Abstract
Introduction
Primary human trophoblasts release a repertoire of extracellular vesicles (EVs). Among them are nano-sized exosomes, which we found to suppress the replication of a wide range of diverse viruses. These exosomes contain trophoblastic microRNAs (miRNAs) that are expressed from the chromosome 19 miRNA cluster and exhibit antiviral properties. Here, we report our investigation of the cargo of placental EVs, focusing on the composition and the antiviral properties of exosomes, microvesicles, and apoptotic blebs.
Methods
We isolated EVs using ultracentrifugation and defined their purity using immunoblotting, electron microscopy, and nanoparticle tracking. We used liquid chromatography-electrospray ionization-mass spectrometry, protein mass spectrometry, and miRNA TaqMan card PCR to examine the phospholipids, proteins, and miRNA cargo of trophoblastic EVs and an in vitro viral infection assay to assess the antiviral properties of EVs.
Results
We found that all three EV types contain a comparable repertoire of miRNA. Interestingly, trophoblastic exosomes harbor a protein and phospholipid profile that is distinct from that of microvesicles or apoptotic blebs. Functionally, trophoblastic exosomes exhibit the highest antiviral activity among the EVs. Consistently, plasma exosomes derived from pregnant women recapitulate the antiviral effect of trophoblastic exosomes derived from in vitro cultures of primary human trophoblasts.
Discussion
When compared to other trophoblastic EVs, exosomes exhibit a unique repertoire of proteins and phospholipids, but not miRNAs, and a potent viral activity. Our work suggests that human trophoblastic EVs may play a key role in maternal-placental-fetal communication.
Keywords: Placenta, trophoblastic vesicles, C19MC, exosomes, viral infection, plasma exosomes
INTRODUCTION
The placenta plays a central role not only in maternal-fetal exchange functions and immunological defense, but also in communicating maternal-fetal signals that are essential for pregnancy health. Accordingly, the human villous trophoblast layer, which includes the syncytiotrophoblast that is directly bathed in maternal blood and the subjacent progenitor cytotrophoblast, regulates the release of these communication messages into the maternal circulation. In contrast, the transmission of trophoblastic biological signals into the fetal circulation may require trafficking through the villous basal membrane and fetal endothelial cells before entering the fetal circulation. In addition to hormones, growth factors, and other signaling proteins and akin to other epithelial cells, trophoblasts release a diverse repertoire of lipid-encapsulated extracellular vesicles (EVs) into the maternal blood, including apoptotic cell-derived EVs (ABs), microvesicles (MVs), and exosomes (EXs) [1–4]. The size of ABs ranges between 1~5 µm, and are produced by deportation of trophoblastic fragments during apoptosis [5]. MVs display a relatively smaller size (100 nm – 1 µm) and are released to the extracellular environment by budding off from the cell membrane [4]. In contrast, EXs (30–200 nm) are generated within multivesicular bodies, which fuse with plasma membrane to release the EXs’ cargo into the intercellular space or maternal blood [6,7]. Human placental EXs derived from trophoblasts and placental non-trophoblastic cells such as placental mesenchymal progenitor cells are involved in cellular adaptation during the course of normal pregnancy, including immuno-modulation and tolerance [8–13], endothelial cell migration [14–16], and proliferation and invasion of extravillous trophoblasts [17]. We have previsouly shown that EXs derived from primary human trophoblast (PHT) cells confer resistance to a broad panel of viruses to non-placental cells that are normally permissive to viral replication [7,18]
In light of their presence in the maternal circulation and their diverse functions at the feto-maternal interface, placental EVs, particularly EXs, have been assessed as potential biomarkers for diagnosis and prognosis of pregnancy disorders such as preterm birth or preeclampsia [3,4]. Indeed, when compared to normal pregnancies, trophoblastic EVs are elevated in the circulation of women with preeclampsia [19–22]. Interestingly, several proteins, including sFlt-1, endoglin, tissue factor, and PAI, all presumed to play a role in the angiogenic and coagulation imbalance of preeclampsia, are present within trophoblastic EVs [23,24], raising the possibility that EVs may contribute to the pathogenesis of preeclampsia [21,25].
Recent data indicate that microRNAs (miRNAs) may have important gene regulatory functions not only in cells that produce them, but also in neighboring or distant cells [26,27]. This unprecedented finding of cell-to-cell communication, mediated by extracellular miRNAs, suggests that extracellular miRNAs play a role in tissue physiology, homeostasis, and disease [7,28,29]. In general, extracellular miRNAs can be found within EVs or in non-vesicular form, bound by proteins such as argounate2 (Ago2), high-density lipoproteins, and nucleophosmin 1 [30–33]. Among the circulating miRNAs associated with pregnancy [34–36], one family of miRNAs, which are expressed from the chromosome 19 miRNA cluster (C19MC), is placenta-specific and highly expressed in the maternal blood throughout pregnancy and sharply diminishes after delivery [37–39]. Moreover, we discovered that C19MC miRNAs are packaged into trophoblastic EXs and, when delivered to non-placental cells, can suppress replication by a wide range of DNA and RNA viruses [7,18].
To systematically investigate the properties and function of trophoblastic EVs, we purified the three major EVs (ABs, MVs, and EXs) from in vitro cultures of term PHT cells derived form healthy pregnancies and characterized their miRNA profiles and phospholipid and protein content. Moreover, we analyzed their antiviral properties using our previously established viral infection assay. Functionally, we found that trophoblastic EXs displayed the most robust antiviral activity among the three trophoblastic EVs tested and that this antiviral effect was recapitulated using total plasma EXs obtained from pregnant women at term.
METHODS
Isolation of trophoblastic EVs from PHT conditioned medium
The collection of placentas used for cell isolation and culture was reviewed and approved by the Institutional Review Board at the University of Pittsburgh. PHT cells were isolated from placentas of uncomplicated pregnancy, labor, and delivery according to our previously published protocol [40]. PHT cells were cultured in 7 × 15-cm plates at 37°C for up to 72 h in complete DMEM medium that contained 1% antibiotics and 10% of either fetal bovine serum (FBS) that had been depleted of bovine EXs by overnight ultracentrifugation at 100,000 g or purchased bovine-EX-depleted FBS (Thermo Fisher Scientific, Waltham, MA). Conditioned medium samples used for vesicle isolation were collected on the third day of culture, when most PHT cells are syncytialized [40]. Approximately 400 ml of PHT conditioned medium (CM) was collected for EV purification using differential centrifugation and OptiPrep (D1556-250 ml, Sigma-Aldrich, St. Louis, MO) continuous gradient ultra-centrifugation. The remaining isolation procedures were performed at 4°C, unless indicated otherwise. We performed serial centrifugation procedures, first at 500 g for 10 min to pellet cell debris, followed by 2,500 g for 20 min to pellet ABs, which were washed three times with 2 ml of PBS and suspended in 50~100 µl of PBS (P5493-1L, RNase-free, Sigma). MVs were pelleted using 12,000 g centrifugation for 30 min, then washed three times with 2 ml of PBS and suspended in 50~100 µl of PBS. To isolate EXs, we filtered AB- and MV-depleted medium through 0.22 µM filter unit (Millipore, Billerica, MA) to exclude any residual particles larger than 200 nm; then the filtrate was concentrated at RT, using a Vivacell 100 filtration unit (100 kDa Mw cut-off, Sartorius, New York, NY). For one Vivacell 100 filtration unit, PHT CM was concentrated to a volume of 5 ml, re-suspended in 9 ml PBS, and centrifuged at 100,000 g overnight. The supernatant was removed, and the pellets were re-suspended in 0.5 ml of PBS and mixed with 1.5 ml of 60% OptiPrep. The mixture (2 ml) was laid at the bottom of the tube and overlaid with 10 ml of 6–40% OptiPrep gradient using a gradient formation chamber and peristaltic pump. After 22 h of OptiPrep gradient ultra-centrifugation at 100,000 g, we withdrew individual fractions from top to bottom and identified the fractions that correspond to trophoblastic EXs by western blotting and NanoSight nanoparticle tracking analysis (NTA) (Malvern Instruments, Westborough, MA). EXs were filtered out of the OptiPrep solution by diluting EXs in PBS and concentrated in a Vivacell 20 filtration unit (100 kDa Mw cut-off, Sartorius). We quantified protein concentration of trophoblastic vesicles, using the Micro BCA method according to the manufacturer’s instructions (Thermo Fisher).
Isolation of EXs from human plasma
The collection of plasma samples, performed during blood testing as a part of routine clinical care, was reviewed and approved by the Institutional Review Board at the University of Pittsburgh. Blood samples (2 ml) were collected in EDTA-treated tubes and centrifuged at 1,000 g for 10 min to remove any blood cells. This was followed by centrifugation at 2,500 g for 20 min to remove platelets. Either fresh or −80°C f rozen plasma was used, as indicated. For EX isolation, frozen plasma was again centrifuged at 2,500 g for 15 min to remove any aggregates. We diluted cleared plasma with PBS (1:1 volume ratio) and filtered the diluted plasma through a 0.2 µM syringe filter (PES membrane, Whatman, Maidstone, UK) to remove particles larger than 0.2 µM. 10 ml of total diluted plasma were loaded into the gravity column that contained 2 ml of gelatin-agarose (G5384, Sigma). We collected the flow-through and repeated loading twice to ensure that fibronectin was bound to the gelatin-agarose [41,42]. We obtained crude EXs by adding PEG6000 (81253, molecular biology grade, Sigma) at the final concentration of 5% (w/v) and incubated the mixture at RT for 10 min. We then centrifuged the mix at 10,000 g at RT for 20 min and suspended pellets in 0.5 ml of PBS. We loaded the crude EX suspension on top of 14 ml of 6%-30% OptiPrep continuous gradient. After overnight 100,000 g OptiPrep gradient ultra-centrifugation, we withdrew individual fractions from top to bottom and identified the fractions that correspond to trophoblastic EXs as described above.
Western blotting
The EVs or cells were lysed in a cell lysis buffer containing 50 mM Tris-HCI, 150 mM NaCI, and 1% Triton X-100, supplemented with protease inhibitors (mini complete Ultra tablets, Roche, Indianapolis, IN) and phosphatase inhibitor cocktail tablets (PhosSTOP, Roche). Lysates were separated on SDS-PAGE and transferred to polyvinylidene fluoride membranes as we previously described [43]. Membranes were immunoblotted with various primary antibodies: rat anti-Ago2 (SAB4200085, final concentration 0.5 µg/ml, Sigma), mouse anti-CD63 (sc-5275, final concentration 0.2 µg/ml, Santa Cruz Biotechnology, Dallas, TX), mouse anti-Syntenin-1 (sc-100336, final concentration 0.1 µg/ml, Santa Cruz Biotechnology), rabbit anti-TSG101 (ab125011, final concentration 0.2 µg/ml, Abcam, Cambridge, MA), Rabbit anti-Calnexin (2433S, dilution 1:1000 of the stock, Cell Signaling, Boston, MA). Each was followed by appropriate horseradish peroxidase secondary antibody. Signals were visualized using SuperSignal West Dura (Thermo Fisher).
NTA analysis of trophoblastic MVs and EXs
MVs or EXs were diluted to an appropriate level (1,000- to 5,000-fold dilution) with 0.1 µM filtered water (W4502, Sigma), which has been validated in our NTA LM-10 system to be particle-free. This dilution ensured that at least 20 particles would be observed in the objective view field per second. The diluted samples were continuously injected into the view field by a syringe pump. Particles were individually recorded and tracked by the high-definition CCD camera for 1 min. Each measurement was repeated three times. Installed NTA particle analysis software was used to analyze all frames containing particles captured by the camera.
Transmission electron microscopy
Isolated trophoblastic EVs were fixed in 2% paraformaldehyde at RT for 10 min and then layered on carbon/Formvar film-coated grids for 20 min. The grids were washed with water and then negatively stained with 1% uranyl acetate for 10 min. EVs were viewed using a JEOL transmission electron microscope JEM 1011 (JEOL, Peabody, MA).
Profiling of EVs’ miRNA
Human miRNA TaqMan A and B cards (Thermo Fisher) were used for analysis of miRNA expression. Total RNA was extracted from purified trophoblastic EVs using a miRNAeasy kit (Qiagen, Valencia, CA). EV RNA concentration was quantified by fluorescence-based measurement with a PicoGreen RNA probe (Thermo Fisher). RNA from trophoblastic vesicles was processed for qPCR-based TaqMan cards, following the manufacturer’s instructions, and analyzed using an Applied Biosystems ViiA 7 sequence detection system. The Ct values of individual miRNAs in the TaqMan A and B cards were retrieved using the preinstalled system software, and differences in miRNA levels between trophoblastic vesicles and the corresponding parental PHT cells were determined using the R package high-throughput quantitative PCR [44]. The Ct values of miRNAs were normalized by the mean of control probes for each card. A moderated t test was used to compare miRNA expression levels (represented as delta Ct value) between trophoblastic vesicles and the corresponding PHT cells [45]. The p values of the moderated t tests were adjusted by the method described by Benjamini and Hochberg to control for false discovery rate [46].
Protein mass spectrometry of trophoblastic EVs
A minimum of 5 µg of purified trophoblastic EVs, derived from the medium of trophoblasts obtained from 2–3 placentas, were solubilized in 0.01% n-dodecyl β-D-maltoside at 4°C overnight. The samples were then processed by the Biomedical Mass Spectrometry Center at the University of Pittsburgh. In-solution tryptic digestion was performed at 37°C overnight with trypsin gold (mass spectrometry grade, Promega, Madison, WI). Digested peptides were analyzed by nano reverse-phase high-performance liquid chromatography (HPLC) interfaced with a linear trap quadrupole-orbitrap Velos mass spectrometer (Thermo Fisher). The tandem mass spectra (MS/MS) were analyzed by the MASCOT search engine (Matrix Science), and identified peptides and proteins were further statistically validated with the Scaffold software. The free Scaffold viewer was downloaded to display a list of identified proteins. High confidence identifications were proteins with (a) protein identification probability of 99% or above, (b) peptide identification probability of 95% or above, and (c) the presence of at least two peptides.
Mass spectrometry for phospholipid analysis
Lipids were extracted from EVs (derived from the medium of trophoblasts obtained from 2–3 placentas) under nitrogen atmosphere, using the Folch procedure [47]. For analysis of phospholipid molecular species, liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis was performed on a Dionex HPLC system (Thermo Fisher Scientific, using the Chromeleon software), consisting of a Dionex UltiMate 3000 mobile phase pump equipped with an UltiMate 3000 degassing unit and UltiMate 3000 autosampler (sampler chamber set at 4 °C). The Dionex HPLC system was coupled to a hybrid quadrupole-orbitrap mass spectrometer, Q-Exactive (Thermo Fisher), with the Xcalibur operating system. The instrument was operated in the negative ion mode (at a voltage 5.0 kV, source temperature was maintained at 150°C). MS spectra were acquired in n egative ion mode using a full-range zoom (400–1800 mass-to-charge ratio [m/z]). The molecular species of phosphatidylcholine and sphingomyelin were detected as a formic acid adducts. Normal phase column separation of phospholipids was performed on a Luna Silica (2) 100 A° column (150 × 1 mm, Phenomenex, Torrance, CA). The analysis was performed using gradient solvent A (hexane:propanol:water, 47: 57:1, v/v) and solvent B (hexane:propanol:water, 47: 57:10, v/v), each containing 5 mM ammonium acetate and 0.01% formic acid. The column was eluted at a flow rate of 0.05 ml/min as follows: 0–3 min, linear gradient, 10–37% solvent B; 3–12.5 min, isocratic at 37% solvent B; 12.5–20 min, linear gradient, 37–100% solvent B; 20–45min, isocratic at 100% solvent B; 45–60 min, isocratic at 10% solvent B. All organic solvents and other biochemical reagents were purchased from Sigma-Aldrich. Analysis of LC-MS data was performed using software package SIEVE ™ for differential analysis (ThemoFisher Scientific).
In vitro viral infection assay
The procedures were performed as we recently described [18]. Briefly, U2OS cells were seeded in 24-well plates before incubation with EVs overnight (16–18 h) at 37°C in DMEM complete medium. The cells were then infected with vesicular stomatitis virus (VSV), at the multiplicity of infection (MOI) equivalent to 0.5–1, for 5 hours, then washed with PBS. RNA was extracted using QIAzol (Qiagen), and VSV infection was quantified by RT-qPCR using VSV primers, as previously described [18].
Statistics
Analysis of qPCR-based TaqMan cards is discussed above. Data were presented as mean values with standard deviations. Statistically significant differences between samples were evaluated by one-way ANOVA (Prism6.0, GraphPad Inc., La Jolla, CA) or using a linear mixed effect model. p < 0.05 was considered statistically significant.
RESULTS
Isolation of EVs from primary human trophoblasts
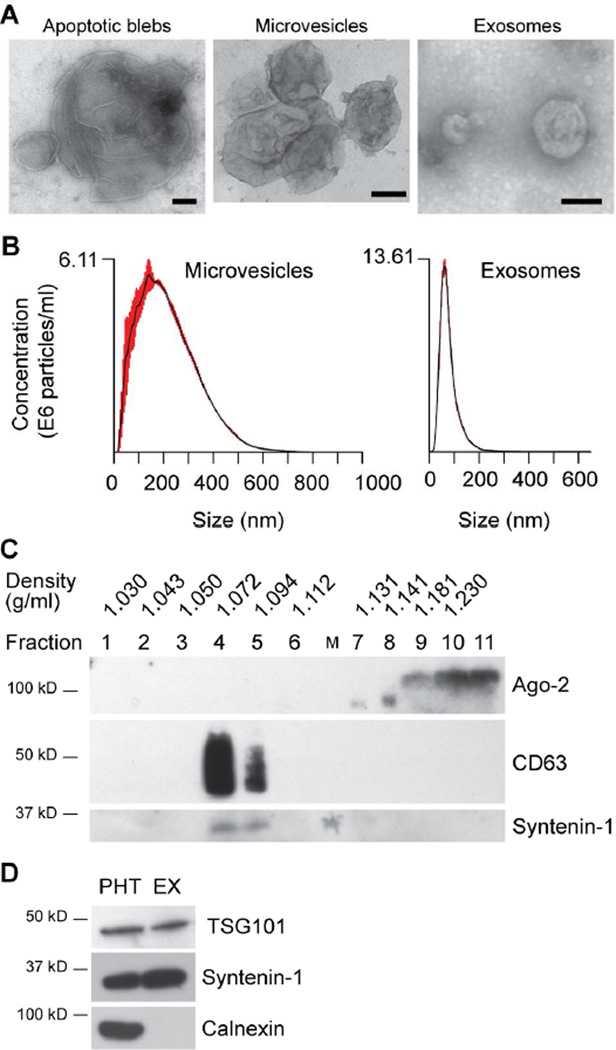
We isolated PHT cells from placentas after uncomplicated term pregnancy and delivery. Conditioned medium (CM) from PHT cells was collected for subsequent fractionation of ABs, MVs, and EXs. Notably, we loaded crude EX samples at the bottom of the continuous 6%-40% OptiPrep gradient rather than overlaying them at the top. This ensured better separation of non-EV proteins (such as Ago2), which were sedimented in 40% OptiPrep after ultracentrifugation, from the EXs migrating at buoyant density ~1.08–1.10 g/ml within the 6%-40% OptiPrep gradient (Fig. 1 and 2A). We confirmed the size distribution of trophoblastic EVs, using nanoparticle tracking analysis (NTA) (Fig. 2B). As expected, EXs exhibited a fairly tight distribution around the median diameter of 80–90 nm, with a somewhat broader distribution of MVs surrounding a peak of 200 nm in diameter. Notably, NTA is designed for measurement of particle size less than 1000 nm, and thus was not used for estimation of AB size. Electron microscopy images confirmed that ABs contained a population of vesicles of approximately 5000 nm, much larger than MVs and EXs, whereas the size of MVs and EXs was consistent with NTA measurement (Fig. 2C). As expected, the purified EXs were immuno-reactive for CD63, syntenin-1, and TSG-101 (Fig. 2A, D), [48–51] and clearly separated from Ago2 proteins (see fractions 4–5 vs fractions 9–11 in Fig. 2A) [30]. Trophoblastic EXs were negative for calnexin, suggesting that endoplasmic reticulum membranes are excluded by our protocol (Fig. 2D). Collectively, our data support our ability to purify the three distinct types of trophoblastic EVs from PHT CM.
Fig. 1. A schematic depiction of our trophoblastic EV isolation workflow.
Fig. 2. Characterization of trophoblastic EVs derived from PHT CM.
(A) A western blotting analysis of key exosomal proteins. Trophoblastic EXs were mainly recovered in the three fractions (see Methods for details), which were immuno-reactive to CD63 and syntenin-1. In contrast, Ago2 protein was only present at the bottom fractions, but absent in the fractions corresponding to trophoblastic EXs. (B) Analysis of trophoblastic EVs using NTA. Note that the X-axis indicates the particle size distribution, and the Y-axis denotes particle concentration. (C) Electron microscopy (EM) images of trophoblastic EVs. EM scale bar: 100 nm. (D) A western blotting analysis of exosomal proteins TSG101 and syntenin-1, compared to PHT cells. Calnexin is a marker of endoplasmic reticulum membranes. An equal amount of total proteins in PHT cells and PHT-derived exosomes were loaded. All the isolation and characterization experiments were performed at least three times.
The distribution of phospholipids in the three trophoblastic EVs
EVs are characteristically enveloped by a phospholipid bilayer. To identify molecular speciation of phospholipids in the three trophoblastic EVs, we performed global phospholipidomic analysis. Overall, we detected 179 individual species of phospholipids in eleven major classes – phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), sphingomyelin (SPH), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidic acid (PA), bis-monoacylglycerophosphate (BMP), cardiolipin (CL), lysophosphatidylcholine (LPC) and lysophosphatidylethanolamine (LPE). While we found multiple minor variations in the content and molecular speciation between the three types of EVs, the major differences were in the highest content of PC in EXs (56.94%), compared to ABs and MVs (19% and 11.7%, respectively, Fig 3). Of note, PC is the most common constitutive phospholipid of stable membrane lipid bilayers. In contrast, phosphatidylethanolamine – known to form non-bilayer hexagonal lipid arrangements – was highest in MVs, had a lower content in ABs and the lowest level in EXs (54%, 33%, 12%, respectively, Fig. 3). The differences between the characteristic EXs phospholipids and those in the other two groups - ABs and MVs - became even more evident when we re-plotted their distribution (Fig. 3, lower panel) between four functionally non-equivalent groups with the propensities to: i) form stable bilayer membrane arrangements (non-charged zwitterionic phospholipids such as PC, PE, Sph, LPC and LPE), ii) contribute to negative charge of the vesicle surface (mono- and di-anionic phospholipids PI, PS, PG, PA, BMP and CL), iii) act as membrane fusogenic agents (PE, PA, BMP, LPC, and LPE), and iv) facilitate the formation of non-bilayer (hexagonal) organizations (PE, PA, and CL). This analysis demonstrated that EXs are composed of phospholipids providing a high level of membrane stability with significantly lower representation by the species known to cause more precarious arrangements. Interestingly, analysis of fatty acid speciation did not reveal higher stability of EXs phospholipids in terms of their poly-unsaturation. The molecular species of PC showed the prominence of stearic, and readily oxidizable linoleic and arachidonic fatty acids (18:0/18:2 and 18:0/20:4 species) in EXs (Supplemental Fig. 1).
Fig 3. Lipidomic analysis of phospholipids in trophoblastic EVs.
The relative fraction of major phospholipids in trophoblastic ABs, MVs, and EXs, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylglycerol (PG), cardiolipin (CL), phosphatidic acid (PA), bis-monoacyl-glycerophosphate (BMP), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE) and sphingomyelin (Sph) species. The lower part of the panel shows the fraction of the depicted phospholipids, defined by their general properties.
The miRNA landscape of trophoblastic EVs
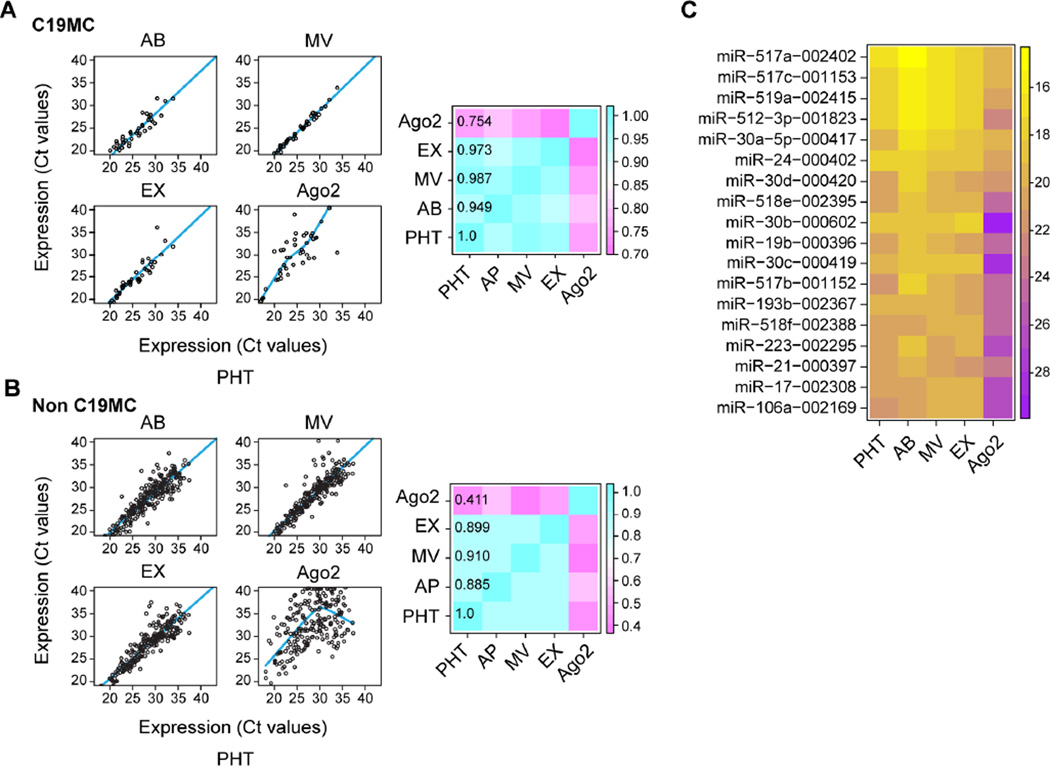
We recently found that C19MC miRNAs, which are largely placenta-specific, are packaged within trophoblastic EXs [39]. To assess the miRNA content of all three main trophoblastic EVs, we used TaqMan card–based RT-qPCR to profile miRNA in trophoblastic EVs. We categorized the 735 mature miRNAs into two subgroups, a C19MC subgroup and a non-C19MC subgroup, and compared their expression in the three EV types to miRNA in the PHT cells from which the EVs were derived and the OptiPrep fraction containing extracellular Ago2 protein. There was a high correlation in the expression of C19MC and non-C19MC miRNA among the three EVs in PHT cells (Fig. 4A–B, right and left panels). In contrast, extracellular, non-vesicular, Ago2-containing miRNAs displayed a significantly different profile, with a lower level of correlation between PHT cells and protein-bound miRNAs (Fig. 4A–B). In addition, compared to the parental PHT cells, the majority of miRNAs in the Ago2 fraction showed significantly decreased expression levels. Together, our data indicate that the three trophoblastic EVs and PHT cells have comparable miRNA contents, distinct from that of the Ago2-positive fraction (Fig. 4C).
Fig. 4. The miRNA landscape of trophoblastic EVs and extracellular Ago2.
(A–B) Scatter plots of C19MC (A) and non-C19MC (B) miRNA profiles, comparing miRNA in each of the EVs and the Ago-2 bound fraction to the miRNA landscape of PHT cells. The heatmap depicts the correlation matrix of miRNA profiles, including three trophoblastic EVs, extracellular Ago2 population, and parental PHT cells. The regression lines for AP, Exo, and MV were generated by ordinary least square linear regression and by locally weighed scatterplot smoothing (LOWESS) for Ago2. (C) Heatmap of the top 20 miRNAs, ascending by the mean Ct values of PHT cells, ABs, MVs, EXs, and extracellular Ago2 population. Trophoblastic EVs and Ago2 population were isolated from three independent PHT cells. Statistical significance was analyzed as described in Methods.
Characterization of EVs’ Proteomics
To examine the repertoire of proteins that characterize the different types of trophoblastic EVs, we extracted proteins from purified trophoblastic EVs and performed mass spectrometry–based proteomics. In total, we identified 1684 proteins in trophoblastic EVs. The list of trophoblastic EV proteins has been deposited in the Data and Specimen Hub (DASH) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We found that the majority of proteins were similar between ABs and MVs. Distinctively, trophoblastic EXs were enriched for surface proteins, such as integrins α6β4 (Table 1) and tetraspanin family members CD9, CD63, and CD81, which characterize non-trophoblastic EXs. Consistent with their early endosome origin and processing within multivesicular bodies, components of the endosomal complexes, known as Endosomal Sorting Complex Required for Transport (ESCRT) [52], including Alix, TSG101, CHMP2A, and CHMP3, were also expressed in trophoblastic EXs. Interestingly, syndecan-1 and its binding partner, syntenin-1, two recently identified EX proteins in breast cancer cell line MCF-7 [48,49], are expressed in trophoblastic EXs. Collectively, these results suggest that trophoblastic EXs share a repertoire of proteins representative of common EX proteins from other cell types. In contrast, and consistent with previous reports [8,12,53,54], we found that four trophoblast-specific proteins, CD276, ERVFRD-1, ERVW-1, and placental alkaline phosphatase (PLAP), were also expressed in trophoblastic EXs. In total, we identified 63 proteins that were exclusively expressed in trophoblastic EXs, but not in the other trophoblastic EVs. Among the 63 EX-exclusive proteins, 38 were identified as harboring membrane-spanning domains on the basis of Ingenuity pathway analysis and the LOCATE database [55], which categorizes protein localization patterns.
Table 1.
Top 10 proteins enriched in ABs, MVs, and EXs, derived from PHT cells
| AB | MV | EX |
|---|---|---|
| Myosin-9/10 | Myosin-9/10 | Integrin beta-4 |
| Dynein 1 heavy chain 1 | Filamin A/B | Integrin alpha-6 |
| Plectin isoform 4 | Plectin isoform 4 | Annexin A2/6 |
| Filamin A/B | Dynein 1 heavy chain 1 | Syntenin-1 |
| Talin-1 | Ras GTPase-activating-like protein IQGAP1 |
Tumor necrosis factor alpha-induced protein 3 |
| Desmoplakin | Talin-1 | Catenin alpha-1 |
| Ras GTPase-activating-like protein IQGAP1 |
Alpha-actinin | Calpain-6 |
| Fatty acid synthase | Desmoplakin | Choline transporter-like protein 2 |
| Alpha-actinin | Annexin A2 | Placental Alkaline phosphatase |
| Spectrin alpha chain | Fatty acid synthase | ADAM10 |
Comparison of antiviral activity of three trophoblastic EVs
We previously showed that trophoblastic EXs could confer viral resistance to non-trophoblastic cells [7,18] and that this effect could be recapitulated, at least in part, by C19MC miRNAs that are packaged within trophoblastic EXs. Because the miRNA cargo of the three trophoblastic EVs was similar, we sought to compare the antiviral properties of the three EVs, using an assay similar to that we previously used, based on assessment of the antiviral effect of PHT EXs on viral infection in non-placental U2OS cells [18,56]. We found that trophoblastic EXs markedly inhibited the production of VSV viral RNA, a measure of viral replication, by approximately 75% (Fig. 5). MVs also attenuated VSV infection in U2OS cells, but to a lesser, statistically insignificant degree. The effect of ABs on viral infection was insignificant. Taken together, these data indicate that, among trophoblastic EVs, EXs exhibit the most potent antiviral activity.
Fig. 5. The effect of trophoblastic EVs on VSV replication in U2OS cells.
Determination of viral replication, assessed by the production of viral RNA using RT-qPCR, as described in Methods. The viral infection assays were repeated using trophoblastic EVs derived from PHT cell prepared from three different placentas. Analysis was performed using one-way ANOVA (Prism 6.0 software). The asterisks indicate statistical significance. * denotes p<0.05 compared to each of the other paradigms.
Comparison of antiviral activity of EXs derived from non-pregnant women and pregnant women
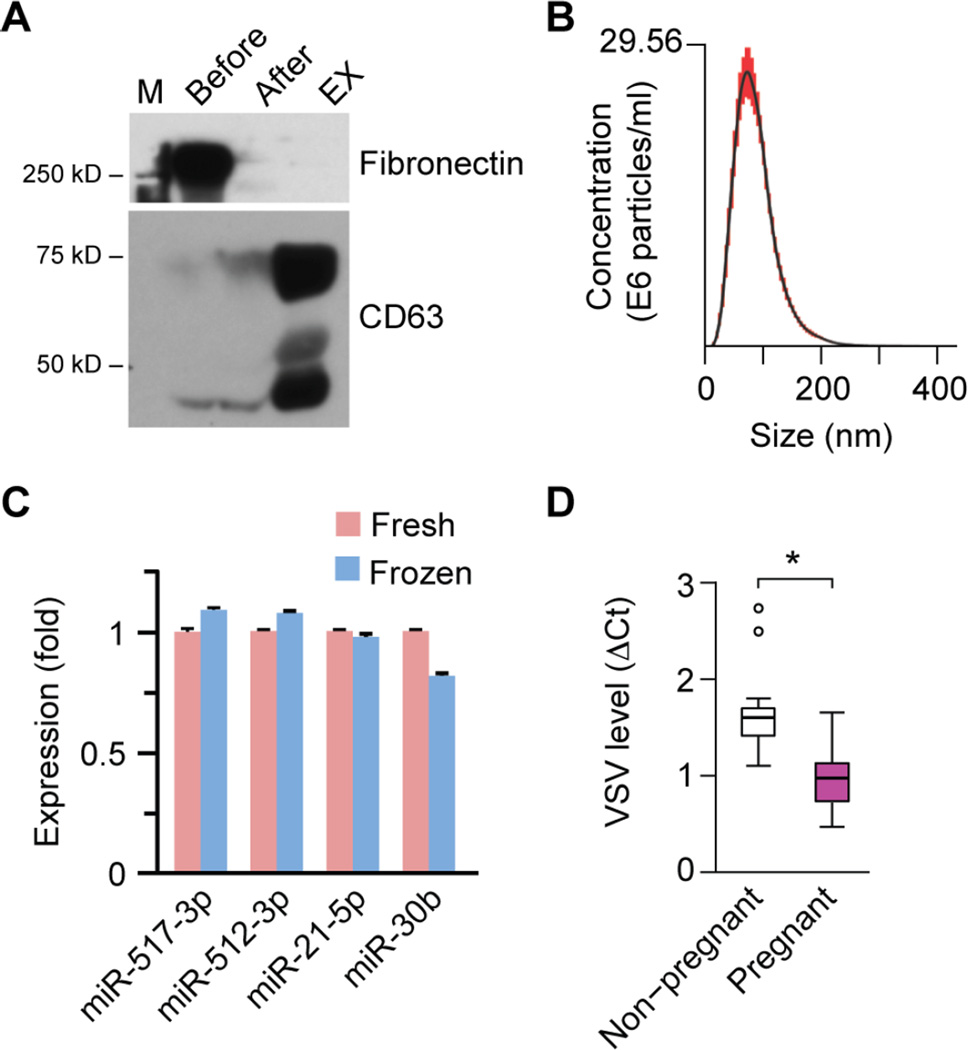
Given the ability of trophoblastic EXs to restrict viral replication in vitro, we posited that EXs purified from the plasma of pregnant women might exhibit an antiviral activity when compared to EXs purified from non-pregnant women. To enhance the purity of EXs derived from plasma, which unlike PHT CM, also contains abundant proteins, such as fibronectin [41,42], we added to our purification methods an additional step in which we used a gelatin-agarose gravity column to specifically remove fibronectin. As shown in Fig. 6A, this additional step led to a marked reduction in plasma fibronectin while retaining the expression of CD63, a prototypical exosomal protein. Notably, for these experiments we used plasma samples that were frozen (−80°C) prior to assay. We therefore verified that EX size distribution and the expression of representative exosomal miRNAs were not influenced by freezing of the plasma samples (Fig. 6B–C), as also shown by others [15]. Assessing the effect of plasma EXs (n=7 for each group) on viral infection, we found that, when compared to EXs from non-pregnant women’s plasma, EXs from pregnant women’s plasma decreased VSV infection of U2OS cells by 20–70% (Fig. 6D).
Fig 6. The effect of EXs derived from plasma of pregnant women on VSV replication in U2OS cells.
(A) A western blotting analysis depicting the removal of fibronectin, performed as a part of gelatin-agarose chromatography for purification of plasma EXs. The human plasma EXs express CD63, a canonical exosome marker. (B) NTA of purified human plasma exosomes, showing the representative distribution of exosomes in the nanometer size range (30~200 nm). (C) The expression of several representative miRNAs in fresh versus frozen plasma (n=3). None of the differences were significant. (D) The effect of plasma EXs on viral replication in U2OS cells, comparing EXs from plasma of pregnant women to EXs from plasma of non-pregnant women. Viral infection was assessed by qPCR measurement of VSV RNA levels (n=7 for each group). Linear mixed effect model was employed to evaluate the statistical difference of plasma EXs of pregnant versus non-pregnant women. *denotes p<0.05.
DISCUSSION
Proper characterization of EV biology requires a well-validated and reproducible vesicle isolation system [57,58]. Inadequate EVs separation that solely relied on differential centrifugation and/or polymer-mediated precipitation might obscure the exact function of individual EVs, likely because of an inconsistent mixture of EVs or co-precipitation of non-vesicular proteins [13,59,60]. These issues are particularly relevant for EX isolation, which has been improved by the use of sucrose or OptiPrep gradient [12,61–63]. We chose to utilize OptiPrep rather than sucrose gradient during ultracentrifugation because of its enhanced resolution [61,64,65]. Furthermore, by loading our crude EX samples at the bottom of the OptiPrep gradient, rather than overlaying them at the top, we improved the separation of non-vesicular proteins, which remained at the bottom, higher density OptiPrep layer. We showed that non-vesicular proteins, such as Ago2 [30], were clearly separated from the EX fractions. We also confirmed the morphology and size distribution of three trophoblastic EVs and thus demonstrated that we were able to isolate the three major trophoblastic EVs.
The diversity membrane lipid composition is known to affect its biophysical properties, and provides a platform for receptor-mediated cellular signaling. Our mass spectrometry screen of phospholipids in trophoblastic EVs revealed that, while the relative proportion of several classes of phospholipid was similar among the three vesicle subtypes, EXs express a higher relative fraction of phosphatidylcholine (PC) and a lower relative fraction of phosphatidylethanolamine (PE) compared to ABs and MVs. While PC represents the major component of stable membrane bilayers, PE typically forms non-bilayer membrane arrangements. PE is also known as a critical factor for fusion [66,67]. Our classification of phospholipids based on their propensity to contribute to the organization of the membrane s bilayer revealed the lowest fraction of fusogenic phospholipids (PE, PA, BMP, LPC, LPE) and the highest fraction of non-charged zwitterionic phospholipids in EXs. Interestingly, PS, which is phospholipid is externalized to the surface of apoptotic cells and during cell fusion, was relatively lower in EXs. Taken together, these data predict that EXs would exhibit greater stability and fewer fusogenic properties than MVs and ABs.
We found that trophoblastic EVs share similar miRNA content, which is comparable to the level in the EV-producing PHT cells. In contrast, the miRNA levels in the non-vesicular, Ago2-associated fraction were different. These findings are intriguing, as the mechanisms of miRNA packaging in EVs remain largely unknown and likely respond to cell-intrinsic and/or -extrinsic signals. For example, stimulation of macrophages with IL-4 causes relocation of a subset of miRNAs between cytoplasmic P-bodies and multivesicular bodies, which will impact EX miRNA content [68]. The similarity among miRNA levels in PHT cells and the EVs does not support the possibility of selective miRNA loading into EVs, yet general conditions that affect cellular homeostasis, such as hypoxia, might impact the EV miRNA landscape [40,69–72]. It is also possible that miRNAs are modified within EVs. For example, a subset of miRNAs that undergo a post-transcriptional uridylation at the 3’ end are highly represented in human urine and B cell– derived EXs, but not in the cognate producer cells [73]. In addition, the RNA-binding protein hnRNPA2B1 in primary T-cells directly recognizes certain miRNAs, such as miR-125a-3p, miR-198, and miR-601, harboring a consensus 4-nucleotide sequence and recruits them to EXs [74]. Collectively, these data suggest that different cell types are capable of sorting discrete cellular miRNAs into EVs. Interestingly, our data point to the presence of hnRNP family members such as hnRNPA2B1 and hnRNPD within trophoblastic EVs and thus suggest that they play a role in miRNA packaging in PHT cells [74]. Lastly, the similar repertoire of miRNA in PHT cells and EVs suggests that trophoblastic EVs, which shield miRNA against degradation [75,76], are better than total plasma or protein-bound plasma miRNA for monitoring trophoblastic miRNA in health and disease [34–36,77,78].
Our observation that different EVs contain a distinct repertoire of proteins suggests that these proteins may influence EV function [79]. Thus far, we have uncovered one relevant function of trophoblastic EVs, an antiviral activity, which was most potent in EXs isolated from PHT conditioned medium. This antiviral effect of EXs was recapitulated using EXs derived from the plasma of pregnant women, but not with those from non-pregnant controls. Because plasma EXs represent a mixture of EXs derived from multiple tissues, not only trophoblasts, it is not surprising that the antiviral effect was markedly weaker than that of isolated EXs. Given that C19MC miRNA are trophoblast-specific and are rarely expressed in other maternal organs, we estimated the percentage of trophoblastic EXs present in whole plasma from the level of C19MC miRNA in PHT-derived EXs relative to that in plasma-derived EXs and found this to be approximately 5%, thus accounting for the weaker antiviral effect of EXs in vivo. As the most potent antiviral effect was observed with EXs, our data suggest that the delivery and/or processing of distinct EV populations may directly contribute to their antiviral properties.
While the function of distinct exosomal proteins remains to be established, it is possible that some of the proteins, and specifically those localized at the vesicle surface, may play a role in vesicle targeting to local or distant cells. Consistent with this possibility, distinctive combinations of integrin subunits, such as α6β4 and αVβ5, were recently shown to target tumor EX metastasis to the lung and liver, respectively [80]. It is also possible the EV cargo is influenced by disease state, as recently suggested in the context of preeclampsia [81–83], as well as the stage of pregnancy and the methodology used for analysis. [84]. Lastly, we note that EV subtypes might not be homogeneous [85]. This underscores the need for further characterization of EV diversity, including a detailed comparison of EVs derived from cytotrophoblasts and syncytiotrophoblasts. Akin to data derived from single cell analysis [86–89], belter definition of EV sub-populations may illuminate EV function in the context of the normal physiology of pregnancy as well as in various disease states and lead to the use of EVs as disease biomarkers.
Supplementary Material
HIGHLIGHTS.
Gradient ultracentrifugation is a useful tool to separate extracellular vesicle subtypes from medium or blood.
When compared to other vesicles, trophoblastic exosomes have a similar repertoire of miRNAs but exhibit a distinct composition of phospholipids and proteins.
Among the three trophoblastic extracellular vesicles, exosomes have the most potent antiviral activity.
Exosomes in blood from pregnant women exhibit a higher antiviral activity compared to exosomes from non-pregnant women.
Acknowledgments
We thank Elena Sadovsky and Judy Ziegler for technical assistance, Lori Rideout for assistance with manuscript preparation, and Bruce Campbell for editing. We also thank Drs. Xuemei Zeng and Nathan Yates from the Biomedical Mass Spectrometry Center of the University of Pittsburgh, which is supported, in part, by NIH P30-CA047904. This work was supported by grants from the 25 Club of Magee-Womens Hospital (YS), the March of Dimes Prematurity Research Center at the University of Pennsylvania (YS), and NIH grants R01AI081759 (CBC), R01HD075665 (CBC and YS), P01HD069316 (YS) and R01HD086325 (YS). In addition, CBC is supported by Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Awards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mincheva-Nilsson L, Baranov V. Placenta-derived exosomes and syncytiotrophoblast microparticles and their role in human reproduction: immune modulation for pregnancy success. Am. J. Reprod. Immunol. 2014;72:440–457. doi: 10.1111/aji.12311. [DOI] [PubMed] [Google Scholar]
- 2.Record M. Intercellular communication by exosomes in placenta: a possible role in cell fusion? Placenta. 2014;35:297–302. doi: 10.1016/j.placenta.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell MD, Peiris HN, Kobayashi M, et al. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015;213:S173–S181. doi: 10.1016/j.ajog.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Tong M, Chamley LW. Placental extracellular vesicles and feto-maternal communication. Cold Spring Harb. Perspect. Med. 2015;5:a023028. doi: 10.1101/cshperspect.a023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp AN, Heazell AE, Crocker IP, et al. Placental apoptosis in health and disease. Am. J. Reprod. Immunol. 2010;64:159–169. doi: 10.1111/j.1600-0897.2010.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang Y, Mouillet JF, Coyne CB, et al. Review: placenta-specific microRNAs in exosomes - good things come in nano-packages. Placenta. 2014;35(Suppl):S69–S73. doi: 10.1016/j.placenta.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kshirsagar SK, Alam SM, Jasti S, et al. Immunomodulatory molecules are released from the first trimester and term placenta via exosomes. Placenta. 2012;33:982–990. doi: 10.1016/j.placenta.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenqvist AC, Nagaeva O, Baranov V, et al. Exosomes secreted by human placenta carry functional Fas ligand and TRAIL molecules and convey apoptosis in activated immune cells, suggesting exosome-mediated immune privilege of the fetus. J. Immunol. 2013;191:5515–5523. doi: 10.4049/jimmunol.1301885. [DOI] [PubMed] [Google Scholar]
- 10.Mincheva-Nilsson L, Baranov V. The role of placental exosomes in reproduction. Am. J. Reprod. Immunol. 2010;63:520–533. doi: 10.1111/j.1600-0897.2010.00822.x. [DOI] [PubMed] [Google Scholar]
- 11.Hedlund M, Stenqvist AC, Nagaeva O, et al. Human placenta expresses and secretes NKG2D ligands via exosomes that down-modulate the cognate receptor expression: evidence for immunosuppressive function. J. Immunol. 2009;183:340–351. doi: 10.4049/jimmunol.0803477. [DOI] [PubMed] [Google Scholar]
- 12.Tolosa JM, Schjenken JE, Clifton VL, et al. The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta. 2012;33:933–941. doi: 10.1016/j.placenta.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Alegre E, Rebmann V, Lemaoult J, et al. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur. J. Immunol. 2013;43:1933–1939. doi: 10.1002/eji.201343318. [DOI] [PubMed] [Google Scholar]
- 14.Gupta AK, Rusterholz C, Huppertz B, et al. A comparative study of the effect of three different syncytiotrophoblast micro-particles preparations on endothelial cells. Placenta. 2005;26:59–66. doi: 10.1016/j.placenta.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Salomon C, Torres MJ, Kobayashi M, et al. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS One. 2014;9:e98667. doi: 10.1371/journal.pone.0098667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salomon C, Ryan J, Sobrevia L, et al. Exosomal signaling during hypoxia mediates microvascular endothelial cell migration and vasculogenesis. PLoS One. 2013;8:e68451. doi: 10.1371/journal.pone.0068451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon C, Kobayashi M, Ashman K, et al. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS One. 2013;8:e79636. doi: 10.1371/journal.pone.0079636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delorme-Axford E, Donker RB, Mouillet JF, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. USA. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redman CW, Tannetta DS, Dragovic RA, et al. Review: Does size matter? Placental debris and the pathophysiology of pre-eclampsia. Placenta. 2012;33(Suppl):S48–S54. doi: 10.1016/j.placenta.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Goswami D, Tannetta DS, Magee LA, et al. Excess syncytiotrophoblast microparticle shedding is a feature of early-onset pre-eclampsia, but not normotensive intrauterine growth restriction. Placenta. 2006;27:56–61. doi: 10.1016/j.placenta.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Redman CW, Sargent IL. Circulating microparticles in normal pregnancy and pre-eclampsia. Placenta. 2008;29(Suppl A):S73–S77. doi: 10.1016/j.placenta.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Knight M, Redman CW, Linton EA, et al. Shedding of syncytiotrophoblast microvilli into the maternal circulation in pre-eclamptic pregnancies. Br. J. Obstet. Gynaecol. 1998;105:632–640. doi: 10.1111/j.1471-0528.1998.tb10178.x. [DOI] [PubMed] [Google Scholar]
- 23.Guller S, Tang Z, Ma YY, et al. Protein composition of microparticles shed from human placenta during placental perfusion: Potential role in angiogenesis and fibrinolysis in preeclampsia. Placenta. 2011;32:63–69. doi: 10.1016/j.placenta.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner C, Tannetta DS, Simms CA, et al. Syncytiotrophoblast microvesicles released from pre-eclampsia placentae exhibit increased tissue factor activity. PLoS One. 2011;6:e26313. doi: 10.1371/journal.pone.0026313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Post JA, Lok CA, Boer K, et al. The functions of microparticles in pre-eclampsia. Semin. Thromb. Hemost. 2011;37:146–152. doi: 10.1055/s-0030-1270342. [DOI] [PubMed] [Google Scholar]
- 26.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, et al. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 2013;97:51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Wu L, Zhou H, Lin H, et al. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143:389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 36.Miura K, Miura S, Yamasaki K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010;56:1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 37.Sadovsky Y, Mouillet JF, Ouyang Y, et al. The function of trophomiRs and other MicroRNAs in the human placenta. Cold Spring Harb. Perspect. Med. 2015;5:a023036. doi: 10.1101/cshperspect.a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo SS, Ishibashi O, Ishikawa G, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 39.Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012;18:417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouillet JF, Chu T, Nelson DM, et al. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuento M, Vaheri A. Purification of fibronectin from human plasma by affinity chromatography under non-denaturing conditions. Biochem. J. 1979;183:331–337. doi: 10.1042/bj1830331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speziale P, Visai L, Rindi S, et al. Purification of human plasma fibronectin using immobilized gelatin and Arg affinity chromatography. Nat. Protoc. 2008;3:525–533. doi: 10.1038/nprot.2008.12. [DOI] [PubMed] [Google Scholar]
- 43.Mishima T, Miner JH, Morizane M, et al. The expression and function of fatty acid transport protein-2 and −4 in the murine placenta. PLoS One. 2011;6:e25865. doi: 10.1371/journal.pone.0025865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dvinge H, Bertone P. HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics. 2009;25:3325–3326. doi: 10.1093/bioinformatics/btp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth GK. In: limma: Linear models for microarray data. Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. New York, NY: Springer New York; 2005. pp. 397–420. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B (Methodological) 1995;57:289–300. [Google Scholar]
- 47.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 48.Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 49.Ghossoub R, Lembo F, Rubio A, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014;5:3477. doi: 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- 50.Escola JM, Kleijmeer MJ, Stoorvogel W, et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 51.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 52.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 53.Vargas A, Zhou S, Ethier-Chiasson M, et al. Syncytin proteins incorporated in placenta exosomes are important for cell uptake and show variation in abundance in serum exosomes from patients with preeclampsia. FASEB J. 2014;28:3703–3719. doi: 10.1096/fj.13-239053. [DOI] [PubMed] [Google Scholar]
- 54.Dragovic RA, Collett GP, Hole P, et al. Isolation of syncytiotrophoblast microvesicles and exosomes and their characterisation by multicolour flow cytometry and fluorescence Nanoparticle Tracking Analysis. Methods. 2015;87:64–74. doi: 10.1016/j.ymeth.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sprenger J, Lynn Fink J, Karunaratne S, et al. LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res. 2008;36:D230–D233. doi: 10.1093/nar/gkm950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bayer A, Delorme-Axford E, Sleigher C, et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am. J. Obstet. Gynecol. 2015;212:71.e71–71.e78. doi: 10.1016/j.ajog.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tkach M, Thery C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 59.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 60.Umezu T, Ohyashiki K, Kuroda M, et al. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 61.Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Cvjetkovic A, Lotvall J, Lasser C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3 Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 64.Cantin R, Diou J, Belanger D, et al. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J. Immunol. Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 65.Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van den Brink-van der Laan E, Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Kasson PM, Pande VS. Control of membrane fusion mechanism by lipid composition: predictions from ensemble molecular dynamics. PLoS Comput. Biol. 2007;3:e220. doi: 10.1371/journal.pcbi.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Squadrito ML, Baer C, Burdet F, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 69.Donker RB, Mouillet JF, Nelson DM, et al. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol. Hum. Reprod. 2007;13:273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- 70.Wu C, So J, Davis-Dusenbery BN, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen J, Xia W, Khotskaya YB, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rampersad R, Nelson DM. Trophoblast biology, responses to hypoxia and placental dysfunction in preeclampsia. Front. Biosci. 2007;12:2447–2456. doi: 10.2741/2246. [DOI] [PubMed] [Google Scholar]
- 73.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 74.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng L, Sharples RA, Scicluna BJ, et al. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koberle V, Pleli T, Schmithals C, et al. Differential stability of cell-free circulatingmicroRNAs: implications for their utilization as biomarkers. PLoS One. 2013;8:e75184. doi: 10.1371/journal.pone.0075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Escudero CA, Herlitz K, Troncoso F, et al. Role of extracellular vesicles and microRNAs on dysfunctional angiogenesis during preeclamptic pregnancies. Front. Physiol. 2016;7:98. doi: 10.3389/fphys.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mouillet JF, Ouyang Y, Coyne CB, et al. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015;213:S163–S172. doi: 10.1016/j.ajog.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tong M, Kleffmann T, Pradhan S, et al. Proteomic characterization of macro-, micro-and nano-extracellular vesicles derived from the same first trimester placenta: relevance for feto-maternal communication. Hum. Reprod. 2016;31:687–699. doi: 10.1093/humrep/dew004. [DOI] [PubMed] [Google Scholar]
- 80.Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baig S, Kothandaraman N, Manikandan J, et al. Proteomic analysis of human placental syncytiotrophoblast microvesicles in preeclampsia. Clin. Proteomics. 2014;11:40. doi: 10.1186/1559-0275-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tannetta D, Mackeen M, Kessler B, et al. OS045. Multi-dimensional protein identification technology analysis of syncytiotrophoblast vesicles released from perfused preeclampsia placentas. Pregnancy Hypertens. 2012;2:201–202. doi: 10.1016/j.preghy.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 83.Li H, Han L, Yang Z, et al. Differential Proteomic analysis of syncytiotrophoblast extracellular vesicles from early-onset severe preeclampsia, using 8-Plex iTRAQ labeling coupled with 2D nano LC-MS/MS. Cell. Physiol. Biochem. 2015;36:1116–1130. doi: 10.1159/000430283. [DOI] [PubMed] [Google Scholar]
- 84.Familari M, Cronqvist T, Masoumi Z, et al. Placenta-derived extracellular vesicles: their cargo and possible functions. Reprod. Fertil. Dev. 2015 doi: 10.1071/RD15143. [DOI] [PubMed] [Google Scholar]
- 85.Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deng Q, Ramskold D, Reinius B, et al. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 87.Shalek AK, Satija R, Shuga J, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 89.Treutlein B, Brownfield DG, Wu AR, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.