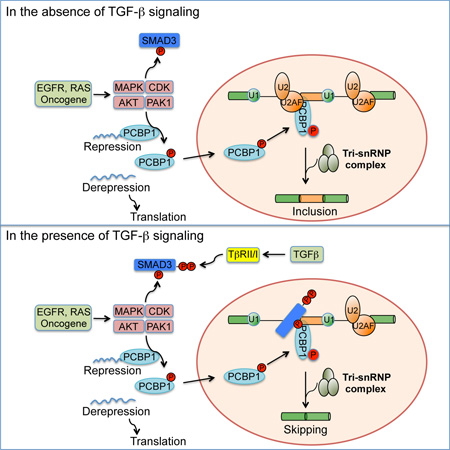
Summary
In advanced stages of cancers, TGF-β promotes tumor progression in conjunction with inputs from receptor tyrosine kinase pathways. However, mechanisms that underpin the signaling cooperation and convert TGF-β from a potent growth inhibitor to a tumor promoter are not fully understood. We report here that TGF-β directly regulates alternative splicing of cancer stem cell marker CD44 through a phosphorylated T179 of SMAD3-mediated interaction with RNA-binding protein PCBP1. We show that TGF-β and EGF respectively induce SMAD3 and PCBP1 to colocalize in SC35 positive nuclear speckles, and the two proteins interact in the variable exon region of CD44 pre-mRNA to inhibit spliceosome assembly in favor of expressing the mesenchymal isoform CD44s over the epithelial isoform CD44E. We further show that the SMAD3-mediated alternative splicing is essential to the tumor-promoting role of TGF-β and has a global influence on protein products of genes instrumental to epithelial to mesenchymal transition and metastasis.
Keywords: TGF-β, SMAD3, PCBP1, alternative splicing, CD44, EMT, metastasis
Graphical Abstract
eTOC
SMADs are known transcription factors of the TGF-β pathway. Tripathi et al report here that SMAD3 also directly regulates alternative splicing through a partnership with RNA binding protein PCBP1 and this regulation is essential to the tumor-promoting role of TGF-β.
Introduction
Transforming growth factor-β (TGF-β) is a potent growth inhibitor of epithelial cells and also a well-known promoter of tumor progression in advanced cancers (Drabsch and ten Dijke, 2012; Ikushima and Miyazono, 2010). The growth inhibitory property of TGF-β stems from its ability to signal through the canonical SMAD-dependent transcription mechanisms to induce cyclin-dependent kinase (CDK) inhibitors and suppress proto-oncogene MYC. On the other hand, the protumorigenic effects of TGF-β rely on the interplay of both SMAD-dependent and SMAD-independent mechanisms with other signaling pathways to elicit epithelial to mesenchymal transition (EMT), cell invasion and metastasis (Derynck et al., 2014; Heldin et al., 2012). However, despite a wealth of information about the context-dependent pathway crosstalks in the literature, it is still not completely clear how the core TGF-β pathway of SMAD signaling mechanism itself adjusts to promote tumor progression.
At the cell surface, TGF-β signals through a complex of membrane-bound type I (TβRI) and type II (TβRII) receptors, which activate signal transducers SMAD2 and SMAD3 by phosphorylation at a consensus SSXS motif in the carboxyl terminus and set forth their movement into the nucleus in association with SMAD4 (Massague, 2012). Within the nucleus, SMADs generally regulate target gene expression in conjunction with other transcription activators and chromatin modifiers. The receptor complex bound to TGF-β also activates MAPKs including ERK, JNK and p38MAPK independent of SMADs, forming the so-called non-SMAD pathways (Zhang, 2009). These MAPKs and CDKs can modulate TGF-β signaling through phosphorylation at T179, S204, and S208 of SMAD3 in the linker region that bridges the two conserved MH1 and MH2 domains (Matsuura et al., 2004; Matsuura et al., 2005). For instance, phosphorylation of T179 of SMAD3 exposes a neighboring PY motif, a docking site for ubiquitin E3 ligase SMURF2 and NEDD4L, causing SMAD3 to be monoubiquitinated and become transcriptionally inept, because this form of SMAD3 can no longer form a complex with SMAD4 nor bind DNA (Gao et al., 2009; Inui et al., 2011; Tang et al., 2011). Commensurate to the complexity in signaling mechanisms, TGF-β uses a number of ways to induce epithelial to mesenchymal transition (EMT) and promote cell invasion and metastasis. In addition to a large number of transcriptional targets, activated receptor complexes have been shown to directly regulate cell-cell contact at tight and adherens junctions (Wang et al., 2003), and translational controls orchestrated by the mTOR complex (Lamouille and Derynck, 2007) and heterogeneous nuclear ribonucleoprotein E1 (hnRNPE1) (Chaudhury et al., 2010).
Here, we report that in a concerted action with inputs from receptor-tyrosine kinase (RTK) pathways, TGF-β induces SMAD3 to form a complex with RNA binding protein poly-(rC) binding protein 1 (PCBP1, also known as hnRNPE1). Using cancer stem cell marker CD44 as a model, we show that the SMAD3-PCBP1 complex regulates alternative splicing via direct RNA binding to allow for expression of protein products of a large number of genes enriched in pathways critical for EMT, cytoskeleton organization, adherens junction signaling and cell migration.
Results
Phosphorylation at T179 switches SMAD3 to a promoter of tumor invasion
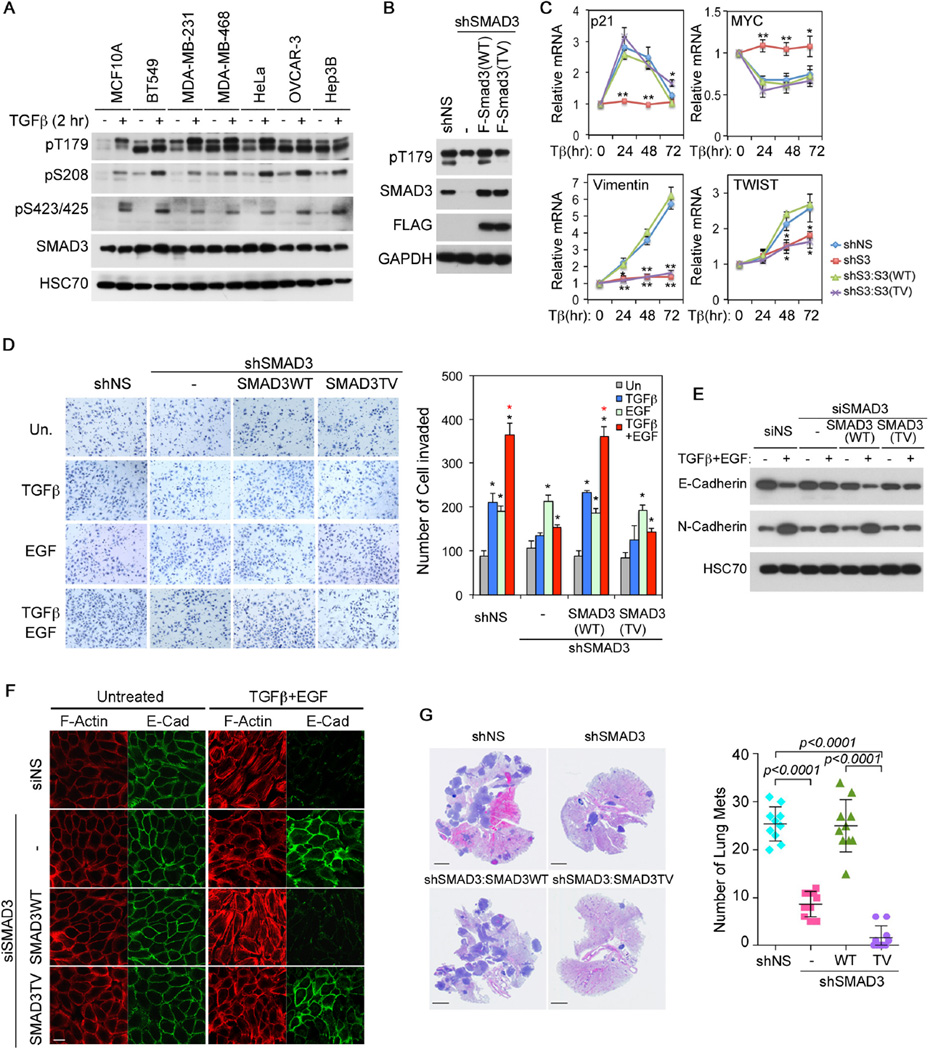
In normal human MCF10A mammary gland epithelial cells and several lines of breast cancer (BT549, MDA-MB-231, MDA-MD-468), cervical cancer (HeLa), ovarian cancer (OVCAR-3) and liver cancer (Hep3B) cells, TGF-β-induced early signaling events of phosphorylation at two C-terminal serine residues as well as T179 and S208 in the linker region of SMAD3 were readily detectable (Figure 1A); however, we noted that the basal level of the linker phosphorylation, particularly the one at T179, was already much higher in the above cancer cells comparing to that in MCF10A cells and in contrast to that of the C-terminal sites (Figure 1A). Previously, we and others showed that T179 phosphorylation inhibits the transcriptional activity of SMAD3 by blocking its ability to form a complex with SMAD4 (Matsuura et al., 2005; Tang et al., 2011; Wang et al., 2009). Thus, it is possible that this phosphorylation event may constitute a contextual adjustment that usurps SMAD3 for cancer cell invasion through noncanonical mechanisms. To test this hypothesis, we generated several pools of HeLa cells in which expression of the endogenous SMAD3 was depleted by shRNA, or depleted and then rescued using vectors that express shSMAD3-resistant, FLAG-tagged, but otherwise wild type SMAD3 (SMAD3(WT)) or the T179V mutant (SMAD3(TV)) (Figure 1B). As expected, TGF-β-induced transcriptional regulation of its direct target genes, including p21 (also known as CDKN1A) and MYC that control growth, was severely compromised in these SMAD3 knockdown cells until SMAD3 expression was rescued by SMAD3(WT) or the mutant SMAD3(TV) (Figures 1C and S1B). Surprisingly, while SMAD3(WT) also restored the transcriptional regulation of two mesenchymal genes, Vimentin and TWIST, which are late response genes during EMT (Tan et al., 2012; Vervoort et al., 2013), the mutant SMAD3(TV) failed to do so (Figure 1C). Since direct test with transcription reporter in SMAD3 knockdown cells indicated that SMAD3(TV) still possesses transcriptional activity (Figure S1A), the differential regulation of mesenchymal genes imposed by T179 phosphorylation of SMAD3 is likely mediated by a mechanism other than transcription.
Figure 1. SMAD3 T179 phosphorylation is required for TGF-β-mediated invasion.
A: Phosphorylation of SMAD3 in normal mammary gland epithelial cell MCF-10A and various cancer cells. The SMAD3 pT179 antibodies recognize both SMAD2 pT220 (upper band) and SMAD3 pT179 (lower band).
B: Generation of stable SMAD3 knockdown HeLa cells and cells in which SMAD3 expression was rescued with shSMAD3-resistant, F-SMAD3(WT) or F-SMAD3(TV) construct.
C: qRT-PCR analyses of p21, MYC, Vimentin and TWIST mRNAs in HeLa cells. Relative levels of expression were normalized to untreated cells. Data are shown as mean ± S.D. (n=3), and statistical significance over shNS cells are indicated. *, p<0.02; **, p<0.002.
D: Invasion assays in HeLa cells carrying various SMAD3 vectors. Quantitation data are shown as mean ± S.D. (n=3). Statistical significance (p<0.02) is indicated for treated versus untreated samples (black *) and for samples treated with TGF-β and EGF versus those with TGF-β alone (red *).
E: E-Cadherin and N-Cadherin expression in MCF10A cells carrying various SMAD3 vectors after TGF-β and EGF treatment for 3 days.
F: Immunofluorescence staining of F-Actin (phalloidin) and E-Cadherin in MCF10A cells after TGF-β and EGF treatment for 4 days. Bar = 20 µm.
G: H&E staining of lung sections from nude mice that were tail-vein injected with various HeLa cells. Bars = 5 mm. Numbers of lung metastasis nodule per animal are shown in the right as mean ± SD (n=10).
See also Figure S1.
TGF-β induction of EMT and cell invasion is known to cooperate with signaling pathways downstream of RTKs (Janda et al., 2002; Lamouille and Derynck, 2007; Wendt et al., 2010). This was recapitulated by mesenchymal gene expression in HeLa cells and EMT experiments in MCF10A cells (Figure S1B–C). The same principle was also demonstrated by invasion assays through the matrigel-coated basement membrane in response to the activation of corresponding pathways by TGF-β and EGF individually or additively by both (Figure 1D). However, the ability of SMAD3-depleted HeLa cells to invade in response to TGF-β was drastically reduced, unless they were rescued by SMAD3(WT) but not SMAD3(TV) (Figure 1D). Similarly, SMAD3(TV) also failed to rescue the ability of SMAD3-depleted MCF10A cells to undergo EMT or invade matrigel in response to TGF-β and EGF treatment (Figures 1E–F, S1D–E). To further substantiate the regulation of T179 phosphorylation on the tumor invasion function of SMAD3 in vivo, we performed the lung metastasis assay in nude mice using the above HeLa cells through tail vein injection. Once again, the mutant SMAD3(TV) failed to restore the 20+ number of histologically confirmed metastasis nodules that were otherwise observed in the lungs receiving the control shNS or SMAD3(WT)-rescued HeLa cells (Figure 1G). These results indicate that phosphorylation at T179 activates a latent function of SMAD3 that likely mediates the tumor invasion function of TGF-β. Since the non-phosphorylatable SMAD3(TV) mutant is still transcriptionally active (Figures 1C and S1A) and even exhibited a stronger role in suppressing tumor formation in xenografts (Figure S1F), our results also suggest that the transcriptional activity of SMAD3 is not sufficient for supporting tumor invasion.
TGF-β and EGF respectively induce SMAD3 and PCBP1 to interact and accumulate in the nucleus
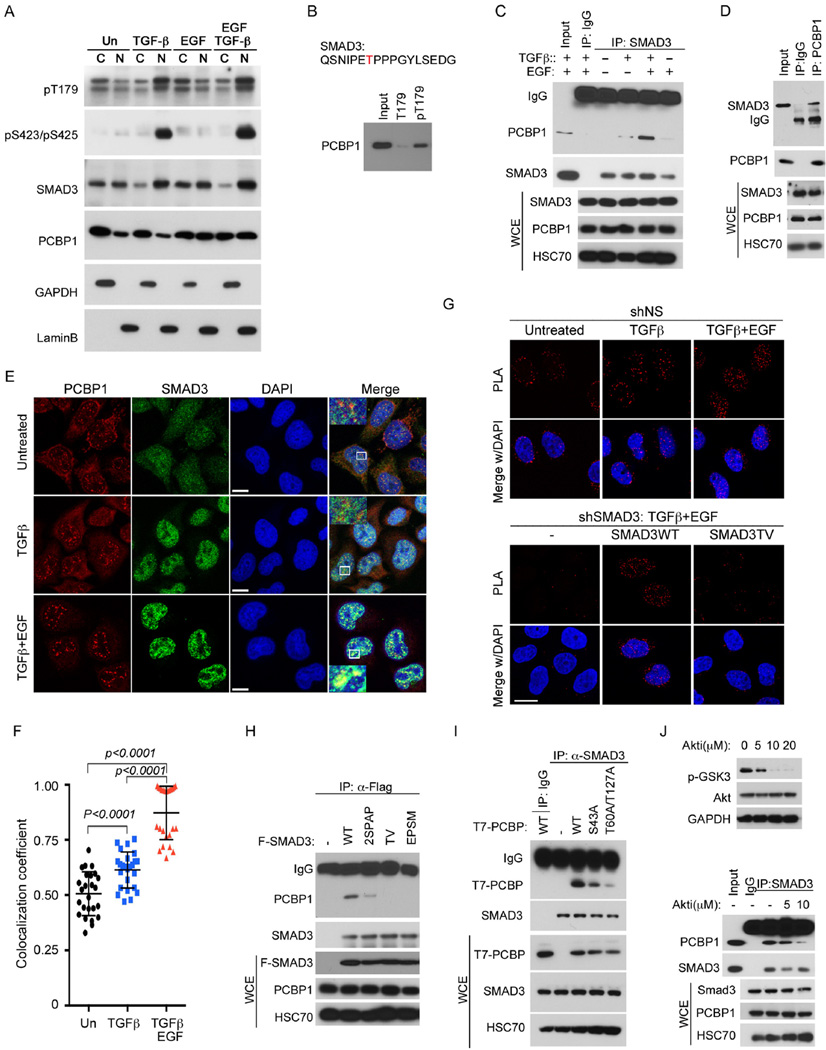
Phosphorylation of SMAD3 at T179 can be induced by noncanonical TGF-β signaling or EGF and its related TGFα via EGFR activation (Gao et al., 2009; Matsuura et al., 2005; Wang et al., 2009) (Figures S2A–C). This form of SMAD3 is primarily present in the cytoplasm of unstimulated cells until it moves into the nucleus upon TGF-β treatment, regardless of the presence of EGF (Figure 2A). To unravel the mechanism of T179-mediated regulation, we sought to identify proteins that specifically recognize pT179 in the nuclear extracts of HeLa cells using a pair of immobilized SMAD3 peptides centered on nonphosphorylated T179 or phosphorylated pT179 (Figure S2D). Among potential interacting proteins, we found NEDD4L, a known binding partner of SMAD3 that recognizes pT179 (Gao et al., 2009), and PCBP1 (Figures S2E and 2B). PCBP1 is an omnipresent protein containing highly conserved triple repeats of the RNA-binding KH domain and known to participate in multiple mRNA processing steps and translational silencing (Ji et al., 2003; Meng et al., 2007). The interaction between endogenous SMAD3 and PCBP1 was confirmed by co-immunoprecipitation in HeLa cells that were treated with TGF-β and/or EGF (Figures 2C–D), and the results also revealed that the highest affinity between SMAD3 and PCBP1 was attained when the cells were treated with both factors (Figure 2C).
Figure 2. Phosphorylation-dependent interaction between SMAD3 and PCBP1.
A: Nuclear and cytosolic distribution of SMAD3 and PCBP1 in HeLa cells treated with indicated growth factors for 2 hr. GAPDH and Lamin B were used as cytoplasmic (C) and nuclear (N) specific protein markers, respectively.
B: Western blot of PCBP1 isolated from HeLa cells nuclear extracts using immobilized SMAD3 T179 or pT179 peptides.
C: Western blots of SMAD3 and PCBP1 following immunoprecipitation (IP) of SMAD3 from whole cell extracts (WCE) of HeLa cells treated with TGF-β and/or EGF for 2 hr.
D: Western blots of SMAD3 and PCBP1 following IP of PCBP1 from WCE of HeLa cells treated with TGF-β and EGF for 2 hr.
E: Immunofluorescence staining of SMAD3 and PCBP1 in HeLa cells treated with TGF-β or TGF-β and EGF for 2 hr. The yellow color is indicative of colocalization. Bar =10 µm.
F: Scatter dot plot of co-localization coefficiency of SMAD3 with PCBP1 from (E). The data are shown as mean ± SD (n=25).
G Proximity ligation assay (PLA) of SMAD3 and PCBP1 in the nucleus of HeLa cells. TGF-β or combined TGF-β and EGF treatments were for 2 hr. Bar = 20 µm.
H: IP-Western analyses of various Flag-tagged SMAD3 mutants to bind endogenous PCBP1 in HeLa cells after treatment with TGF-β and EGF for 2 hr.
I: IP-Western analyses of various T7-tagged PCBP1 mutants to bind endogenous SMAD3 in HeLa cells after treatment with TGF-β and EGF for 2 hr.
J: AKT inhibitor at a concentration (10 µM) that is sufficient to block GSK3β phosphorylation (top) decreased the affinity of SMAD3 to PCBP1 in HeLa cells treated with TGF-β and EGF for 2 hr (bottom).
See also Figure S2.
Previously, EGF or serum-induced phosphorylation on T60 and T127 of PCBP1 was reported to cause the release of PCBP1 from translational repression and render it to be retained in the nucleus (Meng et al., 2007). Indeed, this nuclear enrichment was recapitulated in HeLa cells with EGF treatment, but was not affected by TGF-β (Figures 2A and 2E). Interestingly, SMAD3, which became enriched in the nucleus in response to TGF-β, was found colocalized with PCBP1 in punctate nuclear speckles that are intimately associated with pre-mRNA splicing under the combined TGF-β and EGF treatment (Figures 2E–F). Exogenously expressed GFP-SMAD3 also colocalized with PCBP1 in the nuclear speckles marked by SC35 in TGF-β and EGF treated HeLa cells, but GFP-SMAD3(TV) was never in overlapping with PCBP1, albeit it was broadly present throughout the nucleus (Figure S2F). Proximity ligation assay (PLA) further corroborated the ligand-induced colocalization of PCBP1 with SMAD3 but not SMAD3(TV) in the nucleus (Figures 2G and S2G). Apparently, TGF-β and EGF respectively induce SMAD3 and PCBP1 to accumulate in the nucleus and to interact with each other.
Since the interaction between SMAD3 and PCBP1 is enhanced by simultaneous TGF-β and EGF signaling, both of which are transduced by kinases, we speculated that phosphorylation might regulate the binding as well. To test this, we carried out immunoprecipitation experiments in HeLa cells transfected with phosphorylation site mutants and treated with TGF-β and EGF. The results showed that PCBP1 could bind wild type SMAD3, or weakly the mutant SMAD3 containing S204A and S208A substitution (2SPAP), but the binding to T179V and EPSM (T179V, S204A, S208A, and S213A) mutants was completely abolished (Figure 2H), indicating an absolute requirement of pT179 for binding. PLA experiments also indicated that the mutant SMAD3(TV) does not overlap with endogenous PCBP1 (Figures 2G and S2G). On the other hand, substituting the EGF-induced phosphorylation sites, T60 and T127, of PCBP1 with alanine also abrogated the interaction (Figure 2I). Moreover, mutating the AKT2 site of PCBP1 at S43, whose phosphorylation has the similar effect of releasing PCBP1 from acting on translational repression (Chaudhury et al., 2010), weakened the interaction as well (Figure 2I). Finally, inhibiting AKT kinase activity with a chemical inhibitor, which effectively blocked the phosphorylation of GSK3β, a well-known AKT substrate, and the nuclear localization of PCBP1 (Figure S2H), greatly diminished the TGF-β/EGF-induced interaction between endogenous SMAD3 and PCBP1 (Figure 2J). Because the AKT inhibitor does not affect T179 phosphorylation of SMAD3 (Figure S2A), the effect of AKT must be channeled through PCBP1. In conclusion, the interaction between SMAD3 and PCBP1 relies on phosphorylation at multiple sites on each protein.
TGF-β suppresses epithelial specific splicing of CD44 through SMAD3 and PCBP1
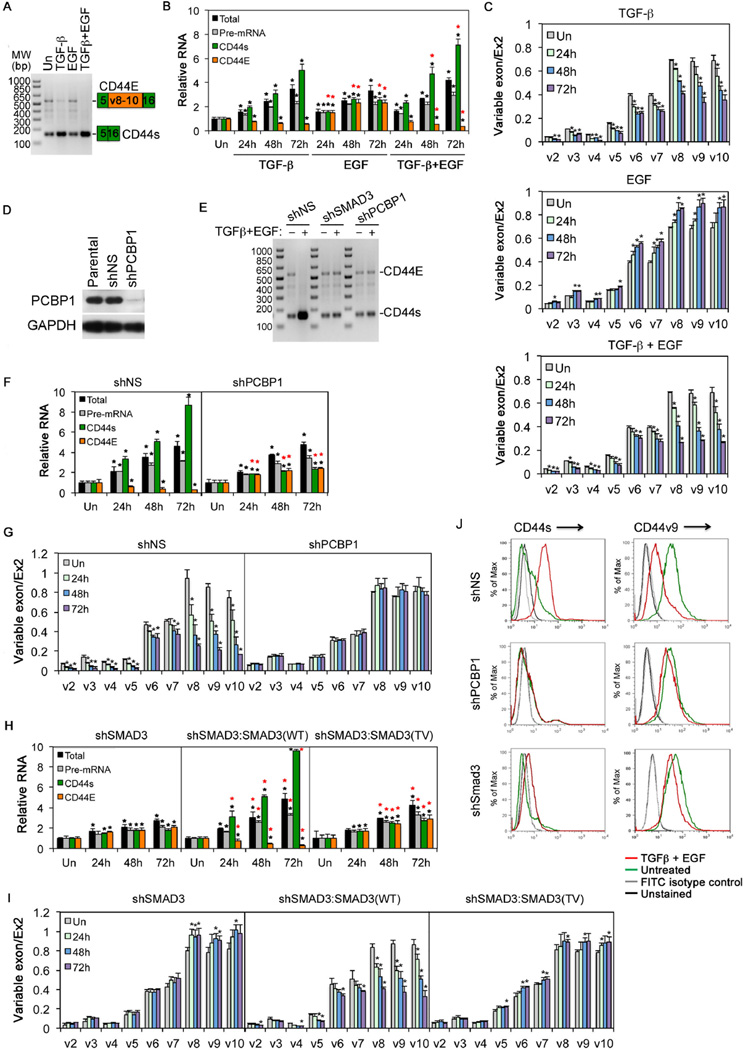
A known PCBP1-regulated splicing target is the CD44 gene, which encodes a family of isoformic glycoproteins with multi-facet functions in cell adhesion, lymphocyte homing, and cancer stem cell signaling (Ponta et al., 2003). All human and mouse CD44 isoforms are generated by alternative splicing of 20 exons (Naor et al., 1997); the shortest isoform devoid of any variable exon, CD44s, is expressed abundantly in most tissues and referred to as the mesenchymal form, whereas other isoforms with a mixed number of variable exons inserted between constant Exon 5 and Exon 16 are restricted to specific tissues (Figure S3A). Normal epithelial cells or epithelium-derived carcinomas preferentially express an isoformic CD44E containing the last 3 variable exons (v8–10). During EMT, production of CD44E is suppressed, while CD44s becomes highly elevated (Biddle et al., 2013; Brown et al., 2011). This switch has been known to play an essential role in EMT (Brown et al., 2011). In HeLa cells, TGF-β treatment, especially in combination with EGF, suppressed the production of the epithelial specific CD44E while promoting the standard isoform CD44s (Figure 3A). To analyze CD44 splicing events in detail, we designed quantitative real time PCR (qRT-PCR) reactions to detect CD44 total, CD44s, CD44E mRNAs, as well as CD44 pre-mRNA (Figure S3A and Table S1). Using this system, we found that while EGF moderately induced all four categories of CD44 RNAs, TGF-β drastically induced CD44s, but selectively suppressed the production of CD44E; this bipartite pattern was even more pronounced when the cells were treated with both factors (Figure 3B). We attributed this regulation to alternative splicing because the levels of CD44 total mRNA and pre-mRNA, used as proxies for transcription and RNA stability, respectively, increased steadily over the time course regardless if the cells were treated with TGF-β, EGF, or both (Figure 3B). In support of this notion, we further measured the usage of each variable exon defined as the ratio of the variable exon in question to the constant Exon 2, and the results showed that exons v6–7 were moderately used and exons v8–10 heavily used in the untreated cells (Figure 3C), which are consistent with the epithelial origin of HeLa cells. However, TGF-β treatment markedly decreased the usage of v6–10 exons (Figure 3C, top panel), whereas EGF enhanced it (Figure 3C, middle panel). More importantly, treating the cells with TGF-β and EGF together further suppressed the usage of the epithelial specific v8–10 exons (Figure 3C, bottom panel), thus confirming a negative regulation of TGF-β on the selection of the alternate variable exons. A similar bipartite pattern of increasing CD44s versus decreasing CD44E with combined TGF-β and EGF or TGF-α treatment was also observed in MCF10A cells (Figure S3B–C), indicating that TGF-β regulation of alternative splicing is not a cell-type specific phenomenon. Of note, unlike that in cancerous HeLa cells, the preference for CD44s over the epithelial CD44E isoform exhibited by TGF-β alone treatment was rather subdued in normal MCF10A cells; this appears to stem out of a requirement to activate RTK signaling pathways (which are not active in normal cells) as evident from the experiments using chemical inhibitors of MEK, CDK and AKT (Figure S3D).
Figure 3. PCBP1 and SMAD3-dependent CD44 isoform switch upon TGF-β and/or EGF treatment.
A: Detection of CD44E and CD44s in HeLa cells in a single RT-PCR reaction using one pair of primers. The growth factor treatment was for 24 hr.
B: qRT-PCR analyses of CD44 total mRNA, pre-mRNA, CD44s, or CD44E mRNAs in HeLa cells using specific primer pairs.
C: qRT-PCR analyses of CD44 variable exon usage in HeLa cells.
D: Western blot showing PCBP1 knockdown in HeLa cells.
E: RT-PCR detection of CD44s and CD44E as in A. HeLa cells were treated with TGF-β and EGF for 72 hr.
F: qRT-PCR analyses of different CD44 RNAs in HeLa cells treated with TGF-β and EGF.
G: qRT-PCR analyses of CD44 variable exon usage in HeLa cells treated with TGF-β and EGF.
H: qRT-PCR analyses of different CD44 RNAs in HeLa cells treated with TGF-β and EGF
I: qRT-PCR analyses of CD44 variable exon usage in HeLa cells treated with TGF-β and EGF.
J: FACS analyses of cell surface expression of CD44 isoforms in HeLa cells. The cells were treated with TGF-β and EGF for 72 hrs.
For qRT-PCR, relative levels of expression were normalized to untreated cells (Un); variable exon usage defined as the ratio between variable exon to standard exon 2. All bars are shown as mean ± SD (n=3). The black * denotes statistical significance (p<0.02) between treated and untreated samples; the red * denotes statistical significance (p<0.02) for samples treated with TGF-β and EGF or EGF versus those with TGF-β alone (B), or for samples expressing shPCBP1 versus those expressing shNS (F), or for samples expressing SMAD3 WT or TV versus those with expressing shSMAD3 (H).
See also Figure S3.
In light of the additive effect of TGF-β and EGF treatment on SMAD3-PCBP1 complex formation and nuclear localization, we reasoned that TGF-β suppression of the alternative use of variable exons is very likely channeled through PCBP1. Indeed, depletion of PCBP1 in HeLa cells diminished the preferred expression of standard CD44s over epithelial CD44E upon the combined TGF-β/EGF treatment (Figures 3D–F) and reversed the suppression of variable exon usage (Figure 3G), while leaving unaffected the levels of CD44 total and pre-mRNAs (Figure 3F). Similar effects were observed in HeLa cells with TGF-β only treatment (Figure S3E). Moreover, depletion of PCBP1 also blunted the EGF-induced increase in variable exon v6–10 usage (Figure S3E). Thus, PCBP1 is an essential factor in TGF-β and EGF-mediated regulation of CD44 alternative splicing.
To assess the role of SMAD3 in the splicing of CD44 variable exons, we quantified the levels of different CD44 RNAs as well as variable exon usage in the SMAD3-depleted HeLa cells. In the absence of SMAD3, the transcription and splicing effects of TGF-β and EGF treatment were greatly diminished (Figure 3E, 3H). The exon usage analysis also showed a complete reversal of the variable exon suppression, particularly v8–10 exons (Figures 3I and S3E). These data indicate that TGF-β-regulated CD44 alternative splicing requires SMAD3. The requirement of SMAD3 and PCBP1 in switching CD44 expression from CD44E to CD44s in response to TGF-β and EGF treatment was also confirmed by cell surface detection of specific CD44 isoforms (Figures 3J and S3F). As with SMAD3 and PCBP1 binding, reintroducing shSMAD3-resistant SMAD3(WT) rescued both transcriptional and splicing responses to TGF-β and EGF treatment, but the SMAD3(TV) mutant rescued only transcriptional response (Figure 3H). Likewise, SMAD3(WT) but not the TV mutant rescued the TGF-β-mediated suppression of variable exon usage (Figures 3I and S3E). These results demonstrate that the activity of SMAD3 in CD44 alternative splicing requires a phosphorylated pT179 and the interaction with PCBP1.
TGF-β/SMAD3 regulates alternative splicing independent of transcription
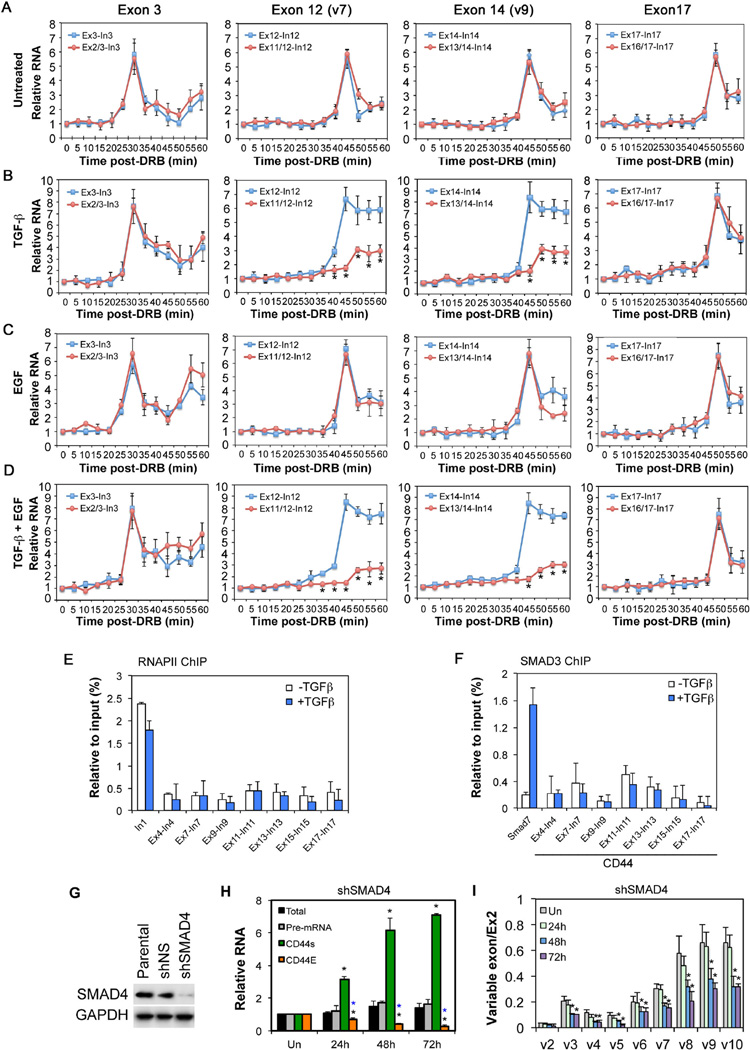
Since RNA splicing occurs co-transcriptionally, transcription regulators such as SMAD3 could theoretically influence exon choice by pausing the RNAPII elongation complex to allow splicing to proceed on alternate exons (Kornblihtt et al., 2013). Alternatively, SMAD3 could act directly on the splicing machinery assembled on pre-mRNAs through PCBP1 without altering the rate of transcription. However, since both TGF-β and EGF signaling also affects CD44 transcription, we designed qRT-PCR reactions capable of resolving these possibilities in real time, and used a reversible transcription inhibitor 5,6-dichlorobenimidazole-1-β-D-ribofuranoside (DRB) to block the transcription elongation such that only the new round of transcription and splicing were examined upon DRB withdrawal (Figures S4A–B). These qRT-PCR reactions employ exon-intron primers to monitor the de novo pre-mRNA synthesis by transcription or a pair of primers from the immediately preceding exon-exon junction and the lagging intron to monitor the production of mature mRNA by splicing at a given exon (Singh and Padgett, 2009) (Figure S4B). This approach would avoid the interference from transcription and would result in the appearance of a peak of new RNA synthesis due to the excision of the lagging intron that contains the intronic primer. We thus examined if TGF-β signaling alters the rate of RNAPII elongation or splicing over several representative exons covering the length of CD44 pre-mRNA. In the case of Exon 3 in CD44, the appearance of the unspliced pre-mRNA peaked at 30 minutes following DRB withdrawal, whereas the pre-mRNA peaks of Exons 12 (v7), 14 (v9), and 17 occurred at 45, 45+, and 50 minutes, respectively (Figure 4A). Taking the genomic distance of these exons into consideration, these measurements were consistent with RNAPII elongation complex moving around 1.7–2.1kb.min−1 reported previously for the CD44 gene (Veloso et al., 2014). In the absence of TGF-β or EGF, the spliced CD44 mRNA products at Exons 3, 12 (v7), 14 (v9), or 17 appeared in congruent kinetic patterns as the unspliced pre-mRNAs (Figure 4A), implying that the variable Exon 12 and 14 were spliced at the same rate as the constant Exon 3 or 17. However, TGF-β upheld the production of the unspliced pre-mRNA after it reached the peak levels while it suppressed the spliced mRNA at variable Exon 12 (v7) and 14 (v9) (Figure 4B). In contrast, the production of pre-mRNA and mature mRNA from the constant Exon 17 in the post-variable region proceeded normally, still reaching the peak 50 minutes after DRB withdrawal (Figure 4B). These results indicated that the RNAPII elongation complex moved through CD44 variable exons without impediment, but the splicing of these alternate exons was blocked in the presence of TGF-β. Once again, EGF alone showed no significant effect on either transcription or splicing (Figure 4C), but TGF-β and EGF together exacerbated the dichotomy between the production of unspliced and spliced RNA at variable exons (Figure 4D).
Figure 4. SMAD3 regulation of CD44 alternative splicing is independent of its transcriptional activity.
A-D: Real time analyses of transcription of nascent RNA (blue line) and splicing (red line) at selective CD44 exons from DRB-synchronized HeLa cells. Data are shown as mean ± SD (n=3), the * denotes statistical significance (p<0.02) between transcription and splicing.
E-F: ChIP analyses of RNAPII (E) or SMAD3 (F) binding at various CD44 genomic regions in HeLa cells treated with TGF-β for 2 hr. Data are shown as mean ± SD (n=3). The SMAD7 promoter was used as a positive control for SMAD3 binding to DNA.
G: Western blot showing SMAD4 knockdown in HeLa cells.
H: qRT-PCR analyses of different CD44 RNAs in shSMAD4-expressing HeLa cells treated with TGF-β and EGF. Data are shown as in Figure 3.
I: qRT-PCR analyses of CD44 variable exon usage in shSMAD4-expressing HeLa cells treated with TGF-β and EGF. Data are shown as in Figure 3.
See also Figure S4.
Consistent with a lack of participation of transcription in CD44 alternative splicing, RNAPII and SMAD3 were not found on the exonic DNA as determined by ChIP assays, nor did TGF-β treatment alter this observation, although RNAPII was found to be enriched near the CD44 transcription start (e.g. Intron 1, Figure 4E), and SMAD3 was enriched at the Smad7 promoter (Figure 4F). Further evidence for excluding transcription in direct regulation of CD44 splicing by SMAD3 was obtained from experiments carried out in SMAD4-knockdown HeLa cells (Figure 4G), in which SMAD4 depletion abrogated the induction of CD44 pre-mRNA and total mRNA by TGF-β and EGF treatment (Figure 4H), as expected from the role of SMAD4 in TGF-β-induced transcriptional responses. However, the production of CD44s continued unabatedly at the expense of CD44E (Figure 4H), and the usage of variable exons continued to be suppressed (Figure 4I).
SMAD3 suppresses CD44 alternative splicing by blocking access of spliceosomal factors to PCBP1
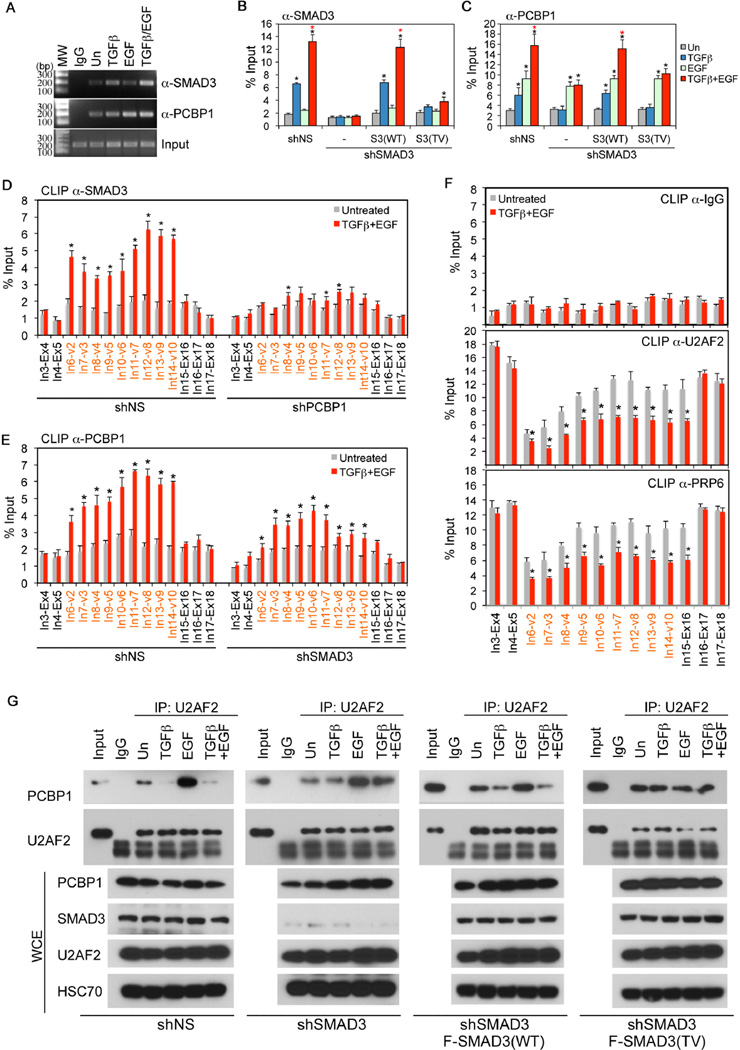
Having demonstrated that the regulation of CD44 splicing by TGF-β signaling is separate from the transcriptional activity of SMAD3, we sought to understand how SMAD3 blocks the selection of CD44 variable exons via the interaction with PCBP1. In the splicing reaction, intron/exon boundaries are recognized through base pairing by U1 and U2 snRNPs in the presence of 5’ and 3’ splice site recognition factors. Alternatively spliced exons often have short and intrinsically weak splice sites; their selections are modulated by various cis-acting sequence elements, RNA binding serine-arginine (SR) factors, and hnRNPs (Kornblihtt et al., 2013). Using RNA-immunoprecipitation (RIP) assay, we found that SMAD3 presented in the same complex of CD44 pre-mRNA and the association was augmented by TGF-β, and additively by simultaneous treatment with both TGF-β and EGF (Figure 5A–B). Likewise, the association of PCBP1 to CD44 pre-mRNA was also enhanced by these two factors (Figure 5A and 5C). Interestingly, although knocking-down SMAD3 showed no effect on EGF-induced interaction of PCBP1 with CD44 pre-mRNA, it blocked the enhancement by TGF-β/EGF (Figure 5C). However, unlike its wild type counterpart, SMAD3(TV) could not be recruited to CD44 pre-mRNA, nor could it augment the association of PCBP1 to CD44 pre-mRNA (Figure 5B–C). In order to determine where SMAD3 and PCBP1 bind to CD44 pre-mRNA, we performed UV cross-linking and immunoprecipitation (CLIP) assay and scanned the intron-exon boundaries spanning CD44 pre-mRNA using qRT-PCR. Results from this experiment revealed that binding of SMAD3 to CD44 pre-mRNA in the variable exon region from Intron 6 to Exon 15 was significantly enhanced by TGF-β/EGF (Figure 5D, left). Knocking-down PCBP1 essentially diminished the binding of SMAD3 to CD44 pre-mRNA to the basal level (Figure 5D, right), suggesting that the interaction of SMAD3 with CD44 pre-mRNA is dependent on PCBP1. This notion is further supported using the RIP assay (Figure S5). Consistent with the idea that SMAD3 and PCBP1 bind CD44 pre-mRNA in the same complex, the binding of PCBP1 in the variable exon region was also enhanced by TGF-β/EGF (Figure 5E, left), and in keeping with the RIP assay result (Figure 5C). Knocking-down SMAD3 partially weakened the PCBP1 binding (Figure 5E, right), confirming the stabilizing effect of SMAD3 on PCBP1 and CD44 pre-mRNA interaction.
Figure 5. Formation of the SMAD3 and PCBP1 complex on variable exon regions of CD44 pre-mRNA precludes spliceosome assembly.
A: RIP assays of SMAD3 and PCBP1 binding to CD44 pre-mRNA in HeLa cells with growth factor treatment for 2 hr.
B-C: qRT-PCR analyses of RIP assay for SMAD3 (B) and PCBP1 (C) in HeLa cells.
D-E: qRT-PCR analyses of CLIP assay for SMAD3 (D) or PCBP1 (E) binding to intron-exon junctions (red denotes variable intron-exon junctions) of CD44 pre-mRNA in HeLa cells.
F: CLIP assays for control non-specific IgG, as well as U2AF2 or PRP6 binding to intron-exon junctions (red denotes variable intron-exon junctions) of CD44 pre-mRNA in HeLa cells.
G: IP-Western analyses of PCBP1 and U2AF2 following IP of U2AF2 from whole cell extracts (WCE) of HeLa cells expressing different vectors and treated with growth factors for 2 hr.
All bars are shown as mean ± SD (n=3). Statistical significance (p<0.02) is indicated for treated (2 hr) versus untreated samples (black *) and samples treated with TGF-β and EGF versus those with TGF-β alone (Red *).
See also Figure S5.
Previously, PCBP1 was postulated to promote the alternative splicing of a CD44 minigene by recruiting U2 snRNP auxiliary factor (U2AF) (Meng et al., 2007). It is possible that the binding of SMAD3 could deny the access of splicing factors to PCBP1, thereby disrupting spliceosome assembly in the variable exon region of CD44 pre-mRNA. To address this possibility, we examined if TGF-β and EGF could reduce the binding of U2AF2 to CD44 pre-mRNA by CLIP. The results indicated that treating cells with TGF-β and EGF indeed reduced U2AF2 binding in variable exon region, but it did not affect their binding in the constant exon region (Figure 5F). Similar results were obtained with the binding of PRP6, a splicing factor required for the assembly of functional spliceosomes with catalytic activity (Kornblihtt et al., 2013) (Figure 5F). Moreover, co-immunoprecipitation experiments showed that U2AF2 was in a complex with PCBP1, and this association was strongly enhanced by EGF, but suppressed by TGF-β and the combined TGF-β/EGF treatment (Figure 5G, left). Depletion of SMAD3 abolished the suppression by TGF-β on the U2AF2 and PCBP1 interaction (Figure 5G, 2nd panel to the left). Importantly, while reintroducing wild type SMAD3(WT) restored the TGF-β-mediated suppression, the SMAD3(TV) mutant showed no such an effect (Figure 5G, two right panels). Taken together, these results indicate that SMAD3 directly regulates the alternative splicing of CD44 by interacting with PCBP1 around variable exons of CD44 pre-mRNA to inhibit the recruitment of the splicing machinery to suboptimal splice sites, thereby promoting a switch in expression of the epithelial isoform to the mesenchymal isoform of CD44.
Alternative splicing of CD44 is critical to the tumor-promoting roles of TGF-β
The chain of events from T179 phosphorylation of SMAD3 leading to PCBP1 binding and to the suppression of CD44 epithelial specific variable exon usage suggests that it might be part of a critical mechanism that converts TGF-β from a tumor suppressor to a tumor promoter. Indeed, in advanced human breast cancers and cancer stem-like cells that have undergone EMT, CD44s expression is markedly upregulated (Biddle et al., 2013; Brown et al., 2011), and moreover, the switching from various epithelial isoforms to the mesenchymal CD44s has been proven essential for cells to undergo EMT and metastasis (Brown et al., 2011; Xu et al., 2014). We also observed the switch from epithelial isoforms to CD44s in two other cell types upon TGFβ/EGF treatment, despite different variable exon preference in these cells (Figure S6A–C). Thus, to determine if the regulation of CD44 alternative splicing is relevant to the tumor-promoting role of TGF-β, we created additional pools of HeLa cells that are devoid of CD44 entirely using shRNAs against constant exons (shCD44), or re-express standard CD44s or an epithelial variable isoform CD44v in the CD44 knockdown background (shCD44 + CD44s, or shCD44 + CD44v), respectively (Figure 6A). We found that removing total CD44 altogether abolished TGF-β/EGF-induced invasion, whereas re-expressing CD44s but not CD44v was sufficient to bring the basal level of invasion to that matching TGF-β/EGF treatment (Figure 6B). These observations are consistent with mesenchymal and metastasis associated protein expression, in which knocking-down total CD44 inhibited induction of Vimentin, CTGF, and MMP2 by TGF-β/EGF (Figure 6C). Re-expressing CD44s but not CD44v in the shCD44 background was sufficient to induce Vimentin and MMP2, albeit slightly less on CTGF (Figure 6C). Consistent with our conclusion, SMAD3(TV) also failed to induce Vimentin and MMP2 expression (Figure S6D). Moreover, CD44s but not CD44v promoted lung metastasis in cells in which total CD44 was depleted (Figure 6D).
Figure 6. The mesenchymal CD44s isoform is required for cell invasion and metastasis.
A: FACS analyses of cell surface expression of different CD44 isoforms in HeLa cells carrying different CD44 expression vectors as indicated. RFI: mean relative fluorescence intensity.
B: Invasion assays of HeLa cells carrying different CD44 expression vectors. Quantification data are shown as mean ± S.D. (n=3), and * denotes statistical significance (p<0.02) against the corresponding shNS control cells.
C: Western analyses of mesenchymal marker Vimentin, CTGF and MMP2 in HeLa cells carrying various CD44 expression vectors. The cells were treated with TGF-β and EGF for 3 days.
D: H&E staining of lung sections from nude mice that were tail vein injected with HeLa cells carrying various CD44 expression vectors. Bar =10 mm. Numbers of lung metastasis nodule per animal are shown in the right as mean ± SD (n=10).
See also Figure S6.
TGF-β regulation of alternative splicing has a global impact on cellular gene expression
To assess the global impact of TGF-β on alternative splicing, we conducted RNA-seq of total RNAs isolated from various pools of HeLa cells that were generated for this study (Table S2). Out of a total 83,257 alternative splicing events detected, 384, representing 287 human genes, were significantly changed (with a cut-off ≥15% exon usage) linked to TGF-β/EGF treatment (Figure 7A, Table S3). The most frequently occurring changes were skipped exon (SE) events, which encompassed 102 included and 137 skipped exons (Figure 7A, Table S4). Other changes were mutually excluded exons (MXE), alternative 5’ or 3’ splice sites (A5SS or A3SS), and retained introns (RI) (Figure 7A). Ingenuity Pathway Analyses indicated that many functional categories instrumental to EMT and cancer cell migration including those regulated by the RHOA, Netrin, actin cytoskeleton, and epithelial adherens junction signaling pathways were enriched in these 287 alternatively spliced genes (Figure 7B, Table S5). When comparing our data on SE events with a published data set from human mammary epithelial cells (HMLE) comprising exons whose splicing patterns were altered during EMT induced by overexpressing TWIST (Shapiro et al., 2011), we identified 23 overlapping SE events between the two data sets (Figure 7C). The absence of a substantial overlap between these two data sets strongly argues that the majority of changes in the splicing pattern that we detected are direct consequence of TGF-β and EGF signaling rather than secondary events associated with EMT or cell morphological changes.
Figure 7. Identification of genome-wide alternative splicing (AS) events in HeLa cells.
A: Pie chart of differentially regulated AS events, total 384 (FDR≤ 5%, Δψ≥ 15%, n=3) between untreated versus TGF-β and EGF treated cells (72hrs). SE: skipped exon; RI: retained intron; MXE: mutually excluded exons; A5SS: alternative 5’ splice site; A3SS: alternative 3’ splice site. The number of exon inclusion and exon skipping SE events is also listed.
B: Ingenuity pathway analyses of affected genes identified in A. Signaling pathways that are associated with EMT and cell movement are significantly enriched. P-value (-log10) is indicated on the X-axis. Cut off (p≤0.02) is shown by a dash line in the graph.
C: Venn diagram of overlapping skipped exon (SE) events between those induced by the combined TGF-β and EGF treatment and by TWIST overexpression. The 163 SE events (Shapiro et al, 2011) were used in comparison because analyzing the original RNA-seq data using our analysis pipeline yielded much smaller number of SE events.
D: Representative RNA-seq reads at the CD44 locus. T+E: cells treated with TGF-β and EGF for 72 hr. Orange rectangle marks the position of the variable exon region.
E: Number of differentially regulated AS events induced by TGF-β and EGF treatment in HeLa cells stably expressing shNS, shPCBP1 or shSMAD3 vector.
F: qRT-PCR validation of nine SE events. Ratios of alternative exon versus constant exon of individual gene are shown as mean ± S.D. (n=3), and * denotes statistical significance (p<0.02) between treated (72 hr) and untreated samples.
See also Figure S7.
The RNA-seq analysis also recapitulated the TGF-β/EGF-induced exclusion of CD44 variable exons revealed through qRT-PCR as indicated by the reduction in the number of variable exon reads (Figure 7D). However, depleting PCBP1 or SMAD3 expression by shRNA nullified the role of the combined ligand treatment, confirming the requirement of PCBP1 and SMAD3 for this regulation (Figure 7D). In fact, the RNA-seq data further showed that PCBP1 was required for most, if not all, of the alternative splicing events regulated by TGF-β/EGF treatment, whereas SMAD3 participated in slightly fewer events (Figure 7E). Conversely, we performed qRT-PCR validation of the RNA-seq results in a cohort of 9 genes, which are crucial to EMT and cancer cell migration. As with CD44 variable exons, the 6 skipped exons within PDGFA, SLK, MAP3K7, KIF13, BAIAP2, and FMNL3, showed varying degrees of sensitivity to TGF-β-induced exon exclusion or EGF-induced exon inclusion, while the 3 included exons (SCRIB, PTEN, and CLSTN1) showed opposite responses (Figures 7F and S7A). However, regardless of the direction and extent of changes that TGF-β or EGF imposed on these alternative splicing events, PCBP1 was invariably required and, so was requirement for SMAD3 in the TGF-β-mediated of alternative splicing (Figure 7F). Nevertheless, SMAD3-independent alternative splicing events do exist (Figure S7B). Since the outcome of the combined TGF-β/EGF treatment was the same as or even stronger than that of TGF-β treatment alone, our results once again indicate that SMAD3 overrides the function of PCBP1.
Discussion
Since their discovery, receptor-specific SMADs and SMAD4 have been known as transcriptional mediators of TGF-β signaling in the nucleus (Derynck et al., 1998). SMAD3-mediated transcription was also reported to regulate splicing indirectly through epithelial splicing regulator protein 1 and 2 (ESRP1 and ESRP2) that were downregulated by TGF-β (Horiguchi et al., 2012; Warzecha et al., 2010). However, the transcriptional activity of SMAD3 is insufficient to regulate alternative splicing, because the SMAD3(TV) mutant is still fully capable of mediating TGF-β-induced downregulation of ESRP1 and ESRP2 (Figure S4C) but failed to act in TGF-β-regulated alternative splicing of CD44 (Figure 3). Our current investigation surprisingly reveals that SMAD3 possesses a previously unappreciated function in alternative splicing, which, unlike in the TGF-β-induced transcription, is independent of the SMAD3-SMAD4 complex that binds DNA of target gene promoters. Instead, after being induced to accumulate in the nucleus, SMAD3 partners with the RNA binding protein PCBP1 for RNA-binding and regulating splicing. Interestingly, SMAD3 was also shown to bind RNA independent of SMAD4 during miRNA processing (Davis et al., 2010).
Previously, PCBP1 was shown to be a translational repressor acting in the cytoplasm through interaction with eukaryotic elongation factor eEF1A1 and TGF-β or EGF treatment disrupted that association and the ability of PCBP1 to repress translation (Chaudhury et al., 2010; Meng et al., 2007). These observations are conspicuously consistent with our data in that the growth factor treatment relieves PCBP1 from its cytoplasmic roles and promotes it to participate in alternative splicing with SMAD3 in the nucleus (see model illustrated in supplemental graphic abstract). Recently, another RNA-binding protein hnRNPM was also reported to promote CD44s expression at the expense of CD44 variable exons during TGF-β-induced EMT (Xu et al., 2014). However, hnRNPM recognizes an intronic GU-rich motif and competes with the epithelial splicing regulator ESRP1 for binding to promote variable exon skipping, whereas PCBP1 recognizes the “poly-(rC) tract” and interacts with U2AF at the 3’ splice site to promote variable exon inclusion. The effect of TGF-β on hnRNPM-mediated alternative splicing is manifested through repressing ESRP1 expression. In contrast, by inducing the pT179-mediated SMAD3-PCBP1 complex in the nucleus, TGF-β disrupts the interaction between PCBP1 and U2AF and directly suppresses the usage of variable exons.
Since T179 phosphorylation, SMAD3 interaction with PCBP1, and the nuclear translocation of PCBP1 are all promoted by the activated RTK signaling, TGF-β is afforded with a capacity to regulate splicing and promote EMT synergistically with RTKs pathways. The requirement of simultaneous inputs from TGF-β and RTK signaling for SMAD3-PCBP1 interaction and splicing regulation has a cancer implication. Oncogenic activation of CDK or MAPKs causes SMAD3 to be phosphorylated at T179, which is essential for SMAD3 to bind PCBP1. However, phosphorylation of T179 by these kinases alone is not sufficient to induce nuclear translocation of SMAD3; TGF-β signaling must be activated as well. On the other hand, RTK signaling pathways such as that of EGFR must be activated to promote the nuclear trafficking of PCBP1 (Figure S2H). This mechanism may explain why tumors that arise from oncogenic mutations in the RTK signaling pathways still resort to TGF-β for the initiation of EMT and metastasis. The unique role of SMAD3 in mediating TGF-β-regulated alternative splicing also has an implication in cancer. In gastrointestinal and pancreatic cancers where the SMAD4 gene is frequently mutated or deleted in advanced stages of tumorigenesis, inactivation of SMAD4 and overexpression of TGF-β both have been shown to be associated with metastasis (Losi et al., 2007; Malkoski and Wang, 2012; Papageorgis et al., 2011). These seemingly paradoxical observations now can be accounted for by the mechanism revealed in our current study; it is possible that in the absence of SMAD4, SMAD3 becomes readily available to participate in the regulation of alternative splicing for the gain in malignant phenotypes. Indeed, a recent report indicates that upregulated SMAD3 promotes EMT and serves as a poor prognostic biomarker even in the absence of SMAD4 in pancreatic ductal adenocarcinoma (Yamazaki et al., 2014).
Experimental Procedures
Detecting splicing events
Primers covering an intronic region were used in qRT-PCR reactions for CD44 pre-mRNA, and primers covering different splicing junctions were used for total CD44, standard CD44s, and epithelial CD44E mRNAs (Table S1). Genome-wide differential alternative splicing events were identified using rMATS tool version 3.0.8. The RNA-Seq data sets have been deposited in GEO with accession number GSE71419.
Invasion and lung metastasis assay
Cell invasion assay was performed using Corning Biocoat™ Growth Factor Reduced Matrigel Invasion Chamber (Corning Life Sciences). Lung metastasis assay was performed using 5–6 weeks old female athymic nu/nu mice after injection of cells through tail vein. All mice were maintained and handled according to protocols approved by the Animal Care and Use Committee of the National Cancer Institute.
RIP, CLIP and ChIP assays
RIP was done using RIP-Assay Kit (MBL International), CLIP was performed with RNACHIP-IT kit (Active Motif) after UV crosslinking and sonication of cells, and ChIP was carried out with an EZ-ChIP Chromatin Immunoprecipitation Kit (Millipore).
Details and other methods are in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
SMAD3 forms a pTl79-dependent complex with PCBP1 on cue of TGF-β and EGFR signaling.
PCBP1 recruits SMAD3 to the alternate exon region of CD44 pre-mRNA.
The SMAD3-PCBP1 complex on CD44 pre-mRNA causes alternate exon exclusion.
Alternative splicing by SMAD3 is essential to the tumor-promoting role of TGF-β.
Acknowledgments
We thank Drs. B. O’Malley and R. Kumar for PCBP1 plasmids, Jing Yang for HMLE cells, Chonghui Cheng for the CD44v plasmid and Fang Liu for SMAD3 pT208 antibodies. This research was supported by the intramural research program of the NTH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.S. and Y.E.Z. conceived the project, V.T. performed most of experiments and analyzed results, K.S. performed peptide pull-down studies, M.Z performed Mass-Spec analyses, X.X performed ChlP experiments, S.G. and J.H. performed bioinformatics analyses of RNA-seq data, R.W. helped with in vivo metastasis assays, V.T. and Y.E.Z wrote the manuscript with input from all authors.
Reference
- Biddle A, Gammon L, Fazil B, Mackenzie IC. CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal transition. PloS one. 2013;8:e57314. doi: 10.1371/journal.pone.0057314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Muthusamy BP, Saeteurn KY. Signaling pathway cooperation in TGF-beta-induced epithelial-mesenchymal transition. Curr Opin Cell Biol. 2014;31:56–66. doi: 10.1016/j.ceb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Drabsch Y, ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer metastasis reviews. 2012;31:553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massague J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS letters. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nature reviews Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Kong J, Liebhaber SA. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol Cell Biol. 2003;23:899–907. doi: 10.1128/MCB.23.3.899-907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi L, Bouzourene H, Benhattar J. Loss of Smad4 expression predicts liver metastasis in human colorectal cancer. Oncology reports. 2007;17:1095–1099. [PubMed] [Google Scholar]
- Malkoski SP, Wang XJ. Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS letters. 2012;586:1984–1992. doi: 10.1016/j.febslet.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Wang G, He D, Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry. 2005;44:12546–12553. doi: 10.1021/bi050560g. [DOI] [PubMed] [Google Scholar]
- Meng Q, Rayala SK, Gururaj AE, Talukder AH, O’Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci U S A. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Advances in cancer research. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS genetics. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nature structural & molecular biology. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EJ, Thuault S, Caja L, Carletti T, Heldin CH, Moustakas A. Regulation of transcription factor Twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J Biol Chem. 2012;287:7134–7145. doi: 10.1074/jbc.M111.291385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Yamashita M, Coussens NP, Tang Y, Wang X, Li C, Deng CX, Cheng SY, Zhang YE. Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. Embo J. 2011;30:4777–4789. doi: 10.1038/emboj.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome research. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort SJ, Lourenco AR, van Boxtel R, Coffer PJ. SOX4 mediates TGF-beta-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PloS one. 2013;8:e53238. doi: 10.1371/journal.pone.0053238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Matsuura I, He D, Liu F. Transforming growth factor-{beta}-inducible phosphorylation of Smad3. J Biol Chem. 2009;284:9663–9673. doi: 10.1074/jbc.M809281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. Embo J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt MK, Smith JA, Schiemann WP. Transforming growth factor-beta-induced epithelial-mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene. 2010;29:6485–6498. doi: 10.1038/onc.2010.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Masugi Y, Effendi K, Tsujikawa H, Hiraoka N, Kitago M, Shinoda M, Itano O, Tanabe M, Kitagawa Y, et al. Upregulated SMAD3 promotes epithelial-mesenchymal transition and predicts poor prognosis in pancreatic ductal adenocarcinoma. Laboratory investigation; a journal of technical methods and pathology. 2014;94:683–691. doi: 10.1038/labinvest.2014.53. [DOI] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.