Abstract
OXA-163 and OXA-48 are closely related class D β-lactamases that exhibit different substrate profiles. OXA-163 hydrolyzes oxyimino-cephalosporins, particularly ceftazidime, while OXA-48 prefers carbapenem substrates. OXA-163 differs from OXA-48 by one substitution (S212D) in the active site β5-strand and a four-amino acid deletion (214-RIEP-217) in the loop connecting the β5 and β6 strands. Although the structure of OXA-48 has been determined, the structure of OXA-163 is unknown. To further understand the basis for their different substrate specificities we performed enzyme kinetic analysis, inhibition assays, x-ray crystallography, and molecular modeling. The results confirm the carbapenemase nature of OXA-48 and the ability of OXA-163 to hydrolyze the oxyimino-cephalosporin ceftazidime. The crystal structure of OXA-163 determined at 1.72-Å resolution reveals an expanded active site compared to OXA-48, which allows the bulky substrate ceftazidime to be accommodated. The structural differences with OXA-48, which cannot hydrolyze ceftazidime, provide a rationale for the change in substrate specificity between the enzymes. OXA-163 also crystallized in another condition that contained iodide. The crystal structure determined at 2.87-Å resolution revealed iodide in the active site accompanied by several significant conformational changes including a distortion of the β5-strand, decarboxylation of Lys73, and distortion of the substrate-binding site. Further studies showed that both OXA-163 and OXA-48 are inhibited in the presence of iodide. In addition, OXA-10, which is not a member of the OXA-48-like family, is also inhibited by iodide. These findings provide a molecular basis for the hydrolysis of ceftazidime by OXA-163 and, more broadly, show how minor sequence changes can profoundly alter the active site configuration and thereby affect the substrate profile of an enzyme.
Keywords: Gram-negative bacteria, antibiotic resistance, β-lactam antibiotics, serine β-lactamases, oxacillinase, carbapenemase
INTRODUCTION
β-Lactams are the most commonly prescribed antibiotics worldwide.1 Therefore, bacterial resistance towards these drugs presents a serious public health threat.2, 3 The effectiveness of β-lactam antibiotics is challenged by the emergence of multi-drug resistant Gram-negative bacteria from the Enterobacteriaceae family such as Klebsiella pneumoniae.4–7 The most common mechanism of β-lactam antibiotic resistance in Gram-negative pathogens is the production of β-lactamases that hydrolyze and inactivate the drugs.8–11 β-Lactamases are divided into four classes (A, B, C and D) based on amino acid sequence homology.8 Class A, C, and D enzymes are serine hydrolases while members of class B are metallo-β-lactamases that are unrelated in sequence and mechanism to the other classes.12, 13 In recent years, β-lactamase variants from classes A and D with the ability to hydrolyze oxyimino-cephalosporins and carbapenems have emerged. This alarming trend is reducing treatment options because oxyimino-cephalosporins are regularly prescribed, while carbapenems are considered last resort options in the treatment of multi-drug resistant infections.14–17
The class D β-lactamases (DBLs) are the most diverse group among serine β-lactamases with certain members possessing less then 20% sequence identity.18, 19 Despite their sequence diversity, DBLs have conserved structural features that are involved in the mechanism of hydrolysis. The mechanism of hydrolysis of β-lactam antibiotics by DBLs involves acylation and deacylation of the active-site serine and features a carboxylated lysine as the general base.20, 21 This reversible lysine modification, which is essential for DBL activity, is proposed to be a spontaneous reaction, facilitated by the hydrophobic environment of the active site and dependent on the protonation state of the lysine and the availability of CO2.17, 20, 22 Another property of DBLs that is not shared by other serine β-lactamases is inhibition by sodium chloride in vitro.18 Most DBLs are fully inhibited at a sodium chloride concentration of 100 mM.23 This property is not fully understood but a tyrosine residue, which is a constituent of a conserved YNG-motif located in the vicinity of the DBL active site, has been implicated.24 Héritier et al. showed that when the active-site tyrosine is substituted with phenylalanine, the resulting mutant is resistant to sodium chloride inhibition. Additionally, the mutant enzyme exhibited weaker activity for all the substrates tested.24 A more recent study suggests that chloride competes with the carboxylation of the lysine and thereby inhibits DBLs by attenuating formation of the general base.25
DBLs are also called oxacillinases (or OXA-enzymes) based on the properties of the first discovered members of this class, which have high catalytic efficiency for the hydrolysis of the semi-synthetic penicillin oxacillin.26 The rapid discovery of DBLs in recent years has led to the description of over four hundred OXA-enzymes.27–30 Based on substrate specificity, they can be classified as narrow-spectrum oxacillinases, extended-spectrum β-lactamases (ESBLs), or carbapenem-hydrolyzing class D β-lactamases (CHDLs).17, 18, 27, 31 Furthermore, to better separate the OXA-enzymes, several subgroups have been formed based on sequence homology. The enzymes within a subgroup differ from each other by one to five amino acids on average and generally have similar kinetic profiles.
OXA-48-like β-lactamases constitute a CHDL subgroup that is widespread in Klebsiella pneumoniae and other Enterobacteriaceae.28, 32 Their emergence represents an alarming development in carbapenem resistance worldwide. OXA-48, which was the first OXA-type carbapenemase isolated from enteric bacteria, has been identified over a decade from a multidrug-resistant K. pneumoniae isolate and it is the most widespread member of this subgroup.28, 33 It has a typical carbapenemase substrate profile with the highest catalytic efficiency for imipenem hydrolysis among all DBLs. However, its activity for oxyimino-cephalosporins is very modest and, in the case of ceftazidime, undetectable.33, 34 Currently, the OXA-48-like subgroup has eleven members and they differ from OXA-48 by few amino-acid substitutions and deletions.32 All of the OXA-48-like enzymes have a similar substrate profile as OXA-48 except for OXA-163.32 The OXA-163 enzyme has a drastically reduced ability to hydrolyze carbapenems, however unlike OXA-48, OXA-163 is able to hydrolyze the oxyimino-cephalosporin ceftazidime.35
OXA-163 is a relatively new β-lactamase that was identified in Klebsiella pneumoniae and Enterobacter cloacae from nosocomial infections.35 OXA-163 differs from OXA-48 by an S212D substitution and a four amino-acid deletion (214-RIEP-217) (OXA-48 numbering).34–36 The S212D substitution is located at the tip of β5 strand and the four amino-acid deletion is located in the loop region between β5 and β6 strands.34 The β5 strand forms one side of the active-site cavity and contains the conserved motif K(S/T)G (residues 208–210) typical for DBLs.34 The loop between β5-β6 beta strands has been suggested to be important for the ability of an OXA-variant to hydrolyze carbapenems and therefore the deletion may impact carbapenem hydrolysis.34, 37–39
The goal of this study was to determine the structural basis for the change in substrate specificity observed in OXA-163 versus OXA-48. The crystal structure of OXA-163 determined at 1.72-Å resolution revealed that the four amino-acid deletion in OXA-163 expands the active-site pocket to accommodate the bulky side chain of ceftazidime, providing a molecular basis for the different substrate profiles of the two enzymes. Additionally, a second structure of OXA-163 was determined at 2.87-Å using crystals from a different crystallization condition. This crystallization buffer contained iodide, which was found in the active site of the enzyme in the structure. Subsequent enzyme inhibition assays indicated that iodide is an inhibitor of both OXA-163 and OXA-48 as well as OXA-10, which is not in the OXA-48-like family. Based on the structural and inhibition analyses, it is proposed that halogen ions inhibit OXA-enzymes by changing the position of key active site residues and inhibiting the formation of the carboxylated lysine, which is essential for the function of all OXA-enzymes.
MATERIALS AND METHODS
Cloning
The blaOXA-163 gene containing the native signal sequence was inserted into the EcoRI site of the pET29a expression vector using T4 DNA ligase (New England Biolabs, Ipswich, MA). The blaOXA-48 gene in the pET29a plasmid was constructed by introducing the D212S substitution and 214-RIEP-217 insertion in blaOXA-163 gene by Quick-change PCR with Pfu Turbo DNA Polymerase (Agilent, Santa Clara, CA). The DNA sequence encoding the mature portion of blaOXA-10 was introduced into pET28a vector using Gibson assembly kit (New England Biolabs) with flanking NdeI and SacI restriction sites that also contained an N-terminal His-tag. DNA sequencing of the entire genes verified the sequence of blaOXA-163, blaOXA-48, and blaOXA-10.
Protein expression and purification
OXA-48 and OXA-163 were expressed following a protocol previously described by Sosa-Peinado et al.40 In brief, cells were grown in 1L LB broth containing 300 mM sorbitol, 2.5 mM betaine, and 30 µg/mL kanamycin to an OD600 of 0.6–0.8 before induction with 0.4 mM IPTG. The culture was then incubated at 23°C for 20 hours with shaking. Afterwards, the culture was centrifuged for 40 minutes at 7,000g and the supernatant was concentrated ten-fold using Vivaflow50 10MWCO (Sartorius, Goettingen, Germany) following dialysis (1:50) overnight at 4°C against buffer containing 20 mM Tris, and 0.4 M NaCl at pH 8.2. The proteins were purified to ~95% homogeneity using a Fast Flow Chelating Sepharose™ (GE Healthcare, Pittsburgh, PA) packed in a column with a 16 mm diameter and 170 mm bed length, loaded with zinc and eluted with a linear gradient of 150 mM imidazole. Purity was determined by SDS-PAGE and protein fractions were concentrated with Vivaspin® Turbo centrifugal filters 10MWCO (Sartorius). After concentration, size-exclusion chromatography using a HiLoad 16/600 Superdex 75 column (GE Healthcare) was performed in 10 mM Tris pH 7.7 and 50 mM NaCl. Size-exclusion chromatography of both OXA-48 and OXA-163 indicated the presence of a dimer.34 Fractions were concentrated and protein concentration was determined by absorbance measurements at 280 nm using an extinction coefficient of 63,940 M−1 cm−1.41
OXA-10 was expressed and purified as follows. 0.5L LB medium supplemented with 30 µg/mL kanamycin was inoculated with 10 mL of overnight culture of E. coli BL21(DE3) carrying the pET28a-blaOXA-10 plasmid. Protein production was induced with 0.5 mM IPTG when the cell culture reached OD600 of 0.8. The culture was then incubated at 30°C for 20 hours. Afterwards, the culture was centrifuged for 40 minutes at 7,000g. The cell pellet was resuspended in 20 mL lysis buffer containing 50 mM phosphate pH7.4, 40 µM MgCl2, and 10 ng/mL DNAse. Cell contents were released using a French press and the lysate was centrifuged at 10,000g for 30 minutes. The supernatant was passed through a HisTrap FF column (GE Healthcare) and the protein was eluted with a linear gradient of 500 mM imidazole. Protein was concentration by buffer exchange using Vivaspin® Turbo centrifugal filters 10MWCO (Sartorius). Protein purity was determined by SDS-PAGE. Protein concentration was determined by absorbance measurements at 280 nm using an extinction coefficient of 47,565 M−1cm−1. The N-terminal His-tag was not removed because steady-state kinetics of the His-OXA-10 enzyme confirmed that activity is not affected by the tag (data not shown).
Enzyme Kinetic Studies
Assays were performed on a DU800 spectrophotometer at 30°C in 50 mM sodium phosphate buffer pH 7.2 supplemented with 15 mM sodium bicarbonate as previously described.42 Substrate hydrolysis was followed at the following wavelengths: 260 nm for ceftazidime (Δε = − 6,900 M−1 cm−1) and cefotaxime (Δε = − 7,500 M−1 cm−1), 262 nm for cephalothin (Δε = − 7,660 M−1 cm−1), 298 nm for meropenem (Δε = − 7,200 M−1 cm−1), 299 nm for imipenem (Δε = − 9, 670 M−1 cm−1), doripenem (Δε = − 11, 540 M−1 cm−1), and nitrocefin (Δε = 20,500 M−1 cm−1). Enzyme kinetics data was analyzed with GraphPad Prism 6 (GraphPad Software, Inc. La Jolla, CA) and fitted to the Michaelis-Menton equation. In the cases when Vmax could not be reached because of a high Km value, the catalytic efficiency (kcat/Km) was calculated by fitting the data in a first-order kinetics equation.43 Inhibition by halogen ions was examined with NaI, NaBr, NaCl or NaF using cephalothin (50 µM for OXA-48 and 12 µM for OXA-10) and 13 µM cefotaxime (OXA-163) as substrates in 50 mM sodium phosphate buffer pH 7.2. The enzymes were preincubated with increasing concentrations of inhibitor for 30 minutes at room temperature and an additional 10 minutes at 30 °C before monitoring hydrolysis. The concentration that reduced hydrolysis by 50% (IC50) was determined and is presented in Table 2.44
Table 2.
IC50 values of OXA-48, OXA-163 and OXA-10 for halogen ions.
| Halide | IC50 (mM)a | ||
|---|---|---|---|
| OXA-48 | OXA-163 | OXA-10 | |
| NaF | 207 ± 11 | 216 ± 20 | 264 ± 20 |
| NaCl | 109 ± 12 | 97 ±13 | 67 ±10 |
| NaBr | 35 ± 7 | 29 ± 9 | 27 ± 6 |
| NaI | 14 ± 5 | 10 ± 4 | 4 ± 2 |
Values and standard deviations were determined from three separate experiments.
Crystallization and data collection
Crystal conditions were screened with 10 mg/mL of concentrated protein using a range of commercially available screens. OXA-163 crystals grew in 0.1M HEPES pH 7.7 and 14% w/v PEG 8, 000. Crystals were cryo-protected with the well solution containing 20% v/v PEG 600 and flash-frozen in liquid nitrogen. A 1.72-Å resolution data set was collected on beamline 5.0.1 of the Berkeley Center for Structural Biology in the context of the Collaborative Crystallography Program. Another crystallization condition was identified containing 0.2 M NaI pH 5.5 and 15% PEG 3, 350. Crystals were cryo-protected with the well solution containing 30% glycerol. A 2.87-Å data set was collected at Baylor College of Medicine on a Rigaku FR-E SuperBright™ High-Brilliance Rotating Anode Generator.
Structure determination and refinement
Diffraction data was processed using the CCP4i suite.45 iMOSFLM was used to process and integrate the images.46 The crystal structures were determined by molecular replacement using OXA-48 (PDB ID 3HBR) as the phasing model with the software MOLREP.47 After one round of REFMAC5 refinement, AutoBuild was used (Phenix suite) followed by manual inspection with COOT.48–50 Several cycles of refinement were performed using phenix.refine (native) and REFMAC5 (with iodide).51, 52 The presence of iodide was confirmed by computing the anomalous difference map using the data set without averaging the Friedel pair intensities (Rmeas 13.3 %). Data collection and refinement statistics are provided in Table 3.
Table 3.
Data collection and refinement statistics.
| Protein | OXA-163 (PDB ID 4S2L) | OXA-163 (PDB ID 4S2M) |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.997 | 1.54 |
| Resolution range (Å) | 38.2 – 1.72 (1.78 – 1.72) | 61.77 – 2.87 (2.98 – 2.87) |
| Space group | P 21 | P 1 |
| Unit cell | ||
| a, b, c (Å) | 44.9, 125.8, 49.7 | 67.6, 68.4, 70.2 |
| α, β, γ (°) | 90.0, 116.8, 90.0 | 62.2, 68.0, 71.6 |
| Unique reflections | 51864 (5184) | 22060 (2111) |
| Multiplicity | 10.4 (10.3) | 8.2 (7.9) |
| Completeness (%) | 99.7 (99.8) | 95.0 (90.3) |
| Mean I/sigma(I) | 52.5 (3.40) | 5.1 (2.0) |
| Wilson B-factor (Å2) | 18.3 | 58.8 |
| Rmerge (%) | 7.7 (53) | 9.3 (32.5) |
| Refinement | ||
| Rwork (Rfree) (%) | 19.5 (23.7) | 20.1 (25.6) |
| Number of non-hydrogen atoms |
4642 | 7752 |
| Protein | 3883 | 7661 |
| Ligands | 20 | |
| Waters | 759 | 71 |
| Protein residues | 474 | 935 |
| r.m.s.d. bond length (Å) | 0.009 | 0.013 |
| r.m.s.d. bond angle (°) | 1.18 | 1.49 |
| Ramachandran favored (%) | 95 | 96 |
| Ramachandran outliers (%) | 0 | 0.33 |
| Average B-factor (Å2) | 22.9 | 55.5 |
| Protein | 21 | 55.6 |
| Ligands | - | 80.9 |
| Waters | 32.8 | 30.4 |
Molecular docking
Docking experiments were performed with cefotaxime (PDB ID: CE3) and ceftazidime. Ceftazidime [CID481173] coordinates were retrieved from the PubChem compound database.53 The substrate molecule was docked into the crystal structure of OXA-48 (PDB ID 3HBR) and OXA-163 (PDB ID 4S2L) using AutoDock Vina (The Scripps Institute, La Jolla, CA).54 Before docking, the proteins were processed by adding polar hydrogen atoms using AutoDockTools. The Lamarckian genetic algorithm was used to generate possible protein-ligand binding conformations.55 The receptor (β-lactamase) was treated as a rigid body, with all possible rotational angles in the substrate. The grid box was centered on the Ser70 residue with the size (22 × 24 × 24 Å) of the box adjusted to cover the entire catalytic site. Docking was carried out with an exhaustiveness of eight.
PDB accession codes and programs used
The OXA-163 structure atomic coordinates were deposited in the Protein Data Bank56 with accession codes 4S2L (native) and 4S2M (with iodide). Alignment and RMSD calculations were performed by SSM procedure using COOT.57 All structural figures were generated with the UCSF Chimera program.58
RESULTS AND DISCUSSION
Enzyme kinetic parameters of OXA-48 and OXA-163 for cephalosporins and carbapenems
To examine if the changes in sequence result in different substrate specificity between OXA-48 and OXA-163, steady-state kinetic parameters were determined for several clinically used cephalosporin and carbapenem substrates as well as the colorimetric cephalosporin, nitrocefin. For OXA-48, the highest catalytic efficiency is observed for nitrocefin hydrolysis (4.0×106 s−1M−1). With regard to clinically-relevant substrates, however, the results confirm that OXA-48 has a preference for carbapenems, with the highest catalytic efficiency (kcat/Km) observed for imipenem hydrolysis (9.0×105 s−1 M−1).33, 34 Compared to imipenem, the OXA-48 kcat/Km value is 53-fold smaller for meropenem and 56-fold smaller for doripenem hydrolysis because of a reduction in the turnover number (kcat). In addition, the early cephalosporin, cephalothin, is hydrolyzed with a slightly higher kcat/Km than the oxyimino-cephalosporin cefotaxime mainly due to a lower Km value. Finally, the steady-state kinetics analysis indicates OXA-48 does not hydrolyze the bulky oxyimino-cephalosporin ceftazidime.33, 59
The substrate profile of OXA-163 is substantially different from that of OXA-48. Unlike OXA-48, OXA-163 has a preference for cephalosporin substrates (Table 1). The highest kcat/Km values were observed for nitrocefin (1.8×106 s−1 M−1), cephalothin (5.0×105 s−1 M−1) and cefotaxime (3.8×105 s−1 M−1). Ceftazidime was hydrolyzed with the lowest kcat/Km (300 s−1M−1) among the cephalosporins tested due to a high Km value. Nevertheless, OXA-163 does hydrolyze ceftazidime. The carbapenemase activity of OXA-163 is attenuated largely due to a reduction in the turnover number while the affinity is very similar to OXA-48 affinity for carbapenems. This leads to a drop of 820-fold in the catalytic efficiency of OXA-163 for imipenem in comparison to OXA-48. However, it is noteworthy that the catalytic efficiencies of OXA-163 are higher for meropenem and doripenem than imipenem, which is opposite to that observed for OXA-48. This change in order of preference is due to a larger reduction in kcat for imipenem hydrolysis compared to meropenem and doripenem in the OXA-163 enzyme (Table 1). Although the reason for this is not known without further structural data, it is possible that the smaller size of imipenem compared to meropenem and doripenem allows it to fit better in the active site of OXA-48, leading to higher turnover of imipenem by OXA-48, which is then lost due to the active site expansion that occurs in OXA-163 as described below.
Table 1.
Steady-state kinetic parameters of OXA-48 and OXA-163 for cephalosporin and carbapenem substrates.
| Substrate | OXA-48 | OXA-163 | ||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (µM) | kcat/Km (s−1M−1) | kcat (s−1) | Km (µM) | kcat/Km (s−1M−1) | |
| Nitrocefin | 143 ± 24 | 36 ± 7 | 4.0 × 106 | 34 ± 2 | 19 ± 8 | 1.8 × 106 |
| Cephalothin | 2.8 ± 0.1 | 140 ± 10 | 2.0 × 104 | 1.7 ± 0.1 | 3.4 ± 0.4 | 5.0 × 105 |
| Cefotaxime | NM | >1000 | 4.7 × 103 | 14 ± 1 | 36 ± 11 | 3.8 × 105 |
| Ceftazidime | ND | - | - | NM | >1000 | 300 |
| Imipenem | 2.7 ± 0.2 | 3.7 ± 0.7 | 9.0 × 105 | 0.004 ± 0.001 | 3.6 ± 1.0 | 1.1 × 103 |
| Meropenem | 0.11 ± 0.01 | 6.0 ± 1.2 | 1.7 × 104 | 0.021 ± 0.001 | 5.3 ± 0.5 | 4.0 × 103 |
| Doripenem | 0.066 ± 0.002 | 4.1 ± 0.6 | 1.6 × 104 | 0.018 ± 0.001 | 5.0 ± 0.4 | 3.6 × 103 |
NM – Not measurable. It was not possible to measure Vmax due to high Km value.
ND – Not detected. No detectable hydrolysis with up to 5 µM enzyme and 500 µM substrate.
In summary, the results confirm the ability of OXA-48 active site to accommodate carbapenem substrates, particularly imipenem, while it is unable to hydrolyze ceftazidime, a bulkier oxyimino-cephalosporin.34, 35 When OXA-48 is transformed to OXA-163 by mutations, it loses high catalytic efficiency for carbapenems, particularly imipenem, but gains the ability to hydrolyze ceftazidime. Therefore, the 214-RIEP-217 deletion and S212D substitution alter the substrate specificity of the enzyme. This finding is consistent with previously published kinetic data showing a significant increase in ceftazidime hydrolysis compared to OXA-48.34, 35 Additionally, two other members of the OXA-48-like β-lactamases, OXA-24760 and OXA-405, which differ from OXA-48 by a four amino-acid deletion, 214-RIEP-218 and 213-TRIE-217, respectively, have significantly lowered activity towards carbapenem substrates compared to OXA-48 (P.N. personal communication).
1.72-Å Crystal Structure of OXA-163
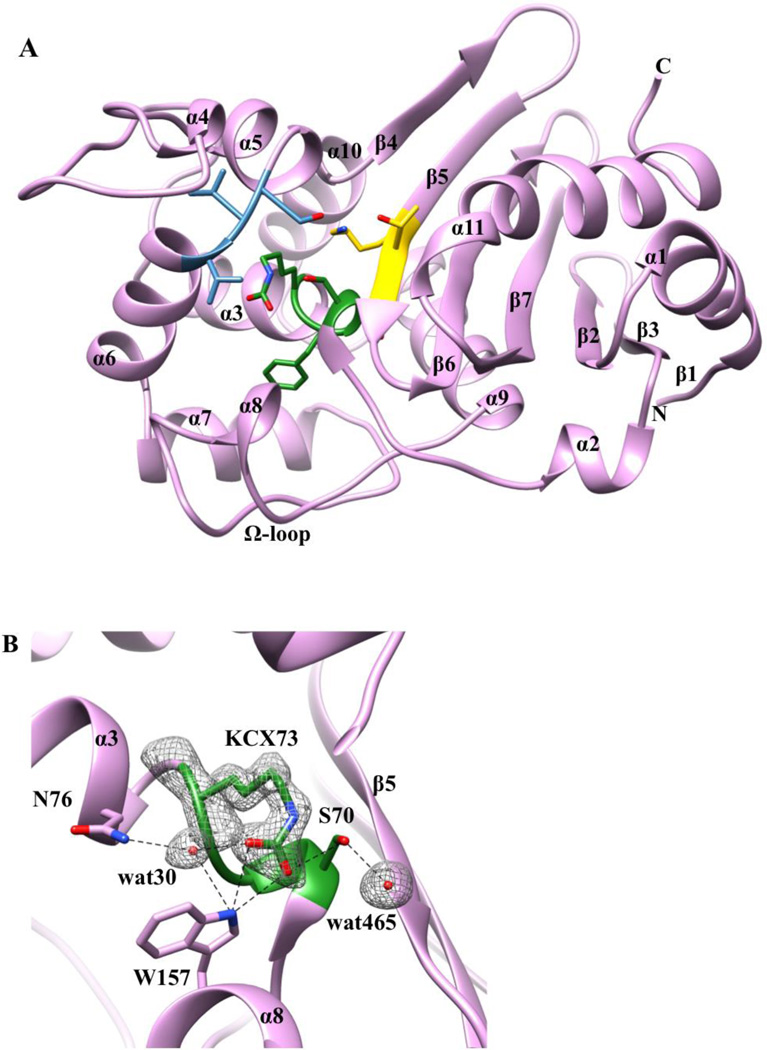
The four amino-acid deletion and S212D substitution transform OXA-48 into OXA-163 and change the substrate profile of the enzyme. To investigate the structural basis for the changes in specificity, the X-ray crystal structure of OXA-163 was determined. The asymmetric unit in the crystal consists of one dimer with the same space group (P21) as the previously published crystal structure of OXA-48.34 The mature OXA-163 enzyme consists of 237 residues and has a two-domain fold typical of DBLs with a globular α-domain and a α/β-domain with the active site located between the two domains (Figure 1A). The active site of OXA-163 forms a groove in the protein surface between the two domains. The clefts of the groove are formed on one side by the β5 strand and the N-terminus of helix α11, and on the other by helices α5, α6 and the Ω-loop (Figure 1A). The three conserved motifs characteristic of the active site of DBLs are outlined in Figure 1A. Motif I consists of the catalytic Ser70 as well as Thr71, Phe72, and the carboxylated Lys73. The carboxylate moiety attached to the εN shows clear density (Figure 1B) in both monomers. Motif II contains residues Ser118, Val119, and Val120 and is located on the loop between the α5 and α6 helices while motif III consists of β5-strand residues Lys208, Thr209, and Gly210. Lys208 interacts with Ser118 from motif II and these residues have been suggested to play a role in substrate binding and proton transfer in the acylation reaction in DBLs.23 Overall, the OXA-163 structure shares a large degree of similarity with the OXA-48 structure34 (Figure 2A) with an RMSD of 0.271 Å for the matching Cα atoms. However, the β5–β6 loop is shorter due to the 214-RIEP-217 deletion that results in an expanded active site cavity compared to OXA-48. Like OXA-48, the quaternary structure of OXA-163 is dimeric, which is not surprising because the residues at the dimerization surface are identical.
Figure 1. Crystal structure of OXA-163.
A) Ribbon representation of OXA-163 with secondary structural elements labeled. The three conserved DBL motifs are shown with their respective residues in a stick model: motif I in green (Ser70, Thr71, Phe72, and Lys73), motif II in blue (Ser118, Val119, and Val120) and motif III in yellow (Lys208, Thr209, Gly210). B) Active site region of OXA-163. The carboxylated side chain of the active-site residue Lys73 is shown together with nearby interacting residues and water molecules. Superimposed is the 2Fo-Fc simulated annealing difference map calculated with phases from the final refined model and contoured at the 2.5σ level. The carboxylate moiety is coordinated by Ser70, Trp157 and, via a water molecule, Asn76. Red spheres represent the two water molecules.
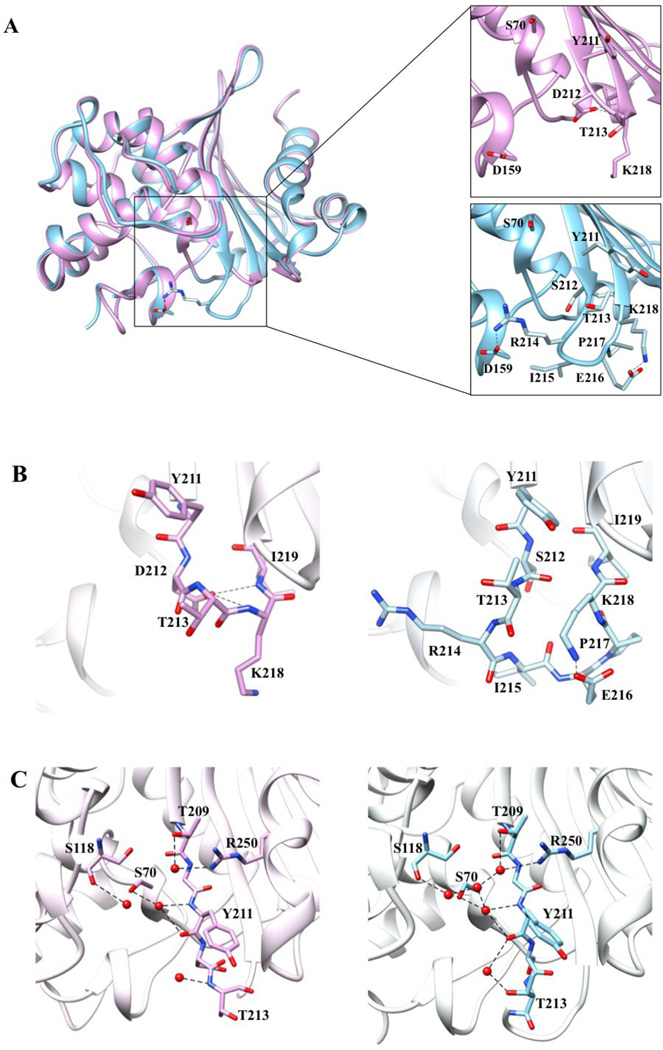
Figure 2. Comparison of the tertiary structures of OXA-48 at 1.9 Å and OXA-163 at 1.72-Å resolution.
A) Cα alignment of OXA-48 (blue) and OXA-163 (lavender). The insets provide a detailed view of the β5–β6 loop region residues represented in stick model with black dashes representing interactions. Arg214 in OXA-48 forms an electrostatic interaction with Asp159 located on the Ω-loop. This interaction confines the bottom boundary of the active-site cavity in OXA-48. In OXA-163 Arg214 is absent and so the interaction is lost, resulting in an expanded active-site cavity. B) Stick representation of the β5–β6 loop region of OXA-48 and OXA-163. The OXA-163 residues that undergo the largest movement compared to OXA-48 are Tyr211, Thr213, and Lys218. The S212D substitution results in newly formed hydrogen bonds between the Asp212 side chain and the -NH main chain of Lys218 and Ile219 represented as black dashed lines. C) Active-site hydrogen bond network in OXA-48 and OXA-163. Several water molecules exhibit altered positions or are missing in OXA-163 compared to OXA-48. Distances of 2.5–3.8 Å are shown as dashed black lines.
A comparison of the active sites of OXA-48 and OXA-163 reveals several differences. i) In OXA-48, the side-chain of Arg214 creates one side of the active site by forming an electrostatic interaction with Asp159 positioned on the Ω-loop (Figure 2A). In OXA-163, Arg214 is within the four amino acid deletion, which eliminates one boundary of the active site and elongates the groove (Figure 2A). Arg214 has been suggested to form electrostatic interactions with carbapenem substrates that facilitate hydrolysis. This could explain the low activity of OXA-163 towards carbapenems.34 Additionally, this expansion of the active site of OXA-163 is consistent with the ability of the enzyme to accommodate a larger substrate such as ceftazidime. The hypothesis that Arg214 contributes to the inability of OXA-48 to accommodate ceftazidime is also supported by the observation that a shorter and uncharged side-chain at position 214 (R to S) results in an increase in the activity of ceftazidime hydrolysis by OXA-232 (an OXA-48-like enzyme).32 ii) The 214-RIEP-217 deletion shortens and alters the conformation of the β5–β6 loop in OXA-163 compared to OXA-48 (Figure 2A). The position and the length of this loop have also been associated with efficient deacylation in carbapenem hydrolysis.34, 39 The altered size and position of the β5–β6 loop in OXA-163 may contribute to the reduced hydrolysis of carbapenems by this enzyme (Table 1). At the same time, this structural change widens the active site of OXA-163 and provides extra space for the oxyimino group of ceftazidime. iii) The 214-RIEP-217 deletion also causes displacement of Thr213, which in part contributes to the enlargement of the active site cavity of OXA-163. The backbone, together with the side chain of Thr213, adopts a different conformation and moves away from the active site (~4 Å) creating a shorter β5–β6 loop (Figure 2A, B). Lys218, which is now adjacent to Thr213, is also displaced further away from the active site (5 Å). iv) The newly introduced aspartate at position 212 is also likely to contribute to the changes in OXA-163 specificity. Compared to Ser212 in OXA-48, the Asp side-chain is pointed towards the β6 strand to form hydrogen bonds with the –NH main chain groups of Lys218 and Ile219 (Figure 2B). This draws the β5 strand main chain closer to the β6 strand and widens the active site wall formed by the β5 strand. v) Finally, several water molecules that are part of a larger interaction network that includes Ser70, Ser118, Thr209, Tyr211, Thr213, and Arg250 in OXA-48 are misplaced or missing in the OXA-163 structure (Figure 2C). This interacting network in OXA-48 was suggested previously to be important in the efficiency of the deacylation reaction for carbapenems.34 This observation is consistent with the large decrease in the turnover number of OXA-163 for carbapenems (Table 1).
In summary, several spatial modifications in the active site of OXA-163 ultimately expand the active site and rearrange the inter-residue interaction network. The larger active site is consistent with improved accommodation of ceftazidime and the loss of critical interactions with carbapenem substrates resulting in an altered substrate profile for OXA-163.
Crystal structure of OXA-163 in the presence of iodide
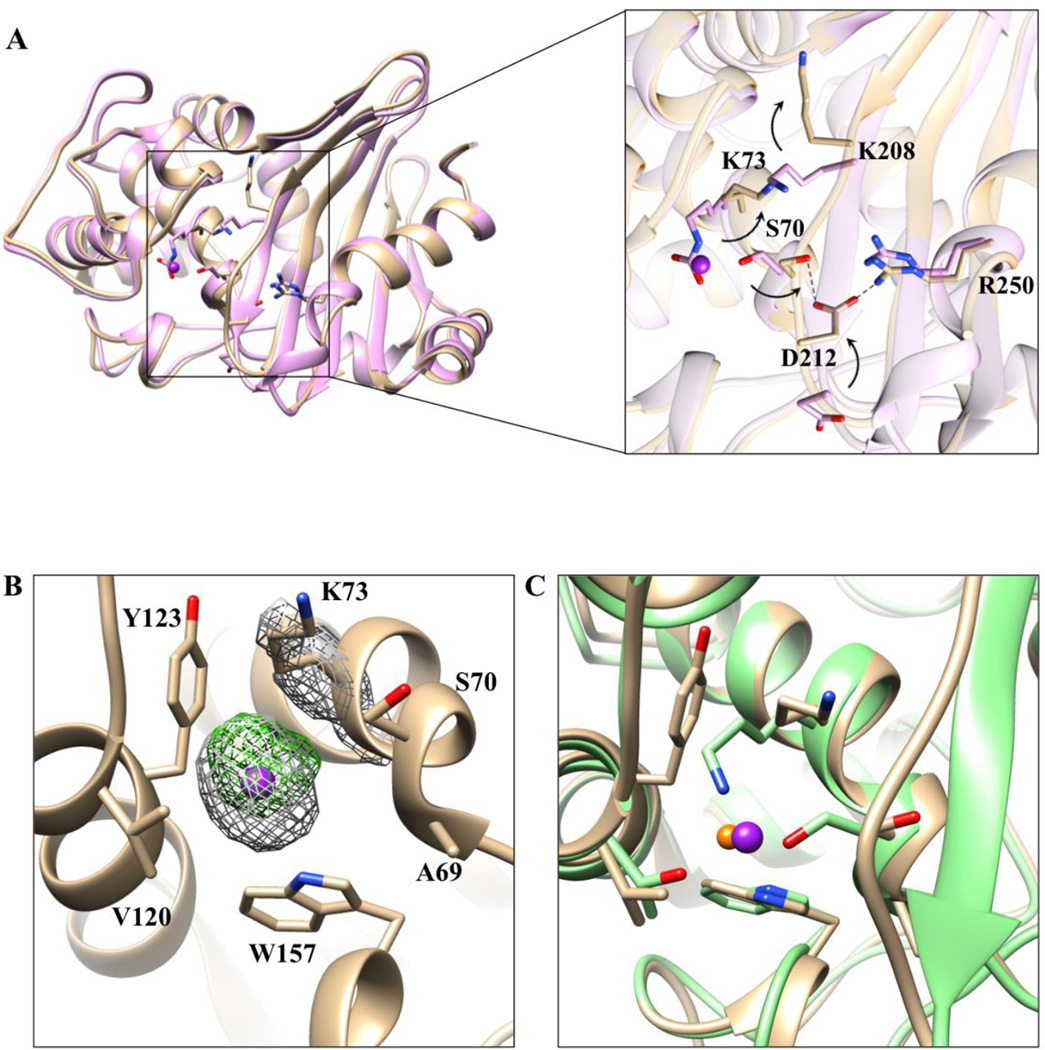
As described above, a second structure of OXA-163 was determined at 2.87-Å from crystals formed in a condition that contained 200 mM sodium iodide. The observed asymmetric unit consists of four protein molecules (two dimers) in a P1 space group. The dimer interface of the structure with iodide is unchanged compared to the native OXA-163 and OXA-48 structures. The 2.87-Å resolution of the structure with iodide clearly reveals electron density of the main chain backbone. An exception was part of the loop (residues 244–247) connecting the β7-strand and the α-helix α11, which did not show density, and was not modeled in the final structure. Some of the solvent exposed side chains, particularly lysines and glutamates were also not visualized. However, the electron density for the active-site lysine 73, which acts as the general base in its carboxylated form20, was clearly resolved and did not show an extension of the side chain indicating the carboxylate group was absent. Instead, an iodide ion was observed in all four molecules in the asymmetric unit at the location where the εN-carboxylate has been observed in other crystal structures. Previously, a chloride ion has been observed at this location in the crystal structures of wild type and a V117T mutant of OXA-10.23, 25
The two OXA-163 structures determined here are very similar with an RMSD of 0.425 Å for their main-chain Cα atoms (Figure 3A). However, the presence of iodide causes a series of rearrangements in the active site, which ultimately leads to distortion of the β5 strand and occlusion of the active-site serine (Figure 3A). In the presence of iodide, Lys73 is not carboxylated and it adopts a conformation in which its side chain points away from the active site and the bound iodide. Similarly, the side chain of Lys208 is also dramatically shifted pointing away from the active site. In fact, the Nζ of Lys73 in the OXA-163 iodide-bound structure occupies the position where the Lys208 Nζ is found in the OXA-163 structure without iodide and in OXA-48 (Figure 3A). Another major change observed in the iodide-bound structure is with respect to Asp212, at the tip of β5-strand, which moves 5Å towards the active site where it hydrogen bonds with Ser70 and engages in electrostatic interactions with Arg250 (Figure 3B). These rearrangements of Lys73, Lys208, and Asp212 along with the newly formed network of interactions involving Ser70-Asp212-Arg250, result in the occlusion of the active site. Previous structural studies of DBLs showed that Arg250 plays an important role in binding and positioning of the carbapenem in the active site of the enzyme.34, 37, 61 The rearrangements in the iodide-bound structure results in a short β5-strand (three residues 204–206), which interacts only with β4-strand. This is in contrast to the OXA-163 structure without iodide, in which the β5-strand is nine residues long (204–212) forming an antiparallel β-sheet involving β4- and β6-strands.
Figure 3. Alignment of the two OXA-163 structures and comparison of chloride and iodide ions in the active site of class D enzymes.
A) 1.72-Å structure (lavender) and 2.87-Å structure with iodide (tan). Iodide is represented as a dark purple sphere with arbitrary radius. The inset provides a more detailed view of the residues (stick model) that undergo large conformational changes due to the presence of the iodide in the active site. In the iodide structure, Asp212 exhibits an altered conformation that results in formation of an electrostatic interaction between with Arg250 (black dashed line). Additionally, the Asp212 residue is within hydrogen bonding distance to the catalytic serine Ser70. The arrows represent the direction of the rearrangements that occur in the iodide structure. B) Iodide and its surrounding environment in the active site. In gray mesh, superimposed, is the 2Fo-Fc simulated annealing difference map calculated with phases from the final refined model and contoured at 3.0 σ level. In green mesh is the anomalous difference map contoured at 5σ. Residues surrounding iodide are labeled and shown in stick model. C) Overlay of OXA-10 (green) inhibited by chloride (orange sphere) and OXA-163 (tan) inhibited by iodide (purple sphere). Iodide is larger than chloride and results in structural rearrangements of the surrounding residues.
The iodide ion found in the active site of OXA-163 is located in a hydrophobic pocket (Figure 3B). It is surrounded mainly by non-polar and aromatic moieties, with the exception of the polar interaction with the εNH of Trp157 (3.7 Å). The hydrophobic interactions include the side-chain hydrocarbons of Lys73 (3.8 Å), the aromatic component of Tyr123 (4.3 Å), the side chain of Val120 (4.5 Å), the aromatic component of Trp157 (4.5 Å), and the main-chain hydrocarbons of Ser70 (4.3 Å) and Ala69 (4.7 Å). The type of displacements observed in the OXA-163 structure with iodide, including alternative side chain conformations and main chain movements, have been observed in several crystal structures that accommodate an iodide ion in proximity to a hydrophobic region.62
Iodide ions have not previously been observed in the active sites of OXA-enzyme crystal structures. However, chloride ions have been identified in the active site of OXA-10.23, 25 The position of the chloride ion in OXA-10 V117T mutant is very similar to the iodide ion in the OXA-163 structure (Figure 3C). However, the iodide occupies a larger volume and results in additional structural displacements in the active site compared to chloride. Also, the interactions of the chloride ion with OXA-10 are exclusively electrostatic while the iodide interactions are overwhelmingly hydrophobic. This is possibly due to the difference in size between the two ion species and their different ability to tolerate a hydrophobic environment. Chloride is smaller and can replace the carboxylate group that is attached to Lys73 and thereby maintain electrostatic interactions with the surrounding residues. On the other hand, iodide is larger and cannot be accommodated in the same location as the carboxylate without expanding the cavity and rearranging the side chains of the close-by residues. Additionally, iodide ions are more tolerant to a hydrophobic environment than chloride ions and are found in hydrophobic patches of proteins.62–64 Tyr141 from the YGN conserved motif that has been implicated in chloride inhibition of OXA-enzymes is 10Å away from the iodide in OXA-163 suggesting it has a minimal role in the binding of the iodide.24
Docking of ceftazidime into the active sites of the OXA-48 and OXA-163
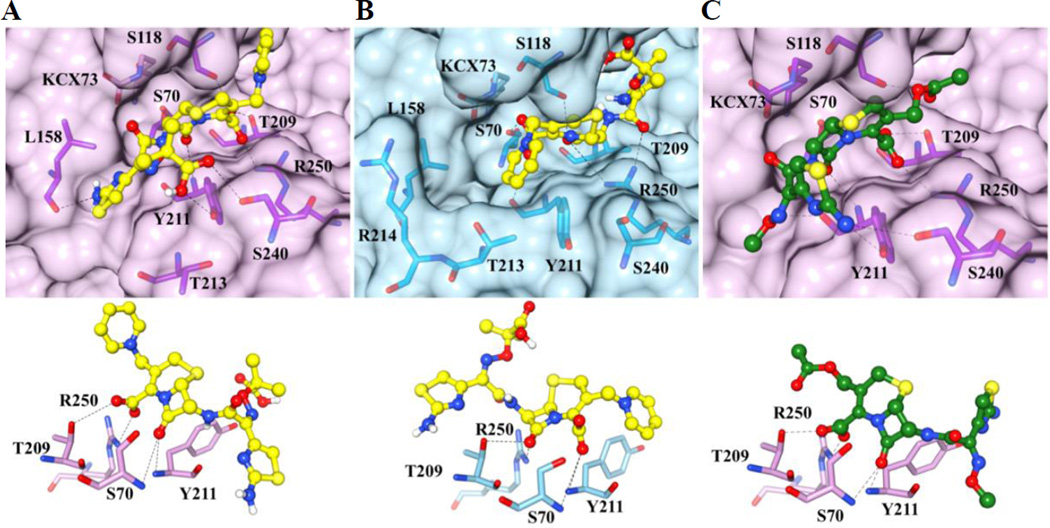
To further investigate why ceftazidime is hydrolyzed by OXA-163 but not OXA-48, molecular docking of ceftazidime was performed using Autodock Vena.54 The protein structures used for docking were OXA-48 (PDB ID 3HBR)34 and the OXA-163 structure without iodide (PDB ID 4S2L). The same constraints were used for both proteins (Materials and Methods). Each docking round gave nine possible conformations, ranked from highest to lowest predicted affinity. It should be noted that all nine conformations of ceftazidime in OXA-48 had the oxyimino-side chain of ceftazidime pointed toward Lys208 and the top of the active site (Figure 4B). The hydrolysis of ceftazidime is not mechanistically feasible from this conformation because the β-lactam carbonyl oxygen is not in close proximity to the NH- main-chain atoms of Ser70 (5.3 Å) and Tyr211 (4.2 Å), which form the oxyanion hole in DBLs. Also, the position of the carboxylate group of ceftazidime does not form electrostatic interactions with Thr209 and Arg250. These residues have been identified previously to form strong electrostatic interactions with the substrate carboxylate in crystal structures of OXA-enzymes.61, 65 The dominance of this catalytically non-productive conformation of ceftazidime in the docking results is due to the narrow active-site cavity of OXA-48 that is confined at the bottom by Arg214 and the longer β5–β6 loop and Thr213. The narrow cavity sterically hinders the formation of a more productive conformation of the oxyimino side chain of ceftazidime.
Figure 4. Docking results of ceftazidime with OXA-163 (lavender) and OXA-48 (light blue) and cefotaxime with OXA-163.
The protein structures are in surface representation and the residues that interact with ceftazidime or cefotaxime are shown in yellow and green stick representation, respectively. The carboxylated Lys73, which is not within interaction distance with the substrate, is also shown and is labeled KCX73. Black dashed lines represent hydrogen bonds. Panel A top: The conformation with the lowest binding energy of ceftazidime for OXA-163 is shown. Panel B top: The conformation of ceftazidime with the lowest binding energy for OXA-48 is shown. In OXA-48, the ceftazidime molecule is flipped compared to the conformation in OXA-163. The docked conformation of ceftazidime in the active site of OXA-48 is not consistent with catalysis. Panel C top: The conformation with the lowest binding energy for OXA-163 with cefotaxime is shown. The bottom panels show the oxyanion hole formed by Ser70 and Tyr211 and the interaction between the carboxylate group of ceftazidime or ceftotaxime with Thr209 and Arg250. These interactions are altered in the docked conformation of ceftazidime in OXA-48 (panel B bottom) such that the β-lactam carbonyl interacts with Thr209 and Arg250 while Ser70 and Tyr211 interact with the carboxylate moiety.
In contrast, the highest affinity binding conformation predicted for ceftazidime bound to OXA-163 has the substrate in an orientation where the oxyimino-side chain occupies the bottom portion of the active-site cavity (Figure 4A). Importantly, the carbonyl oxygen of the β-lactam ring of ceftazidime is hydrogen-bonded to the main chain nitrogens of Ser70 and Tyr211 to form the oxyanion hole so that the substrate is in a catalytically competent conformation. The carboxylated Lys73 is on the other side of Ser70 and is not within an interaction distance with the substrate. In addition, the carboxylic acid moiety of the oxyimino group is within hydrogen bond distance to Oη of tyrosine 211 and Oγ of serine 240. These interactions turn the imino-thiazole ring toward the Ω-loop where it interacts with the main-chain CO of leucine 158. This flip in the conformation of ceftazidime in the active site of OXA-163, compared to the conformation in OXA-48, is allowed by the expanded cavity resulting from the absence of Arg214 side-chain and the shorter β5–β6 loop. The inability of ceftazidime to be docked in the active site of OXA-48 in an orientation that is catalytically feasible is consistent with the hypothesis that the active-site cavity of OXA-48 is too small to fit the bulky ceftazidime substrate.
OXA-163 also exhibits an increased kcat/Km for hydrolysis of the oxyimino-cephalosporin cefotaxime compare to the OXA-48 enzyme (Table 1). This is due to a large decrease in Km from >1000 µM for OXA-48 to 36 µM for OXA-163. The Km of cefotaxime for OXA-163 is also much lower than that observed for ceftazidime. Molecular docking was therefore performed with OXA-163 with cefotaxime for comparison with the ceftazidime docking results. The docking results for show that cefotaxime fits in the active site in a catalytically productive pose with the β-lactam carbonyl oxygen present in the oxyanion hole as observed for ceftazidime (Figure 4C). However, cefotaxime is embedded deeper in the active site cavity and also has the aminothiazole ring flipped 180 degrees pointing towards the solvent compared to ceftazidime. Although there is no X-ray structural information available to examine the molecular details of OXA-163 binding with cefotaxime and ceftazidime, the docking studies suggest cefotaxime can assume a more buried position in the active site, which may explain the lower Km observed for cefotaxime hydrolysis by OXA-163 (Table 1).
Halogen ion inhibition of OXA-enzymes
The observation of an iodide ion and the associated rearrangement of key active site residues as well as the absence of carboxylated Lys73 in the crystal structure of OXA-163 in the presence of iodide suggest that iodide ions inhibit the activity of the enzyme. Considering the relative ability of iodide versus the other halogens to enter a hydrophobic hydration shell66 and the hydrophobic environment of the active site of DBLs, we predict that iodide would be the most potent inhibitor among halogen ions. In order to test these ideas, OXA-48 and OXA-163 were examined for halogen ion inhibition. The substrates cephalothin (OXA-48) and cefotaxime (OXA-163) were used to measure the activity of the enzymes in the presence of halogen ions. It was found that halogen ions inhibit both OXA-48 and OXA-163 with similar potency (Table 2). The IC50 depends on the size of the ion with larger ions being better inhibitors. The crystal structure of OXA-163 with iodide in the active site shows that iodide is not in hydrogen bond proximity to any of the active-site residues except the εNH of Trp157 (Figure 3B). The inhibition assay results are consistent with the ability of the halide ion to be accommodated in a hydrophobic environment since the polar nature of the halogens decreases with increasing size.67
To test whether halogens inhibit other Class D enzymes that are not members of the OXA-48-like family, inhibition assays were performed with OXA-10 β-lactamase. OXA-10 has been studied extensively and it was the first class D β-lactamase with a determined X-ray structure.23 OXA-10 shares 46% sequence identity with OXA-48 and it is a narrow spectrum β-lactamase with high catalytic efficiency for penicillins and early-cephalosporins.33, 39 As indicated in Table 2, the activity of OXA-10 is inhibited by halogens with similar potency in the same order as for OXA-48 and OXA-163 where larger ions are better inhibitors. Additionally, the iodide IC50 for OXA-10 is 4.2 mM, which is approximately 2.5-fold more potent compared to both OXA-48 and OXA-163.
Taken together the inhibition studies suggest that halogen inhibition might be a property of all Class D β-lactamases. Although the IC50s are in low mM range (for iodide), the inhibition of carboxylation of Lys73 by halogens may provide a starting point in the development of new β-lactamase inhibitors given the fact that carboxylation of the active-site lysine is an absolute requirement for the function of all class D β-lactamases.
CONCLUSIONS
As multi-drug resistant infections are on the rise, carbapenems represent one of the last-resort antibiotics to treat these infections.16 Carbapenem-hydrolyzing class D β-lactamases (CHDLs) inactivate carbapenems and confer resistance to these drugs. One common CHDL that is widespread in clinics is OXA-48.28, 33 OXA-163 is a variant of OXA-48 with attenuated catalytic efficiency for carbapenems that has gained the ability to hydrolyze ceftazidime, expanding the ability of this class of enzymes to hydrolyze β-lactams.32, 35 Our structural data suggests that in OXA-163 an enlargement of the active-site cavity occurs that allows this enzyme to accommodate ceftazidime. The enlargement of the active site provides a molecular basis for the distinct substrate profile of these two closely related enzymes and, more broadly, shows that minor sequence variations can profoundly alter the active site of an enzyme. Lastly, we found that OXA-enzymes are inhibited by halogen ions, with iodide being the most potent inhibitor. The structure of OXA-163 in the presence of iodide shows that it changes the position of key active site residues and prevents carboxylation of Lys73. This information may be utilized in the future development of class D inhibitors considering that the carboxylation of Lys73 is essential for the function of these enzymes.
Acknowledgments
We thank Hiram F. Gilbert for discussions and comments on the manuscript.
Funding Sources
This work was supported by NIH grant AI32956 to TP. BVVP acknowledges support from Robert Welch Foundation (Q1279). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH1123. VS is supported by training grant T32 AI55449 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 2.McKenna M. Antibiotic resistance: the last resort. Nature. 2013;499:394–396. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Garbati MA, Al Godhair AI. The growing resistance of Klebsiella pneumoniae; the need to expand our antibiogram: case report and review of the literature. Afr J Infect Dis. 2013;7:8–10. doi: 10.4314/ajid.v7i1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti Infect Ther. 2014;12:565–580. doi: 10.1586/14787210.2014.902306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Martinez L, Gonzalez-Lopez JJ. Carbapenemases in Enterobacteriaceae: Types and molecular epidemiology. Enferm Infecc Microbiol Clin. 2014;32(Suppl 4):4–9. doi: 10.1016/S0213-005X(14)70168-5. [DOI] [PubMed] [Google Scholar]
- 8.Gutkind GO, Di Conza J, Power P, Radice M. beta-lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr Pharm Des. 2013;19:164–208. [PubMed] [Google Scholar]
- 9.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice LB. Mechanisms of resistance and clinical relevance of resistance to beta-lactams, glycopeptides, and fluoroquinolones. Mayo Clin Proc. 2012;87:198–208. doi: 10.1016/j.mayocp.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Therrien C, Levesque RC. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol Rev. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 12.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 13.Bush K. Metallo-beta-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 14.Gniadkowski M. Evolution of extended-spectrum beta-lactamases by mutation. Clin Microbiol Infect. 2008;14(Suppl 1):11–32. doi: 10.1111/j.1469-0691.2007.01854.x. [DOI] [PubMed] [Google Scholar]
- 15.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard DA, Bonomo RA, Powers RA. Class D beta-lactamases: a reappraisal after five decades. Acc Chem Res. 2013;46:2407–2415. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahiri SD, Mangani S, Jahic H, Benvenuti M, Durand-Reville TF, De Luca F, Ehmann DE, Rossolini GM, Alm RA, Docquier JD. Molecular Basis of Selective Inhibition and Slow Reversibility of Avibactam against Class D Carbapenemases: A Structure-Guided Study of OXA-24 and OXA-48. ACS Chem Biol. 2014 doi: 10.1021/cb500703p. [DOI] [PubMed] [Google Scholar]
- 20.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc Natl Acad Sci U S A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Cross JB, Vreven T, Meroueh SO, Mobashery S, Schlegel HB. Lysine carboxylation in proteins: OXA-10 beta-lactamase. Proteins. 2005;61:246–257. doi: 10.1002/prot.20596. [DOI] [PubMed] [Google Scholar]
- 22.Baurin S, Vercheval L, Bouillenne F, Falzone C, Brans A, Jacquamet L, Ferrer JL, Sauvage E, Dehareng D, Frere JM, Charlier P, Galleni M, Kerff F. Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases. Biochemistry. 2009;48:11252–11263. doi: 10.1021/bi901548c. [DOI] [PubMed] [Google Scholar]
- 23.Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D beta-lactamase OXA-10. Nat Struct Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 24.Heritier C, Poirel L, Aubert D, Nordmann P. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob Agents Chemother. 2003;47:268–273. doi: 10.1128/AAC.47.1.268-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vercheval L, Bauvois C, di Paolo A, Borel F, Ferrer JL, Sauvage E, Matagne A, Frere JM, Charlier P, Galleni M, Kerff F. Three factors that modulate the activity of class D beta-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J. 2010;432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- 26.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bush K. Proliferation and significance of clinically relevant beta-lactamases. Ann N Y Acad Sci. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 28.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 29.Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 30.Kamolvit W, Derrington P, Paterson DL, Sidjabat HE. A Case of IMP-4-, OXA-421-, OXA-96-, and CARB-2-Producing Acinetobacter pittii Sequence Type 119 in Australia. J Clin Microbiol. 2015;53:727–730. doi: 10.1128/JCM.02726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother. 2009;64(Suppl 1):i29–i36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- 32.Oueslati S, Nordmann P, Poirel L. Heterogeneous hydrolytic features for OXA-48-like beta-lactamases. J Antimicrob Chemother. 2015 doi: 10.1093/jac/dku524. [DOI] [PubMed] [Google Scholar]
- 33.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. OXA-163, an OXA-48-related class D beta-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelaziz MO, Bonura C, Aleo A, El-Domany RA, Fasciana T, Mammina C. OXA-163-producing Klebsiella pneumoniae in Cairo, Egypt, in 2009 and 2010. J Clin Microbiol. 2012;50:2489–2491. doi: 10.1128/JCM.06710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith CA, Antunes NT, Toth M, Vakulenko SB. Crystal structure of carbapenemase OXA-58 from Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58:2135–2143. doi: 10.1128/AAC.01983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maveyraud L, Golemi D, Kotra LP, Tranier S, Vakulenko S, Mobashery S, Samama JP. Insights into class D beta-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8:1289–1298. doi: 10.1016/s0969-2126(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 39.De Luca F, Benvenuti M, Carboni F, Pozzi C, Rossolini GM, Mangani S, Docquier JD. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D beta-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci U S A. 2011;108:18424–18429. doi: 10.1073/pnas.1110530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosa-Peinado A, Mustafi D, Makinen MW. Overexpression and biosynthetic deuterium enrichment of TEM-1 beta-lactamase for structural characterization by magnetic resonance methods. Protein Expr Purif. 2000;19:235–245. doi: 10.1006/prep.2000.1243. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E, H C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 42.Stojanoski V, Chow DC, Hu L, Sankaran B, Gilbert HF, Prasad BV, Palzkill T. A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis. J Biol Chem. 2015;290:10382–10394. doi: 10.1074/jbc.M114.633438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahl RC. The calculation of initial velocity from product progress curves when [S] << Km. Anal Biochem. 1994;219:383–384. doi: 10.1006/abio.1994.1284. [DOI] [PubMed] [Google Scholar]
- 44.Poirel L, Girlich D, Naas T, Nordmann P. OXA-28, an extended-spectrum variant of OXA-10 beta-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob Agents Chemother. 2001;45:447–453. doi: 10.1128/AAC.45.2.447-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 53.Bolton E, W Y, Thiessen PA, Bryant SH. Annual Reports in Computational Chemistry. Chapter 12. Washington, DC: American Chemical Society; 2008. PubChem: Integrated Platform of Small Molecules and Biological Activities. [Google Scholar]
- 54.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated Docking Using a Lamarckian Genetic Algorithm and and Empirical Binding Free Energy Function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 56.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF, G T, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;13:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Docquier JD, Benvenuti M, Calderone V, Giuliani F, Kapetis D, De Luca F, Rossolini GM, Mangani S. Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob Agents Chemother. 2010;54:2167–2174. doi: 10.1128/AAC.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomez S, Pasteran F, Faccone D, Bettiol M, Veliz O, De Belder D, Rapoport M, Gatti B, Petroni A, Corso A. Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae. Clin Microbiol Infect. 2013;19:E233–E235. doi: 10.1111/1469-0691.12142. [DOI] [PubMed] [Google Scholar]
- 61.Pernot L, Frenois F, Rybkine T, L'Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. Crystal structures of the class D beta-lactamase OXA-13 in the native form and in complex with meropenem. J Mol Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 62.Abendroth J, Gardberg AS, Robinson JI, Christensen JS, Staker BL, Myler PJ, Stewart LJ, Edwards TE. SAD phasing using iodide ions in a high-throughput structural genomics environment. J Struct Funct Genomics. 2011;12:83–95. doi: 10.1007/s10969-011-9101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibb CL, Gibb BC. Anion binding to hydrophobic concavity is central to the salting-in effects of Hofmeister chaotropes. J Am Chem Soc. 2011;133:7344–7347. doi: 10.1021/ja202308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fox JM, Kang K, Sherman W, Heroux A, Sastry GM, Baghbanzadeh M, Lockett MR, Whitesides GM. Interactions between Hofmeister Anions and the Binding Pocket of a Protein. J Am Chem Soc. 2015;137:3859–3866. doi: 10.1021/jacs.5b00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider KD, Karpen ME, Bonomo RA, Leonard DA, Powers RA. The 1.4 A crystal structure of the class D beta-lactamase OXA-1 complexed with doripenem. Biochemistry. 2009;48:11840–11847. doi: 10.1021/bi901690r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rankin BM, Ben-Amotz D. Expulsion of ions from hydrophobic hydration shells. J Am Chem Soc. 2013;135:8818–8821. doi: 10.1021/ja4036303. [DOI] [PubMed] [Google Scholar]
- 67.Politzer P, Murray JS, Bulat FA. Average local ionization energy: A review. J Mol Model. 2010;16:1731–1742. doi: 10.1007/s00894-010-0709-5. [DOI] [PubMed] [Google Scholar]