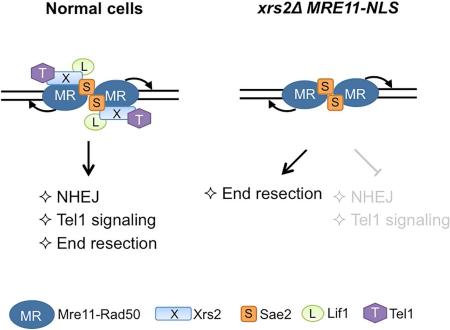
SUMMARY
The Mre11-Rad50-Xrs2/Nbs1 (MRX/N) complex orchestrates the cellular response to DSBs through its structural, enzymatic and signaling roles. Xrs2/Nbs1 is essential for nuclear translocation of Mre11, but its role as a component of the complex is not well defined. Here we demonstrate that nuclear localization of Mre11 (Mre11-NLS) is able to bypass several functions of Xrs2, including DNA end resection, meiosis, hairpin resolution and cellular resistance to clastogens. Using purified components, we show the MR complex has equivalent activity to MRX in cleavage of protein-blocked DNA ends. Although Xrs2 physically interacts with Sae2, we found that end resection in its absence remains Sae2 dependent in vivo and in vitro. MRE11-NLS was unable to rescue the xrs2Δ defects in Tel1/ATM kinase signaling and non-homologous end joining, consistent with the role of Xrs2 as a chaperone and adaptor protein coordinating interactions between the MR complex and other repair proteins.
Graphical Abstract
INTRODUCTION
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that must be accurately repaired to avoid genomic instability and cell death. DSBs arise during DNA replication and can be induced by genotoxic agents, such as ionizing radiation and topoisomerase inhibitors. Although accidental DSBs pose a threat to genome integrity, DSBs are important intermediates during meiosis and lymphocyte development to drive genetic diversity (Lam and Keeney, 2015; Schatz and Swanson, 2011). The two main mechanisms to repair DSBs are non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ directly ligates DNA ends with little or no sequence homology (Chiruvella et al., 2013), whereas HR requires a homologous duplex as a template, usually the sister chromatid, resulting in restoration of the original sequence (Jasin and Rothstein, 2013).
The Mre11-Rad50-Xrs2/Nbs1 (MRX/N) complex plays multiple roles in the DNA damage response, including sensing and repair of DSBs, activation of the Tel1/ATM kinase and telomere length homeostasis (Stracker and Petrini, 2011). Hypomorphic mutations of human MRN complex components are associated with Nijmegen breakage syndrome and Ataxia telangiectasia-like disorder, which are characterized by cellular radiosensitivity, immune deficiency and cancer proneness, similar to individuals with Ataxia telangiectasia (A-T) (Carney et al., 1998; Stewart et al., 1999; Stracker and Petrini, 2011). Mre11 and Rad50 are evolutionarily conserved and form an ATP-regulated nuclease involved in processing hairpin-capped and protein-bound DNA ends. Xrs2/Nbs1 is less conserved than Mre11 and Rad50, and has only been identified in eukaryotes (Stracker and Petrini, 2011). Xrs2/Nbs1 is required for nuclear localization of Mre11 and has a number of protein-protein interaction motifs, suggesting it functions as a chaperone and scaffold (Carney et al., 1998; Tsukamoto et al., 2005). The C-terminal region of Xrs2/Nbs1 contains Mre11 and Tel1/ATM binding motifs (Falck et al., 2005; Nakada et al., 2003; Tsukamoto et al., 2005; You et al., 2005). Mutation of residues within the Mre11 interaction motif that prevent binding to Mre11 confers a phenotype indistinguishable from xrs2Δ and mre11Δ null mutations in Saccharomyces cerevisiae (budding yeast) (Tsukamoto et al., 2005). Schizosaccharomyces pombe Nbs1 binds to sequences in the N-terminal region of Mre11, including an insertion loop between nuclease motifs II and III that is absent from bacterial and archaeal proteins (Schiller et al., 2012). The insertion loop extends the Mre11 dimer interface and Nbs1 binding is suggested to stabilize Mre11 (Schiller et al., 2012). Deletion of the Tel1 interaction motif at the C-terminus of Xrs2 (xrs2-11) results in a phenotype similar to the tel1Δ mutant, including a defect in DNA damage signaling and short telomeres (Nakada et al., 2003). The N-terminal region of Xrs2/Nbs1 contains a forkhead associated (FHA) domain, which binds to phosphorylated Sae2/Ctp1/CtIP and Lif1/Xrcc4 (Chen et al., 2001; Liang et al., 2015; Lloyd et al., 2009; Matsuzaki et al., 2008; Palmbos et al., 2008; Wang et al., 2013; Williams et al., 2009).
MRX/N is essential for NHEJ in budding yeast, and plays a supporting role in classical and alternative NHEJ in mammalian cells (Boulton and Jackson, 1998; Dinkelmann et al., 2009; Moore and Haber, 1996; Rass et al., 2009; Xie et al., 2009). In vitro, MRX tethers DNA ends and stimulates their ligation by the DNA ligase IV complex (Dnl4-Lif1) (Chen et al., 2001). In addition, MRX/N plays a conserved role in HR by promoting degradation of the 5’-terminated strands in a process referred to as end resection. The resulting 3’ single-strand tailed intermediates are essential intermediates for Rad51 or Dmc1-catalyzed strand invasion to initiate homology-directed repair. MRX acts with Sae2 to initiate 5’-3’ end resection by endonucleolytic cleavage internal to protein-blocked 5’ ends (Cannavo and Cejka, 2014; Garcia et al., 2011; Neale et al., 2005). Mutations that eliminate the Mre11 nuclease activity (e.g., mre11-H125N), or sae2Δ, prevent resection initiation at protein-blocked or hairpin-capped ends (reviewed by (Mimitou and Symington, 2009)). Besides its catalytic function, the MRX complex also recruits Exo1 and Sgs1-Dna2 nucleases, which catalyze extensive resection (Mimitou and Symington, 2010; Shim et al., 2010). The mre11-H125N and sae2Δ mutants show a mild resection defect at DSB ends with no covalent modification because in the absence of Mre11 endonuclease activity, Exo1 and Sgs1-Dna2 can initiate resection, albeit with some delay (Mimitou and Symington, 2009). However, mre11Δ, rad50Δ, and xrs2Δ mutants show a more severe resection defect, presumably because the absence of the complex attenuates recruitment of Exo1, Sgs1 and Dna2 to DSBs (Shim et al., 2010).
A previous study identified the Mre11 interaction domain of Xrs2 and demonstrated an important role for Xrs2 in nuclear localization of Mre11 (Schiller et al., 2012; Tsukamoto et al., 2005). In addition, it was shown that nuclear localization of Mre11 via fusion to the Gal4 DNA binding domain (GBD) could partially bypass the requirement for Xrs2 in DNA repair. However, the mechanism by which nuclear Mre11 mediates DNA repair in the absence of Xrs2 was not investigated. Here we show that localization of Mre11 to the nucleus suppresses the slow growth, DNA damage sensitivity and meiotic defects of the xrs2Δ mutant by restoring Mre11 nuclease and Sae2-dependent end resection. However, Xrs2 is crucial for NHEJ and Tel1 signaling functions of the MRX complex.
RESULTS
Nuclear Mre11 rescues xrs2Δ slow growth and genotoxin sensitivity
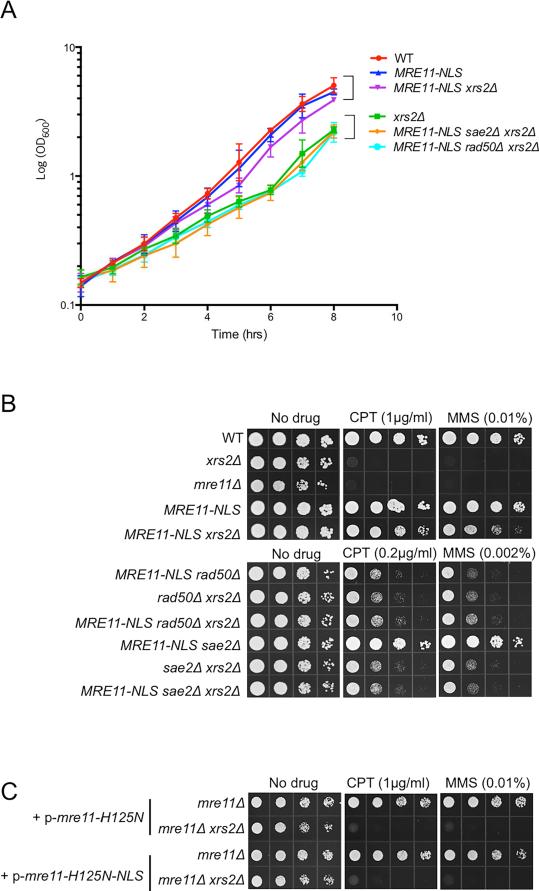
To investigate the contribution of Xrs2 to MR activities, independently of Mre11 nuclear translocation, we fused a nuclear localization signal (NLS) to the C-terminus of Mre11 expressed from the endogenous locus and assayed for complementation of xrs2Δ defects. The steady-state protein level of Mre11-NLS was about two-fold lower than Mre11 in XRS2 and xrs2Δ backgrounds (Fig S1A). Nevertheless, expression of MRE11-NLS suppressed the growth defect of xrs2Δ and this suppression was completely dependent on RAD50 and SAE2 (Fig 1A). A previous study had shown that expression of Mre11-GBD restores partial resistance to methyl methane sulfonate (MMS) to the xrs2Δ mutant (Tsukamoto et al., 2005). We found that MRE11-NLS partially suppresses the camptothecin (CPT) and MMS sensitivity of the xrs2Δ mutant in a RAD50-dependent manner, indicating that Xrs2 is dispensable for DNA repair once Mre11 and Rad50 are present in the nucleus (Fig 1B). Furthermore, Sae2 and Mre11 nuclease are required for DNA damage resistance in the MRE11-NLS xrs2Δ context, even though the sae2Δ and mre11-H125N single mutants are not sensitive to the same low dose of CPT or MMS (Fig 1B, C), indicating that the nuclease activity of Mre11 is essential for DNA repair in the absence of Xrs2.
Figure 1. MRE11-NLS rescues xrs2Δ in growth and sensitivity to DNA damaging agents.
A. Growth curves representing cell concentration measured by OD600 at the indicated time-points. Error bars indicate standard deviation (s.d.) from 3 trials. B. Ten-fold serial dilutions of the indicated strains were spotted onto rich medium without drug, or medium containing CPT or MMS at the indicated concentrations. WT refers to wild type. C. Ten-fold serial dilutions of indicated strains transformed with indicated plasmids were spotted onto selective plates containing 1μg/ml CPT or 0.01% MMS. See also Fig S1.
Because the suppression by MRE11-NLS is incomplete, and the protein is expressed at lower steady-state level than Mre11, we over-expressed MRE11-NLS using a 2-micron plasmid construct but did not see further suppression of the DNA damage sensitivity of the xrs2Δ mutant (Fig S1B, C). Furthermore, the MRE11-NLS strain showed equivalent DNA damage resistance to the MRE11 parental strain, even at high MMS concentration, indicating that the reduced protein level is sufficient for normal activity (Fig S1D).
Xrs2 is required for Tel1 activation and NHEJ
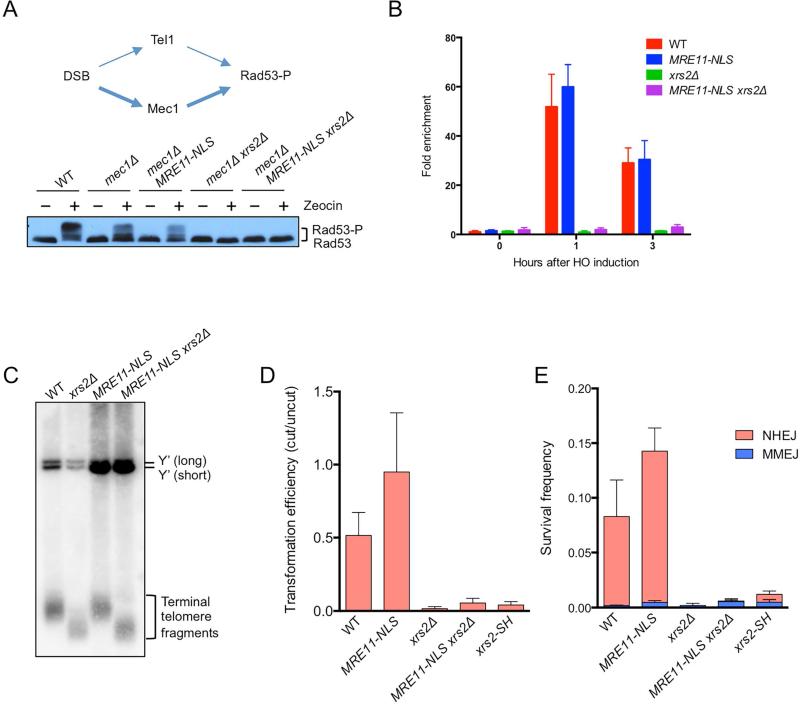
Xrs2/Nbs1 is the only component of the complex known to directly interact with Tel1/ATM (Falck et al., 2005; Nakada et al., 2003). To test whether this physical interaction is required for Tel1 activation, phosphorylation of the downstream effector kinase, Rad53, was measured following acute zeocin treatment. The strains used contain mec1Δ and sml1Δ mutations to eliminate the main pathway for Rad53 activation (the sml1Δ mutation is required to suppress lethality caused by mec1Δ) (Gobbini et al., 2013; Zhao et al., 1998). As expected, there was no activation of Rad53 in the mec1Δ xrs2Δ mutant, and MRE11-NLS was unable to rescue this defect (Fig 2A). Consistent with the lack of Tel1 activation, Tel1 was not recruited to a site-specific DSB in xrs2Δ MRE11-NLS cells (Fig 2B). We also showed that MRE11-NLS does not restore normal telomere length to the xrs2Δ mutant, in agreement with a previous study (Fig 2C) (Tsukamoto et al., 2005). These data indicate that Tel1 binding to the MR complex through its interaction with Xrs2 is necessary for Tel1-dependent checkpoint signaling and telomere maintenance functions.
Figure 2. Xrs2 is required for end joining and Tel1 signaling functions of the MRX complex.
A. Model of Rad53 phosphorylation (Rad53-P) in response to DNA damage. Tel1/ATM responds to MRX/N bound DSBs, whereas Mec1/ATR is activated by RPA bound to the ssDNA formed at resected DSBs. Western blot analysis showing Rad53-P in response to zeocin treatment in the indicated strains. B. ChIP-qPCR for HA-Tel1 0.2 kb from the DSB. Error bars indicate s.d. (n=3). C. Genomic DNA of indicated strains was digested with XhoI and separated on a 1% agarose gel. The DNA fragments were transferred to a nylon membrane and hybridized with a Y’ element probe. Y’ long and Y’ short refers to the two classes of subtelomeric Y’ repeats. D. Transformation efficiency of BamHI-digested linear plasmid DNA measured by the plasmid-ligation assay. Transformation efficiency was calculated as a ratio of the number of transformants with BamHI-digested pRS414 DNA to that with undigested DNA. Error bars indicate s.d. (n=4). E. Frequency of chromosomal NHEJ. Error bars indicate s.d. (n=3). See also Fig S2.
In budding yeast, the MRX complex is essential for NHEJ (Boulton and Jackson, 1998; Moore and Haber, 1996). To determine whether Xrs2 is directly involved in facilitating NHEJ, we used a plasmid-ligation assay. A self-replicating plasmid linearized with BamHI is efficiently recircularized by NHEJ when transformed into yeast cells (Boulton and Jackson, 1998). Repair was completely eliminated in the xrs2Δ mutant and expression of MRE11-NLS did not complement this defect, indicating that Xrs2 has a direct role in promoting NHEJ (Fig 2D). The FHA domain of Xrs2/Nbs1 was previously shown to directly interact with Lif1/Xrcc4 in vitro (Chen et al., 2001; Matsuzaki et al., 2008; Palmbos et al., 2008); thus defective Lif1 binding could contribute to the xrs2Δ NHEJ defect. Consistent with this hypothesis, ablating Lif1 interaction by mutation of conserved residues within the Xrs2 FHA domain (xrs2-SH mutant) reduced NHEJ by 10-fold (Fig 2D). To verify these results in a chromosomal context we also determined the efficiency of repair using a substrate with an inverted duplication of I-SceI cut sites that measures imprecise NHEJ (Deng et al., 2014). Because I-SceI is continuously expressed, the NHEJ products recovered are inaccurate and mostly utilize a 2 bp microhomology within the 4-bp 3’ overhangs produced by I-SceI cleavage (Deng et al., 2014). Minor classes of end joining products are also recovered that utilize microhomologies flanking the DSB. Consistent with the plasmid assay, MRE11-NLS was unable to suppress the xrs2Δ NHEJ defect and the xrs2-SH mutant displayed a similar defect to that seen using the plasmid assay (Fig 2E).
Since the xrs2Δ MRE11-NLS strain is deficient for Tel1 signaling and NHEJ we asked whether combining these defects results in DNA damage sensitivity. The lif1Δ tel1Δ double mutant was more resistant to CPT and MMS than the xrs2Δ MRE11-NLS strain indicating that loss of NHEJ and Tel1 signaling is not responsible for the residual DNA damage sensitivity of xrs2Δ MRE11-NLS cells (Fig S2).
Rescue of the xrs2Δ resection defect by MRE11-NLS
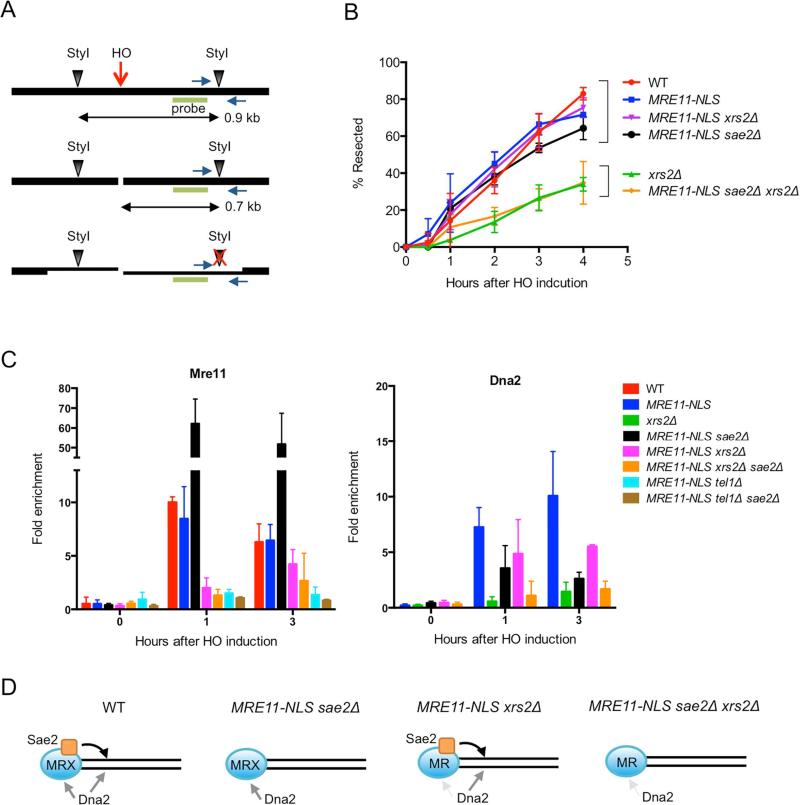
The MRX complex promotes HR by initiating end resection. To analyze resection activity of the MR complex in the absence of Xrs2, we monitored formation of ssDNA following DSB formation at the MAT locus. The strains used express HO from the galactose-inducible GAL1 promoter, allowing synchronous DSB formation, and the HML and HMR loci were deleted to prevent repair by gene conversion. As resection proceeds, the StyI restriction enzyme site located 0.7 kb distal to the DSB becomes single-stranded and resistant to digestion, which results in disappearance of the 0.7 kb restriction fragment over time (Fig 3A). Resection was also measured by the accumulation of StyI-resistant ssDNA by real-time PCR (Zierhut and Diffley, 2008). Kinetic analysis of resection using both assays revealed that expression of MRE11-NLS restores resection to the wild-type level in the xrs2Δ mutant (Fig 3B, Fig S3A, B). In the absence of Sae2, the suppression was completely abolished, indicating that Sae2 is still critical for resection in the absence of Xrs2 (Fig 3B, Fig S3B). Because Tel1 signaling is defective in xrs2Δ derivatives, we tested resection in the sae2Δ tel1Δ MRE11-NLS strain and found it was reduced relative to sae2Δ MRE11-NLS, but not to the same extent as observed for sae2Δ xrs2Δ MRE11-NLS cells indicating a requirement for Xrs2 independent of Tel1 (Fig S3B).
Figure 3. Xrs2 is dispensable for end resection of HO-induced DSBs.
A. Representation of the MAT locus used to measure end resection after introduction of a targeted DSB. Green bar shows the location of the probe used for hybridization and blue arrows show primers used for real-time PCR. B. Quantification of the southern blot data shown in Supplementary Fig 3A. Error bars indicate s.d. (n=3). C. ChIP-qPCR for Mre11 and Dna2-Myc 1 kb from the DSB. Error bars indicate s.d. (n=3). D. Model for Sae2-dependent end resection in xrs2Δ MRE11-NLS cells. In WT cells, Dna2 is recruited via MRX and also by MRX-Sae2 clipping, which creates a substrate for RPA. Dna2 is recruited by MRX in sae2Δ MRE11-NLS cells and can bypass the need for Sae2 to initiate resection. Less Mre11 is recruited to ends in the xrs2Δ MRE11-NLS strain, but Sae2 clipping can still initiate resection and Dna2 loading. In the absence of Sae2 and Xrs2, MR poorly recruits Dna2 due to no clipping and reduced stabilization of Mre11 at ends. See also Fig S3.
To determine whether Mre11-NLS is recruited normally to DSBs we measured Mre11 binding to sequences 1 kb from the HO cut site by chromatin immunoprecipitation (ChIP). While the enrichment of Mre11 at the DSB was comparable in MRE11 and MRE11-NLS cells, less Mre11-NLS was retained at the DSB in the xrs2Δ background at the 1 hr time point (P<0.05) (Fig 3C). Since Tel1 signaling is abrogated in the xrs2Δ MRE11-NLS mutant, and Tel1 is required for retention of Mre11 at DSBs (Gobbini et al., 2015), we considered the possibility that the Mre11 localization defect in xrs2Δ MRE11-NLS cells could be due to loss of Tel1 recruitment. Indeed, we found that Mre11 enrichment at the HO-induced DSB was the same in MRE11-NLS xrs2Δ and MRE11-NLS cells tel1Δ (Fig 3C). Note that resection is at almost the wild-type level in MRE11-NLS xrs2Δ and MRE11-NLS tel1Δ xrs2Δ cells, indicating that Mre11 localization to DSBs is not limiting for resection initiation and loss of Tel1 signaling does not impair Sae2 activity (Fig S3B). We observed greater enrichment of Mre11-NLS in the sae2Δ mutant as compared with SAE2 cells, consistent with previous studies (Clerici et al., 2006; Langerak et al., 2011; Lisby et al., 2004), and this was dependent on Xrs2 and Tel1 (P<0.05) (Fig 3C).
Previous studies implicate Sgs1-Dna2 in resection initiation when Sae2 or the Mre11 nuclease is absent (Bonetti et al., 2015; Budd and Campbell, 2009; Ferrari et al., 2015; Mimitou and Symington, 2010; Shim et al., 2010); thus, the dependence of resection on Sae2 in the MRE11-NLS xrs2Δ strain could be because the Sgs1-Dna2 mechanism is disabled. If this were the case, we would predict similar resection products in exo1Δ MRE11-NLS xrs2Δ and exo1Δsgs1Δ mutants. We did not detect the characteristic end clipped products in exo1Δ xrs2ΔMRE11-NLS cells that are observed when Exo1 and Sgs1 are absent indicating that Sgs1-Dna2 is active, at least when Sae2 is present (Fig S3C). A previous study showed that Sgs1-Dna2 recruitment to DSBs is MRX dependent (Shim et al., 2010). Therefore, the decreased enrichment of Mre11 at DSBs in the MRE11-NLS sae2Δ xrs2Δ mutant could potentially result in reduced Sgs1-Dna2 recruitment and explain the decreased end resection relative to MRE11-NLS sae2Δ cells. Interestingly, we found the level of Dna2 enrichment 1 kb from the DSB was not significantly decreased in the MRE11-NLS xrs2Δ strain, despite the decrease in Mre11, whereas Dna2 enrichment was barely above background in MRE11-NLS sae2Δ xrs2Δ cells (Fig 3C). The decreased Dna2 binding in the MRE11-NLS sae2Δ mutant could be due to delayed resection initiation or Rad9 inhibition (Ferrari et al., 2015; Gobbini et al., 2015). We suggest that there are normally two modes of Sgs1-Dna2 recruitment to DSBs: via MRX interaction and by RPA interaction after MRX-Sae2 dependent cleavage creates ssDNA (Fig 3D) (Chen et al., 2013; Shim et al., 2010). Although Mre11 binding is reduced in MRE11-NLS xrs2Δ cells, resection still initiates by MR-Sae2 dependent cleavage and Dna2 is recruited to the substrate generated. When Sae2 is absent, Mre11 accumulates at DSBs facilitating Sgs1-Dna2 recruitment and resection initiation. However, reduced Mre11 localization to DSBs and absence of clipping in the MRE11-NLS sae2Δ xrs2Δ strain results in diminished Sgs1-Dna2 activity.
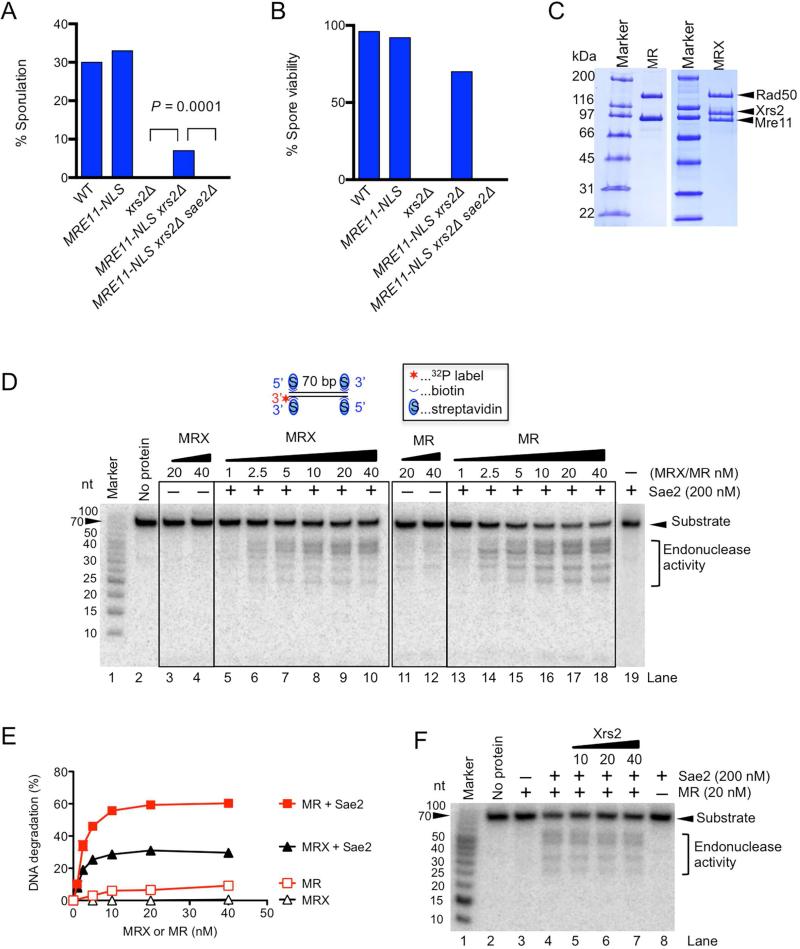
Xrs2 is dispensable for meiosis
The Spo11 protein creates DSBs at meiotic recombination hotspots by covalent linkage to the 5’ ends at break sites and is then removed by endonucleolytic cleavage releasing Spo11 attached to a short oligonucleotide (Neale et al., 2005). mre11Δ, rad50Δ, and xrs2Δ diploids fail to generate meiosis-specific DSBs and do not progress through meiosis in the W303 strain background, while mre11-H125N and sae2Δ diploids are able to form meiotic DSBs, but are unable to remove Spo11 from ends and consequently arrest during meiotic prophase with unrepaired DSBs (Mimitou and Symington, 2009). To determine the requirement for Xrs2 during meiosis, homozygous diploid strains were generated and grown under conditions to induce sporulation. Around 30% of wild-type cells sporulated and 97% of the dissected ascospores were viable; similar values were obtained for the MRE11-NLS homozygous diploid (Fig 4A). We found that expression of MRE11-NLS partially rescued the sporulation defect of xrs2Δ (7% of cells formed tetrads), and, remarkably, 70% of the spores were viable indicating formation and repair of meiosis-specific DSBs (Fig 4B). Restoration of sporulation in the MRE11-NLS xrs2Δ strain was dependent on SAE2, consistent with the Sae2 requirement to process Spo11-bound ends (Fig 4A, B).
Figure 4. Xrs2 is not required for Sae2- and Mre11-Rad50-dependent endonuclease activity in the vicinity of protein-blocked DNA ends.
A. Sporulation percentage determined by counting cells that contain three or four visible spores out of at least 700 total cells counted. B. Spore viability determined by dissection of asci and counting spores germinating to give visible colonies. No fewer than 50 asci were dissected for each strain. C. Recombinant Mre11-Rad50 (MR) and Mre11-Rad50-Xrs2 (MRX) used in this study. D. Nuclease assay with Mre11-Rad50 (MR), Mre11-Rad50-Xrs2 (MRX) and Sae2, as indicated. E. Quantitation of data from panel D. Error bars indicate SEM (n=2). F. The effect of Xrs2 on the endonuclease activity of Mre11-Rad50 in the presence of Sae2, as indicated. See also Fig S4
The MR complex is competent for Sae2-dependent resection initiation in vitro
Previously, Mre11 within the MRX complex was found to possess a Sae2-stimulated endonuclease activity on dsDNA in the vicinity of protein-blocked DNA ends (Cannavo and Cejka, 2014). This reaction is believed to recapitulate the initial steps in DNA end resection that require the Mre11 nuclease. To determine whether Xrs2 is also required for dsDNA clipping by Mre11, we prepared recombinant MR complex and compared its activity with the MRX heterotrimer (Fig 4C, D). We used a DNA substrate with fully blocked DNA ends to prevent the Mre11 3’-5’ exonuclease activity. We found that both MRX and MR complexes possess endonuclease activities that are strongly stimulated by Sae2 (Fig. 4D). The MR complex appeared to be even more active than MRX (Fig 4E), however this may be due to different preparation procedures and specific activities of MR and MRX, respectively. The addition of recombinant Xrs2 did not have a significant inhibitory effect on the MR and Sae2-dependent cleavage of dsDNA (Fig 4F). Very similar results were obtained with a DNA substrate containing a single protein block, which allows both endonuclease as well as exonuclease activities of MR/MRX (Fig S4A, B). Therefore, Mre11 dependent cleavage in the vicinity of protein-blocked DNA in vitro requires Sae2, but not Xrs2, in agreement with the in vivo data.
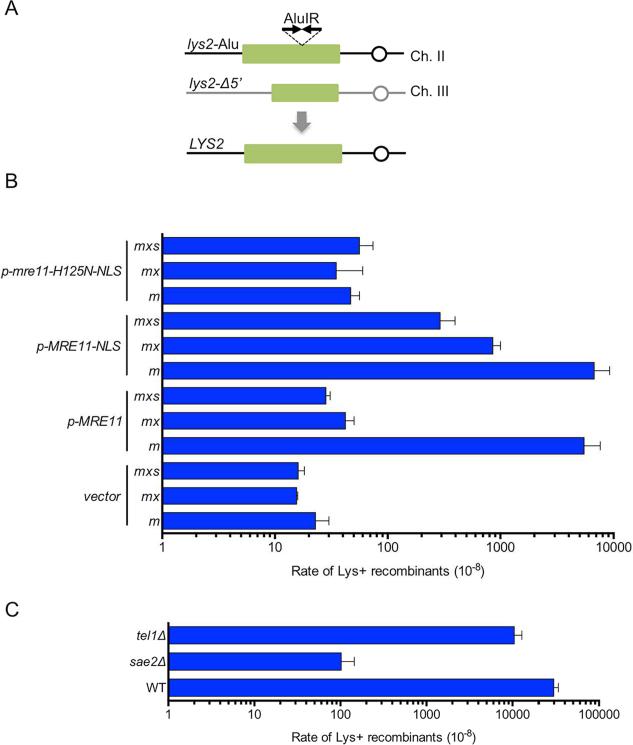
Sae2-dependent and independent hairpin resolution
The MRX complex and Sae2 also play an important role in resolving hairpin-capped DNA ends. Hairpin resolution in MRE11-NLS derivatives was measured using a genetic assay developed by Lobachev and colleagues (Fig 5A) (Lobachev et al., 2002). Inverted Alu elements inserted in the lys2 gene stimulate ectopic recombination with a truncated lys2 gene by ~1000-fold relative to a strain with a direct repeat of Alu elements inserted at the same site in lys2, and this stimulation is largely dependent on the MRX complex, the Mre11 nuclease and Sae2 (Lobachev et al., 2002). The inverted repeats are thought to extrude to form a hairpin or cruciform that is cleaved by an unknown nuclease to form a hairpin-capped end, or to form a foldback structure following resection of a nearby DSB, which is then opened by MRX-Sae2 and stimulates recombination generating Lys+ cells. Expression of MRE11-NLS increased the recombination rate of the xrs2Δ mutant by 20-fold (P = 0.01) (Fig. 5B). Because the steady-state protein level of Mre11-NLS is reduced and could be the cause of the partial suppression, MRE11-NLS was overexpressed using a plasmid construct; however, this did not restore recombination to the wild-type rate (Fig. S5A).
Figure 5. Hairpin resolution is independent of Xrs2 and partially Sae2-independent.
A. Cartoon representation of the lys2-AluIR ectopic recombination assay. B. Recombination frequencies of strains with the lys2-AluIR and lys2-Δ5’ ectopic recombination reporter system. The rate of Lys+ recombinants was derived from the median recombination frequency determined from eight different isolates of each strain. Error bars indicate s.d. (n=3). m, mre11Δ; mx, mre11Δ xrs2Δ; mxs, mre11Δ xrs2Δ sae2Δ. C. Recombination frequencies of sae2Δ and tel1Δ derivatives. Error bars indicate s.d. (n=3). See also Fig S5.
Interestingly, expression of MRE11-NLS in the sae2Δ xrs2Δ background enhanced the recombination rate by 10-fold (P = 0.04) compared to expression of MRE11. The increased rate of Lys+ recombinants observed in the sae2Δ mutant was dependent on the Mre11 nuclease activity indicating that it is the Mre11 nuclease, and not Sae2, that promotes hairpin cleavage. Previous studies have demonstrated structure-selective nuclease activity for Sae2, and postulated a direct role for Sae2 in cleaving hairpin structures (Lengsfeld et al., 2007); by contrast, our data indicate that hairpin cleavage is catalyzed by Mre11 nuclease. Our data also suggest the possibility that Sae2 is required to relieve an inhibitory effect of Xrs2 on hairpin cleavage in vivo because the rate of Lys+ recombinants when MRE11-NLS was expressed in the mre11Δ sae2Δ background was lower than in the mre11Δ sae2Δ xrs2Δ mutant (Fig S5B). These data show a separation of Sae2 function in regulating Mre11 nuclease activity, and are consistent with the finding that MR and MRX/N complexes cleave DNA hairpins in vitro independently of Sae2/CtIP (Cannavo and Cejka, 2014; Paull and Gellert, 1999; Trujillo and Sung, 2001).
Previous studies have shown redundancy between Mec1 and Tel1 for damage-induced Sae2 phosphorylation (Baroni et al., 2004). Since Tel1 signaling is abrogated in xrs2Δ MRE11-NLS cells, we tested whether the tel1Δ mutation results in decreased hairpin opening (Fig 5C). The tel1Δ mutant exhibited a small (P=0.0017) decrease in formation of Lys+ recombinants, but the frequency was 100-fold higher than the sae2Δ mutant indicating that Tel1-mediated phosphorylation of Sae2 is not required for hairpin opening. Similarly, the tel1Δ diploid exhibits close to normal sporulation and spore viability (Carballo et al., 2008), in contrast to the sporulation-defective sae2Δ diploid.
DISCUSSION
Previous studies have shown that Xrs2/Nbs1 is a flexible scaffold that binds to several DNA repair proteins, including Mre11, Tel1/ATM, Sae2/Ctp1/CtIP and Lif1 (Liang et al., 2015; Lloyd et al., 2009; Matsuzaki et al., 2008; Nakada et al., 2003; Palmbos et al., 2008; Tsukamoto et al., 2005; Williams et al., 2009). Xrs2 binding to Mre11 is required for its nuclear localization raising the question of the contribution of Xrs2 to MR functions once the complex is in the correct cellular compartment. Here we show that localizing Mre11 to the nucleus in the absence of Xrs2 restores functions associated with MR nuclease activity including DNA end resection, hairpin resolution and meiotic recombination. However, MRE11-NLS is unable to rescue the end joining and Tel1 activation defects of the xrs2Δ mutant indicating an essential role for Xrs2 in these processes.
Mre11 and Rad50 are conserved in all domains of life and together form an ATP-regulated nuclease involved in DNA end processing. In Escherichia coli, the main function of SbcCD (Rad50-Mre11) is to resolve hairpin-capped ends formed by closely spaced inverted repeats and the complex has no known function in 5’-3’ end resection (Connelly and Leach, 1996; Eykelenboom et al., 2008). MRX shares the hairpin cleavage activity with SbcCD and in addition facilitates 5’-3’ end resection, either directly by endonucleolytic cleavage internal to protein-blocked DNA ends or indirectly by recruitment of Exo1 and Dna2 nucleases (Cannavo and Cejka, 2014; Lobachev et al., 2002; Shim et al., 2010). Our data show that Xrs2 is not essential for hairpin resolution or end resection of HO and Spo11-induced DSBs consistent with MR comprising the core nuclease activity and the finding that Xrs2 is absent from bacteria and archaea.
DNA damage resistance and meiosis were not restored to wild type levels, and this could be due to either a direct role for Xrs2 in promoting MR activity or reduced localization of Mre11-NLS to DSBs in the absence of Xrs2. Previous studies have shown Xrs2/Nbs1 binds to DNA, and to the Mre11 latching loop, potentially stabilizing the Mre11 dimer at ends (Paull and Gellert, 1999; Schiller et al., 2012; Trujillo et al., 2003). Mre11 retention was equally reduced in the MRE11-NLS xrs2Δ and MRE11-NLS tel1Δ mutants suggesting the xrs2Δ defect could result from failure to recruit Tel1 (Gobbini et al., 2015). However, it is important to note that tel1Δ mutants are proficient for end resection, hairpin resolution and meiosis indicating that the reduction in Mre11 retention at DNA ends is of no consequence for these Mre11-dependent functions (Carballo et al., 2008; Iwasaki et al., 2016): thus, Xrs2 must contribute in some way to Mre11 activity, independent of the role of Tel1 stabilization of Mre11 at ends.
The sporulation and spore viability defects of the xrs2Δ diploid were partially rescued by MRE11-NLS indicating that Xrs2 is not essential for meiotic DSB formation and subsequent Spo11 removal. Our data contradict an earlier study in which it was shown that expression of GBD-Mre11 (nuclear localized) from a plasmid failed to suppress the meiotic defect of the xrs2Δ mutant, even though it was able to partially complement MMS sensitivity (Tsukamoto et al., 2005). There are several possible explanations for this discrepancy. First, we created a chromosomal version of MRE11-NLS instead of expressing it from a plasmid and the plasmid might have been unstable during meiosis. Second, we used a different strain background to the one used by Tsukomoto et al (Tsukamoto et al., 2005). In S. pombe, the MRN complex is dispensable for meiotic DSB formation, but is required for Spo11 removal (Milman et al., 2009; Young et al., 2004). The nbs1Δ mutant displays a temperature-sensitive defect for Spo11 clipping and meiotic recombination indicating a separation of MR nuclease activity from Nbs1 (Milman et al., 2009). The finding that Xrs2 is not integral to MR nuclease activity explains why no separation-of-function alleles of XRS2 have been identified in genetic screens for mutants proficient for meiotic DSB formation but deficient for Spo11 release, in contrast to MRE11 and RAD50 (Alani et al., 1990; McKee and Kleckner, 1997; Nairz and Klein, 1997; Prinz et al., 1997).
Surprisingly, we found that end resection by the MR complex retains the requirement for Sae2 even though the FHA domain of Xrs2/Nbs1 was thought to recruit Sae2/Ctp1 to sites of DNA damage (Liang et al., 2015; Lloyd et al., 2009; Williams et al., 2009). A recent study found no defect in Sae2 recruitment to an HO-induced DSB in the xrs2-SH mutant (Iwasaki et al., 2016), and a weak interaction between purified Sae2 and Mre11 was detected by co-immunoprecipitation (Cannavo and Cejka, 2014), potentially accounting for Sae2-dependent end resection. Resection is only mildly impacted by the sae2Δ mutation in budding yeast, yet resection in MRE11-NLS sae2Δ xrs2Δ cells was reduced to the same extent as in the absence of the MRX complex, suggesting that resection initiation by Sgs1-Dna2 is compromised. Consistent with this idea, we found reduced localization of Dna2 to DSBs in MRE11-NLS sae2Δxrs2Δ cells.
In contrast to MR-catalyzed end resection, which is Sae2 dependent, we found a partial restoration of hairpin opening by Mre11-NLS in the absence of Sae2 and Xrs2. Expression of Mre11-NLS in cells lacking only Sae2 did not restore hairpin opening suggesting Sae2 is required to overcome an inhibitory role of Xrs2. In vitro, MR can cleave hairpin structures independently of Sae2, raising the question of why hairpin cleavage in vivo requires Sae2. We speculate that in normal cells, MRX and Sae2 promote cleavage of hairpin-capped ends at a distance from the end, analogous to protein-bound DSBs. Xrs2 has been shown to bind to branched-DNA structures in vitro and could potentially shield the ssDNA region of a DNA hairpin from MR cleavage (Trujillo et al., 2003). By this model, MR endonuclease could cleave the exposed ssDNA at the hairpin without Sae2 only when Xrs2 is absent, whereas MRX cleavage would require Sae2. As noted above, E. coli SbcCD cleaves hairpins in vivo independently of Xrs2 or Sae2-like functions and has no role in end resection. We suggest that the acquisition of end resection by the MR complex in eukaryotes coincided with evolution of Sae2, a regulatory subunit to coordinate end resection with the cell cycle (Huertas et al., 2008). How Sae2 converts the Mre11 endonuclease from being ssDNA specific to clipping the 5’ terminated strand of duplex DNA is currently unknown.
EXPERIMENTAL PROCEDURES
Media, growth conditions, yeast strains and plasmids
Media and growth conditions were as described previously (Amberg, 2005). CPT or MMS was added to SC or YPD medium at the indicated concentrations. Yeast cells were grown in 30°C and all the experiments were carried out with log-phase cells. For DNA damage sensitivity assays, ten-fold serial dilutions of cells were spotted on solid medium with the indicated drug concentration and plates were incubated for 3-4 days. The yeast strains used are listed in Supplementary Table 1. Details of strain and plasmid constructions are provided in Supplementary data.
Physical monitoring of end resection and ChIP-qPCR
Yeast cells were grown in YP medium containing 2% lactate (YPL) to log phase and were arrested in G2/M phase with the addition of nocodazole (15μg/ml) to the medium. Then, galactose was added to a final concentration of 2% for HO induction. Cells were collected at 30-60 min intervals after HO induction for genomic DNA extraction or chromatin isolation. Southern blot and real-time PCR of resection intermediates were performed as described previously (Mimitou and Symington, 2008). Real-time PCR assay was performed as described with primers flanking the StyI site located 0.7 kb away from the HO cut site (Zierhut and Diffley, 2008). A control primer pair was used to amplify a region on chromosome XV that does not contain StyI sites. The PCR reaction program and calculation of the fraction of DNA resected were done in the same manner as described (Chen et al., 2013). The mean of three independent experiments is presented. The ChIP assays were performed as described using anti-HA (ab9110), anti-Mre11 (Chen et al., 2015) or anti-cMyc 9E10 (Santa Cruz Biotechnology). qPCRs were carried out by the SYBR green system using primer pairs complementary to DNA sequences located 1 kb or 200 bp from the HO-cut site at MAT (DSB) and to DNA sequences located 66 kb from MAT (control). Fold enrichment was calculated by 2ΔΔCq, where ΔΔCq= (Cq(IP, control) – Cq(input, control)) – (Cq(IP, DSB) – Cq(input, DSB)).
Recombinant proteins, DNA substrates and nuclease assays
Recombinant Mre11-Rad50-Xrs2 was purified as a complex using baculoviruses coding for his-tagged Mre11, FLAG-tagged Xrs2 and untagged Rad50 as described previously (Cannavo and Cejka, 2014). Mre11-Rad50 heterodimer was prepared using his-tagged Mre11 and untagged Rad50 constructs, Xrs2 was purified using FLAG-tagged Xrs2 (see Supplementary Data). Recombinant Sae2, DNA substrates and nuclease assays were used as described previously (Cannavo and Cejka, 2014).
In vivo hairpin opening assay
The rate of Lys+ recombinants was derived from the median recombination frequency determined from eight different isolates of each strain as described (Lobachev et al., 2002). Three trials were performed and the mean recombination rate was calculated.
End joining assays
Yeast cells were transformed with 250ng of BamHI-digested or undigested plasmid pRS414 (ARS-CEN, TRP1). Transformation efficiency was calculated as a ratio of the number of transformants with digested plasmid DNA to that with undigested plasmid DNA. The chromosomal end-joining assay was performed as described (Deng et al., 2014). Three trials were performed for each strain, and the mean frequency was calculated.
Western blot analysis and Rad53 phosphorylation assay
Protein extracts for western blot analysis were prepared by TCA precipitation. Anti-Mre11 (Krogh et al., 2005), anti-Rad53 (gift from M. Foiani), and anti-Rfa1 (Agrisera) were used for western blot analysis. For checkpoint activation, cells were collected after one hour in a media containing 500mg/ml of zeocin (Invitrogen).
Supplementary Material
Xrs2 is not required for nuclease-associated functions of the Mre11-Rad50 complex
Xrs2-independent end resection requires Sae2
Tel1 recruitment to DSBs requires Xrs2
Xrs2 is essential for canonical non-homologous end joining
ACKNOWLEDGEMENTS
We thank S. Oh and S. Deng for assistance with strain construction, K. Lobachev. K. Sugimoto and K.P. Hopfner for gifts of yeast strains and plasmids, M. Foiani for Rad53 antibodies, and W.K. Holloman for critical discussion of the data. This study was supported by grants from the National Institutes of Health (R01GM041784 and P01CA174653 to L.S.S) and the Swiss National Science Foundation (PP00P3_159323 to P.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.O. performed the experiments shown in Fig 1, Fig 2, Fig 3, Fig 4A,B, Fig 5B and Supplementary Figs 1, 2, 3 and 5. A.A.-Z. constructed the MRE11-NLS strain and performed pilot experiments of growth and end resection, and the data presented in Fig 4C. E.C and P.C. performed the experiments shown in Fig 4C, D, E, F and Supplementary Fig 4. All authors contributed to the experimental design, and writing the manuscript.
REFERENCES
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold spring Harbor Laboratory Press; 2005. [Google Scholar]
- Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Molecular and cellular biology. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Villa M, Gobbini E, Cassani C, Tedeschi G, Longhese MP. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO reports. 2015;16:351–361. doi: 10.15252/embr.201439764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PloS one. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Chen H, Donnianni RA, Handa N, Deng SK, Oh J, Timashev LA, Kowalczykowski SC, Symington LS. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc Natl Acad Sci U S A. 2015;112:E1880–1887. doi: 10.1073/pnas.1503331112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Molecular cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Molecular cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harbor perspectives in biology. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO reports. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Leach DR. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes to cells : devoted to molecular & cellular mechanisms. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eykelenboom JK, Blackwood JK, Okely E, Leach DR. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Molecular cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Ferrari M, Dibitetto D, De Gregorio G, Eapen VV, Rawal CC, Lazzaro F, Tsabar M, Marini F, Haber JE, Pellicioli A. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS genetics. 2015;11:e1004928. doi: 10.1371/journal.pgen.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbini E, Cesena D, Galbiati A, Lockhart A, Longhese MP. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair. 2013;12:791–799. doi: 10.1016/j.dnarep.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Gobbini E, Villa M, Gnugnoli M, Menin L, Clerici M, Longhese MP. Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity. PLoS genetics. 2015;11:e1005685. doi: 10.1371/journal.pgen.1005685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki D, Hayashihara K, Shima H, Higashide M, Terasawa M, Gasser SM, Shinohara M. The MRX Complex Ensures NHEJ Fidelity through Multiple Pathways Including Xrs2-FHA-Dependent Tel1 Activation. PLoS genetics. 2016;12:e1005942. doi: 10.1371/journal.pgen.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, Symington LS. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics. 2005;171:1561–1570. doi: 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harbor perspectives in biology. 2015;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS genetics. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Molecular cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Suhandynata RT, Zhou H. Phosphorylation of Sae2 Mediates Forkhead-associated (FHA) Domain-specific Interaction and Regulates Its DNA Repair Function. J Biol Chem. 2015;290:10751–10763. doi: 10.1074/jbc.M114.625293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Shinohara A, Shinohara M. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics. 2008;179:213–225. doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Molecular and cellular biology. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K, Klein F. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics. 2008;180:1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annual review of genetics. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell. 2005;16:597–608. doi: 10.1091/mbc.E04-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS genetics. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Molecular and cellular biology. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Molecular cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.