Abstract
Ensuring the appropriate spatial-temporal control of protein abundance requires careful control of transcript levels. This process is regulated at many steps, including the rate at which transcripts decay. microRNAs (miRNAs) and RNA Binding Proteins (RBPs) represent two important regulators of transcript degradation. We review here recent literature that suggests these two regulators of transcript decay may functionally interact. Some studies have reported an excess of miRNA binding sites surrounding the positions at which RBPs bind. Experimental reports focusing on a particular transcript have identified instances in which RBPs and miRNAs compete for the same target sites, and instances in which the binding of a RBP makes a miRNA recognition site more accessible to the RISC complex. Further, miRNAs and RBPs use similar enzymes for degradation of target transcripts and the degradation of the target transcripts occurs in similar subcellular compartments. In addition to miRNA-RBP interactions involving transcript decay, RBPs have also been reported to facilitate the processing of pri-miRNAs to their final form. We summarize here several possible mechanisms through which miRNA-RBP interactions may occur.
Keywords: AU-rich elements, microRNAs, Pumilio, RNA binding proteins, transcript decay
INTRODUCTION
mRNA transcripts are subjected to a complex array of regulatory controls as they are processed into proteins. These regulatory steps may occur at the level of mRNA splicing, polyadenylation, transport, and stabilization. The ultimate goal of such regulation is the correct spatial and temporal distribution of encoded proteins. Transcript degradation, the focus of this review, is utilized extensively during development to eliminate maternal transcripts at the maternal-to-zygotic transition [1, 2]. In adult tissues, transcript degradation is employed to define cell lineages [3], and to rapidly eliminate transcripts in signaling pathways that could be deleterious if continuously expressed [4]. Inappropriate protein expression resulting from deficiencies in transcript degradation mechanisms may contribute to developmental abnormalities and the progression of cancer [5]. Many aspects of mRNA regulation are controlled by RBPs, and there are many types of proteins that bind RNA. We focus here on the class of RBPs that recognize and bind to sequence elements in the transcript’s 3’ UTR, thereby affecting the transcript’s stability [6]. Transcript stability and translatability can also be regulated by miRNAs, short 21-23 nucleotide RNAs that regulate targeted mRNA transcripts [7-10]. RBPs and miRNAs recognize specific recognition sequences in the transcript RNA indicated by the nucleotide sequence and/or secondary structure [11, 12]. While RBPs and miRNAs have generally been considered distinct transcript degradation pathways, it is becoming clearer that RBPs can regulate miRNA activity through synergistic activity at the level of transcript decay, and by affecting the miRNAs themselves. We briefly summarize the known mechanism of action of miRNAs and RBPs. We then describe evidence indicating that RBPs and miRNAs have a functional interaction. We consider several possible mechanisms in which RBPs and miRNAs might promote or antagonize each other’s effects, and the evidence for each. We summarize with suggestions for future areas of research.
INTRODUCTION TO RBPs
RBPs can regulate many aspects of RNA processing, localization, export, and stability by binding to recognition sequences within 3’ UTRs. As an example, trans-acting RBPs ensure that there is coordinated regulation of the transcripts that encode proteins involved in iron metabolism. Iron-responsive RBPs are regulated by iron levels. These proteins interact with iron-responsive elements—conserved hairpins that form distinctive secondary structures in untranslated portions of target transcripts [13]. Binding of the iron-responsive RBPs results in changes in transcript stability or translation rate. Other families of RBPs include the Pumilio or PUF family [14], and the adenosine-uracil rich element (ARE) binding family [15]. The Pumilio protein in Drosophila is essential for axis formation and stem cell maintenance [14]. The ARE, UAUUUAU, is present in many signaling transcripts including cytokines, growth factors and oncogenes [16]. Among the proteins that can bind to AREs are HuR/ELAV [6], Tristetrapolin (TTP) [17] and FXR1 [18]. Some of these ARE-binding proteins (ARE-BPs) promote degradation of the target transcript, while others, like the HuR family of proteins [19, 20], can cause stabilization of the targeted message [6].
The short and degenerate recognition sites for RBPs, and the difficulty in defining RBP occupancy in vivo, has hindered attempts to clearly define RBP recognition sites and the transcripts they regulate. Immunoprecipitation of RBPs as part of RBP-RNA complexes, followed by detection of the bound sequences by microarrays or next generation sequencing, a procedure called RIP-chip or RIP-Seq, respectively represents a method for better defining the sequences bound by specific RBPs [21, 22]. More recently introduced methods allow the definitive localization of the recognized binding sites. With photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP), photoactive thiouridine is incorporated into RNA and then crosslinked with ultraviolet light [23]. T to C conversions in the portion of the cDNA derived from the protected cross-linked regions allow for a definitive indication of the specific nucleotides bound by the RBP [23-25].
INTRODUCTION TO miRNAs
In addition to RBPs, miRNAs are also important regulators of transcript decay. miRNAs have been shown to play a central role in the regulation of a wide variety of biological processes [26]. Many miRNAs display tissue-specific or development time point-specific expression patterns [27-29]. Dysregulation of miRNA pathways has been implicated in human disorders including cancer, heart disease and neurological disorders [30-33]. In the latest version of MiRBase, version 18, there are 1527 unique human miRNAs, and over 60% of human protein-coding genes are conserved targets of miRNAs [34]. miRNAs are initially transcribed as primary miRNAs (pri-miRNAs) by RNA Pol II. The pri-miRNA contains a hairpin precursor that includes the sequence that will be the ultimate miRNA. pri-miRNAs are processed to precursor miRNAs (pre-miRNAs) by the Microprocessor complex of Drosha/DGCR8 [35-37]. The pre-miRNAs are then cleaved by the RNase III enzyme Dicer to generate mature miRNAs [38, 39]. Mature miRNAs direct the RNA-induced silencing complex (RISC), including the catalytic component Argonaute (Ago), to messenger RNAs with partially complementary sequences [40]. Recent studies using microarrays and ribosome profiling to measure the effects of miRNAs have revealed that miRNAs act predominantly to decrease levels of target mRNAs [41].
miRNAs generally bind their targets through 6-8mer matches to the miRNA “seed” region [8-10]. There may also be imperfect matching throughout the rest of the miRNA [8-10, 42]. There is selective pressure to conserve seed-matched targets, including not only the stronger 8mer seed matches, but also the weaker 6mer seed matches [34], which tend to effect minimal changes in target RNA or protein levels [43-46]. In addition to a stronger seed match, some of the other parameters that lead to enhanced miRNA activity include increased AU content in the surrounding sequence, proximity to other miRNA recognition sites, and avoidance of the middle of 3’ UTRs [46].
EVIDENCE FOR AN INTERACTION BETWEEN miRNAs AND RBPs
RBPs and miRNAs have generally been viewed as two distinct mechanisms to regulate transcript abundance. There are several reasons to hypothesize that these two mechanisms might act coordinately. miRNA recognition sites are most prevalent and most effective in 3’ UTRs, the regions of the transcript that are also targeted by RBPs [46, 47]. The increased effectiveness of 3’ UTR recognition sites could reflect several factors, including the passing of ribosomes across coding regions. Indeed, there is experimental evidence indicating that translating ribosomes can impede miRNA function. When the same miRNA targeting site was moved from the coding region to the 3’ UTR by shifting the location of the stop codon, it was more effective in the 3’ UTR [48]. Mutating specific base pairs upstream of a miRNA site so that they now encode rare codons caused the translation machinery to pause, and made the miRNA binding sites more effective [48]. In addition to the absence of passing ribosomes, the preference for 3’ UTRs could also reflect proximity to the polyA tail, which is a critical regulator of transcript stability, or to the cap once the transcript is circularized [49]. Thus, one of several possible explanations for the improved efficacy of miRNA binding sites in 3’ UTRs is that 3’ UTRs also contain recognition sites for RBPs.
Another line of indirect evidence that supports a possible role of RBPs in miRNA function is that the sequences surrounding a given recognition site also make an important contribution to the effectiveness of the site [46]. Indeed, sometimes the sequences surrounding a miRNA recognition site are conserved along with the site itself [46, 50, 51]. These findings suggest the importance of flanking sites, and could reflect either a contribution of accessibility [52], or that these sequences are conserved because they represent a docking point for RBPs [51].
More direct evidence for a potential interaction between miRNAs and RBPs derives from the observation that miRNA binding sites and RBP binding sites are present on the same 3’ UTRs, and in some cases are in close proximity to each other. In a study of HuR binding sites using RIP-chip and PAR-CLIP, there was extensive colocalization of HuR and Ago binding sites [24]. Over 75% of mRNAs with 3’ UTR Ago binding sites also contained 3’ UTR HuR binding sites [24]. An enrichment for miRNA recognition sites has also been reported in the sequences surrounding Pumilio recognition sites identified based on immunoprecipitation with antibodies to Pumilio [53].
Finally, miRNAs and RBPs share mechanisms of action. Both affect target transcripts by causing deadenylation, decapping, and transcript degradation by exonucleolytic enzymes [54]. The CCR4 deadenylase complex and the decapping enzymes Dcp1/2 have been implicated in both AU-rich element binding protein-mediated decay and miRNA-mediated decay [43, 55, 56]. Further, both miRNAs [57, 58] and RBPs [59, 60] have been localized to cytoplasmic P bodies, sites within the cell that may be involved in mRNA decay [54].
GENOMEWIDE ANALYSES IMPLICATING AN INTERACTION BETWEEN RBPs AND miRNAs
Researchers analyzing large scale miRNA overexpression microarrays have identified putative RBP recognition sites as sequence motifs associated with miRNA function. Jacobsen and colleagues transfected miRNAs into cells and monitored gene expression changes with microarrays [61]. U-rich motifs previously identified as binding sites for the HuD RBP were associated with downregulation in response to transfection with small RNAs. In another study, a subset of the AU-rich elements correlated with changes in mRNA expression after transfection with either miR-1 or miR-124 in HeLa cells [62]. Thus, the transcripts that are actually downregulated in response to overexpression of miRNAs tend to also contain recognition sites for ARE-BPs.
One concern in evaluating these studies is that the actual sequences bound by miRNAs and RBPs tend to be AU-rich. Thus, the apparent colocalization of miRNA and RBP binding sites could be a consequence of the localization of both types of sites in AU-rich regions of 3’ UTRs, rather than a signal that the two types of sequences are interacting with each other functionally. Further, there may be portions of 3’ UTRs that are more important for regulating transcript decay. If so, RBP recognition sites and miRNA recognition sites might both tend to be present in such regions. These possible confounding factors make it difficult to assess the functional importance of colocalization of miRNA and RBP recognition sites.
EXPERIMENTAL MODELS TO TEST THE INTERACTION BETWEEN RBPs AND miRNAs
Several model systems have been developed to experimentally test for a functional interaction between miRNA and RBP recognition sites. Didiano and Hobert analyzed the ability of the endogenous C. elegans miRNA lsy-6 to regulate the activity of its target cog-1 in the neurons in which it is typically active [63]. The authors found that lsy-6 recognition sites in the cog-1 3’ UTR may not be active when transferred to a different 3’ UTR, indicating the importance of the surrounding sequence. Further, mutation of the nucleotides within the seed had a minimal effect on the efficacy of miRNA binding sites within permissive contexts. A specific sequence flanking a particular lsy-6 binding site was shown to be important for miRNA regulation. The sequence contained a possible recognition site for Pumilio, but deleting that recognition site had no effect on 3’ UTR regulation. Thus, while the authors discovered that the sequences surrounding miRNA binding sites are important for their efficacy, it was unclear whether this reflects a role for these sequences for the docking of RBPs or in regulating the local secondary structure.
Sun and colleagues performed a similar set of experiments in a different reporter system, and arrived at a different conclusion [64]. They performed reporter assays to determine critical regions within the RhoB 3’ UTR for the efficacy of two miR-223 binding sites. One site was more important than the other based on site-directed mutagenesis, and yet was not the site predicted to have stronger binding. They demonstrated that the distinction lay in the presence of specific sequences upstream of the more effective binding site, and these sequences included binding sites for AU-rich elements. The simple presence of As and Us was not as strong a signal as known RBP recognition sites. Further, the directionality mattered, as the RBP sequences could promote miRNA-mediated repression of reporter activity when present upstream, but not downstream, of miR-223 sites or recognition sites for other miRNAs. Directionality in the relative position of the miRNA binding sites and RBP recognition sites was also noted for the Pumilio RBP. While Pumilio binding sites were enriched in the sequences both upstream and downstream of the miRNA binding site, the downstream enrichment was stronger [53].
POSSIBLE MODELS FOR RBP-miRNA INTERACTION: RBPs MAKE miRNA SITES MORE ACCESSIBLE
Previous studies have highlighted the accessibility of miRNA binding sites as an important factor in their functionality [52]. A role for RBPs in making miRNA target sites more accessible has been reported for an interaction between Pumilio and miR-410 [65]. Leibovich and colleagues identified motifs that are enriched in the least accessible miRNA targets. Most motifs were GC-rich, but one AU-rich motif was identified, and it resembled the Pumilio binding site. By analyzing gene expression data, they discovered that high expression of both Pumilio and miR-410 had a greater suppressive effect on targets with binding sites that are present in poorly accessible positions within 3’ UTRs. The results support a model in which, for Pumilio and miR-410 specifically, Pumilio binding increases the accessibility of specific targets to the RISC complex.
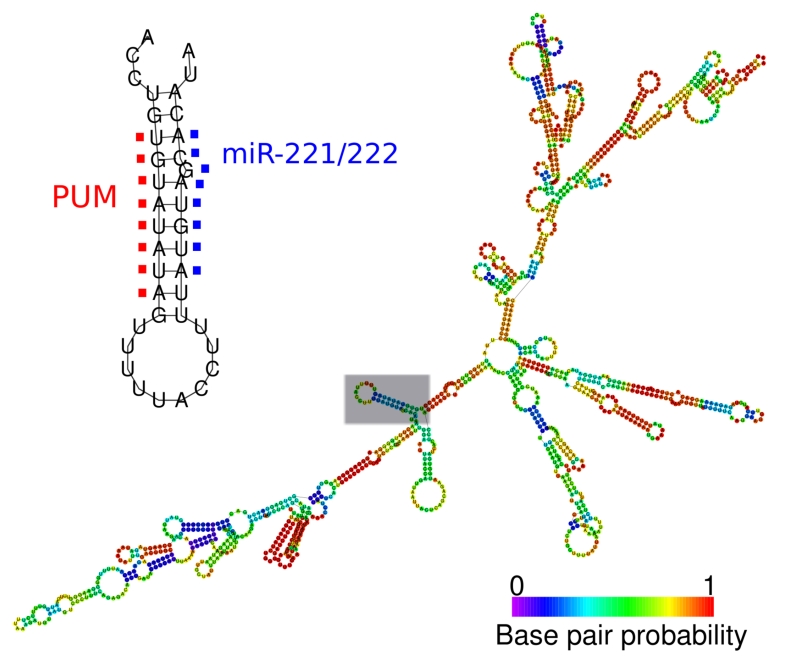
A similar conclusion that RBPs can make transcripts more accessible to the RISC complex was drawn for the case of the Pumilio binding protein and miR-221/222 [5]. miR-221 and miR-222 are regulators of p27Kip1, a cyclin-dependent kinase inhibitor that contributes to the establishment of proliferative quiescence [66]. In quiescent cells, p27Kip1 accumulates despite high miR-221/222 levels. Kedde and colleagues revealed that in quiescent cells, Pumilio is not phosphorylated and therefore not active on the p27Kip1 3’ UTR [5]. Upon growth factor stimulation, Pumilio becomes phosphorylated, binds the p27Kip1 3’ UTR and causes a change in local conformation that facilitates its association with miR-221/222. The authors introduced an RNA molecule with both donor and acceptor fluorophores. With this system, the lifetime of acceptor fluorescence is proportional to the distance between the donor and acceptor fluorophores, and indicates the openness of the structure. The lifetime acceptor fluorescence was longer in proliferating than quiescent cells because of the more open conformation conferred by Pumilio binding (Fig. 1). Knockdown of Pumilio in cycling cells eliminated the decrease in transcript lifetime. Thus, in proliferating cells, Pumilio binding makes the miR-221/222 sites accessible and results in down-regulation of p27Kip1 levels, allowing proliferation to proceed. In this study, measurements of RNA secondary structure in cells permitted a clear demonstration of a role for Pumilio in affecting miRNA site accessibility.
Fig. (1).
Local secondary structure of PUM and miR-221/222 binding sites.
PUM and miR-221/222 binding sites form a stem loop secondary structure by binding to each other on the 3’UTR of the p27Kip1 transcript. Binding of Pumilio is necessary to increase the accessibility of miRNA binding sites. The Pumilio and miR-221/222 recognition sites are shown in detail and the entire 3’ UTR, folded with RNAfold [101], is shown. The probability that a base is paired with other nucleotides is indicated with the color scheme shown.
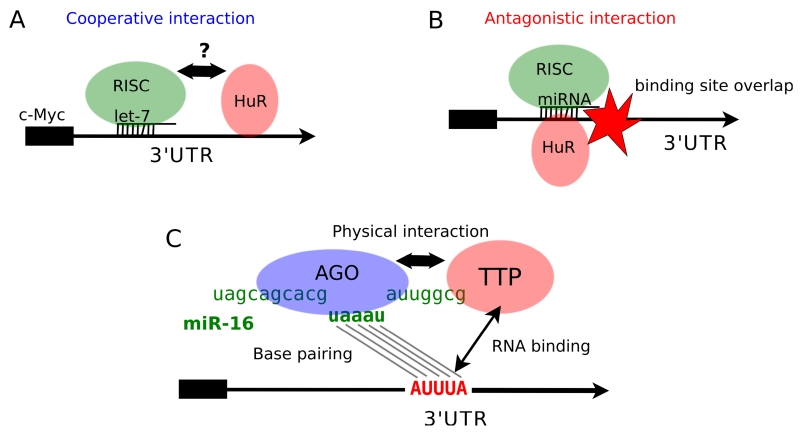
A similar model was proposed for the case of HuR and let-7 binding to the c-Myc 3’ UTR [67]. The RBP HuR was found to reduce c-Myc expression by complexing with the 3’ UTR in a position adjacent to a let-7 binding site. The activity of let-7 was dependent on HuR, and the activity of HuR depended on let-7. The authors discovered that HuR binding promoted the association of let-7 with the c-Myc mRNA and therefore proposed a model in which HuR binding changes the local RNA conformation, unmasking the let-7 recognition site. However, there is no further evidence for this structural model, thus more experiments are still needed to understand the relationship between HuR and let-7 in this model system (Fig. 2A).
Fig. (2).
Models of RBP-miRNA combinatorial binding on mRNA 3’UTRs.
Three examples of RBP-miRNA interaction with the proposed mechanisms are shown. Gene 3’UTR regions are indicated with black arrows. (A) HuR and let-7 bind closely to each on the c-Myc 3’UTR and promote the effect of miRNA-mediated repression. (B) HuR binding competes with miRNA binding sites and alleviates the effect of miRNA repression. (C) TTP physically interacts with AGO. TTP binds ARE elements on the mRNA 3’UTRs, which facilitates sequence pairing between the 3’UTR ARE and parts of the miR-16 mature sequence.
POSSIBLE MODELS FOR RBP-miRNA INTERACTION: RBPs AND AGO DIRECTLY INTERACT
There is also evidence that RBPs and the RISC complex can have a direct physical interaction. In an early study, Caudy and colleagues demonstrated that the RISC complex in Drosophila contains two proteins that associate with AU-rich elements: PAI-RBP1 [68] and FMRP/FXR1 [69]. Reduction of FXR1 in Drosophila S2 cells resulted in less efficient RNAi silencing. The FMRP protein is encoded in a portion of the genome that is silenced in Fragile X syndrome [70]. A mutation in FMRP observed in Fragile X syndrome affected the ability of the protein to associate with RISC [69]. Further work confirmed that FXR1 associates with Ago2 in mammalian cells as well [71]. Subsequent proteomic analysis of Ago-binding proteins in human cells revealed AU-rich element binding proteins FMR, FXR and HuR [72]. In two Drosophila models, the phenotypic functional effects of FMRP were reduced in strains in which expression of Ago proteins was deficient [71]. The discovery of a functional interaction between FXR1 and Ago2 resulted in suggestions that either RBPs are targeted to substrates by the RNAi complex [69], or that FXR1 scans for the G-quartet previously reported to be a recognition site for FXR1 [73], and then recruits RISC [71]. Caudy and colleagues suggested that different RISC component RBPs could be present in a tissue-specific or development-specific manner, allowing for flexibility in the regulation of target genes in different ways, including transcript decay or translation initiation.
Jing and colleagues proposed a similar, but not identical, model as a result of their investigations of TNF-α mRNA [74]. They performed a RNAi-based screen in Drosophila cells and discovered that Dicer and Ago proteins are required for the rapid decay of ARE-containing 3’ UTRs. Thus, their results suggest that the miRNA machinery is central to the ability of AU-rich element binding proteins to induce rapid transcript turnover. Further, their results specifically implicated the miRNA miR-16 in ARE-mediated transcript turnover. miR-16 contains the sequence UAAAUAUU, which is complementary to the AU-rich element binding site (Fig. 2C). For miR-16 mediated degradation of the TNF-α mRNA, the specific ARE binding protein TTP was required. A physical interaction between TTP and the RISC complex was discovered, and proposed to be an important mechanism for miR-16-mediated decay. The authors suggest a model in which TTP binds to an ARE and, when a RISC-containing miR-16 is nearby, the miRNA and ARE interact through complementary base pairing and mediate decay.
POSSIBLE MODELS FOR RBP-miRNA INTERACTION: COMPETITION FOR THE SAME BINDING POSITION
In addition to data indicating that miRNAs and RBPs collaborate to mediate transcript decay, there are also reports of competition between miRNAs and RBPs for the same sequence, resulting in an antagonistic effect (Fig. 2B) [51, 75, 76]. Competition for binding sites has mostly focused on RBPs of the HuR/ELAV family, the family of proteins that stabilize transcripts rather than promote target degradation (with the exception of [67]). One report focused on Dnd1, a RBP required for germ cell survival and migration in zebrafish [77] that has a conserved RNA binding domain similar to that of ELAV/Hu family members. Dnd1 was discovered to bind to target mRNAs through stretches of uridines, and thereby prevent access of miR-430-loaded RISC complexes to the target [51]. This Dnd1-mediated antagonism of miR-430 activity was active in zebrafish germ cells, and represents a mechanism whereby specific sets of miRNA-mRNA interactions can be inhibited in a cell type-specific manner. Loss of Dnd1 can result in reduced numbers of germ cells, sterility and germ cell cancers [51, 78]. The authors suggest two possible mechanisms for Dnd1 action: Dnd1 binding may result in a secondary structure for the target RNA that makes miRNA recognition sites inaccessible. Alternatively, binding of Dnd1 could result in a subcellular localization for the transcript that makes it inaccessible to the RISC complex.
As another example, downregulation of the cationic amino acid transporter 1 (CAT-1) mRNA by miR-122 was inhibited by stress, and the derepression required binding of HuR to the 3’ UTR [75]. The derepression required a specific region of the 3’ UTR that bound HuR, and was accompanied by relocalization of CAT-1 mRNA from P bodies, subcellular structures that are sites of transcript decay, to polysomes. Further, antibodies against Ago2 immunoprecipitated CAT-1 mRNA from the cytoplasm of stressed cells, indicating that HuR likely remains bound to the transcript, but prevents miRNPs from acting as effective repressors.
As a final example, a competition between miRNAs and RBPs for the same binding site has also been identified for a coding region miRNA recognition site. While less common and less effective than 3’ UTR sites, miRNAs can also target coding region recognition sites [47, 79-81]. Degradation of the βTrCP1 ubiquitin ligase is dependent on miR-183-mediated binding to a recognition site within the coding region [75]. The RBP coding region determinant binding protein (CRD-BP) was shown to bind to the coding region of βTrCP1 and compete with miR-183. CRD-BP thereby results in stabilization of the transcript and the protein.
Competition between RBPs and miRNAs was also addressed in two different studies performing PAR-CLIP with antibodies to HuR to assess the relationship between HuR and miRNAs. Lebedeva and colleagues concluded that HuR and miRNA binding sites were frequently near each other in 3’ UTRs, but usually did not overlap [25]. In fact, the HuR binding pattern across 3’ UTRs was essentially the opposite of miRNA recognition sites. HuR binding was almost uniform across the 3’ UTR, but was depleted near the stop codon and the polyadenylation site, while binding of Ago proteins was enriched in these two regions [82]. They conclude that miRNA seeds tend to be present within 20 nucleotides of HuR binding sites, and that this preference may represent the preference of miRNA recognition sites for AU-rich regions.
Mukerjee and colleagues, upon performing a similar analysis, came to a different conclusion. They discovered that there was a strong signal for HuR and miRNA binding sites to be very close together and even overlapping [24]. When they analyzed the effects of miRNA knockdown on mRNA expression, transcripts with overlap between the miRNA site and the HuR site were significantly less strongly upregulated upon miRNA depletion than transcripts with nonoverlapping sites, or with a miRNA recognition site and no HuR binding site. The authors concluded that these overlapping sites represent instances in which the stabilizing effect of HuR out-competes the destabilizing effect of miRNA-mediated transcript decay through a competition for the same or nearby base pairs (Fig. 2B).
POSSIBLE MODELS FOR RBP-miRNA INTERACTION: CONVERSION FROM REPRESSION TO ACTIVATION
An alternative and intriguing model for RBP-miRNA interaction was described by Vasudevan and Steitz [18]. They discovered that the TNFα ARE mediates translational induction upon cell cycle arrest of HEK293 cells and THP-1 monocytes. Purification of S1-aptamer tagged mRNP complexes revealed that FXR1 and Ago2 are associated with the TNFα ARE only in serum-starved cells, and that both FXR1 and Ago2 activities are important for translational activation upon serum withdrawal and other forms of cell cycle arrest [18]. Upon growth arrest, insoluble foci of FXR1/Ago2 decreased, and these same two proteins became associated with ARE-containing mRNP complexes. A human miRNA miR-369-3 that contains a seed sequence that recognizes the TNFα ARE, was found to direct the association of FXR1 and Ago2 to the TNFα 3’ UTR upon cell cycle arrest [83]. The authors further showed that other miRNAs, including the miRNA let-7, can also induce translational upregulation of target mRNAs in cell cycle-arrested cells [83]. A similar mode of miRNA-mediated translational induction was also discovered in G0-arrested Xenopus leavis oocytes, but not in oocytes matured with progesterone [84]. Translational activation in the oocytes also depended upon FXR1 and Ago, as oocytes depleted of either factor failed to show translational activation. The results support a model in which, upon cell cycle withdrawal, new RBPs and miRNAs may be recruited to ARE-containing sites, and these new components can mediate translational activation rather than repression.
RBPs AFFECT miRNA BIOGENESIS: FACILITATING PROCESSING
Many miRNAs display cell- and tissue-specific regulatory patterns [27-29]. Some of these regulatory events occur at the level of transcription of the pri-miRNA, and some of the same transcription factors that control the expression of genes also regulate the expression of miRNAs [85]. In addition, the process through which pri-miRNAs are converted to pre-miRNAs and to the mature form of the transcript can also be regulated, and RBPs have been implicated in this form of regulation. The human immunodeficiency virus transactivating response RNA-binding protein (TRBP), a protein that binds the stem-loop of the HIV trans-activating response region, is an integral component of the Dicer-containing complex that prepares pre-miRNAs for loading into the RISC complex [86, 87]. Dicer, TRBP and the catalytic component of RISC, Ago2, are present in a ternary complex. The presence of TRBP is important for the recruitment of Ago2 and miRNA activity, and thus TRBP represents an important platform for RISC assembly [86]. Truncating mutations in the TRBP protein result in defective miRNA processing, and have been identified in carcinomas with microsatellite instability [88]. Restoration of TRBP protein levels improved miRNA activity and suppressed tumor growth and tumorigenicity.
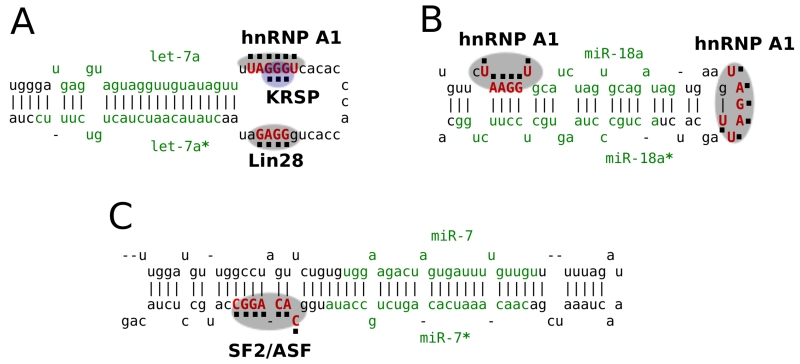
Further studies indicated a role for other RBPs in the processing of specific classes of miRNAs, also through binding of the terminal loop. KH-type splicing regulatory protein (KSRP), for instance, binds AU-rich elements in 3’ UTRs and acts as a mediator of decay [89, 90]. KSRP was found to be a component of both the Drosha and Dicer complexes. KSRP interacts directly with a GGG triplet in the terminal loop of let-7a and promotes the processing of pre-let-7a to the mature form [91]. KSRP also exhibited activity toward several other miRNAs, but not all of the miRNAs tested. KSRP knockdown reduced expression of mature let-7a and upregulated cell proliferation. Thus, KSRP, a protein previously identified as an ARE-BP, is also an important component of the pathway that forms miRNAs by binding the terminal loop of immature miRNAs and promoting their processing (Fig. 3A).
Fig. (3).
RBPs regulate miRNA biogenesis.
Mature miRNA sequences and the star miRNA sequence are indicated in green. RBP binding sites are indicated in red. The specific binding position of each RBP motif is highlighted with dots. (A) pre-miRNA hairpin loop of let-7a. (B) pri-miR-18a. (C) pri-miR-7.
Another RBP, heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), can also bind the terminal loop region of let-7a [92]. hnRNP A1 binds to a UAGGG[AU] sequence in the terminal loop of pri-let-7a and inhibits processing by Drosha. Depletion of hnRNP A1 resulted in increased pri-let-7a processing. hnRNP A1 interfered with the binding of KSRP (Fig. 3A), and thus, hnRNP A1 functions as a negative regulator of pre-let-7a biogenesis through a competition with KSRP. In contrast, hnRNP A1 can also act positively on miRNA biogenesis by binding to the terminal loop region of miR-18a (Fig. 3B). In this case, hnRNP A1 binding creates a more favorable cleavage site for Drosha and actually promotes formation of the pre-miRNA [93]. The results suggest that the terminal loop of miRNAs represents a position at which there can be a competition between multiple proteins, some of which promote biogenesis while others inhibit it, and that the same RBP may promote or inhibit miRNA processing depending on the terminal loop sequence and its conformation.
Besides binding to the terminal loop region of a pre-miRNA sequence, RBPs may also bind other parts of the pri-miRNA sequence to affect their biogenesis. Splicing factor SF2/ASF preferentially binds to the intronic pri-miR-7-1 sequence to facilitate the Drosha cleavage step in miR-7 maturation [94]. A SF2/ASF binding site was identified on the 3’ end of the pri-miR-7 stem (Fig. 3C), and a functional miR-7 binding site was also identified on the 3’ UTR of SF2/ASF to repress its translation [94]. Thus, the SF2/ASF splicing factor and miR-7 form a negative feedback loop.
RBPs AFFECT miRNA BIOGENESIS: IMPACT THROUGH MODIFICATION OF miRNA 3’ ENDS
RBPs can also regulate miRNA biogenesis by modifying miRNA 3’ ends. As an example, in embryonic stem cells, processing of the let-7 family of miRNAs is regulated by the RBP Lin28 [95]. Human Lin28 interacts with a GGAG sequence motif in the terminal loop region of let-7. Lin28 can recruit terminal uridylyltransferase 4, which adds terminal uridines to pre-let-7. The uridylated pre-let-7 fails Dicer processing and is degraded [96-98] (Fig. 3A). Inhibiting let-7 biogenesis may be a key constituent of pluripotency as Lin28 is essential for proper germ cell development in mice and overexpression of Lin28 is associated with human germ cell tumors [99]. Further, Lin28 is one of the proteins that, in concert with others, can contribute to the creation of induced pluripotent stem cells from differentiated precursors [100].
CONCLUDING THOUGHTS AND SUGGESTIONS FOR FUTURE RESEARCH
Transcript decay is an important mechanism for the regulation of protein abundance in the creation of intracellular polarization, in the establishment and maintenance of cell fates and identities, and in the transduction of intracellular and extracellular signals. Mechanisms that affect transcript decay rates can potentially be used to alter cellular protein content and distribution. Defects in these pathways can result in inappropriate protein expression and contribute to developmental abnormalities and disease. We describe here two different pathways for controlling transcript decay rates: miRNAs and RBPs. We specifically explore the ways in which these two regulators interact with each other.
Both global computational analyses and experimental manipulation of a specific system have supported a possible interaction between RBPs and miRNAs. Some specific examples are summarized in Table 1. While both approaches can provide evidence to support such a model, each is limited. In global computational analyses, each putative binding site cannot be directly verified, and thus some sites may be incorrect. Experimentation for individual RBP-miRNA interactions can provide more direct and definitive evidence for a specific interaction, but is limited to the small number of instances that can be directly investigated. For this reason, it has been difficult to assess the impact of a possible synergistic mode of action on the global role of miRNAs. Novel approaches that bridge this gap would be valuable.
Table 1. Summary of selected reports of miRNA-RBP interaction.
| miRNA | RBP | Targeted Transcript | Mechanism | References |
|---|---|---|---|---|
| Cooperative Binding | ||||
| miR-221/222 | PUM | p27 3′UTR | open up secondary structure | [5] |
| miR-410 | PUM | open up secondary structure | [65] | |
| miR-1, miR-124 | ARE | [62] | ||
| miR-223 | ARE | RhoB 3′UTR | Upstream ARE promotes miRNA site activity | [64] |
| let-7 | HuR | c-Myc 3′UTR | [67] | |
| miR-16 | TTP | TNF-α 3′UTR | ARE-BP TTP physically interacts with AGO, and miR-16 sequence could pair up with ARE |
[74] |
| let-7 | Puf-9 | hbl-1 | [102] | |
| Antagonistic Binding | ||||
| miR-443 | Dnd1 | Nanos1, TDRD7 3′UTR | [51] | |
| miR-122 | HuR | CAT-1 3′UTR | [75] | |
| miR-183 | CRD-BP | βTrCP1 coding region | [76] | |
| Biogenesis | ||||
| let-7a | KSRP | pre-let-7a terminal loop | promote pre-miRNA processing | [91] |
| let-7a | hnRNP A1 | pre-let-7a terminal loop | compete with KRSP binding to repress pre-miRNA processing | [92] |
| miR-18a | hnRNP A1 | pri-miR-18a terminal loop | promote Drosha cleavage | [93] |
| miR-7 | SF2/ASF | pri-miR-7 stem | promote Drosha cleavage | [94] |
| let-7a | Lin28 | pre-let-7a terminal loop | recruit TUT4 which uridylates pre-let-7a for degradation | [97] |
There are many other important avenues of research that remain open. While there are a large number of RBPs (~800 for human) catalogued in the Gene Ontology database, the activity of relatively few of them is understood. Further studies with RIP-CLIP or PAR-CLIP to identify the binding motifs and the regulated transcripts for RBPs would be helpful in determining the role of different RBPs in different contexts.
Further application of other approaches used in a small number of instances would also be valuable. Kedde and colleagues used a FRET approach to clearly define a role for the Pumilio protein in the conformation of the p27Kip1 3’ UTR [5]. Application of this or similar techniques to other 3’ UTRs could help to clarify how general this mechanism is, and thus the extent to which RBPs modify the accessibility of transcripts to the miRNA machinery. Vasudevan developed an in vivo formaldehyde crosslinking approach to “freeze” and purify aptamer-tagged mRNA complexes [18]. Further applications of this approach could provide valuable information about the association of miRNA components with AU-rich sequence elements more broadly. Didiano and Hobert performed the critical experiment of testing whether mutating putative RBP recognition sites and introducing or eliminating AU-rich sequences affected miRNA effectiveness [63]. Other similar experiments could help to clarify whether putative RNA-binding sites are truly docking sites for an RBP or reflect changes in RNA secondary structure. The studies of Didiano and Hobert also emphasize the importance of model systems in which miRNA-target interactions can be studied in the cells in which they normally occur, and without the introduction of high levels of exogenous miRNAs.
Further investigation of the mechanisms by which RBPs and miRNAs achieve their effects on transcript decay would also be beneficial. While it is understood that binding of RBPs results in the subcellular movement of transcripts between polysomes and P bodies, the basis for this is still unclear. Further, how some RBPs facilitate transcript decay, while others promote transcript stability, is also unclear. A combination of genetic and biochemical methods that allow for monitoring of the localization of specific target transcripts under conditions in which particular components of the transcript processing machinery are inactivated or modified could help to clarify the molecular basis for RBP function.
If miRNAs and RBPs can in some cases promote, and in other cases oppose, each other, this represents new opportunities for regulation [50]. RBPs that affect transcript stability can represent a rapid mechanism of regulation. Cell-type, cell-fate or signal-induced changes in the levels of RBPs could regulate a subset of miRNA targets, and would therefore provide modularity to the regulation achieved by miRNAs. A better understanding of this process could provide valuable insight into the mechanisms through which cells perform specific physiological functions.
ACKNOWLEDGEMENTS
Declared none.
ABBREVIATIONS
- Ago
Argonaute
- ARE
Adenosine-uracil rich element
- ARE-BP
Adenosine-uracil rich element-binding protein
- CAT-1
Cationic amino acid transporter 1
- CRD-BP
Coding region determinant binding protein
- hnRNP A1
Heterogeneous nuclear ribonucleoprotein A1
- KSRP
KH-type splicing regulatory protein
- miRNA
microRNA
- RBP
RNA binding protein
- PAR-CLIP
Photoactivatable ribonucleoside crosslinking and immunoprecipitation
- pre-miRNA
Precursor-miRNA
- pri-miRNA
Primary miRNA
- RIP-chip
Ribonucleoprotein immunoprecipiation followed by microarray
- RIP-Seq
Ribonucleoprotein immunoprecipitation followed by next generation sequencing
- RISC
RNA-induced silencing complex
- TRBP
Transactivating response RNA-binding protein
- TTP
Tristetrapolin
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- [1].Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.03.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- [2].Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–7. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- [3].Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- [4].Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley interdisciplinary reviews. RNA. 2010;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–20. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- [6].Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- [9].Krek A, Grun D, Poy MN. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- [10].Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–59. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ray D, Kazan H, Chan ET, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–70. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- [13].Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–97. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- [14].Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 2002;18:150–7. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- [15].Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–46. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- [17].Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–5. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- [18].Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–18. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–30. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–95. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–7. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- [22].Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97:14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mukherjee N, Corcoran DL, Nusbaum JD, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lebedeva S, Jens M, Theil K, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- [26].Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- [27].Wienholds E, Kloosterman WP, Miska E, et al. microRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- [28].Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- [29].Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Negrini M, Nicoloso MS, Calin GA. microRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470–9. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- [31].Latronico MV, Condorelli G. microRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–29. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- [32].Hebert SS. Putative role of MicroRNA-regulated pathways in comorbid neurological and cardiovascular disorders. Cardiovasc Psychiatry Neurol. 2009;2009:849519. doi: 10.1155/2009/849519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- [36].Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- [37].Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- [39].Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- [40].Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- [41].Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- [43].Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–7. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- [45].Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. microRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Forman JJ, Coller HA. The code within the code: MicroRNAs target coding regions. Cell Cycle. 2010;9:1533–41. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–50. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–40. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- [50].Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- [51].Kedde M, Strasser MJ, Boldajipour B, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–86. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- [52].Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- [53].Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PloS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–94. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–61. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. microRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- [59].Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Stoecklin G, Stubbs T, Kedersha N, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–24. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–9. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sun G, Li H, Rossi JJ. Sequence context outside the target region influences the effectiveness of miR-223 target sites in the RhoB 3′UTR. Nucleic Acids Res. 2010;38:239–52. doi: 10.1093/nar/gkp870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Leibovich L, Mandel-Gutfreund Y, Yakhini Z. A structural-based statistical approach suggests a cooperative activity of PUM1 and miR-410 in human 3′-untranslated regions. Silence. 2010;1:17. doi: 10.1186/1758-907X-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- [67].Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plas-minogen activator inhibitor mRNA. J Biol Chem. 2001;276:3341–7. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- [69].Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–6. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–91. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- [71].Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- [72].Hock J, Weinmann L, Ender C, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–60. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–99. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- [74].Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- [75].Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- [76].Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–6. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Weidinger G, Stebler J, Slanchev K, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–34. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- [78].Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest. 2010;40:370–4. doi: 10.1111/j.1365-2362.2010.02279.x. [DOI] [PubMed] [Google Scholar]
- [79].Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. microRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- [81].Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- [84].Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs) Proc Natl Acad Sci U S A. 2011;108:8281–6. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- [86].Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang HW, Noland C, Siridechadilok B, et al. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–53. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [89].Garcia-Mayoral MF, Hollingworth D, et al. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–98. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- [90].Gherzi R, Lee KY, Briata P, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–83. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- [91].Trabucchi M, Briata P, Garcia-Mayoral M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wu H, Sun S, Tu K, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- [96].Lehrbach NJ, Miska EA. Regulation of pre-miRNA Processing. Adv Exp Med Biol. 2011;700:67–75. doi: 10.1007/978-1-4419-7823-3_7. [DOI] [PubMed] [Google Scholar]
- [97].Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- [98].Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- [99].West JA, Viswanathan SR, Yabuuchi A, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–13. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- [101].Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M. Fast folding and comparison of RNA secondary structures. Monatshefte f Chemie. 1994;125:167–88. [Google Scholar]
- [102].Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–63. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]