddcc mutants but is increased in drm2, drm1drm2 and ddcmt3 (Fig. 6d). This indicates that DRM1 and CMT2 may function redundantly at these regions.
Figure 6. ROS1 antagonizes RdDM-independent DNA methylation.
a. IGB display of DNA methylation levels at shared hyper DMRs between nrpd1 and ros1-4. DNA methylation levels of cytosines were indicated with the heights of vertical bars on each track.
b–d. The 1026 genomic regions with increased DNA methylation in both ros1 and nrpd1 mutants were used. Box plots of CG (b), CHG (c), and CHH (d) methylation level changes (mutant-WT) of these regions were shown in different mutants. ddcmt2 is drm1drm2cmt2 triple mutant, ddcmt3 is drm1drm2cmt3 triple mutant and ddcc is drm1drm2cmt2cmt3 quadruple mutant. (*p-value<1e-10, one sample one-tailed Student’s t-test).
At these loci, mCHG and mCHH levels are increased in met1 mutant plants (Fig. 6c, d). This may be caused by the reduction in ROS1 expression in met1 mutants 34, such that the mCHG and mCHH methylation by CMT3, CMT2 and DRM1 could not be removed by ROS1. These results suggested that ROS1 antagonizes CMT3-, CMT2-, DRM1-, and MET1-mediated DNA methylation, which are independent of DRM2, the major DNA methyltransferase in the RdDM pathway (Supplementary Fig. 6c).
We examined siRNA levels at these 1026 loci in wild type and nrpd1, and found that 24-nt siRNAs accumulate at the loci in the wild type, but are lost in nrpd1 mutant plants (Supplementary Fig. 6d). Since the siRNAs but not DNA methylation at these loci are dependent on Pol IV, the siRNAs at these loci would not be required for the methylation. This is consistent with a recent study showing that a reduction of siRNA levels in RdDM mutants does not substantially reduce CMT2-dependent CHH methylation 33.
In summary, our study revealed that, besides RdDM, ROS1 can antagonize DNA methylation mediated by MET1, DRM1 and CMTs in an siRNA-independent manner.
Discussion
Among the four proteins in the ROS1/Demeter family in Arabidopsis, ROS1 is the major DNA demethylase in vegetative tissues. In this study, we showed that genome-wide, ROS1 preferentially targets TEs that are close to protein coding genes (Fig. 1b). We also showed that the sequences just outside the boarders of ROS1-targeted TEs have increased DNA methylation in ros1 mutants (Fig. 1c), suggesting that ROS1 prevents the spreading of DNA methylation from highly methylated TEs. Consistently, Yamamuro et al. reported that ROS1 is required for the expression of the EPF2 gene by preventing the spreading of methylation from a TE near the promoter of EPF2 18. In addition, ROS1 family demethylases can positively regulate fungal pathogen responsive genes via demethylating TEs located in or near their promoters 19. Together with these previous studies, our data support that ROS1 is involved in the regulation of gene expression by preventing DNA methylation spreading from nearby TEs.
H3K18Ac is an active histone mark correlated with transcriptional activation 35. We found that ROS1 targets are positively associated with H3K18Ac (Fig. 2a–c), supporting our previous work showing that IDM1, an H3K18/23 acetyltransferase, can create a permissive chromatin environment important for ROS1 to access target loci 14. ROS1 targets were also found to be enriched with H3K27me3, but depleted of H3K27me and H3K9me2, in contrast to general TEs (Fig. 2a–c). This is consistent with a previous finding that there was a strong correlation between H3K18Ac and H3K27me3 in Arabidopsis 36, and is also consistent with findings in mammals that DNA demethylation process is coupled with decreased H3K9me2 and increased H3K27me3 37.
Consistent with previously observed antagonism between ROS1 and RdDM, ROS1-targeted TEs display decreased DNA methylation in nrpd1 mutants (Fig. 1c). We hypothesized that ROS1-mediated DNA demethylation may be so strong at some loci that methylation does not accumulate in the wild type at these regions. These potential RdDM targets could not be identified by comparing RdDM mutants with wild type plants; however, in this study we discovered over two thousand of these “type II” RdDM targets by comparing ros1 and ros1/nrpd1 mutants. These RdDM targets have eluded previous attempts of RdDM target identification. Our discovery of type II loci suggests that the number of RdDM targets has been greatly underestimated.
Overall siRNA enrichment and Pol IV occupancy were lower at type II targets compared to type I targets. More than half of the type II loci do not have any siRNA reads, and we did not observe any significant Pol IV enrichment at type II loci. The type II loci may be better targeted by RdDM in the ros1 mutant background. We observed increased siRNA levels at type II loci in ros1-4 mutant, indicating that RdDM becomes stronger in ros1 mutant plants. It is possible that demethylation or occupancy by ROS1 at these loci in wild type plants limits RdDM accessibility, thus leading to weak RdDM in the wild type, and that this inhibition of RdDM in the wild type is alleviated by ros1 mutation. ROS1 has been shown to have a similar binding affinity to both methylated and non-methylated DNA through a Lysine-rich Domain at the N terminus 38. Thus, it is possible that ROS1 antagonizes RdDM not only by removal of DNA methylation, but also by preventing the access of RdDM machinery to the target loci. In the future, it will be interesting to compare Pol IV occupancy at these loci in wild type and ros1 mutant plants to further investigate this possibility.
It is well known that ROS1 expression is dramatically reduced in RdDM mutants 23. Our results suggested that the reduction in ROS1 expression in nrpd1 mutant plants induces DNA hypermethylation at over a thousand genomic regions. The DNA hypermethylation in RdDM mutants must also be caused by some RdDM-independent DNA methylation pathways. Our analysis suggested that four DNA methylases including DRM1, CMT2, CMT3, and MET1 contribute to the hypermethylation in RdDM mutants. This finding implies that ROS1 can also antagonize RdDM-independent DNA methylation. Interestingly, we noticed that the nrpd1 mutant has slightly increased DNA methylation at type II RdDM loci compared to the wild type (Fig. 3c): 198/2319 type II loci overlap with the 1026 hyper DMRs in nrpd1 mutant. Thus, RdDM-independent DNA methylation may compensate to methylate DNA at some type II loci when RdDM is lacking. Our findings suggest that the fine tuning of the plant methylome is complex and involves interactions between DNA methylation mediated by RdDM and RdDM-independent mechanisms, and DNA demethylation mediated by ROS1 family demethylases.
Methods
Plant materials
Mutants including ros1-4, nrpd1-3 (SALK_128428), ros1/nrpd1 double mutant and nrpe1-11 (SALK_029919) are in the Col-0 background. ros1-4 and nrpd1-3 were crossed to generate ros1/nrpd1 double mutant. ros1-1, nrpd1(C24) and ros1-1/nrpd1 (C24) are mutants of C24 ecotype.
Seeds were stratified for 2–3 d at 4 °C before being sown on 1/2 MS plates containing 2% (wt/vol) sucrose and 0.7% (wt/vol) agar. All of the plants were grown under long day conditions at 22 °C.
Whole genome bisulfite sequencing and analysis
DNA was extracted from 1 gram of 14-day-old seedlings using the Plant DNeasy Maxi Kit from Qiagen. And 5 μg of gDNA was used for library construction using Illumina’s standard DNA methylation analyses protocol and the TruSeq DNA sample preparation kit. The samples in Col-0 background were sequenced in the Genomics Core Facilities of the Shanghai Center for Plant Stress Biology, SIBS, CAS (Shanghai, China) with Illumina HiSeq2500. The samples in C24 background were sequenced in the Biosciences Core Laboratory of King Abdullah University of Science & Technology (KAUST) with Illumina HiSeq2000.
For Col-0 background data analysis, low quality sequences (q < 20) were trimmed using trim in BRAT-BW 39, and clean reads were mapped to the TAIR10 genome using BRAT-BW and allowing two mismatches. To remove potential PCR duplicates, the remove-dupl command of BRAT-BW was used. DNA hypomethylated regions were identified according to Ausin et al. 40 with minor modification. In brief, only cytosines with 4X coverage in all libraries in the same background were considered. A sliding-window approach with a 200-bp window sliding at 50-bp intervals was used to identify DMRs. Fisher’s exact test was performed for methylated versus unmethylated cytosines for each context, within each window, with FDRs estimated using a Benjamini–Hochberg adjustment of Fisher’s p-values calculated in the R environment. Windows with an FDR ≤ 0.05 were considered for further analysis, and windows within 100 bp of each other were merged to larger regions. Regions were then adjusted to shrink to the first and last differentially methylated cytosines (DMCs). A cytosine was considered DMC if it showed at least a two-fold change in methylation percentage in the mutant. The regions were then filtered to include only those with at least 10 DMCs and with at least a twofold change in arithmetic mean of methylation percentage of all cytosine.
For C24 data, clean reads were mapped to a pseudo-C24 genome using BRAT-BW allowing two mismatches. We used public data set of ros1-1/nrpd1(C24) double mutant in C24 background 41 to analyze type II RdDM targets. The pseudo-C24 genome was generated through the replacement of SNPs in the Col-0 genome with C24 variants (http://1001genomes.org/data/MPI/MPISchneeberger2011/releases/current//C24/Marker/C24.SNPs.TAIR9.txt).
TE border analysis
The analysis was according to previously described method 42: ros1-4 hyper DMR associated TEs were aligned at the 5′ end or the 3′ end. We discarded from the analysis 250 bp from the end opposite to the one used for alignment to avoid averaging the edges of shorter TEs with the middles of longer sequences.
Histone feature analysis
Histone features were analyzed according to a previously described method 4 with a minor modification: Briefly, the public data used for the analysis were downloaded from GEO (Accession No: GSE28398) 36. The reads were aligned to TAIR10 using Bowtie 43 allowing 3 mismatches. Only reads that were uniquely mapped to the genome were retained for the downstream analysis. To generate the relative histone signal distribution in the flanking 5-kb region of the mid-point of DMRs, the whole region (10050 bp long) was divided into 201 bins with a size of 50 bp and the 101th bin aligning at the middle point of each DMR. The number of depth in each of the 201 bins was summed. The relative histone modification signal (y axis) in each of the 201 bins was defined as: n( Histone_modification ) × N( Input )/[N( Histone_modification ) × n( Input )] where n is the sum of depth of the corresponding library in each bin and N is the number of mapped reads of the corresponding library.
For box plots, DMRs were considered as the 1050 bp region from the DMR mid-point (+/−10 bins plus the mid-bin). In each region, the relative histone modification signal was calculated as above. The box plots were generated in R using function “boxplot” with parameter “range = 1.5, outline = F, notch = T”. The p-values were calculated in R using function “wilcox.test”.
Small RNA analysis
Small RNA samples were prepared from 14-day-old seedlings. The analysis pipeline was according to Zhang et al. 44.
Pol IV ChIP-seq analysis
The data sets we used are from published paper 29. According to this paper, WT is pure wild-type plants without any transgenes. nrpd1/NRPD1-3xFLAG is nrpd1 mutant with NRPD1-3xFLAG transgene.
Data access
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE83802. Previously published data, including whole genome sequencing data and ChIP-seq data, used in this study were listed in Supplementary Table 2.
Supplementary Material
a. Chromosomal distribution of hyper DMRs in ros1-4 and ros1-1 mutants.
b. Methylation changes in ros1 mutants relative to wild type at ros1 hyper DMRs are shown in the kernel density plots. Changes in mCG, mCHG and mCHH are shown separately.
c. Methylation levels of ros1 hyper DMRs in ros1 mutants and their corresponding wild types. Left two panels show hyper DMRs in both ros1-4 and ros1-1. Middle two panels show hyper DMRs in only ros1-4 but not ros1-1. Right two panels show hyper DMRs in only ros1-1 but not ros1-4.
a. Association of different histone modifications surrounding the mid-point of ros1-4 hyper DMRs, all TEs and simulated regions. Similar with general TEs, ros1-4 hyper DMRs are negatively associated with H3K9Ac, H3K4me2, H3K36me2, H3K36me3, and H3K4me3.
b. Box plots display of the results in Supplementary Fig. 2a (*p-value < 1e-15, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Supplementary Fig. 2b. For each histone mark, simulated regions, TEs and ros1-4 DMRs were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.
d. Box plot display of histone modifications of different types of ros1-4 hyper DMRs and the corresponding simulation regions (*p-value < 1e-6, one-tailed Wilcoxon rank sum test).
a. Methylation levels of two type I RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1. Whole genome bisulfite sequencing data are shown for two type I RdDM targets. The region on the left is the type of RdDM target shown in the upper panel of Fig. 3a, while the region on the right is the type of RdDM targets shown in the middle panel of Fig. 3b.
b. Heat maps showing CG, CHG and CHH methylation levels of all type I RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1.
c. Methylation levels of type II RdDM targets. Methylation levels of two type II RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1 were shown in left panels. Methylation levels of two type II RdDM targets in C24, nrpd1 (C24), ros1-1, and ros1-1/nrpd1 (C24 background) were shown in right panels.
d. Heat maps showing CG, CHG and CHH methylation levels of all type II RdDM target in Col-0, nrpd1, ros1-4, ros1/nrpd1.
e. Heat maps and box plots showing 24-nt siRNA abundance of type I (left panel) and type II (right panel) RdDM targets in Col-0 and nrpd1 (*p-value < 1e-7, paired two-sample t-test).
a. Association of different histone modifications at regions surrounding the mid-points of type I and type II targets and simulation regions.
b. Box plots showing the same results as in Supplementary Fig. 4a. (*p-value < 0.005, significantly lower than that of simulation, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Supplementary Fig. 4b. For each histone mark, simulated regions, type I and type II RdDM targets were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.
d. Box plots showing histone features of different categories of type I and type II RdDM targets (*p-value < 1e-6, one-tailed Wilcoxon rank sum test; NS, not significant).
e. Venn diagrams showing relationships among different groups of hyper and hypo DMRs.
a. Heat maps showing CG, CHG and CHH methylation levels of nrpd1 hyper DMRs in Col-0, nrpd1 and ros1-4.
b. Heat maps showing CG, CHG and CHH methylation levels of nrpe1 hyper DMRs in Col-0, nrpe1 and ros1-4.
a. Chromosomal distribution of shared hyper DMRs between nrpd1 and ros1-4.
b. Box plots display of different histone modifications surrounding 1026 shared hyper DMRs, all ros1-4 DMRs and simulated regions (*p-value < 0.0005, one-tailed Wilcoxon rank sum test).
c. Diagram showing that the 1026 shared hyper DMRs are regulated by ROS1, RdDM, and RdDM-independent DNA methylation pathways in wild type, and might be also regulated by DMLs-mediated demethylation pathways.
d. Heat map showing 24-nt siRNA abundance of the 1026 shared hyper DMRs in Col-0 and nrpd1.
Table of the percentiles of the length of ROS1-targeted TEs and all TEs.
Table of previously published data used in this study.
Acknowledgments
This work was supported by National Institutes of Health Grant R01GM070795 and by the Chinese Academy of Sciences (to J.-K. Z.).
Footnotes
Author contributions
J.-K.Z, Z.L and K. T. designed the study, interpreted the data and wrote the manuscript. K.T. and Z.L. did the bioinformatics analysis. H.Z. performed sequencing experiments.
Competing interests
The authors declare no competing financial interests.
References
- 1.Collier J. Epigenetic regulation of the bacterial cell cycle. Curr Opin Microbiol. 2009;12:722–729. doi: 10.1016/j.mib.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 2.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D-L, et al. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 2015 doi: 10.1038/cr.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhai J, et al. A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell. 2015;163:445–455. doi: 10.1016/j.cell.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye R, et al. A Dicer-Independent Route for Biogenesis of siRNAs that Direct DNA Methylation in Arabidopsis. Mol Cell. 2016;61:222–235. doi: 10.1016/j.molcel.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins T, et al. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife. 2015;4:e09591. doi: 10.7554/eLife.09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 9.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis [corrected] PLoS Genet. 2015;11:e1004905. doi: 10.1371/journal.pgen.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Macías MI, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Duan CG, Zhu X, Qian W, Zhu JK. A DNA ligase required for active DNA demethylation and genomic imprinting in Arabidopsis. Cell Res. 2015;25:757–760. doi: 10.1038/cr.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.lang Z, et al. The Methyl-CpG-Binding Protein MBD7 Facilitates Active DNA Demethylation to Limit DNA Hyper-Methylation and Transcriptional Gene Silencing. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, et al. Methyl-CpG-Binding Domain Protein MBD7 Is Required for Active DNA Demethylation in Arabidopsis. Plant Physiol. 2015;167:905–914. doi: 10.1104/pp.114.252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 18.Yamamuro C, et al. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le TN, et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014;15:458. doi: 10.1186/s13059-014-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007;145:1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei M, et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA. 2015;112:3553–3557. doi: 10.1073/pnas.1502279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams BP, Pignatta D, Henikoff S, Gehring M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 2015;11:e1005142. doi: 10.1371/journal.pgen.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Zhu JK. Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol. 2012;77:161–173. doi: 10.1101/sqb.2012.77.014936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He XJ, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Eskandarian HA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 36.Luo C, et al. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013;73:77–90. doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- 37.Rose CM, van den Driesche S, Meehan RR, Drake AJ. Epigenetic reprogramming: preparing the epigenome for the next generation. Biochem Soc Trans. 2013;41:809–814. doi: 10.1042/BST20120356. [DOI] [PubMed] [Google Scholar]
- 38.Ponferrada-Marín MI, Martínez-Macías MI, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of Repressor of Silencing 1 (ROS1) and facilitates demethylation in long substrates. J Biol Chem. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris EY, Ponts N, Le Roch KG, Lonardi S. BRAT-BW: efficient and accurate mapping of bisulfite-treated reads. Bioinformatics. 2012;28:1795–1796. doi: 10.1093/bioinformatics/bts264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausin I, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang CF, et al. A Pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013;9:e1003779. doi: 10.1371/journal.pgen.1003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol Cell. 2014;54:418–430. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. Chromosomal distribution of hyper DMRs in ros1-4 and ros1-1 mutants.
b. Methylation changes in ros1 mutants relative to wild type at ros1 hyper DMRs are shown in the kernel density plots. Changes in mCG, mCHG and mCHH are shown separately.
c. Methylation levels of ros1 hyper DMRs in ros1 mutants and their corresponding wild types. Left two panels show hyper DMRs in both ros1-4 and ros1-1. Middle two panels show hyper DMRs in only ros1-4 but not ros1-1. Right two panels show hyper DMRs in only ros1-1 but not ros1-4.
a. Association of different histone modifications surrounding the mid-point of ros1-4 hyper DMRs, all TEs and simulated regions. Similar with general TEs, ros1-4 hyper DMRs are negatively associated with H3K9Ac, H3K4me2, H3K36me2, H3K36me3, and H3K4me3.
b. Box plots display of the results in Supplementary Fig. 2a (*p-value < 1e-15, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Supplementary Fig. 2b. For each histone mark, simulated regions, TEs and ros1-4 DMRs were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.
d. Box plot display of histone modifications of different types of ros1-4 hyper DMRs and the corresponding simulation regions (*p-value < 1e-6, one-tailed Wilcoxon rank sum test).
a. Methylation levels of two type I RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1. Whole genome bisulfite sequencing data are shown for two type I RdDM targets. The region on the left is the type of RdDM target shown in the upper panel of Fig. 3a, while the region on the right is the type of RdDM targets shown in the middle panel of Fig. 3b.
b. Heat maps showing CG, CHG and CHH methylation levels of all type I RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1.
c. Methylation levels of type II RdDM targets. Methylation levels of two type II RdDM targets in Col-0, nrpd1, ros1-4, and ros1/nrpd1 were shown in left panels. Methylation levels of two type II RdDM targets in C24, nrpd1 (C24), ros1-1, and ros1-1/nrpd1 (C24 background) were shown in right panels.
d. Heat maps showing CG, CHG and CHH methylation levels of all type II RdDM target in Col-0, nrpd1, ros1-4, ros1/nrpd1.
e. Heat maps and box plots showing 24-nt siRNA abundance of type I (left panel) and type II (right panel) RdDM targets in Col-0 and nrpd1 (*p-value < 1e-7, paired two-sample t-test).
a. Association of different histone modifications at regions surrounding the mid-points of type I and type II targets and simulation regions.
b. Box plots showing the same results as in Supplementary Fig. 4a. (*p-value < 0.005, significantly lower than that of simulation, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Supplementary Fig. 4b. For each histone mark, simulated regions, type I and type II RdDM targets were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.
d. Box plots showing histone features of different categories of type I and type II RdDM targets (*p-value < 1e-6, one-tailed Wilcoxon rank sum test; NS, not significant).
e. Venn diagrams showing relationships among different groups of hyper and hypo DMRs.
a. Heat maps showing CG, CHG and CHH methylation levels of nrpd1 hyper DMRs in Col-0, nrpd1 and ros1-4.
b. Heat maps showing CG, CHG and CHH methylation levels of nrpe1 hyper DMRs in Col-0, nrpe1 and ros1-4.
a. Chromosomal distribution of shared hyper DMRs between nrpd1 and ros1-4.
b. Box plots display of different histone modifications surrounding 1026 shared hyper DMRs, all ros1-4 DMRs and simulated regions (*p-value < 0.0005, one-tailed Wilcoxon rank sum test).
c. Diagram showing that the 1026 shared hyper DMRs are regulated by ROS1, RdDM, and RdDM-independent DNA methylation pathways in wild type, and might be also regulated by DMLs-mediated demethylation pathways.
d. Heat map showing 24-nt siRNA abundance of the 1026 shared hyper DMRs in Col-0 and nrpd1.
Table of the percentiles of the length of ROS1-targeted TEs and all TEs.
Table of previously published data used in this study.