SUMMARY
For many years, a connection between circadian clocks and cancer has been postulated. Here, we describe an unexpected function for the circadian repressor CRY2 as a component of an FBXL3-containing E3 ligase that recruits T58-phosphorylated c-MYC for ubiquitylation. c-MYC is a critical regulator of cell proliferation; T58 is central in a phosphodegron long recognized as a hotspot for mutation in cancer. This site is also targeted by FBXW7, though the full machinery responsible for its turnover has remained obscure. CRY1 cannot substitute for CRY2 in promoting c-MYC degradation; their unique functions may explain prior conflicting reports that have fueled uncertainty about the relationship between clocks and cancer. Thus, we demonstrate that c-MYC is a target of CRY2-dependent protein turnover, suggesting a molecular mechanism for circadian control of cell growth and a new paradigm for circadian protein degradation.
eTOC blurb
Circadian disruption increases the risk of many types of cancer. Huber et al. demonstrate that the circadian clock protein CRY2 recruits T58-phosphorylated c-MYC to SCFFBXL3, thus promoting its ubiquitination and degradation. This unexpected function of CRY2 may contribute to circadian protection from tumorigenesis.

INTRODUCTION
Mice harboring genetically disrupted clock function (Fu et al., 2002; Geyfman et al., 2012; Janich et al., 2011; Lee et al., 2010; Ozturk et al., 2009) or subjected to experimental jet lag (Papagiannakopoulos et al., 2016; Van Dycke et al., 2015) exhibit altered rates of tumor formation. Analysis of epidemiological studies spurred the World Health Organization to designate circadian disruption as a probable carcinogen (Straif et al., 2007). While several molecular connections have been suggested (Gaddameedhi et al., 2011; Gotoh et al., 2015; Gotoh et al., 2014; Papp et al., 2015; Unsal-Kacmaz et al., 2007; Unsal-Kacmaz et al., 2005), the relationship between clocks and cancer is not well understood and remains controversial (West and Bechtold, 2015). Mammalian clocks involve a transcriptional feedback loop (Partch et al., 2014): CLOCK and BMAL1 drive expression of period (PER1-3) and cryptochrome (CRY1,2) repressors, which inhibit CLOCK:BMAL1, resulting in oscillating transcription of thousands of genes (Panda et al., 2002; Storch et al., 2002). Ubiquitylation of CRY1/2 by SCFFBXL3 is important for setting clock speed (Busino et al., 2007; Godinho et al., 2007; Siepka et al., 2007).
c-MYC plays a critical role in cell proliferation, and its levels are tightly controlled by regulated protein degradation (Farrell and Sears, 2014). When c-MYC is phosphorylated on T58, it is targeted by SCFFBXW7 for ubiquitylation and proteasomal degradation (Popov et al., 2007; Welcker et al., 2004; Yada et al., 2004). However, FBXW7 preferentially interacts with doubly phosphorylated substrates (Hao et al., 2007; Welcker and Clurman, 2008) and in some contexts, mutation of T58 to a non-phosphorylatable residue stabilizes c-MYC more than ablation of SCFFBXW7 does (Chakraborty and Tansey, 2009; Popov et al., 2007; Salghetti et al., 1999). It has therefore been suggested that one or more additional E3 ligase(s) also stimulate(s) the proteolysis of c-MYC phosphorylated on T58 (Thomas and Tansey, 2011). Understanding the mechanisms leading to destruction of T58-phosphorylated c-MYC is important since the sequence surrounding c-MYC T58 has long been recognized as a hotspot of mutations in human cancer (Bhatia et al., 1993; Bhatia et al., 1994). Moreover, disrupted turnover of singly T58-phosphorylated MYC is associated with MYC stabilization and tumorigenesis (Malempati et al., 2006). Here, we demonstrate that CRY2 and SCFFBXL3 drive proteolytic turnover of T58-phosphorylated c-MYC via binding of phospho-T58 to CRY2 near its binding interface with FBXL3. These findings suggest that CRY2-driven cycles of c-MYC turnover represent a previously unappreciated mode of clock output that impacts cancer susceptibility due to circadian disruption.
RESULTS
Loss of Cry2 enhances proliferation and transformation
To examine the roles of CRY1 and CRY2 in growth control, we performed proliferation and transformation assays using five independently isolated sets of primary mouse fibroblasts derived from wildtype, Cry1−/−, and Cry2−/− littermates. We infected three independently derived primary mouse embryonic fibroblast (MEF) cell lines and two fibroblast cell lines derived from adult skin biopsies of each genotype with viruses expressing the oncogenes c-MYC or HRASV12, or shRNA targeting the tumor suppressor P53. To avoid selection of clonal lines that may not accurately reflect the effects of Cry1/2 deletion, we characterized heterogeneous pools of virally transduced cells subjected to antibiotic selection for less than two weeks. In all contexts, Cry2 deficiency enhanced proliferation and colony formation (Figures 1A–G), indicating surprisingly widespread cooperative effects with multiple oncogenic changes. Similarly, wildtype fibroblasts stably expressing shRNA targeting Cry2 were more highly proliferative than those expressing control sequences or shRNA targeting Cry1 (Figure 1B). Mostly, loss of Cry1 had little or no effect on proliferation or transformation; however, in the context of P53 depletion, cells lacking Cry1 were less sensitive to transformation (Figures 1D and 1G). This is consistent with the increased survival of Cry1/2-deficient mice in a P53−/− background (Ozturk et al., 2009) and suggests that CRY1- and CRY2-specific functions may explain the different effects on tumor formation in previous studies of tumorigenesis in Cry1−/−;Cry2−/− mice (Fu and Kettner, 2013; Lee et al., 2010). Loss of Cry2 was not sufficient to transform primary cells and Cry2−/− cells did not robustly form colonies in soft agar with a single oncogenic insult (P53 depletion or expression of either HRASV12 or c-MYC; not shown), indicating that CRY2 is not a tumor suppressor per se.
Figure 1. CRY2 deletion enhances cell growth and transformation. See also Figure S1.
Proliferation (A–F) and colony formation in 2-dimensional culture (G) in primary MEFs of indicated genotypes subjected to 3T3 protocol (Spont. Imm.) or stably expressing the indicated plasmids. (H) mRNA (left) and protein (right) measured by immunoblot (IB) or quantitative PCR (qPCR) from MEFs of the indicated genotypes stably overexpressing c-MYC. (I–K) Proteins measured by IB from adult skin fibroblasts (I–J) or MEFs (K) of the indicated genotypes stably expressing the indicated plasmids. (L–N) Endogenous mRNA (L) and proteins (M) detected by qPCR or IB in MEFs of the indicated genotypes and harvested at the indicated times (CT: hours after circadian synchronization). (N) Quantitation of the IB data shown in (M). In (A–G), data represent the mean ± s.d. for triplicate samples (A–F) or a typical result (G) from a representative experiment of at least 3 experiments of each type performed in 3 sets of MEFs derived from littermate animals. Similar results were obtained using 2 sets of adult skin fibroblasts derived from littermate animals.
Loss of Cry2 stabilizes c-MYC
Enhanced proliferation and colony formation of Cry2−/− cells expressing either c-MYC or shRNA targeting P53 suggests that the loss of CRY2 impacts multiple pathways that control cell growth. For example, cooperation with c-MYC indicates a defect in P53 function, while cooperation with P53 depletion could be explained by enhanced MYC activity. In assessing the efficiency of viral oncogene expression, we found that while overexpressed human c-MYC mRNA is somewhat (~2.2-fold) elevated in both Cry1−/− and Cry2−/− cells compared to wildtype controls, c-MYC protein abundance is highly increased specifically in Cry2−/− cells (Figure 1H), suggesting that the loss of Cry2 alters c-MYC post-transcriptional regulation. Upon further examination, we found that overexpression of human c-MYC repressed the endogenous c-Myc transcript in wildtype and Cry2−/− fibroblasts as expected (Figure S1A). Strikingly, overexpressed human c-MYC and especially endogenous mouse c-MYC protein levels are elevated in Cry2−/− cells compared to controls (Figure 1I). Since elevated c-MYC protein could contribute to the enhanced colony formation observed in Cry2−/− cells upon depletion of P53 or overexpression of mutant RAS, we measured endogenous c-MYC protein in those cells as well as in cells expressing shRNA targeting P19Arf (an upstream regulator of P53). In every case, c-MYC is increased in Cry2−/− cells compared to controls (Figures 1J and 1K). High levels of c-MYC induce apoptosis via P53, so defective P53 signaling can enable MYC protein accumulation. Increased c-MYC in Cry2−/− cells expressing shRNA targeting P53 or P19Arf cannot be attributed to such a defect. Taken together, these data indicate that CRY2 regulates c-MYC protein levels.
In primary MEFs following the induction of synchronized circadian rhythms by dexamethasone (Balsalobre et al., 2000), endogenous c-Myc mRNA is rhythmically expressed, consistent with an earlier report (Fu et al., 2002), and its peak is ~1.7-fold higher in Cry2−/− cells (Figure 1L). Strikingly, c-MYC protein exhibits a high amplitude circadian rhythm, and peak c-MYC protein is much higher in Cry2−/− compared to wildtype cells (Figures 1M, 1N, S1B). Thus, c-MYC protein levels are elevated in Cry2-deficient cells regardless of the endogenous or viral promoter elements controlling its transcription, strongly suggesting that CRY2 modulates post-transcriptional regulation of c-MYC. Metabolic tracer analysis of primary MEFs revealed changes consistent with enhanced MYC and increased proliferative and transformative capacity in Cry2−/− primary MEFs (Figures S1C–G). Elevated levels of endogenous c-MYC protein in Cry2−/− cells could explain the observed cooperativity between CRY2 loss and HRASV12 expression or P53 depletion; other changes in Cry2−/− cells probably contribute to the increase in transformation by c-MYC in Cry2−/− cells compared to wildtype controls. Here, we focus on understanding how CRY2 regulates c-MYC protein levels.
Post-transcriptionally enhanced c-MYC protein levels could reflect increased production and/or decreased decay. The incorporation of 35S-methionine into c-MYC was slightly elevated in Cry2−/− cells compared to controls, similar to the ~2-fold change in c-Myc mRNA, suggesting that translation of c-Myc is not specifically increased in the absence of CRY2 (Figure S2A). To determine whether c-MYC protein half-life is increased in Cry2−/− cells, we examined c-MYC protein turnover after addition of the translation inhibitor cycloheximide (CHX) in wildtype and Cry2−/− embryonic and adult fibroblasts (Figures 2A–C, S2B, and S2C). Consistent with previous studies, c-MYC has a very short half-life in wildtype cells as evidenced by the rapid turnover within 15–20 minutes. In Cry2−/− cells, c-MYC is stabilized in the presence of CHX. Notably, re-introducing CRY2 in these cells decreased the half-life of endogenous c-MYC (Figures 2B, 2C, S2B, and S2C). Thus, CRY2 regulates the half-life of endogenous c-MYC.
Figure 2. CRY2 deletion increases c-MYC stability. See also Figure S2, Table S1 and Table S2.
(A–C) Endogenous proteins detected by IB in MEFs of the indicated genotypes expressing the indicated plasmids and harvested at the indicated times (CHX: minutes after cycloheximide). (B) Samples from time 0 for all cell lines shown in (A) loaded together. (D,E) Transcripts measured by qPCR in primary MEFs of the indicated genotypes at the indicated times after synchronization of circadian rhythms with dexamethasone treatment. Data represent the mean ± s.d. for three biological replicates each measured in triplicate. (F) Heat-map from RNA sequencing in primary MEFs at the indicated times after circadian synchronization. Colors represent high (red) to low (blue) expression. Gene names and expression values are provided in Supplemental Table S2.
Because CRY2 can repress transcription and targets many unique sites in chromatin independent of other circadian factors (Koike et al., 2012), we hypothesized that it could alter c-MYC stability via transcription of one or more effectors. However, quantitative PCR showed no alterations of transcripts encoding regulators of c-MYC degradation that would explain the observed stabilization of c-MYC (Figure 2D). Deep sequencing of RNA isolated from wildtype and Cry2−/− primary MEFs over a full circadian cycle revealed 865 transcripts that were altered by the loss of CRY2 (FDR <0.01; Supplemental Table S1), but no obvious explanation for c-MYC stabilization. Notably, core circadian transcripts oscillate in Cry2−/− cells (Figure 2E), because CRY1 can support circadian rhythms in the absence of CRY2 (Ukai-Tadenuma et al., 2011). Although prolonged elevation of c-MYC can lead to general amplification of transcription (Lin et al., 2012; Nie et al., 2012), transcripts that are likely to be directly regulated by c-MYC (Sabo et al., 2014; Walz et al., 2014) are overrepresented among those that are significantly altered by deletion of Cry2 (~14% of MYC targets identified in (Sabo et al., 2014) vs ~4% of the genome are altered by Cry2 deletion; Figures S2D and S2E). Furthermore, Gene Set Enrichment Analysis (Subramanian et al., 2007) detected significantly enriched expression of a Hallmark set of c-MYC target genes (Liberzon et al., 2015) in Cry2−/− cells (Figures 2F, S2F, Supplemental Table S2), suggesting that the highly elevated c-MYC protein observed in Cry2−/− cells is transcriptionally active.
CRY2 and FBXL3 cooperatively bind c-MYC
Since transcriptional changes did not appear to explain the greatly increased c-MYC stability in Cry2−/− cells, we investigated other ways that CRY2 could affect c-MYC. Expression of CRY2 did not prevent interaction of c-MYC with MAX, FBXW7 or βTRCP (Figures S3A–C). Surprisingly, overexpressed c-MYC co-purifies with overexpressed CRY2 (Figure 3A) and this interaction requires the N-terminal regulatory domain of c-MYC (Figures 3B, 3C). Since CRY2 associates with the SCF E3 ligase substrate receptor FBXL3, we considered the possibility that SCFFBXL3 acts on c-MYC. Although previous studies have not identified c-MYC in FBXL3-containing protein complexes (Busino et al., 2007), mass spectrometry often produces false negative results, especially for F-box proteins, which stimulate the degradation of their substrate partners (Yumimoto et al., 2012). Proteins of very low abundance like c-MYC are most likely to be missed. Indeed, we found that FBXL3 can interact with c-MYC (Figure 3D), and CRY2 enhances the FBXL3-MYC interaction (Figure 3E). We also observed interactions between CRY2, FBXL3, and c-MYC when all three were produced by in vitro transcription and translation (Figure S3D), suggesting that they interact directly.
Figure 3. c-MYC forms a trimeric complex with CRY2 and FBXL3. See also Figure S3.
(A,B,D,E,H–J) Proteins detected by IB following FLAG, HA, or V5 IP from nuclear and cytoplasmic fractions or whole cell lysates from 293T cells expressing the indicated plasmids. (C) Schematic representation of the wildtype and amino- or carboxy-terminal deletion mutants of c-MYC used in experiments shown in Figures 3B and 3C. (F) Superposition of crystal structures from Protein Data Bank accession numbers 1LDK (SKP1-SKP2-CUL1-RBX1), 2OVR (SKP1-FBW7-CYCEDEGN), 2AST (SKP1-SKP2-CKS1-p27KIP1), and 4I6J (SKP1-FBXL3-CRY2). (G) Proteins detected by IB in whole cell lysates of wildtype MEFs expressing the indicated viral shRNA.
Aligning SKP1 in the crystal structures of SKP1-CRY2-FBXL3 (Xing et al., 2013), SKP1-SKP2-CUL1-RBX1 (Zheng et al., 2002), SKP1-FBXW7-CYCLIN E (Hao et al., 2007), and SKP1-SKP2-CKS1-p27KIP1(Hao et al., 2005) reveals that the orientation of CRY2 relative to FBXL3 overlaps with that of the adaptor CKS1 relative to SKP2/FBXL1 and leaves a large open space between CRY2, FBXL3, and the RBX1 active site, including the position occupied by the CYCLIN E and p27KIP1 degron peptides (Figures 3F and S3E). These findings led us to hypothesize that in addition to being a substrate for SCFFBXL3, CRY2 could act as a co-factor to recruit additional substrates, including c-MYC, to SCFFBXL3. Thus, the observed stabilization of c-MYC in the absence of CRY2 may reflect a loss of SCFFBXL3-mediated ubiquitylation of c-MYC. Importantly, c-MYC protein was greatly increased in fibroblasts expressing either of two shRNAs targeting Fbxl3 (Figures 3G and S3F), similar to what we observed in Cry2-deficient cells and consistent with a role for a CRY2-FBXL3 heterodimer in ubiquitylation of c-MYC.
Our model leads to a prediction that mutations in CRY2 or FBXL3 that reduce their interaction with each other would reduce their interaction with c-MYC. Several point mutations in CRY2 that disrupt its association with FBXL3 without altering interactions with CLOCK, BMAL1 or PER2 have been described (Xing et al., 2013). The interaction of those mutants with c-MYC is abolished (Figure 3H). In addition, mutation of the FBXL3 C-terminal tryptophan, which decreases its interaction with CRY2 (Xing et al., 2013), also reduces the interactions of CRY2 and FBXL3 with c-MYC (Figure 3I). To further examine the ability of CRY2, FBXL3, and c-MYC to form a trimeric complex, we serially immunoprecipitated overexpressed CRY2 and FBXL3 and observed progressive enrichment of c-MYC (Figures 3J and S3G).
CRY2-FBXL3 promotes c-MYC ubiquitylation
We measured ubiquitylation of overexpressed c-MYC in the presence or absence of overexpressed CRY2 and FBXL3 and found that CRY2 and FBXL3 together stimulate c-MYC ubiquitylation (Figures 4A, S4A and S4B). Deletion of the F-box (ΔF) in FBXL3 prevents its association with SKP1 and CUL1 and thus prevents formation of the active SCFFBXL3 complex. Consistent with the observed loss of CUL1 binding, though FBXL3ΔF interacts with CRY2 and c-MYC, it did not promote c-MYC ubiquitylation. Mutation of the FBXL3 C-terminal tryptophan, which inhibits FBXL3-CRY2 interaction, reduced the recruitment of c-MYC to FBXL3 and blocked its ubiquitylation (Figures S4A and S4B). As reported by others (Yumimoto et al., 2013), we found that CRY2 promotes the association of FBXL3 with CUL1 (Figures S4B, S4C), further supporting the importance of CRY2 as a scaffold for formation of a fully active SCFFBXL3 complex to promote ubiquitylation and turnover of c-MYC.
Figure 4. CRY2 is an essential component of SCFFBXL3-driven c-MYC ubiquitylation. See also Figure S4.
Proteins detected by IB following HA IP (A) or in whole cell lysates (B,C,G,H) from 293T cells expressing the indicated plasmids or from primary MEFs of the indicated genotypes expressing the indicated plasmids (I–L) and following treatment with cycloheximide (CHX) for the indicated times (C,G,I,K). (A) Monoclonal antibody (Mono Ab) was used to detect c-MYC (E) Proteins detected by autoradiography after a 35S-Methionine Pulse-Chase from 293T cells expressing the indicated plasmids. (D,F) Quantitation of the IB data (D) or the autoradiography data (F) showed in (C) and (E) respectively. Data represent the mean ± s.d. for triplicate samples. In (D,F) * P<0.05, ** P<0.01 vs. c-MYC control by 2-way ANOVA. In (H,J,L) samples from time 0 for cell lines shown in (G,I,K) were loaded together for comparison. In (A) cells were treated with MG132 for 4 hours prior to lysis.
We examined the contributions of CRY2 and FBXL3 to c-MYC protein turnover in cells overexpressing c-MYC and either FBXW7 or FBXL3 and CRY2. The steady state level of overexpressed c-MYC was decreased by overexpression of CRY2 and FBXL3 to a similar extent as overexpression of the established c-MYC SCF substrate adaptor FBXW7 (Figures 4B and S4D). The half-life of overexpressed c-MYC is much longer than that of the endogenous protein likely due to saturation of endogenous systems involved in c-MYC degradation. Using either cycloheximide treatment or 35S-Methionine labeling to measure c-MYC protein turnover, we observed a similar increase in c-MYC turnover with expression of either FBXW7 or FBXL3 and CRY2 (Figures 4C–F). The effects of FBXL3 and CRY2 on MYC half-life are reduced by mutations (FBXL3W428A or CRY2F428D) that disrupt the CRY2-FBXL3 interaction (Figures 4G and 4H), further supporting our hypothesis that the CRY2-FBXL3 heterodimer promotes c-MYC ubiquitylation.
To examine the effects of CRY2 and SCFFBXL3 on the half-life of endogenous c-MYC, we used shRNA knockdown of Fbxl3 and of Fbxw7 in primary embryonic and adult fibroblasts. Decreasing FBXW7 stabilizes c-MYC in both wildtype and Cry2−/− cells, while depleting FBXL3 stabilizes c-MYC in wildtype but not in Cry2−/− cells (Figures 4I–L and S4E–F), demonstrating that CRY2 is required for FBXL3- but not FBXW7-dependent effects on c-MYC stability. Finally, we examined the effect of Fbxl3 disruption in the context of complete genetic deletion of Fbxw7 using Cre-mediated recombination in primary fibroblasts. Depletion of Fbxl3 increased c-MYC protein stability regardless of the presence or absence of Fbxw7 (Figures S4G–I), demonstrating that these two F-box SCF substrate adaptors represent independent pathways for destabilizing c-MYC.
CRY2-FBXL3 binds c-MYC phospho-T58
Phosphorylation of T58 is a primary determinant of c-MYC stability (Gregory et al., 2003; Malempati et al., 2006; Salghetti et al., 1999; Yada et al., 2004). A recent structure of CRY2 bound to a small molecule revealed a conserved, ordered phosphate-binding loop (P-loop) on the surface of CRY2 (Nangle et al., 2013), which has also been observed in a conserved region of photolyases (Hitomi et al., 2009), though no phosphorylated partners have been identified. Aligning this structure with our model, we found that this P-loop would be located near the CRY2-FBXL3 interface (Figure 5A), suggesting that c-MYC interaction with CRY2-FBXL3 could be regulated by phosphorylation. Since phosphorylation of T58 is a well-established driver of c-MYC proteolysis and because phosphorylation of T58 occurs only after phosphorylation of S62, we examined whether either modification alters the interaction of c-MYC with CRY2-FBXL3. Mutation of T58 and/or S62 to a nonphosphorylatable amino acid greatly reduced the interaction between CRY2 and c-MYC (Figures 5B and S5A) but had little effect on the interaction between c-MYC and FBXL3 when CRY2 was not overexpressed (Figure 5C).
Figure 5. SCFFBXL3+CRY2 interacts specifically with T58-phosphorylated c-MYC. See also Figure S5.
(A) Top: Phosphate-binding loop (P-LOOP, magenta) at the interface of CRY2 (blue) and FBXL3 (red). Middle: Superposition of crystal structures from Protein Data Bank accession numbers 1LDK (SKP1-SKP2-CUL1-RBX1), 4I6J (SKP1-FBXL3-CRY2), and 4MLP (CRY2-KL001) with the CRY2 P-LOOP highlighted in magenta. Bottom: 90 degree rotation of the above image. (B,C) Proteins detected by IB following FLAG IP from 293T cells expressing the indicated plasmids. (D,E) Proteins detected by IB following streptavidin affinity purification after incubation of purified Flag-CRY2 and/or Flag-FBXL3 with the indicated biotinylated peptides. (F,J) Proteins detected by IB in whole cell lysates from 293T cells expressing the indicated plasmids and harvested at the indicated times (CHX: minutes after cycloheximide). (H) Proteins detected by autoradiography after a 35S-Methionine Pulse-Chase from 293T cells expressing the indicated plasmids. (G,I) Quantitation of the IB data (G) or the autoradiography data (I) showed in (F) and (H) respectively. Data represent the mean ± s.d. for triplicate samples. * P<0.05 vs. c-MYCT58A control by 2-way ANOVA.
To investigate whether phosphorylated T58 interacts directly with CRY2-FBXL3, we used biotinylated synthetic peptides derived from the sequence of c-MYC surrounding either phosphorylated (P) or non-phosphorylated (NP) threonine at the position corresponding to T58, or a doubly phosphorylated peptide (DP) corresponding to c-MYC phosphorylated on both T58 and S62. While FBXL3 interacts weakly with all three peptides (Figure 5D, middle IPs), CRY2 interacts preferentially with the peptide containing phospho-T58 (Figure 5D, left IPs). When purified CRY2 and FBXL3 are combined, the resulting heterodimer exhibits strongly phospho-specific binding to the c-MYC derived peptide phosphorylated only on the residue corresponding to T58 (Figure 5D, right IPs). Mutation of the reported CRY2 P-loop decreases binding to the T58-phosphorylated c-MYC peptide in vitro and to overexpressed c-MYC in cells (Figures 5E, S5B–D), suggesting that this phospho-specific interaction involves direct binding between phosphorylated T58 and the P-loop on the surface of CRY2.
We also examined the role of the CRY2 P-loop and c-MYC T58 phosphorylation in CRY2 and FBXL3 driven effects on c-MYC protein turnover. While overexpression of CRY2 and FBXL3 or of FBXW7 decreases the stability of wildtype c-MYC (Figures 4C–F), the same manipulations do not significantly increase turnover of c-MYCT58A (Figures 5F–I and S5E). Mutation of the CRY2 P-loop also decreases the impact of CRY2 and FBXL3 on c-MYC turnover (Figure 5J), supporting the idea that phosphorylation of T58 enhances the interaction of c-MYC with CRY2 and FBXL3 by binding the CRY2 P-loop. Lending further support to the model that CRY2 and FBXL3 drive degradation of T58-phosphorylated c-MYC, c-MYCT58A is much less efficiently ubiquitylated than wildtype c-MYC when FBXL3 and CRY2 are overexpressed (Figure S5F).
A role for CRY2-FBXL3-MYC in tumors
To investigate whether this model may have relevance for human cancer, in which c-MYC is a well-established major driver, we examined the relationship between CRY2 and c-MYC in human tumor-derived cells. By ranking 1,059 cell lines in the Cancer Cell Line Encyclopedia (CCLE) by CRY2 mRNA expression (Supplemental Table S3), we selected several among those with the lowest CRY2 for further examination. Exogenous expression of wildtype CRY2 reduced c-MYC protein, proliferation, and anchorage-independent growth in SW480 colon cancer and A549 lung cancer cells (Figures 6A–D), suggesting that CRY2 may be rate-limiting for c-MYC degradation in tumors harboring low CRY2 expression. Importantly, a CRY2 mutant incapable of binding FBXL3 did not significantly reduce c-MYC protein in these cells (Figures 6A and 6B). Furthermore, we observed a striking and significant correlation of reduced CRY2 expression in tumor samples compared to normal controls for a wide variety of human tissue types (Figure 6E), while no such consistent pattern of altered expression was observed for CRY1 (Figure 6F). In bone and lung tumors, FBXL3 is lower than in controls (Figure 6G).
Figure 6. Low CRY2 expression enhances growth of human cancer cell lines. See also Table S3.
Proteins detected by IB (A), proliferation (C), and colony formation in soft agar (D) in SW480 and A549 cells expressing the indicated plasmids. (B) Quantitation of the IB data shown in (A). (E–G) CRY2, CRY1, and FBXL3 expression in human tumor vs. normal samples.
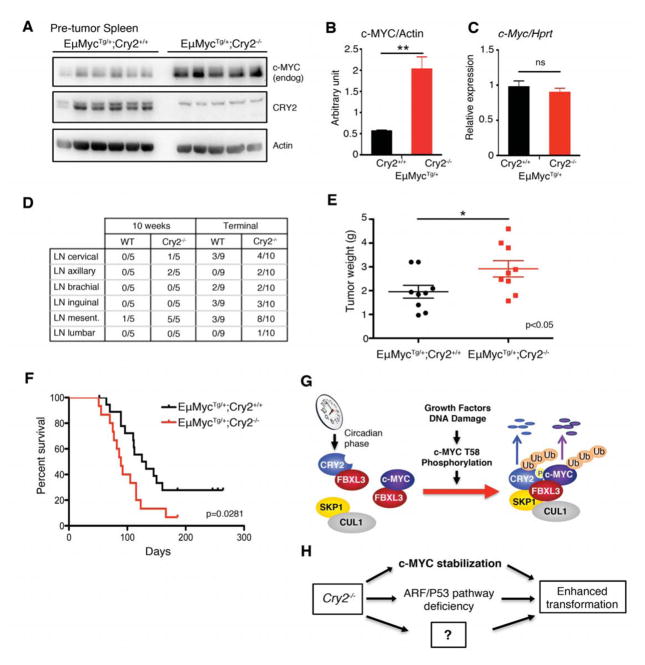
To experimentally test the potential for CRY2 to alter c-MYC in vivo, we bred Cry2−/− mice with mice expressing elevated c-MYC in lymphoid cells (Adams et al., 1985), which develop lymphomas after 8 weeks of age. Pre-tumor spleen samples taken from 6-week-old EμMycTg/+;Cry2−/− mice contain higher levels of c-MYC protein than those from EμMycTg/+;Cry2+/+ littermates (Figures 7A and 7B), while c-Myc mRNA is unchanged (Figure 7C), supporting the hypothesis that deletion of CRY2 enhances c-MYC via a post-transcriptional mechanism in vivo. At the same 6-week-old pre-tumoral stage, we detected small numbers of lymphocytes in Giemsa-stained blood smears from EμMycTg/+;Cry2−/− mice while blood smears from control littermates did not contain detectable circulating lymphocytes (Figure S6A).
Figure 7. CRY2 deletion enhances MYC-driven lymphoma in vivo. See also Figure S6.
(A) Proteins detected by IB in spleen samples taken from 6-week-old EμMyc+/−;Cry2+/+ and EμMyc+/−;Cry2−/− littermates. (B) Quantitation of proteins from WB in (A). (C) mRNA expression measured by qPCR in spleen samples used in (A). (D) Numbers of solid tumors observed in the indicated locations in EμMyc+/−;Cry2+/+ and EμMyc+/−;Cry2−/− littermates of the indicated ages or at the time of sacrifice necessitated by advanced disease progression. LN, lymph node. (E,F) Total combined tumor weight (E) and survival (F) at time of sacrifice due to advanced disease progression in EμMyc+/−;Cry2+/+ (black, N=19) and EμMyc+/−;Cry2−/− (red, N=15) littermates. In (B,C) data represent the mean ± s.d. of samples from 5 mice for which IB data is shown in (A). In (B,E) * P<0.05, ** P<0.01 by t-test. In (F), P value is from log-rank calculations. (G) Schematic model for the role of CRY2 as a cofactor for SCFFBXL3-driven ubiquitylation of phosphorylated c-MYC. (H) Deletion of Cry2 may contribute to cellular transformation in a pleiotropic manner.
All 10-week-old EμMyc mice lacking CRY2 had grossly visible tumors in the mesenteric lymph nodes while EμMycTg/+;Cry2+/+ littermates were mostly tumor-free (Figure 7D), suggesting accelerated lymphoma development in the absence of CRY2. Eventually, EμMycTg/+;Cry2−/− mice succumbed earlier and with an overall greater tumor burden than EμMycTg/+;Cry2+/+ littermates (Figures 7D, 7E, 7F, and S6B). We did not detect spontaneous tumors in either wildtype or Cry2−/− mice without the EμMyc transgene. Taken together, our findings indicate that CRY2 protects cells from transformation via pleiotropic effects, including destabilizing c-MYC via SCFFBXL3 and also via MYC-independent pathways to enable enhanced MYC-driven transformation in cells and in vivo (Figures 7G and 7H).
DISCUSSION
Here we report that the circadian clock component CRY2 cooperates with the E3 substrate receptor FBXL3 to degrade target proteins in a phospho-specific manner, revealing a mechanism for mammalian circadian output via CRY-driven cycles of proteasomal degradation. Our findings suggest that CRY2 and FBXL3 contribute to limiting tumor formation by promoting the turnover of c-MYC. While CRY has never before been proposed to act as a physical component of an E3 ligase, Drosophila melanogaster dCRY is genetically required for light-induced degradation of dTIM via the E3 ligase JETLAG (Koh et al., 2006), suggesting that some aspects of the function that we describe here may be conserved in other systems. In addition, a recent study found that mammalian CRY1 can stimulate MDM2-driven ubiquitylation and degradation of FOXO1 (Jang et al., 2016), which may involve a related mechanism, though MDM2 is not part of an SCF complex. We anticipate that our findings will provide the basis for future identification of additional substrates recruited to SCFFBXL3 and/or the related CRY-binding E3 ligase SCFFBXL21 (Hirano et al., 2013; Yoo et al., 2013) by CRY1 and/or CRY2.
Here, we describe the first demonstrated target of mammalian CRY2-dependent protein turnover: the oncoprotein c-MYC. Because the sequence surrounding c-MYC T58, which binds to CRY2, is conserved in N-MYC, L-MYC and the c-MYC cleavage product MYC-NICK (Conacci-Sorrell et al., 2010), these may also be targeted by CRY2-SCFFBXL3, though they were not examined in this study. Since ubiquitylation and proteasome activity have been shown to regulate CLOCK and BMAL1 activation of target genes (Luo et al.; Stratmann et al., 2012; Tamayo et al., 2015), the novel function of CRY proteins described here may play a role in circadian clock function. Intriguingly, c-MYC can interfere with CLOCK and BMAL1-dependent transactivation (Altman et al., 2015), so CRY2-driven MYC destabilization could reinforce circadian clock amplitude.
We recently demonstrated that CRY1 and CRY2 have divergent roles in modulating transcription in response to DNA damage (Papp et al., 2015). Reduced degradation of c-MYC in response to DNA damage could contribute to the loss of p21 induction in Cry2−/− cells. The unique functions of CRY1 and CRY2 in this pathway may explain why Cry1−/−;Cry2−/− mice are protected from tumor development in the context of P53 deletion (Ozturk et al., 2009), yet are more susceptible to radiation-induced tumor formation (Lee et al., 2010), while loss of CRY2 alone enhances susceptibility to transformation in our study. Disruption of circadian rhythms likely contributes to enhanced cancer susceptibility via multiple mechanisms (Kang et al., 2010; Kemp et al., 2010; Lee et al., 2013; Lee and Sancar, 2011a, b; Masri et al., 2013; Masri et al., 2015; Unsal-Kacmaz et al., 2007; Unsal-Kacmaz et al., 2005) – indeed, the cooperation between CRY2 deficiency and multiple oncogenes in proliferation and transformation assays (Figures 1A–G) suggests that CRY2 itself may have additional roles in suppressing these effects. The observed cooperativity between CRY2 loss and c-MYC overexpression indicates that Cry2−/− cells harbor a defect in P19ARF and/or P53 function (Sherr, 2006, 2012). PER2 has been implicated in stabilizing P53 by opposing its MDM2-mediated ubiquitylation (Gotoh et al., 2015; Gotoh et al., 2014) and has been suggested to function as a tumor suppressor in some contexts (Chen-Goodspeed and Lee, 2007; Lee, 2006; Papagiannakopoulos et al., 2016). CRY2 deficiency could influence P19ARF/P53 by altering PER2 expression, localization or stability. Further studies will be required to understand how the loss of CRY2 impacts P53 and how PER2 and CRY2 independently and/or synergistically modulate tumor formation.
Repeated transient inactivation of c-MYC has been shown to suppress even established tumors while allowing normal tissue proliferative functions to proceed (Soucek et al., 2008; Soucek et al., 2013); perhaps CRY2-driven circadian proteolysis of c-MYC provides analogous daily cycles of c-MYC action and degradation in a physiological context. While c-MYC protein remains rhythmic in Cry2−/− cells (Figures 1M and S1B), perhaps due to rhythmic mRNA expression, its elevated expression overall prevents this prolonged daily absence of c-MYC protein. Increased cancer susceptibility in shift workers and in animals subjected to circadian disruption may be due in part to elevated c-MYC driven by loss of CRY2 and FBXL3-coordinated regulation, since circadian disruption would alter the dynamics of CRY2 expression. Notably, Cry2-deficient mice and cells maintain robust circadian rhythms (Khan et al., 2012; Thresher et al., 1998) while clearly exhibiting enhanced susceptibility to transformation, indicating that neither behavioral nor transcriptional rhythms must be globally disrupted for increased cancer susceptibility in the context of circadian disruption.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293T (293T), A549 and SW480 cells were purchased from the American Type Culture Collection (ATCC). Three different series of MEFs were isolated from E15.5 embryos. Two different series of adult skin fibroblasts were prepared from ear biopsies of adult mice of the indicated genotypes. All MEFs and adult ear fibroblasts (AEFs) were used as primary (passaged no more than 10 times and grown in 3% oxygen) or spontaneously immortalized. More detailed descriptions of all procedures are available in Supplemental Experimental Procedures.
Generation of Stable Cell Lines
Lentiviral shRNAs were produced by transient transfection in 293T cells. Beginning 48 hours after viral transduction, infected cells were cultured in selection media for 2 days to 2 weeks.
Proliferation and Transformation Assays
For proliferation analysis, cells were seeded in 6-well plates at 5,000 cells/well and counted every 2 days for 6 to 10 days. For colony formation, 1,000 cells were plated in 10cm plates and grown for 2 to 3 weeks prior to staining. For anchorage independent growth, MEFs infected with virus expressing c-MYC/shP53 were selected for 2 to 3 days and suspended in complete medium containing 0.4% low melting agarose and plated in 6-well plates at a density of 20,000 cells per well, onto solidified 0.8% agarose. Cells were grown for 2 to 3 weeks prior to staining with MTT.
Immunoprecipitation and Western Blotting were performed using standard protocols.
Pulse-Chase Labeling Experiment
Lysates from transfected 293T cells were incubated with Pulse medium containing 35S-Methionine. The labeling was stopped by replacing the Pulse medium with regular 293T cell culture medium and cells were collected at the indicated time points.
Peptide Binding Assay
Flag-Fbxl3 and Flag-Cry2 were separately immunoprecipitated from transfected 293T cells and eluted with 3XFLAG peptide. After quantification, equal amounts of Flag-Fbxl3 and/or wildtype or mutant Flag-Cry2 were combined with biotinylated synthetic peptides as indicated and incubated at 30°C for 30 minutes while shaking. Streptavidin beads were added, incubated overnight at 4°C, and washed 3 times.
Sequential IP
Lysates from transfected 293T cells were incubated with anti-Flag M2 agarose beads overnight at 4°C, washed 3 times, and incubated in 3X FLAG peptide. The elution was collected and subsequently incubated with anti-HA agarose beads for 2 hours at 4°C. The anti-HA beads were washed 3 times and boiled with sample buffer.
Quantitative RT-PCR
RNA was extracted from MEFs using standard protocols. cDNA was prepared using QScript cDNA Supermix and analyzed for gene expression using quantitative real-time PCR with iQ SYBR Green Supermix. Primer sequences are available in Supplemental Experimental Procedures.
Isotope Labeling
Following dexamethasone-induced synchronization, cells were cultured in glucose free DMEM (Sigma) containing 15% FBS and 25 mM [U-13C6] glucose for 10 hours. Polar metabolites and total fatty acids were extracted, derivatized and analyzed as previously described (Metallo et al., 2012; Vacanti et al., 2014).
RNA Sequencing
100bp reads were generated by the HISeq Analyzer 2000. The Genome Analyzer Pipeline Software was used to perform the early data analysis. For mRNA-Seq, TopHat v2.0.13 with Bowtie2 was used to align to mm10 genome.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) is a computational method (Subramanian et al., 2007) that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes). RNA sequencing output for wildtype and Cry2−/− cells were used as input for GSEA comparing the two genotypes (WT vs. Cry2−/− regardless of circadian time).
Structure Modeling
Molecular graphics and analyses were performed with the UCSF Chimera package (Pettersen et al., 2004). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Tissue Expression Profile
To investigate the expression profile of CRY2 in human normal and cancer samples, publicly available microarray data were compiled, normalized and re-analyzed by Novartis bioinformatics group. Original data were extracted from GEO database, CCLE database and TCGA database.
Mice
Cry2−/− mice were shared by Dr. Aziz Sancar (Thresher et al., 1998); EμMyc+/− mice (Adams et al., 1985) were purchased from Jackson laboratories. Cry2+/− females were mated to EμMyc+/− males to generate EμMyc+/−;Cry2+/− mice. EμMyc+/−;Cry2+/− males were mated to Cry2+/− females to generate EμMyc+/−;Cry2+/+ and EμMyc+/−;Cry2−/− mice used for analysis. Mice showing any signs of systemic illness were sacrificed and necropsied. All animal care and treatments were in accordance with The Scripps Research Institute guidelines for the care and use of animals.
Nuclear Extract from Tissue
Nuclear extracts from spleen were prepared using NUN procedure as described (Lavery and Schibler, 1993).
Data Analysis and Statistics
All experiments were repeated at least three times and results are presented either as one representative experiment or as an average ± s.d. Statistical analyses were done using two-tailed student’s t-test or with two-way ANOVA. Time-dependent survival in mouse experiments was represented with Kaplan-Meir methods and significance was evaluated with the log-rank test.
Supplementary Material
Highlights.
Loss of CRY2 stabilizes c-MYC and enhances cellular transformation.
CRY2 can function as a co-factor for the SCF substrate adaptor FBXL3.
c-MYC phosphorylated on threonine 58 (T58) interacts with CRY2.
SCFFBXL3+CRY2 promotes the ubiquitylation and turnover of c-MYC.
Acknowledgments
This work was supported by grants from the National Institutes of Health to K.A.L. (DK090188 and DK097164), and C.M.M. (CA188652), Searle Scholars awards to K.A.L. and C.M.M. from the Kinship Foundation, a cancer research scholar award to K.A.L. from the Sidney Kimmel Cancer Research Foundation, a grant to K.A.L. from the Lung Cancer Research Foundation, and by fellowships from the Swedish Research Council to E.H. and the Deutsche Forschungsgemeinschaft and the American Heart Association (15POST22510020) to S.D.J. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). We thank Reuben Shaw, Clodagh O’Shea, Michael Hemann, Thales Papagiannakopoulos, Luke Wiseman, Eric Bennett, Brenda Schulman, Daniel Scott, Megan Afetian, Drew Duglan, Megan Vaughan, Colby Sandate, Jamie Williamson, Michael and Louise McHeyzer-Williams, and Eros Lazzerini-Denchi for helpful discussions, sharing reagents or equipment, and/or critical reading of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
A.-L.H. performed the majority of experiments, analyzed data, made figures and contributed to writing the paper. S.J.P., A.B.C., E.H., S.D.J., A.K., and M.N. performed experiments. M.W. and C.M.M. designed, performed, and supervised metabolic labeling experiments and analysis. Z.L. performed bioinformatics analyses (Tables S1 and S3; Figures 6D–F); K.A.L. conceived and supervised the study, performed structural modeling and gene set enrichment analysis, analyzed data, made figures, and wrote the paper. All authors edited and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha-Hicks P, et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell metabolism. 2015 doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nature genetics. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- Bhatia K, Spangler G, Gaidano G, Hamdy N, Dalla-Favera R, Magrath I. Mutations in the coding region of c-myc occur frequently in acquired immunodeficiency syndrome-associated lymphomas. Blood. 1994;84:883–888. [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Chakraborty AA, Tansey WP. Inference of cell cycle-dependent proteolysis by laser scanning cytometry. Experimental cell research. 2009;315:1772–1778. doi: 10.1016/j.yexcr.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Goodspeed M, Lee CC. Tumor suppression and circadian function. Journal of biological rhythms. 2007;22:291–298. doi: 10.1177/0748730407303387. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AS, Sears RC. MYC degradation. Cold Spring Harbor perspectives in medicine. 2014:4. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Kettner NM. The circadian clock in cancer development and therapy. Progress in molecular biology and translational science. 2013;119:221–282. doi: 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Vila-Caballer M, Liu J, Schiffhauer S, Finkielstein CV. Association of the circadian factor Period 2 to p53 influences p53’s function in DNA-damage signaling. Molecular biology of the cell. 2015;26:359–372. doi: 10.1091/mbc.E14-05-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Vila-Caballer M, Santos CS, Liu J, Yang J, Finkielstein CV. The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Molecular biology of the cell. 2014;25:3081–3093. doi: 10.1091/mbc.E14-05-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. The Journal of biological chemistry. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Molecular cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6962–6967. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Lee GY, Selby CP, Lee G, Jeon YG, Lee JH, Cheng KK, Titchenell P, Birnbaum MJ, Xu A, et al. SREBP1c-CRY1 signalling represses hepatic glucose production by promoting FOXO1 degradation during refeeding. Nature communications. 2016;7:12180. doi: 10.1038/ncomms12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos-Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, Cordeiro-Stone M, Kaufmann WK, Abraham RT, Sancar A, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. The Journal of biological chemistry. 2010;285:16562–16571. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SK, Xu H, Ukai-Tadenuma M, Burton B, Wang Y, Ueda HR, Liu AC. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. The Journal of biological chemistry. 2012;287:25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes & development. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- Lee JH, Gaddameedhi S, Ozturk N, Ye R, Sancar A. DNA damage-specific control of cell death by cryptochrome in p53-mutant ras-transformed cells. Cancer research. 2013;73:785–791. doi: 10.1158/0008-5472.CAN-12-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Sancar A. Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:10668–10672. doi: 10.1073/pnas.1106284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Sancar A. Regulation of apoptosis by the circadian clock through NF-kappaB signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108:12036–12041. doi: 10.1073/pnas.1108125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PloS one. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li Y, Tang CH, Abruzzi KC, Rodriguez J, Pescatore S, Rosbash M. CLOCK deubiquitylation by USP8 inhibits CLK/CYC transcription in Drosophila. Genes & development. 26:2536–2549. doi: 10.1101/gad.200584.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, Sears RC. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20:1572–1581. doi: 10.1038/sj.leu.2404317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Cervantes M, Sassone-Corsi P. The circadian clock and cell cycle: interconnected biological circuits. Current opinion in cell biology. 2013;25:730–734. doi: 10.1016/j.ceb.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Kinouchi K, Sassone-Corsi P. Circadian clocks, epigenetics, and cancer. Current opinion in oncology. 2015;27:50–56. doi: 10.1097/CCO.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 2013;23:1417–1419. doi: 10.1038/cr.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell metabolism. 2016 doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp SJ, Huber AL, Jordan SD, Kriebs A, Nguyen M, Moresco JJ, Yates JR, Lamia KA. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife. 2015:4. doi: 10.7554/eLife.04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends in cell biology. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Popov N, Herold S, Llamazares M, Schulein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell cycle. 2007;6:2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. The EMBO journal. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nature reviews Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley interdisciplinary reviews. Developmental biology. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes & development. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Benbrahim-Tallaa L, Cogliano V. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8:1065–1066. doi: 10.1016/S1470-2045(07)70373-X. [DOI] [PubMed] [Google Scholar]
- Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Molecular cell. 2012;48:277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Tamayo AG, Duong HA, Robles MS, Mann M, Weitz CJ. Histone monoubiquitination by Clock-Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nature structural & molecular biology. 2015;22:759–766. doi: 10.1038/nsmb.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Advances in cancer research. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Molecular and cellular biology. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Molecular and cellular biology. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacanti NM, Divakaruni AS, Green CR, Parker SJ, Henry RR, Ciaraldi TP, Murphy AN, Metallo CM. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Molecular cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dycke KC, Rodenburg W, van Oostrom CT, van Kerkhof LW, Pennings JL, Roenneberg T, van Steeg H, van der Horst GT. Chronically Alternating Light Cycles Increase Breast Cancer Risk in Mice. Current biology: CB. 2015;25:1932–1937. doi: 10.1016/j.cub.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature reviews Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Bechtold DA. The cost of circadian desynchrony: Evidence, insights and open questions. BioEssays: news and reviews in molecular, cellular and developmental biology. 2015;37:777–788. doi: 10.1002/bies.201400173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumimoto K, Matsumoto M, Oyamada K, Moroishi T, Nakayama KI. Comprehensive identification of substrates for F-box proteins by differential proteomics analysis. Journal of proteome research. 2012;11:3175–3185. doi: 10.1021/pr201216u. [DOI] [PubMed] [Google Scholar]
- Yumimoto K, Muneoka T, Tsuboi T, Nakayama KI. Substrate binding promotes formation of the Skp1-Cul1-Fbxl3 (SCF(Fbxl3)) protein complex. The Journal of biological chemistry. 2013;288:32766–32776. doi: 10.1074/jbc.M113.511303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.