Summary
The affinity cryo-electron microscopy (cryo-EM) approach has been explored in recent years to simplify and/or improve the sample preparation for cryo-EM, which can bring previously challenging specimens such as those of low abundance and/or unpurified ones within reach of the cryo-EM technique. Despite the demonstrated successes for solving structures to low to intermediate resolutions, the lack of near-atomic structures using this approach has led to a common perception of affinity cryo-EM as a niche technique incapable of reaching high resolutions. Here, we report a ~2.6 Å structure solved using the antibody-based affinity grid approach with low concentration Tulane virus purified from a low-yield cell culture system that has been challenging to standard cryo-EM grid preparation. Quantitative analyses of the structure indicate data and reconstruction quality comparable to conventional grid preparation method using samples at high concentration.
Keywords: antibody-based affinity grid, affinity cryo-electron microscopy, single particle 3D reconstruction, Tulane Virus
Graphical abstract
E-TOC
The antibody-based affinity cryo-EM approach alleviates the required sample concentration of cryo-EM by 2–3 orders of magnitude, and makes low-abundance/yield specimens accessible to cryo-EM. Yu et al. have solved a low-yield, medium-sized virus to 2.6 Å and demonstrated the capability of affinity cryo-EM for near-atomic structural characterization.
Introduction
The remarkably improved image quality provided by direct electron detectors, together with other hardware and software advances, have resulted in an explosion of near-atomic resolution structures determined by single particle cryo-EM (Bai et al., 2015; Banerjee et al., 2016; Bartesaghi et al., 2015; Campbell et al., 2015; Grant and Grigorieff, 2015; Merk et al., 2016; Wang et al., 2014). Furthermore, the recent breakthrough in the Volta phase plate technique (Danev and Baumeister, 2016; Khoshouei et al., 2016) holds great promise to extend single particle cryo-EM to more samples with molecular masses below 100 KDa. Since microscopes, detectors, and image processing algorithms are no longer bottlenecks, the major hurdles for most cryo-EM projects have now shifted to sample grid preparation, which usually consists of a multi-step sample purification process and the subsequent preparation of a thin film of frozen-hydrated sample on TEM grids (Grassucci et al., 2007). Further innovation in cryo-EM sample grid preparation is of great significance for developing single particle cryo-EM into a routine structural biology tool.
The affinity cryo-EM approach that modifies TEM grids with an extra affinity layer to immobilize, purify, and concentrate target samples possesses a great potential to simplify and improve the cryo-EM grid preparation for a broader range of specimens, such as those of low yield and unpurified samples (Glaeser, 2015; Taylor and Glaeser, 2008; Yu et al., 2016a). Multiple affinity cryo-EM approaches based on different types of affinity layers including functionalized lipid layer (Azubel et al., 2004; Benjamin et al., 2016; Kelly et al., 2008; Medalia et al., 2002), 2D crystals of streptavidin (Han et al., 2012), antibody layer (Yu et al., 2014) and chemically functionalized carbon surface (Llaguno et al., 2014) have been reported. The affinity cryo-EM method will provide various advantages. First, it will enable single particle cryo-EM studies of low-concentration samples that are often encountered due to low abundance in natural sources, low yield of expression systems, or safety concerns for highly contagious/dangerous pathogens. Secondly, it will allow direct isolation of target particles from crude extracts via a specific interaction (Kelly et al., 2008; Yu et al., 2014), which combines sample purification with grid setup, simplifying cryo-EM grid preparation into a single-step process. Moreover, better sample integrity could potentially be obtained for labile macromolecular complexes with affinity cryo-EM approaches by avoiding multiple biochemical purification steps. Finally, immobilization of particles to an affinity layer can reduce particle diffusion and minimize potential sample damages at the air-water interface during and after sample blotting (Taylor and Glaeser, 2008). Altogether, these benefits of affinity cryo-EM will potentially bring more samples within reach of single particle cryo-EM, and also provide a more convenient, higher throughput, and mild cryo-EM grid preparation method.
Despite these promising properties of affinity cryo-EM approaches, there is currently no widespread use of such technique primarily due to the lack of high resolution structures solved using affinity cryo-EM. Multiple 3-D reconstructions have been reported with affinity cryo-EM approaches (Han et al., 2012; Kelly et al., 2008; Llaguno et al., 2014; Yu et al., 2014; Zhang et al., 2015). These structures were, however, all limited to low to intermediate resolutions and no near-atomic reconstructions have been obtained. As a step forward, here we report a demonstration case of using the antibody-based affinity grid approach to solve a medium-sized (~40 nm) viral particle to sub-3 Å.
Tulane virus (TV) is a monkey calicivirus in the recovirus genus of the caliciviridae and serves as a surrogate for human norovirus studies (Wei et al., 2008; Yu et al., 2013). Due to the low yield, the purified TV particles from cell cultures were often at a concentration of ~1010–11 particles/ml, which is too low for conventional cryo-EM studies that usually need ~1014 particles/ml or higher for most viral samples. In this work, we have used this challenging low-concentration TV sample to explore the potential of using antibody-based affinity grid for high-resolution cryo-EM. First, TV particles at a low sample concentration were concentrated to higher surface density via the anti-TV antibody affinity layer on the grid prior to sample freezing. Secondly, different cryo-EM grid freezing conditions were screened and optimized to achieve an optimal freezing where particles of interest immobilized on the antibody-affinity layer were frozen in vitreous ice with a thickness slightly larger than the particle size. In particular, we have adopted a Beer-Lambert law-based ice thickness estimation method, which can provide immediate feedback for freezing optimization. With such optimization, we have obtained high quality cryo-EM data for TV particles and a ~2.6 Å structure of TV was solved using the antibody-based affinity cryo-EM approach. This demonstrates the ability of affinity cryo-EM approach for solving sub-3 Å structures of medium-sized particles of high symmetry. Further optimization of the affinity cryo-EM approach and testing with particles of smaller molecular mass and lower symmetry will be performed in the future.
Results
Sample freezing optimization of TV on the antibody-based affinity grid
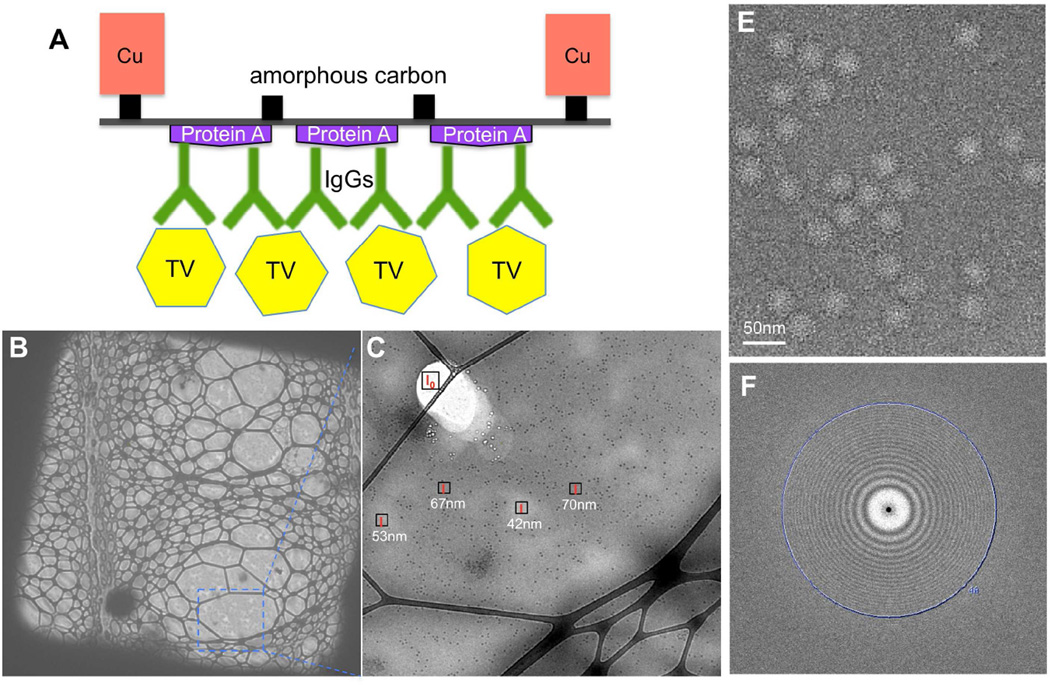
TEM grids were coated with anti-TV IgGs aided by protein A as described previously (Yu et al., 2016a, 2014). As expected, TV particles at a low sample concentration (~1010–11 particles/ml, ~2 µg/ml or less) were immobilized and concentrated efficiently on the anti-TV antibody-coated TEM grid to a level allowing for cryo-EM grid preparation (Figure S1A, B). In addition, after such on-grid purification/concentration, better sample purity was also achieved as indicated by the removing of contaminating vesicles in the original TV sample (Figure S1A). For data collection, cryo-EM sample of TV on the antibody-coated affinity grid was prepared using perforated carbon grids layered with thin continuous carbon as depicted in Figure 1A. The extra affinity layer brings more background noise to cryo-EM images relative to standard cryo-EM grids, which cannot be computationally removed in most affinity cryo-EM approaches (Kelly et al., 2008; Llaguno et al., 2014; Yu et al., 2014) and is essentially the major reason for the common concern about applying affinity cryo-EM for high-resolution cryo-EM reconstruction. On the other hand, however, presence of the affinity layer immobilizes particles and reduces particle-air interface collisions (Taylor and Glaeser, 2008), which thereby allows much thinner vitreous ice to embed the sample particles. Theoretically, reducing the ice thickness by ~30 nm could balance out the extra electron scattering by an affinity layer formed by protein A and IgGs (~13 nm in thickness) on the carbon film (~3 nm) (Vulović et al., 2013). Therefore, to pursue the best possible affinity cryo-EM images, freezing conditions were screened for an optimal ice thickness that is slightly larger than the size of TV particle (~40 nm).
Figure 1. Antibody-based affinity cryo-EM of TV.
(A) Schematic diagram of TV immobilized on the antibody-based affinity grid. For clear visualization, it was not drawn to scale. (B–C) Affinity cryo-EM micrographs of TV frozen using the optimized condition at 500× (B) and 3,800× (C) magnifications. Local ice thicknesses were estimated using the Beer-Lambert law based method. (E) A representative motion-corrected micrograph. (F) The 2-D power spectrum of the micrograph in (E). The position of 4 Å is labeled. See also Figure S1.
Visual evaluation of ice thickness in the range of tens to hundreds of nanometers (e.g., 50–150 nm), however, is challenging. Quantitative estimation of ice thickness therefore would be necessary. We have adopted a simple ice thickness measurement method based on the Beer-Lambert law (Yan et al., 2015) considering its convenience and on-line measurement. Briefly, the ice thickness (t) can be approximated from the ratio of image intensity of a melted area (I0) and an area with ice (I), and the inelastic mean free path of electrons in vitreous ice (λin) using the equation (Figure S1C, D). Mean image intensities (I0 and I) can be measured directly using the DigitalMicrograph software (Gatan, Inc.) controlling the camera, which allows convenient on-line measurement of ice thickness during sample screening. As shown by a demonstration case with phage T7 (Figure S1E–L), the quantitative estimation of ice thicknesses with the the Beer-Lambert law-based method allowed successful optimization of ice thickness with obviously improved image contrast. Similarly, a sample freezing condition producing vitreous ice with optimal thickness of ~40–70 nm was obtained for TV particles on the anti-TV antibody-coated grid (Figure 1B, C), and applied for following data collection. Cryo-EM movies of TV were collected automatically by using Leginon (Carragher et al., 2000) to control the FEI Titan Krios microscope and the Gatan K2 Summit direct electron detector. For most movies, signals at 4 Å or higher resolutions were recovered after motion correction (Figure 1E, F).
Single particle 3-D reconstruction of TV and quality evaluation
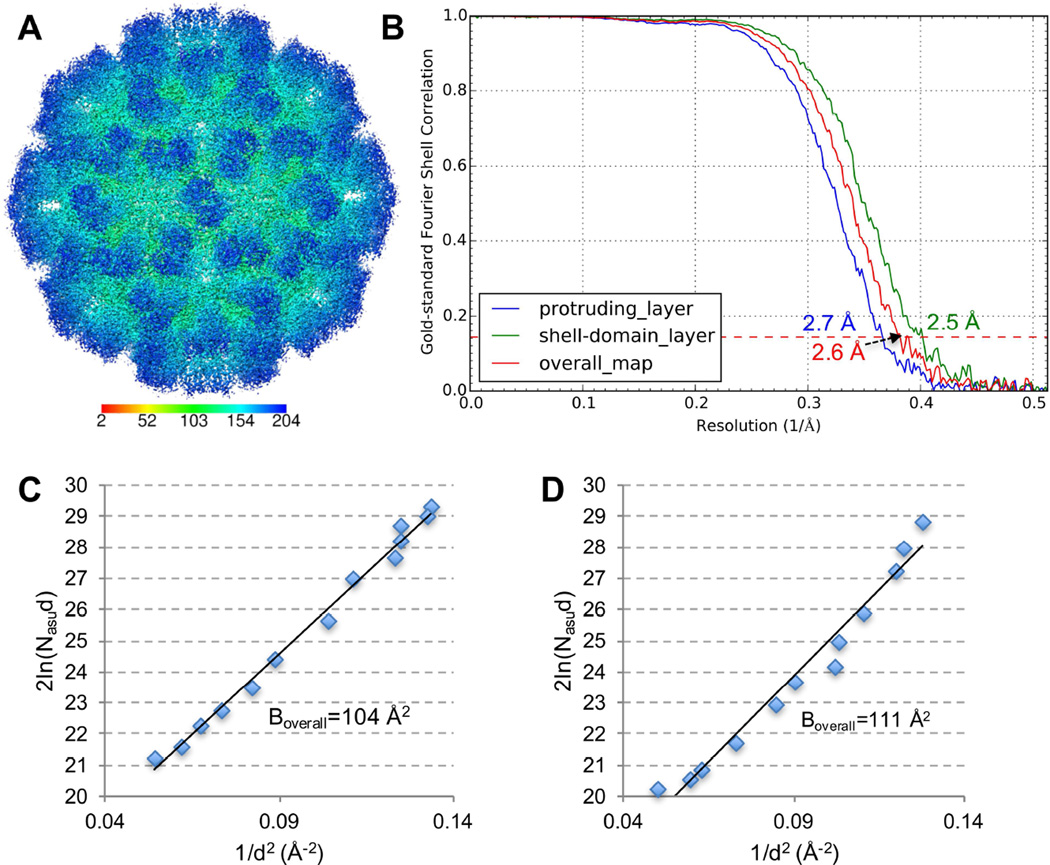
Single particle analysis was performed using jspr program (Guo and Jiang, 2014). The truly independent image processing strategy was adopted, in which the whole dataset was split into two halves and processed completely independently (Guo and Jiang, 2014; Liu et al., 2016). Based on the 0.143 cutoff criterion of gold-standard Fourier shell correlation (FSC) curve, a final 3-D reconstruction of TV at an overall resolution of 2.6 Å was obtained from ~14K particles (Figure 2A, B). As shown in Figure 2A, TV structure contains two major density layers, the basal shell layer (green-colored) and the protruding layer (blue-colored). Conformational flexibility has been suggested for the protruding layer of TV and TV-related structures (Chen et al., 2006; Katpally et al., 2010; Prasad, 1999; Yu et al., 2013). To evaluate the resolutions of the two layers, FSC curves were calculated for the shell layer and the protruding layer, respectively. As expected, the shell layer with an estimated resolution of ~2.5 Å was better resolved than the more flexible protruding layer (~2.7 Å) (Figure 2B). Analysis of the determined Euler angles of TV particles revealed a uniform coverage of the angle space (Figure S2), suggesting that no view preference was introduced by the antibody affinity layer.
Figure 2. Single particle 3-D reconstruction of TV using affinity cryo-EM data.
(A) Surface representation of TV radially colored according to the provided color key. (B) Gold-standard FSC curves of the overall map (red), the protruding layer (blue) and the shell layer (green). The position of the 0.143 FSC cutoff for resolution estimation is labeled with a dashed line. (C, D) 2ln(Nasud) vs 1/d2 is plotted for Boverall estimation of the affinity cryo-EM data of TV (C) and the cryo-EM dataset of 20S proteasome (EMPIAR-10025) re-processed using jspr (D), respectively. See also Figure S2 and S3.
To further evaluate the quality of this affinity cryo-EM TV dataset and 3-D reconstruction, its overall B factor (Boverall) was approximated. Boverall, which reflects the composite effect of sample integrity and homogeneity, ice thickness, imaging quality and computing errors, is a good indicator of cryo-EM data and 3-D reconstruction quality (Liu et al., 2007; Rosenthal and Richard, 2003). Based on the linear relationship between 2ln(Nasud) and 1/d2, in which Nasu is the number of asymmetric units used for the 3-D reconstruction and d is the resolution, Boverall is equal to the slope of the linear fitting (Rosenthal and Richard, 2003). As shown in Figure 2C, the 2ln(Nasud)-1/d2 plot of this affinity cryo-EM TV dataset showed an apparent linear relationship and yielded a Boverall of 104 Å2. This Boverall is smaller than previously reported Boverall for structures determined at 3.2 to 7 Å resolutions (Danev and Baumeister, 2016; Liu et al., 2007; Wang et al., 2014), which further confirms the excellent overall quality of this TV dataset. Moreover, we reprocessed a published cryo-EM dataset of 20S proteasome (Campbell et al., 2015) in the same way as for TV using jspr and obtained a 3-D reconstruction of ~2.7 Å (Figure S3). The 20S proteasome data was collected under similar imaging conditions as TV (i.e., Titan Krios and Gatan K2 Summit) but using standard holey grids and samples. The plot of 2ln(Nasud)-1/d2 for the 20S proteasome yielded a Boverall of 111 Å (Figure 2D) that is comparable to the Boverall of the affinity cryo-EM TV dataset. This comparison suggests that the antibody-based affinity grid approach can not only provide cryo-EM grids of similar quality (if not better) as the standard cryo-EM grid preparation approach, but also provide distinct advantages for those samples beyond reach of standard single particle cryo-EM.
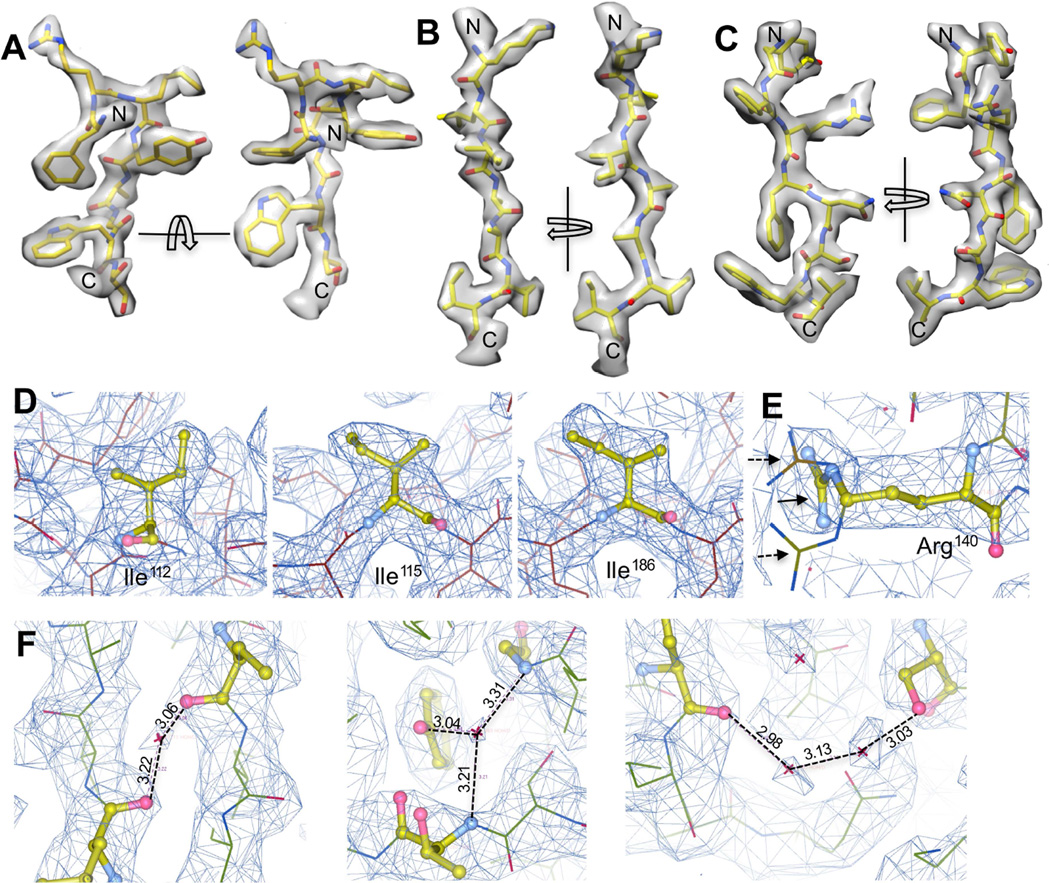
With our ~2.6 Å 3-D reconstruction of TV, many high-resolution structural features were resolved (Figure 3). Figure 3 shows three representative regions of TV density map including a short α-helix followed by a loop (Figure 3A) and two β-strands (Figure 3B, C), in which side chain densities can be easily recognized for most residues. Both hydroxyl groups in Tyr residues and the backbone carbonyl oxygen atoms are distinguishable (Figure 3A–C). In addition, this high-quality map of TV also allowed unambiguous assignment of rotameric conformations of most side chains. For example, different rotamers of Ile residues were unambiguously identified based on the density (Figure 3D) and the correct rotamers were distinguishable from multiple similar rotameric states (Figure 3E). Many ordered water molecules were also observed to form hydrogen bonds with neighboring residues or other water molecules (Figure 3F).
Figure 3. Atomic structural features resolved in the ~2.6 Å map of TV.
(A) Densities for a short α-helix followed by a short loop. (B, C) Densities for two β-strands. (D) Multiple rotameric states identified for Ile residues. (E) shows one example of distinguishing the correct rotamer (solid arrow) from alternative rotameric states (dashed arrow) for residue Arg140. (F) Water molecules resolved in TV map. Water molecules were only assigned to densities present in both maps generated using the two half datasets. Hydrogen bonds were shown in dashed lines with the bond length labeled.
Discussion and Conclusion
Single particle cryo-EM, which in general requires less concentrated samples than X-ray crystallography, has enabled structural studies of many macromolecular complexes refractory to crystallization. However, there are still many specimens such as toxic cellular complexes and some pathogens (e.g., TV used in this work) cannot be purified to the required sample concentrations by standard cryo-EM. The affinity cryo-EM approaches that enrich sample particles on the grid surface from low volume concentration have alleviated the required sample concentration by 2–3 orders of magnitude (or possibly higher with stronger affinity) (Kelly et al., 2008; Yu et al., 2014), and can provide a solution to those challenging specimens.
Inspired by the traditional diagnostic immuno-EM, we have established an antibody-based affinity cryo-EM approach, which can specifically isolate and concentrate target particles in the presence of contaminating proteins (Yu et al., 2014). The feasibility of preparing cryo-EM sample with the antibody-based affinity grid has been demonstrated with multiple low resolution structures (Yu et al., 2016a, 2014). Here, we have significantly extended its high resolution potential by solving a ~2.6 Å structure of TV using the antibody-based affinity cryo-EM approach.
In order to minimize the reduction of image contrast by the antibody-layer, we have carefully optimized the freezing condition (e.g., blotting time, humidity) for minimal ice thickness. A Beer-Lambert law-based ice thickness estimation approach (Yan et al., 2015) has been applied during sample screening to quantitatively evaluate the potential for further ice minimization. As such ice thickness estimation with the Beer-Lambert law-based method can be easily performed by melting a small region of the grid (similar to the sacrificed area for focusing during low-dose imaging), taking an image and simple calculation (Figure S1C, D), it provides a convenient and quick feedback for freezing optimization. An optimal condition that allowed the immobilized particles on the antibody-affinity layer embedded in vitreous ice with a thickness slightly larger than the particle size, was obtained and reproduced with different samples.
With this work, we have demonstrated the feasibility of using the antibody-based affinity cryo-EM approach for sub-3 Å structures, suggesting the potential of using affinity cryo-EM approach as a more efficient and robust way of grid preparation for high-resolution cryo-EM. With antibodies commercially available for a vast number of samples, the antibody-based affinity cryo-EM method can potentially revolutionize cryo-EM sample preparation for a broader spectrum of native specimens (at least for most viral samples as demonstrated in this work). Further optimization of the antibody-based affinity cryo-EM approach (Yu et al., 2016a) and demonstration for high resolution structure determination of smaller or low symmetry protein complexes will be performed in the near future for more general usage of this approach in cryo-EM.
Experimental Procedures
Purification of TV
TV was purified as described previously with some modifications (Yu et al., 2013). Briefly, LLC-MK2 cells (ATCC, Manassas, VA) grown to 80–90% confluency were inoculated with TV at a multiplicity of infection (MOI) of 0.2. Cells were collected after 72 hours and subject to three freeze-thaw cycles to release progeny TV. After clarifying by centrifugation at 13,776× g to remove cell debris, the supernatant was centrifuged at 28,000 rpm (SW28 rotor, Beckman, Danvers, MA). The pellet was suspended and further clarified by 20% (w/v) sucrose cushion ultracentrifugation at 28,000 rpm for 4 hours. Finally, the pellet was suspended and applied to a CsCl density gradient centrifugation at 288,000× g for 45 h (SW41Ti rotor, Beckman, Danvers, MA). The fractions containing TV virion were collected. Plaque assay was performed and revealed a titer of ~108 plaque forming units (PFU)/ml. Sample was dialyzed against 1×PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM phosphate, pH 7.4) for use.
Preparation of antibody-coated TEM grids
The amorphous carbon layer of TEM grids was coated with antibodies as described before (Yu et al., 2016a, 2014). The protein A-aided antibody coating method was applied in this work. Briefly, homemade Formvar/carbon grids or commercial grids layered with lacey carbon and ultrathin continuous carbon (e.g., Ted Pella #01824) were glow-discharge cleaned for ~40 s at 15 mA current and 0.2–0.15 Torr vacuum using a Ladd vacuum evaporator (Ladd Research, Williston, VT). 5 µl of 50 µg/ml Protein A (Sigma Aldrich) was applied to the cleaned carbon surface and incubated for ~10–15 min at room temperature. After removing the excess liquids, 5 µl of diluted antiserum was quickly applied to the grid and incubated for 15–20 min in a humid chamber at ambient temperature. The residual liquid and unbound antibody were blotted away, and the grid was subjected to 30s washing with 100–200 µl of 1×PBS buffer (pH 7.4).
Ice thickness estimation
Cryo-EM grids of frozen sample under different conditions were checked using a Philips CM200 microscope with a 1K CCD camera. The overall quality of the frozen grids were first visually evaluated as usual (Grassucci et al., 2007); For those promising conditions, the ice thickness was quantitatively estimated using the thickness estimation method based on Beer-Lambert law (Malis et al., 1988; Yan et al., 2015). More specifically, after melting the ice at a corner of the vitreous ice film, a low-magnification image (e.g., 3,800×) was taken covering both the region of interest and the melted area using a CCD camera. Beam intensities without (I0) and with (I) ice scattering, and inelastic mean free path of electrons in vitreous ice (λin) determine the ice thickness: . Since I0 and I can be measured directly using the DigitalMicrograph software (Gatan, Inc.) controlling the camera during sample screening, it provides immediate feedback for the ice thickness optimization. λin of 300 nm was used for the microscope operated at 200 KV as estimated in the references (Vulovic et al., 2013; Yan et al., 2015).
Affinity cryo-EM grid preparation and data collection of TV
TEM grids layered with lacey carbon support and a continuous ultrathin carbon (Ted Pella #01824) were coated with anti-TV antiserum (1:10 diluted, (Yu et al., 2014)) as described above. 3 µ1 of the purified TV sample was applied to the grid-coated with anti-TV antibody, and incubated in a humid chamber at 4 °C for ~30 min. Due to the limited amount of purified TV sample, only 3 µl sample was applied per grid. The grid was washed for ~2 min by floating on a drop of 200 µl cold l×PBS, pH7.4, and plunge-frozen using a Gatan Cp3 (or FEI Vitrobot) cryo-plunger. Different cryo-EM grid freezing conditions (e.g., blotting time, humidity) were attempted and evaluated visually complemented by quantitative ice thickness estimation as aforementioned. A freezing condition (8s, ~60% humidity) leading to vitreous ice of ~60 nm on average for TV was identified. A dataset of ~1,000 movies was collected using FEI Titan Krios TEM operating at 300 KV and liquid nitrogen temperature, and recorded with the Gatan K2 Summit direct electron detector at super-resolution movie mode (5 frames/second) at 225,00 × nominal magnification (0.65 Å/super-pixel). A dose rate of 8 electrons/physical pixel/second and a total exposure of 5 seconds resulted in a total dose of ~21 e/Å2 for the collected images.
Single particle 3-D reconstruction using jspr
A script motionCorrect.py that runs the dosefgpu_driftcorr program (Li et al., 2013) in batch mode was used to determine motions among raw movie frames (https://github.com/jianglab/motioncorr). ~20,000 particles were selected from motion-corrected movie sums using RELION automatic particle picking module (Scheres, 2012). The jspr program batchboxer.py (Guo and Jiang, 2014; Liu et al., 2016) was used to extract the particles from raw movie frames based on the particle coordinates files, the frame shift parameters determined by motionCorrect.py, and the total electron dose. The extracted particles were both motion-corrected and dose-weighted adopting the radiation damage compensation strategy described in reference (Wang et al., 2014). The particle images were resampled from 0.650 to 0.975 Å/pixel using the Fourier cropping method. The jspr program fitctf2.py (Jiang et al., 2012) was used to determine the initial CTF parameters from the extracted particles. The estimation and correction of astigmatism aberration was performed later during iterative refinement using the aligner refineAstigmatism in jspr (Guo and Jiang, 2014; Liu et al., 2016). The whole dataset was then split into two half datasets, even and odd subsets, which were processed independently, including constructions of de novo initial models and subsequent refinements as described before (Guo and Jiang, 2014; Liu et al., 2016). Three correct de novo initial models were obtained using the random model method (Guo and Jiang, 2014; Liu et al., 2016) for both the “even” and “odd” halves, respectively, which were used as initial references to determine the Euler/Center parameters of all particles. This resulted in three sets of Euler/center parameters for each particle, based on which particles with consensus Euler/Center parameters were selected for further refinement. In the jspr program (Guo and Jiang, 2014), in addition to Euler/Center searching, many potential modulations present in 2-D images are also formulated as 2-D alignment tasks (often referred to as high-order alignments), including micrograph and per-particle defocus, astigmatism aberration, overall scale variance among 2-D images, elliptic magnification distortion, and phase error from residual coma (Guo and Jiang, 2014; Liu et al., 2016; Yu et al., 2016b). Initial refinement was performed with only the Euler/Center parameters. After the convergence of the orientation/center refinement at ~3.6 Å, high-order aligners were gradually added to the iterative refinement loop to determine and correct those image artifacts as aforementioned, which all together improved the quality of the 3-D reconstruction to ~2.6 Å. The 0.143 cutoff for gold-standard Fourier Shell Correlation (FSC) curve (Rosenthal and Richard, 2003) was used to estimate the resolution of reconstruction with “even” and “odd” maps soft-masked using EMAN2 (Tang et al., 2007) adaptive masking processor (mask.auto3d). The final map of TV was reconstructed from 14,154 particles by pooling the two half datasets. The final map was sharpened using the 1D structure factor of TV fitted from the images, and a low pass filter derived from the final FSC curve to boost the high-resolution Fourier amplitudes, and to suppress the high-frequency noise, respectively (Guo and Jiang, 2014; Rosenthal and Richard, 2003).
Estimation of overall B factor (Boverall) of cryo-EM data and reconstruction
Different numbers of particles were randomly selected from final refinement particle sets. New 3-D reconstructions were performed and the resolutions were estimated based on gold-standard FSC curves. The number of particles for 3-D reconstructions was then plotted against the corresponding achievable resolutions. Specifically, the 2ln(Nasud) as a function of 1/d2 was plotted, in which Nasu is the number of asymmetric units and d is the achievable resolution of 3-D reconstructions. Based on the mathematical formulation of the relationship of Nasu, d and the Boverall (Figure 11 in (Rosenthal and Richard, 2003)), Boverall can be approximated as the slope of the linear fitting of 2ln(Nasud) and 1/d2.
Supplementary Material
Highlights.
2.6 Å Tulane virus structure solved using antibody-based affinity cryo-EM approach
Specimens at low sample concentrations (e.g., 2 µg/ml) applicable to cryo-EM study
Cryo-EM sample freezing optimization with quantitative ice thickness estimation
Acknowledgments
This work was supported by NIH grant (1R01AI111095). Both negative staining and cryo-EM images were taken at Purdue Cryo-EM Facility. We thank Dr. Philip Serwer for providing the bacteriophage T7 antiserum, and Frank Vago for proofreading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers:
The final density map has been deposited in the Electron Microscopy Databank with accession code EMD-8252.
Author Contributions:
Conceptualization, W.J., and G.Y.; Methodology, G.Y. and W.J.; Investigation, G.Y., K.L., and P.H.; Writing, G.Y., X.J., and W.J.
References
- Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol. Cell. 2004;15:833–839. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SHW. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4 doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016 doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, Subramaniam S. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin CJ, Wright KJ, Seok-Hee H, Kyle K, Guimei Y, Ruchika B, Fei G, Stauffacher CV, Wen J, Thompson DH. Nonfouling NTA-PEG-Based TEM Grid Coatings for Selective Capture of Histidine-Tagged Protein Targets from Cell Lysates. Langmuir. 2016;32:551–559. doi: 10.1021/acs.langmuir.5b03445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife. 2015;4 doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B, Bridget C, Nick K, David K, Milligan RA, Potter CS, James P, Amy R. Leginon: An Automated System for Acquisition of Images from Vitreous Ice Specimens. J. Struct. Biol. 2000;132:33–45. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- Chen R, Neill JD, Estes MK, Prasad BVV. X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity. Proceedings of the National Academy of Sciences. 2006;103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Baumeister W. Cryo-EM single particle analysis with the Volta phase plate. Elife. 2016:5. doi: 10.7554/eLife.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser RM. How good can cryo-EM become? Nat. Methods. 2015;13:28–32. doi: 10.1038/nmeth.3695. [DOI] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Joachim F. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Jiang W. Single Particle Cryo-electron Microscopy and 3-D Reconstruction of Viruses. In: Kuo J, editor. Electron Microscopy, Methods in Molecular Biology. Humana Press; 2014. pp. 401–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B-G, Bong-Gyoon H, Walton RW, Amos S, Peter H, Stubbs MT, Yannone SM, Pablo A, Ming D, Glaeser RM. Electron microscopy of biotinylated protein complexes bound to streptavidin monolayer crystals. J. Struct. Biol. 2012;180:249–253. doi: 10.1016/j.jsb.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Jiang W, Guo F, Liu Z. A graph theory method for determination of cryo-EM image focuses. J. Struct. Biol. 2012;180:343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katpally U, Voss NR, Cavazza T, Taube S, Rubin JR, Young VL, Stuckey J, Ward VK, Virgin HW, 4th, Wobus CE, Smith TJ. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J. Virol. 2010;84:5836–5841. doi: 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: A rapid method for isolating protein complexes for single-particle electron microscopy. Proceedings of the National Academy of Sciences. 2008;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshouei M, Radjainia M, Phillips AJ, Gerrard JA, Mitra AK, Plitzko JM, Baumeister W, Danev R. Volta phase plate cryo-EM of the small protein complex Prx3. Nat. Commun. 2016;7:10534. doi: 10.1038/ncomms10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xiangan L, Wen J, Joanita J, Wah C. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-path Simulated Annealing optimization algorithm. J. Struct. Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Guo F, Wang F, Li TC, Jiang W. 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles. Structure. 2016;24:319–328. doi: 10.1016/j.str.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaguno MC, Hui X, Liang S, Nian H, Hong Z, Qinghua L, Qiu-Xing J. Chemically functionalized carbon films for single molecule imaging. J. Struct. Biol. 2014;185:405–417. doi: 10.1016/j.jsb.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malis T, Cheng SC, Egerton RF. EELS log-ratio technique for specimen-thickness measurement in the TEM. J. Electron Microsc. Tech. 1988;8:193–200. doi: 10.1002/jemt.1060080206. [DOI] [PubMed] [Google Scholar]
- Medalia O, Typke D, Hegerl R, Angenitzki M, Sperling J, Sperling R. Cryoelectron microscopy and cryoelectron tomography of the nuclear pre-mRNA processing machine. J. Struct. Biol. 2002;138:74–84. doi: 10.1016/s1047-8477(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, Subramaniam S. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 2016 doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV. X-ray Crystallographic Structure of the Norwalk Virus Capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Richard H. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulović M, Miloš V, Ravelli RBG, van Vliet LJ, Koster AJ, Ivan L, Uwe L, Hans R, Ozan Ö, Bernd R. Image formation modeling in cryo-electron microscopy. J. Struct. Biol. 2013;183:19–32. doi: 10.1016/j.jsb.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, Chen D-H, Liu X, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 2014;5:4808. doi: 10.1038/ncomms5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Farkas T, Sestak K, Jiang X. Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J. Virol. 2008;82:11429–11436. doi: 10.1128/JVI.00696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Edwards TJ, Pankratz LM, Kuhn RJ, Lanman JK, Liu J, Jiang W. Simultaneous determination of sample thickness, tilt, and electron mean free path using tomographic tilt images based on Beer-Lambert law. J. Struct. Biol. 2015;192:287–296. doi: 10.1016/j.jsb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Li K, Jiang W. Antibody-based affinity cryo-EM grid. Methods. 2016a doi: 10.1016/j.ymeth.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Li K, Liu Y, Chen Z, Wang Z, Yan R, Klose T, Tang L, Jiang W. An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J. Struct. Biol. 2016b;195:207–215. doi: 10.1016/j.jsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Vago F, Zhang D, Snyder JE, Yan R, Zhang C, Benjamin C, Jiang X, Kuhn RJ, Serwer P, Thompson DH, Jiang W. Single-step antibody-based affinity cryo-electron microscopy for imaging and structural analysis of macromolecular assemblies. J. Struct. Biol. 2014;187:1–9. doi: 10.1016/j.jsb.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Zhang D, Guo F, Tan M, Jiang X, Jiang W. Cryo-EM Structure of a Novel Calicivirus, Tulane Virus. PLoS One. 2013;8:e59817. doi: 10.1371/journal.pone.0059817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Vago F, Guo F, Liu Z, Yu G, Serwer P, Jiang W. Affinity Cryo-Electron Microscopy Studies of Viral Particles Captured Directly from Cell Culture. Microsc. Microanal. 2015;21:547–548. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.