SUMMARY
The oral cavity is a dynamic environment characterized by hundreds of bacterial species, saliva, and an influx of nutrients and metal ions such as copper. While there is a physiologic level of copper in the saliva, the oral cavity is often challenged with an influx of copper ions. At high concentrations copper is toxic and must therefore be strictly regulated by pathogens in order to persist and cause disease. The cariogenic pathogen Streptococcus mutans manages excess copper using the copYAZ operon that encodes a negative DNA-binding repressor (CopY), the P1-ATPase copper exporter (CopA), and the copper chaperone (CopZ). These hypothetical roles of the copYAZ operon in regulation and copper transport to receptors led us to investigate their contribution to S. mutans virulent properties.
Mutants defective in the copper chaperone CopZ, but not CopY or CopA, were impaired in biofilm formation and competitiveness against commensal streptococci. Characterization of the CopZ mutant biofilm revealed a decreased secretion of glucosyltransferases and reduced expression of mutacin genes. These data suggest that the function of copZ on biofilm and competitiveness is independent of copper resistance and CopZ is a global regulator for biofilm and other virulence factors. Further characterization of CopZ may lead to the identification of new biofilm pathways.
Keywords: copYAZ, Glucan matrix, Dental caries, Mutacin
INTRODUCTION
Streptococcus mutans is an oral pathogen associated with the development of dental caries. Crucial to S. mutans cariogenicity is the ability to form a tenacious, well-structured, three-dimensional biofilm. To establish a cariogenic biofilm and consequently cause disease, S. mutans must first manage to survive environmental stress encountered within the oral cavity which harbors antagonistic bacteria, but also host-derived compounds or antimicrobials and metal ions such as copper. In the oral cavity S. mutans is continuously exposed to copper, however, S. mutans has been reported to be resistant to copper at high concentrations (i.e. approximately 64 mg/L), in comparison to bacteria that dwell in niches that contain no copper (Vats & Lee 2001). The physiologic concentration of copper in saliva ranges from 0.2 to 7.05 mg/L (Duggal et al. 1991), however, copper concentrations in the oral cavity can fluctuate depending on intake of dietary copper and the presence of metal alloy dental restorative materials which may leak copper ions into the surrounding milieu (Orstavik 1985). Under physiologic conditions copper is used as a cofactor for enzymes such as cytochrome C oxidase (Karlin 1993). At elevated concentrations however, abundant free copper is toxic and commonly used as an antimicrobial against S. mutans (Drake et al. 1993; Evans et al. 1986). Therefore, it is necessary for S. mutans to regulate excess copper to survive and consequently establish a cariogenic biofilm.
In S. mutans UA159, copper resistance is mediated by operon copYAZ (Ajdic et al. 2002; Vats & Lee 2001). Although sensitivity to copper increases without copYAZ, functional annotation of S. mutans CopY, CopA, and CopZ is based on extensive studies of the Enterococcus hirae copYABZ operon (Solioz & Stoyanov 2003). The S. mutans copYAZ operon is predicted to encode a negative repressor (CopY) that allows for transcription upon copper transfer from direct binding with CopZ, a P1-ATPase whose sole function is to export excess copper ions out of the cell for copper resistance (CopA), and a small protein that tightly binds and delivers copper to CopY to presumably positively regulate copYAZ (CopZ) (Cobine et al. 1999; Vats & Lee 2001). Due to the function of CopYAZ to recognize, bind, and transport copper, and the connection between copper and virulence in other bacterial pathogens (Baker et al. 2010; Cobine &Wickramasinghe &Harrison &Weber &Solioz &Dameron 1999; Wolschendorf et al. 2011), we sought to characterize the role of CopYAZ in S. mutans virulence traits and competitiveness.
In this study, we determined copper chaperone CopZ indeed binds to copper and established a role for CopZ in the regulation of S. mutans virulence factors. Distinct from copY or copA mutants, S. mutans copper chaperone mutants were defective in biofilm formation and competitiveness against commensal streptococci. Elucidation of the new role of copZ could identify novel regulatory pathways in S. mutans.
METHODS
Bacterial strains and cultures
All strains and plasmids used in this study are listed in Table 1. Streptococcus mutans UA159 (wild-type, WT UA159), serotype C, was used as the model organism. Single gene mutants of copY (ΔcopY), copA (ΔcopA), and copZ (ΔcopZ) were made through allelic exchange with a kanamycin cassette as described (Lau et al. 2002; Wen et al. 2015). In brief, the gene and flanking regions were amplified and inserted into pGEM T-easy vector (Promega, Madison, WI). Since the copY and copA genes overlap, ΔcopY and ΔcopA were created by replacing domains 1–123aa and 2–742aa, respectively, with a kanamycin cassette. In contrast, the copZ gene is downstream of both copY and copA, therefore, the intact gene of ΔcopZ (1–67aa) was replaced by a kanamycin cassette (Vats & Lee 2001). Transformants were recovered following the pGEM protocol. In brief, transformants were screened on X-gal and IPTG plates. White colonies were chosen and checked for insertion by PCR. pGEM plasmids containing the amplified regions were extracted and used for inverse PCR with primers listed in Table 2. Inverse PCR products and the kanamycin cassette were digested with KpnI. Digested products were ligated and resulting plasmids were transformed into E. coli. Plasmids containing the kanamycin cassette in place of the gene of interest were transformed into S. mutans UA159 to replace the kanamycin cassette with the gene in the chromosome. Primers used to make mutants are listed in Table 2. All mutants were examined by PCR for correct placement of the kanamycin cassette in the chromosome. For the ΔcopZ complement strain (ΔcopZ/copZpVPT), ΔcopZ was transformed with a shuttle plasmid, pVPT(erythromycin resistance), expressing copZ. All S. mutans strains, Streptococcus gordonii DL1, and Streptococcus sanguinis SK36 were grown statically in Todd-Hewitt media (THB) (BD Biosciences, Franklin Lakes, NJ) at 37°C under 5% CO2.
Table1.
Strains and plasmids used in this study
| Strain or Plasmid | Description | Source |
|---|---|---|
| WT UA159 | Streptococcus mutans UA159 | This study |
| ΔcopY | S. mutans UA159 with in frame replacement with a kanamycin cassette | This study |
| ΔcopA | S. mutans UA159 with in frame replacement with a kanamycin cassette | This study |
| ΔcopZ | S. mutans UA159 with in frame replacement with a kanamycin cassette | This study |
| ΔcopZ/copZpVPT | S. mutans UA159 ΔcopZ transformed with pVPT encoding copZ | This study |
| S. gordonii | Streptococcus gordonii DL1 | This study |
| S. sanguinis | Streptococcus sanguinis SK36 | This study |
| pET28a-hisSUMO | Plasmid optimal for expressing soluble proteins with a His-tag in E. coli BL21 Gold (DE3) | This study |
| pVPT | E. coli-Streptococcus shuttle vector and expression plasmid (erythromycin) | Zhou et al. (2008) |
| copZpVPT | S. mutans UA159 copZ gene cloned into pVPT for expression of copZ | This study |
Table 2.
Primers used to make mutants or constructs
| Constructs | Primers used (5’–3’) Restriction enzyme sites are underlined |
|---|---|
| copY mutant | Amplify gene + flanking region:
|
| copA mutant | Amplify gene + flanking region:
|
| copZ mutant | Amplify gene + flanking region:
|
| ∆copZ/copZpVPT | Amplify gene with added BspHI & BamHI sites for insertion:
|
| copZpET28a-hisSUMO | Amplify gene with added BamHI & XhoI sites for insertion:
|
Biofilm assay
Biofilms were formed similar to Liu et al. (2011). In brief, overnight cultures were diluted into fresh THB and grown to exponential phase. Cultures were diluted to O.D.470 0.01 into 10mL of pre-warmed chemically-defined biofilm media containing 1% sucrose as the carbohydrate source (Loo et al. 2000). Doubled inoculum of ΔcopZ was added to biofilm media to create a biofilm with biomass similar to wild-type, labelled as ΔcopZ(double) in Fig.3B to compensate for decreased biomass of ΔcopZ. Biofilms were grown statically for 18 hours in 96-well polystyrene plates. To measure biomass loosely adhered cells were gently washed off and the biofilm was stained with 0.1% crystal violet for 15 min and solubilized in 30% acetic acid. Biomass was quantified using O.D.562.
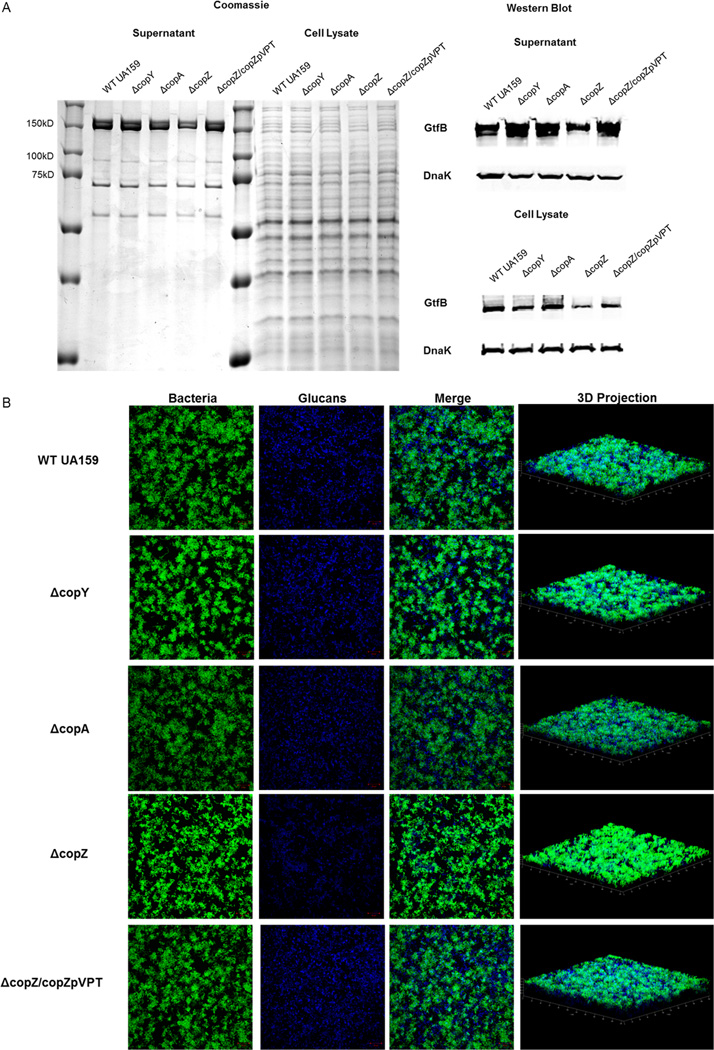
Figure 3. CopZ mutant is deficient in Gtf protein levels and production of glucan matrix.
(A) Protein profiling of S. mutans. Proteins were extracted from supernatants and cells of WT UA159, ΔcopY, ΔcopA, ΔcopZ, and ΔcopZ/copZpVPT cultures grown to the same OD470. Extracted proteins from supernatants and cell lysates were run on SDS-PAGE gels and stained with coomassie (left) or probed with the GtfB or DnaK (for loading control) antibodies for western blot (right). (B) Biofilms of S. mutans. Biofilms of WT UA159, ΔcopY, ΔcopA, ΔcopZ, and ΔcopZ/copZpVPT were grown for 18 hours. All images are maximum intensity projections or Three-dimensional projections of bacteria (green) and glucan matrix (blue). Biomass of biofilms imaged by CLSM is plotted. Results are representative of three independent experiments.
Protein Expression & Isothermal Titration Calorimetry
The copZ (SMU_427) coding region from S. mutans UA159 was cloned into the pET28a-hisSUMO vector in E. coli BL21 Gold (DE3) cells to make the copZpET28a-hisSUMO expression plasmid. Primers used to make the construct are listed in Table 2. Recombinant protein hisSUMO-CopZ was expressed and purified similar to Zhang et al. (2014). For Isothermal Titration Calorimetry, 1mM CopZ and 10mM of CuSO4, MgSO4, ZnSO4, or NiSO4 were each diluted in water and were loaded on a Microcal Automated Isothermal Titration Calorimetry ITC 200 in the Center for Biophysical Sciences and Engineering, UAB. The sample cell was filled with 400uL CopZ protein and the injection syringe was loaded with 150uL metal solution. Data was collected from 25 injections at 20°C, and analyzed using Origin 7 Microcal Software (OriginLab, Northampton, MA).
Protein extraction
Five milliliter cultures of each strain were grown to the same OD470 in THB. Cells were pelleted and the supernatants were mixed with 1.5mL of 100% Molecular Grade Ethanol (Fisher Scientific, Pittsburgh, PA) and frozen overnight at −80°C. Supernatants were thawed out, spun down, and pellets containing proteins were collected. Equal amounts of cells were lysed by vortexing with glass beads and boiled for 10 min at 95°C. Proteins were separated on SDS-PAGE gels for Coomassie staining and western blotting.
Inhibition assay
Inhibition assays were performed similar to the interference assays described in Scoffield & Wu (2015). S. mutans strains were grown to early exponential phase and plated on THB agar plates. Plates were incubated overnight at 37°C under 5% CO2. Streptococcus sanguinis or Streptococcus gordonii cultures in early exponential phase were spotted immediately adjacent to the established S. mutans colony and incubated overnight. Inhibition of S. sanguinis or S. gordonii growth was visually observed and imaged using Gel Logic 100 Imaging System.
Confocal Laser Scanning Microscopy
For confocal laser scanning microscopy (CLSM), cultures were diluted in biofilm media+1% sucrose containing 500nM dextran-conjugated cascade (Ex: 400/Em: 420, Molecular Probes, Invitrogen, Life Technologies, Grand Island, NY). Dextran-conjugated dye has been shown to help visualize the extracellular polysaccharide matrix in S. mutans (Koo et al. 2010; Peng et al. 2015). Biofilms were grown on ibiTreat 8 well µ-slides (ibidi #80826, Martinsried, Germany) for imaging. After 18 hours, biofilms were gently dip washed 3X in sterile 1XPBS. Bacteria in biofilms were visualized with 1 µM Syto9 green (Ex: 485/Em: 495, Molecular Probes, Invitrogen, Life Technologies, Grand Island, NY). Biofilms were imaged at 63X magnification using a Zeiss Laser Scanning Microscope LSM 710 Confocal Microscope at the UAB High Resolution Imaging Shared Facility.
Qualitative Real Time PCR
RNA was extracted from late exponential phase cultures using the Direct-zol kit (Zymo Research, Irvine, CA). Residual DNA was digested using RQ1 DNase (Promega). RNA was purified with the miniRNAeasy kit (Qiagen, Venlo, Limberg), and converted into cDNA using the iScript cDNA Synthesis kit (Bio-rad, Hercules, CA). cDNA was then used for qRT-PCR with iQ SYBR Green Supermix (Bio-rad). Primers used are shown in Table 3.
Table 3.
qRT-PCR Primers
| Gene Name | Primer Sequences (5’–3’) |
|---|---|
| nlmA | F-ATGGATACACAGGCATTTC R-TATGGGGTAACAAGAGTCC |
| nlmB | F-TGTCAGAAGTTTTTGGTGGA R-AGCACATCCAGCAAGAATA |
| nlmC | F-AGCATATGGACCAAGAAATC R-ACGTAATGGATAATGAAGCAC |
| nlmD | F-GAGGGTGGTGGTATGATTAGATGTG R-TCCAGACCAGCCTCCTAAAGC |
| comC | F-ACGAATTAGAGATTATCATTGGCGG R-CCCAAAGCTTGTGTAAAACTTCTGT |
| comD | F-TGATTGCTGTTACGATGGTG R-AAGTCAGAACTGGCAACAGG |
| comE | F-TCATACTGCCGTAGAATTCA R-AAGAATGGTCAATCAGAGGA |
| gtfB | F-ACACTTTCGGGTGGCTTG R-GCTTAGATGTCACTTCGGTTG |
| brpA | F-TACAGCATCAGTTGAGCCCG R-ACCTTGCTGATGACCTCACG |
Statistical analysis
All experiments were repeated three times. Biomass and qRT-PCR results are presented as mean ± SEM and statistical significance determined by one-way ANOVA. Samples were considered statistically significant if the difference has a p-value of <0.05. Analysis of biomass and gene expression data was done using GraphPad Prism 6 (GraphPad Software, San Diego, CA).
RESULTS
CopZ plays a role in proper biofilm formation
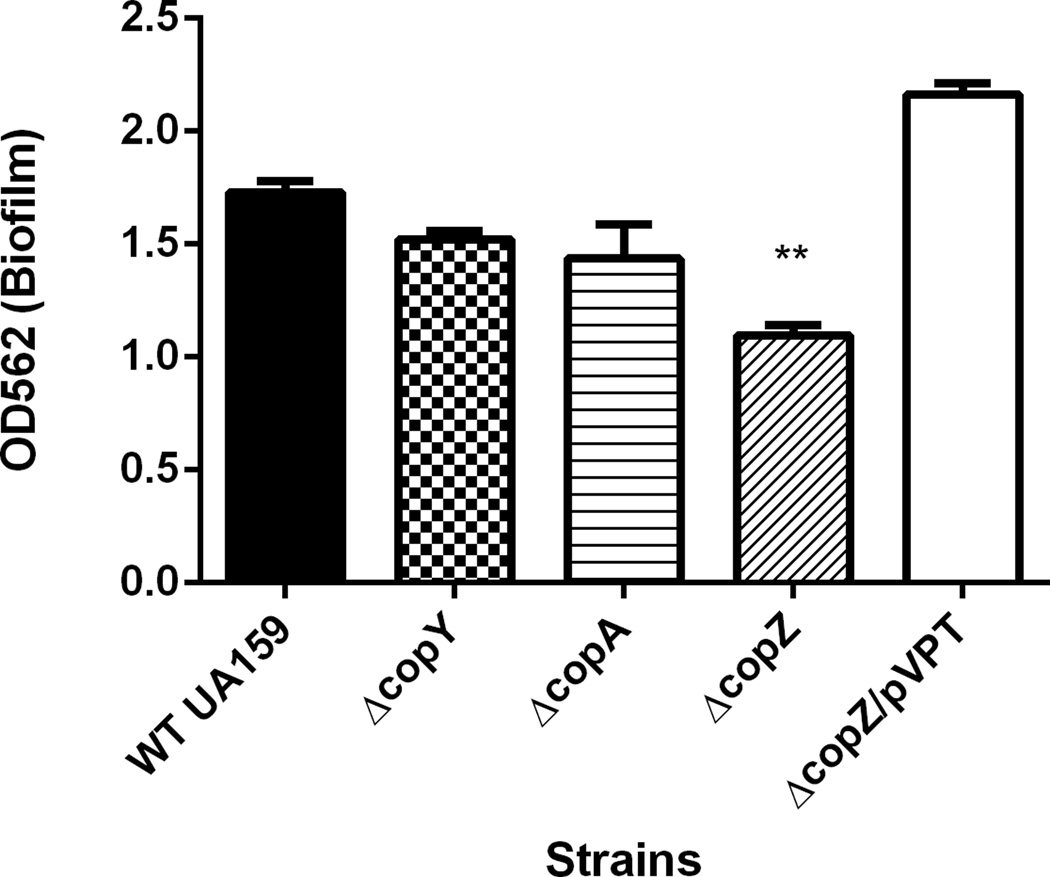
Recently, the intact copYAZ operon of Streptococcus mutans has been implicated in cariogenic biofilm properties such as acid tolerance and competence (Singh et al. 2015). To investigate which gene(s) of the copYAZ operon could be attributed to biofilm formation, we constructed single-gene mutants (ΔcopY, ΔcopA, and ΔcopZ). Mutants were allowed to form biofilms in biofilm media containing 1% sucrose for 18 hours. At the end of 18 hours, biomass was quantitated by crystal violet staining. Compared to the parent strain (WT UA159), the ΔcopZ biofilm had significantly less biomass, while loss of copY and copA did not affect biofilm formation (p<0.05, Fig. 1). Proper biofilm formation could be restored by complementation of copZ (Fig. 1, ΔcopZ/copZpVPT). All strains grew similarly in biofilm media for 18 hours, suggesting the biofilm defect of ΔcopZ was not due to reduced cell growth (data not shown).
Figure 1. S. mutans UA159 copZ mutant is defective in biofilm formation.
Single-gene mutants of copYAZ and the parent strain were allowed to form biofilms for 18 hours in biofilm media supplemented with 1% sucrose. Biomass was quantitated by crystal violet staining and reading at O.D.562. Complement strain of ΔcopZ restored biofilm formation (ΔcopZ/copZpVPT). **=p<0.01. Data represents three biological replicates.
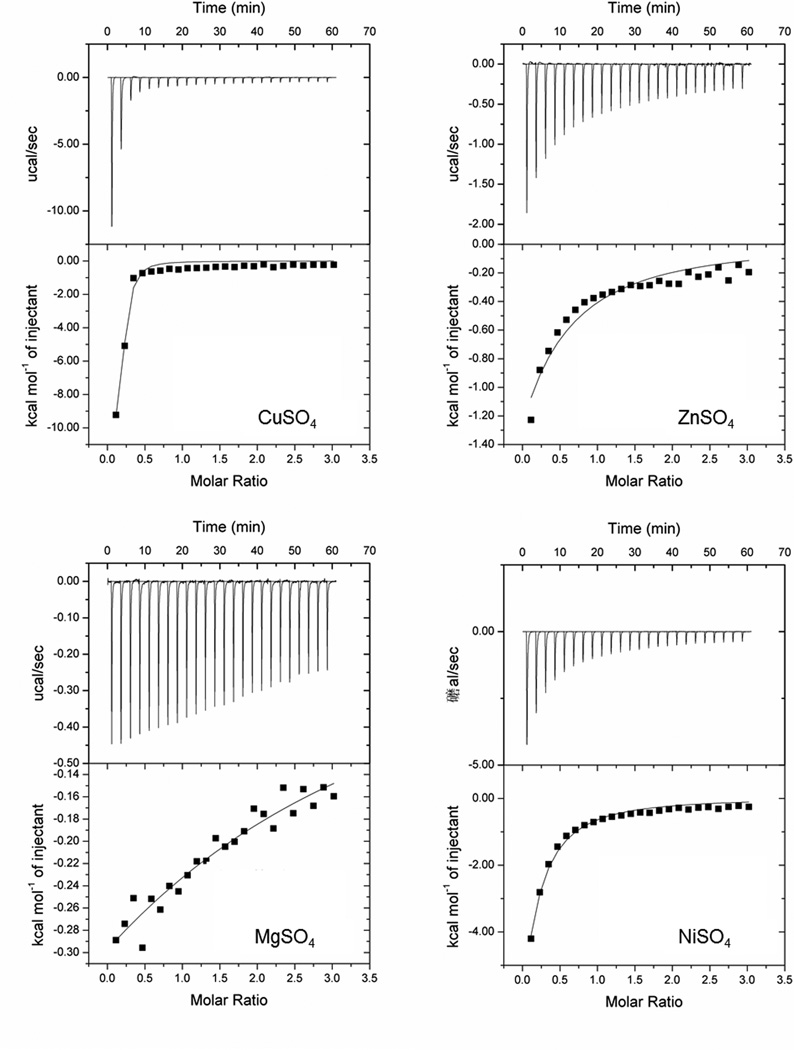
We first wanted to demonstrate the function of S. mutans CopZ as a copper-binding chaperone. Copper chaperones display strong and specific binding to copper (Kihlken et al. 2002; Urvoas et al. 2004). Purified CopZ protein and divalent cations CuSO4, ZnSO4, MgSO4, and NiSO4 were used for isothermal titration calorimetry (ITC). CopZ bound copper with high affinity, as evidenced by the best fit line (Fig. 2). In contrast, while there seems to be some binding activity of CopZ to ZnSO4, MgSO4, and NiSO4, this binding occurred at a much lower affinity. This data demonstrates the selective binding of CopZ and copper and validates the function of CopZ as a copper chaperone.
Figure 2. CopZ affinity for Copper.
ITC curves of purified CopZ protein and free copper, zinc, magnesium, or nickel binding.
We reasoned that a mutation in copZ may result in an altered protein profile, which could potentially be involved in regulating biofilm formation. To investigate this, we examined intracellular and secreted protein patterns in cells and culture media, respectively, of WT UA159, ΔcopZ, and ΔcopZ/copZpVPT. Extracted proteins were separated on an SDS-PAGE gel for comparison. The top two bands (approximately 150kDa) were reduced in the copZ mutant secreted protein profile. Since these bands correlated with the size of glucosyltransferases (Gtfs), the bands were probed with a GtfB antibody (Nakano & Kuramitsu 1992). ΔcopY and ΔcopA protein profiles were similar to the parent strain. In contrast, there were drastically less Gtfs in the cell and secreted into the supernatant of ΔcopZ (Fig. 3A). Gtf production and secretion were restored in by complementation (ΔcopZ/copZpVPT, Fig. 3A). Gtfs, particularly GtfB and GtfC, extracellularly metabolize sucrose into mostly insoluble glucans which are essential for providing a rigid scaffold for the development of a tenacious, cariogenic biofilm (Bowen & Koo 2011; Nakano & Kuramitsu 1992). To determine whether decrease in secreted Gtf translated to the production of less glucan matrix, we examined production of the glucan matrix in biofilms using a dextran-conjugated cascade dye with confocal laser scanning microscopy (CLSM). Biofilms were grown identically to those depicted in Fig.1 except with added dextran-conjugated cascade dye and on an 8-well slide for optimal CLSM imaging. Under these conditions, we visualized composition and architecture of the biofilms with CLSM. We compared the WT UA159 to the ΔcopY, ΔcopA, and ΔcopZ mutant biofilms. Eighteen-hour biofilms of the WT UA159, ΔcopY, and ΔcopA displayed similar biofilm structure of aggregates of bacteria encased or surrounded by glucan matrix (First and fourth rows, Fig. 3B) (Xiao et al. 2012). The ΔcopZ biofilm produced less glucan matrix which led to bacterial cells being more exposed, and therefore, seemed to exhibit higher intensity of fluorescence (Fig. 3B). Three-dimensional panels reflect thickness observed in Fig. 1 (last column, Fig. 3B). To demonstrate ΔcopZ biofilm phenotypes are not biomass dependent, we doubled the inoculum of ΔcopZ bacteria added to biofilm media prior to biofilm development to compensate for differences in biomass (ΔcopZ(double)) and then examined biofilms by CLSM (Fig. S1). At 18 hours, the ΔcopZ(double) biomass was similar to WT UA159 and ΔcopZ/copZpVPT (bar graph, Fig. S1). Since both ΔcopZ and ΔcopZ(double) exhibited reduced glucan matrix and compared to the parent and complement strains, these phenotypes observed in ΔcopZ were therefore not due to reduced biomass. These data suggest copZ, but not copY or copA, plays an important role in proper biofilm formation in a Gtf-dependent manner.
S. mutans competitiveness dependent on CopZ
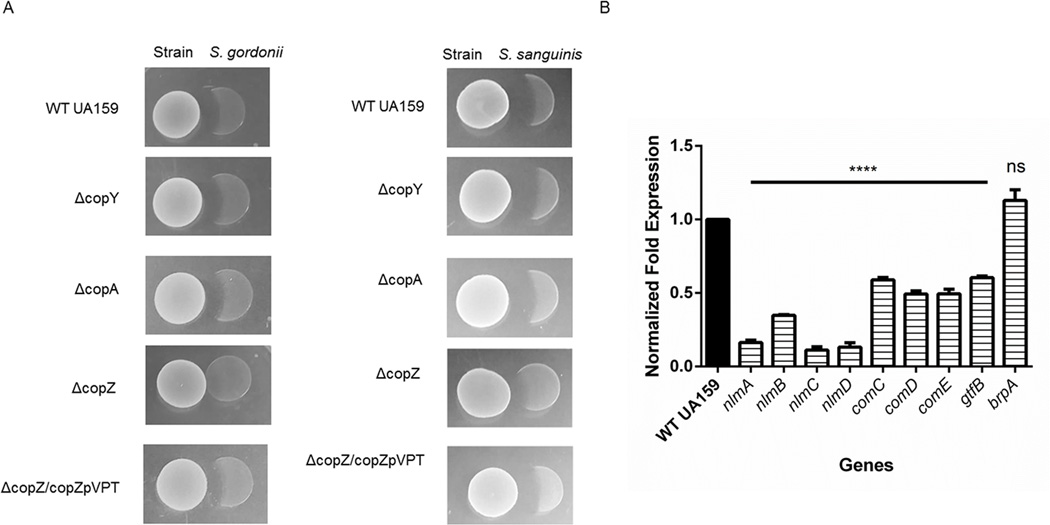
To persist in the oral cavity, S. mutans must out-compete commensal bacteria that produce detrimental hydrogen peroxide or other antimicrobials. As a defense mechanism, S. mutans secretes bacteriocins-antimicrobial peptides that inhibit growth of commensal species such as Streptococcus sanguinis and Streptococcus gordonii (Hossain & Biswas 2011; Kreth et al. 2008). Because the copZ mutant biofilm was defective in biomass, composition, and architecture, we suspected that the mutant would not be able to effectively inhibit commensal species. To determine whether ΔcopZ could inhibit commensal species, strains were analyzed using an inhibition assay. Strains WT UA159, ΔcopY, ΔcopA, ΔcopZ, and ΔcopZ/copZpVPT were inoculated on THB plates first. Liquid cultures of commensal species were then plated immediately adjacent to the S. mutans strains. After 18 hours incubation, growth inhibition of commensal species was observed. Strains ΔcopY and ΔcopA inhibited S. gordonii and S. sanguinis similar to the parent strain. ΔcopZ showed reduced ability to inhibit S. gordonii or S. sanguinis, albeit the effect on S. sanguinis was modest while inhibition of S. gordonii was completely abolished (Fig. 4A). This effect was restored by complementation (ΔcopZ/copZpVPT, Fig. 4A).
Figure 4. CopZ mutant is defective in inhibiting commensal Streptococci.
(A) Plate inhibition assays. WT UA159, ΔcopY, ΔcopA, ΔcopZ, and ΔcopZ/copZpVPT were inoculated first. Commensals S. gordonii and S. sanguinis were plated second and growth inhibition was assessed the following day. (B) Expression of mutacin genes is reduced in the CopZ mutant. RNA was extracted from late exponential phase cultures of WT UA159 and ΔcopZ. Expression of mutacin genes were analyzed by qRT-PCR and normalized by comparison to the expression of 16S. Results are representative of three biological replicates. ****=p-value≤0.0001.
S. mutans UA159 secretes three bacteriocins, mutacin IV,mutacin V, and mutacin VI (Hossain & Biswas 2011; Merritt & Qi 2012). To validate the inhibition assay, we examined the expression of genes nlmA and nlmB (mutacin IV), nlmC (mutacin V), and nlmD (mutacin VI) in the copZ mutant. Expression of all four genes was significantly downregulated in ΔcopZ compared to the parent strain (Fig. 4B). Since the com system has been recently implicated in mediating mutacin production, we checked expression of comCDE (Reck et al. 2015). All com genes were downregulated in ΔcopZ (Fig.4B). The expression for gtfB was downregulated in ΔcopZ, further demonstrating the decrease in Gtfs and glucans observed in ΔcopZ only (Fig. 3A & 3B). Because Biofilm Regulatory Protein A, BrpA, has been associated with biofilm, we examined the expression of brpA in ΔcopZ. Notably, brpA expression in ΔcopZ looked similar to WT UA159, suggesting that the impact of copZ on S. mutans biofilms and competitiveness is BrpA-independent. Collectively, these data suggest that CopZ plays a critical role in the expression of mutacins and consequently inhibition of commensal Streptococci.
DISCUSSION
Copper plays a critical role in bacterial pathogenesis, survival, stress response, and biofilm formation (Baker &Sitthisak &Sengupta &Johnson &Jayaswal & Morrissey 2010; Mitrakul et al. 2004; Shafeeq et al. 2011; Solioz et al. 2010; Solioz & Stoyanov 2003; Wolschendorf &Ackart &Shrestha &Hascall-Dove &Nolan &Lamichhane &Wang &Bossmann &Basaraba & Niederweis 2011). The cariogenic pathogen, S. mutans survives copper challenge by inducing the copYAZ operon and mediating copper resistance (Singh &Senadheera &Levesque & Cvitkovitch 2015; Vats & Lee 2001). While copYAZ counterparts in other pathogenic organisms have been shown to be necessary for virulence expression, the individual copYAZ genes in S. mutans have not been investigated for their potential roles in bacterial fitness and virulence traits. In this study, we demonstrate the importance of copper chaperone CopZ in S. mutans biofilm formation and competitiveness. Unlike ΔcopY and ΔcopA biofilms, the ΔcopZ biofilm had significantly decreased biomass. The defective biofilm of the copZ mutant was in part attributed to decreased production and secretion of biofilm-essential enzymes, GtfB and GtfC. The copZ mutant biofilm characterized with CLSM revealed reduced glucan matrix and exposed bacterial cells in contrast to the microcolonies enveloped by glucans in biofilms formed by WT UA159, ΔcopY, ΔcopA, and ΔcopZ/copZpVPT. Overexpression of CopZ in the complement strain (ΔcopZ/copZpVPT) resulted in increased biomass and robust glucan matrix formation, further demonstrating the crucial role of CopZ for biofilm formation. In addition, the copZ mutant was impaired in competitiveness against oral commensal species S. sanguinis and S. gordonii. Essential to competitiveness of S. mutans UA159 is the expression and secretion of mutacins. These mutacins are critical for S. mutans UA159 to out-compete commensal species in order to establish a biofilm in the oral cavity. The copZ mutant exhibited impaired ability to inhibit growth of commensal species S. gordonii and S. sanguinis. This result was further corroborated by our qRT-PCR data showing decreased expression of S. mutans UA159 mutacin IV,V, and VI genes.
Reduced competitiveness of the copZ mutant cannot be attributed to reduced competence observed in the copYAZ mutant since recent studies suggest that mutacin secretion is independent of competence (Reck &Tomasch & Wagner-Dobler 2015; Singh &Senadheera &Levesque & Cvitkovitch 2015). Mutacin transcription and synthesis has instead been shown to be regulated by ComCDE (Reck &Tomasch & Wagner-Dobler 2015). Indeed we demonstrate reduced expression of mutacin IV,V, VI and comCDE genes in ΔcopZ. This study first demonstrated the role of CopZ in competitiveness of S. mutans against commensal oral species and uncovered the underlying mechanism.
The copYAZ operon has previously been characterized for its role in copper resistance in Streptococcus mutans. S. mutans encounters an influx of copper from diet and metal dental restorations. Under copper stress, S. mutans induces the copYAZ operon for resistance against copper toxicity (Vats & Lee 2001). The copper chaperone CopZ is unique compared to its operon counterparts; CopY, a negative repressor of copYAZ, and CopA, a P1-ATPase copper exporter. In the study, we also demonstrated CopZ tightly binds copper, which provided the first experimental evidence that CopZ is a copper chaperone in S. mutans. Due to the apparent difference among the copYAZ components, we investigated their contribution to S. mutans virulence traits.
CopZ is a copper chaperone, previously characterized for its importance in copper homeostasis in other organisms. In Enterococcus hirae, CopZ has been shown to bind and transport copper to CopY for transcription initiation of the copYAZ operon (Cobine &Wickramasinghe &Harrison &Weber &Solioz & Dameron 1999). Since CopZ can bind and transport copper to regulate copYAZ, it is possible that CopZ may help regulate other processes. Beyond copper transport, copZ has been shown to affect biofilm detachment but unnecessary for biofilm formation in oral commensal Streptococcus gordonii (Mitrakul &Loo & Hughes & Ganeshkumar 2004). In S. mutans, the copYAZ operon has been implicated in oxidative stress and gtfB transcription (Singh &Senadheera &Levesque & Cvitkovitch 2015). In this study, we demonstrate that CopZ is critical for biofilm formation by Gtfs and competitiveness by mutacins. The impact of copZ on S. mutans biofilm formation and competitiveness are, to the best of our knowledge, unique to a copper chaperone. The phenotypes of the copZ mutant are independent of copY and copA, implicating the importance of CopZ in S. mutans virulence traits. The function of CopZ to bind and transport copper, and its unique role in S. mutans biofilm formation and competitiveness suggests CopZ may have an independent role aside from copper resistance. Further investigation into the mechanism of copZ on biofilm formation and competitiveness may lead to the identification of novel virulence-regulatory pathways.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants NIH/NIDCR F31 DE024041 and R01 DE022350.
Footnotes
Additional Supporting information may be found in the online version of this article:
Figure S1. CopZ biofilm phenotype is not due to reduced biomass.
REFERENCES
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Sitthisak S, Sengupta M, Johnson M, Jayaswal RK, Morrissey JA. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol. 2010;76:150–160. doi: 10.1128/AEM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P, Wickramasinghe WA, Harrison MD, Weber T, Solioz M, Dameron CT. The Enterococcus hirae copper chaperone CopZ delivers coppeRI) to the CopY repressor. FEBS Lett. 1999;445:27–30. doi: 10.1016/s0014-5793(99)00091-5. [DOI] [PubMed] [Google Scholar]
- Drake DR, Grigsby W, Cardenzana A, Dunkerson D. Synergistic, growth-inhibitory effects of chlorhexidine and copper combinations on Streptococcus mutans, Actinomyces viscosus, and Actinomyces naeslundii. J Dent Res. 1993;72:524–528. doi: 10.1177/00220345930720020901. [DOI] [PubMed] [Google Scholar]
- Duggal MS, Chawla HS, Curzon ME. A study of the relationship between trace elements in saliva and dental caries in children. Arch Oral Biol. 1991;36:881–884. doi: 10.1016/0003-9969(91)90118-e. [DOI] [PubMed] [Google Scholar]
- Evans SL, Tolbert C, Arceneaux JE, Byers BR. Enhanced toxicity of copper for Streptococcus mutans under anaerobic conditions. Antimicrob Agents Chemother. 1986;29:342–343. doi: 10.1128/aac.29.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol. 2011;77:2428–2434. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin KD. Metalloenzymes, structural motifs, and inorganic models. Science. 1993;261:701–708. doi: 10.1126/science.7688141. [DOI] [PubMed] [Google Scholar]
- Kihlken MA, Leech AP, Le Brun NE. Copper-mediated dimerization of CopZ, a predicted copper chaperone from Bacillus subtilis. Biochem J. 2002;368:729–739. doi: 10.1042/BJ20021036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Liu C, Worthington RJ, Melander C, Wu H. A new small molecule specifically inhibits the cariogenic bacterium Streptococcus mutans in multispecies biofilms. Antimicrob Agents Chemother. 2011;55:2679–2687. doi: 10.1128/AAC.01496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. 2012;27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrakul K, Loo CY, Hughes CV, Ganeshkumar N. Role of a Streptococcus gordonii copper-transport operon, copYAZ, in biofilm detachment. Oral Microbiol Immunol. 2004;19:395–402. doi: 10.1111/j.1399-302x.2004.00176.x. [DOI] [PubMed] [Google Scholar]
- Nakano YJ, Kuramitsu HK. Mechanism of Streptococcus mutans glucosyltransferases: hybrid-enzyme analysis. J Bacteriol. 1992;174:5639–5646. doi: 10.1128/jb.174.17.5639-5646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orstavik D. Antibacterial properties of and element release from some dental amalgams. Acta Odontol Scand. 1985;43:231–239. doi: 10.3109/00016358509046503. [DOI] [PubMed] [Google Scholar]
- Peng X, Zhang Y, Bai G, Zhou X, Wu H. Cyclic di-AMP mediates biofilm formation. Mol Microbiol. 2015 doi: 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M, Tomasch J, Wagner-Dobler I. The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in Streptococcus mutans. PLoS Genet. 2015;11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoffield JA, Wu H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun. 2015;83:101–107. doi: 10.1128/IAI.02396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafeeq S, Yesilkaya H, Kloosterman TG, et al. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- Singh K, Senadheera DB, Levesque CM, Cvitkovitch DG. The copYAZ Operon Functions in Copper Efflux, Biofilm Formation, Genetic Transformation, and Stress Tolerance in Streptococcus mutans. J Bacteriol. 2015;197:2545–2557. doi: 10.1128/JB.02433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solioz M, Abicht HK, Mermod M, Mancini S. Response of gram-positive bacteria to copper stress. J Biol Inorg Chem. 2010;15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Urvoas A, Moutiez M, Estienne C, Couprie J, Mintz E, Le Clainche L. Metal-binding stoichiometry and selectivity of the copper chaperone CopZ from Enterococcus hirae. Eur J Biochem. 2004;271:993–1003. doi: 10.1111/j.1432-1033.2004.04001.x. [DOI] [PubMed] [Google Scholar]
- Vats N, Lee SF. Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiology. 2001;147:653–662. doi: 10.1099/00221287-147-3-653. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Bitoun JP, Liao S. PBP1a-deficiency causes major defects in cell division, growth and biofilm formation by Streptococcus mutans. PLoS One. 2015;10:e0124319. doi: 10.1371/journal.pone.0124319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschendorf F, Ackart D, Shrestha TB, et al. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu F, Yang T, et al. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat Commun. 2014;5:4339. doi: 10.1038/ncomms5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Fives-Taylor P, Wu H. The utility of affinity-tags for detection of a streptococcal protein from a variety of streptococcal species. J Microbiol Methods. 2008;72:249–256. doi: 10.1016/j.mimet.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.