Abstract
At fertilization, the highly condensed and transcriptionally inert chromatin of the spermatozoa becomes remodelled into the decondensed and transcriptionally competent chromatin of the male pronucleus. The chromatin initially becomes dispersed and then transiently recondenses into a small mass upon entry into the ooplasm. This morphological change is coincident with and likely dependent on the replacement of the sperm-specific protamines by oocyte-supplied histones and the organization of the chromatin into nucleosomes. The chromatin then extensively decondenses within the male pronucleus and acquires many of the proteins that are associated with the maternal chromatin. Nonetheless, the paternal chromatin manifests distinct characteristics, including transient hyperacetylation of histone H4, increased transcription of endogenous and microinjected genes, and replication-independent demethylation of DNA. Sperm chromatin remodelling is controlled by an oocyte activity that appears during meiotic maturation and disappears approximately 3 h after activation (release from metaphase II arrest), and which requires factors associated with the germinal vesicle of the oocyte. The molecular components of this activity remain largely unknown. In frogs, nucleoplasmin is required to assemble histones H2A and H2B onto the paternal chromatin. Evidence is presented that related proteins may perform similar functions in mammals. Identifying the mechanisms that underlie sperm chromatin remodelling at fertilization may be relevant for understanding reprogramming of somatic cell nuclei after transfer into oocytes.
Fertilization achieves two objectives: it re-initiates the cell cycle of the dormant egg, and it produces an embryo that contains one functional copy each of the maternal and paternal genomes. The second objective is ensured through a variety of mechanisms, including the block to polyspermy, the completion of the second meiotic division in the oocyte, and the biochemical remodelling of the sperm chromatin to form the male pronucleus. Remodelling of the paternal chromatin at fertilization is necessary because although the paternal genome is required for embryonic development, the DNA of the mature spermatozoa is compacted and transcriptionally inactive. The process through which this remodelling occurs is reviewed here.
Biparental androgenetic and gynogenetic mammalian embryos produced by pronuclear transplantation arrest development at mid-gestation (McGrath and Solter, 1984; Surani et al., 1984). These well-known experiments of McGrath and Solter (1984) and Surani et al. (1984) established that normal mammalian development requires transcription from both parental genomes. In mice, transcription by RNA polymerase II is first observed in the late one-cell embryo, initially from the paternal pronucleus (Bouniol et al., 1995; Aoki et al., 1997), and is followed by the major activation of the embryonic genome at the two-cell stage (Flach et al., 1982). Likewise, transcripts of Y-linked genes Zfy and Sry are present in one-cell human zygotes (Ao et al., 1994), and expression from paternal copies of autosomal genes is observed at the time of activation of the human embryonic genome, the four-cell stage (Taylor et al., 1997). Thus, in the embryo, the paternal genome is transcribed from the earliest stages of embryogenesis and is necessary for successful development.
Sperm chromatin before fertilization
The chromatin of mature mammalian spermatozoa differs markedly in composition and structure from somatic chromatin. The DNA is not associated with histones or arranged in nucleosomes. Instead, it is complexed with sperm-specific basic proteins in a crystalline-like structure that, being sixfold more compact than metaphase chromosomes, is the most highly condensed chromatin in eukaryotes. The predominant chromatin-associated protein is protamine P1: a small (50 amino acid), highly basic protein possessing an arginine-rich central domain (reviewed in Wouters-Tyrou et al., 1998). Mice and humans are exceptional among mammals in that the spermatozoa also contain a second protamine, P2. The Balhorn model posits that the arginine-rich core of the protamine lies lengthwise in the minor groove of the DNA double helix, filling one helical turn (Balhorn, 1982). This neutralizes the negative charge of the DNA phosphate backbone, allowing close alignment of adjacent DNA strands to form a sheet-like arrangement. Inter-protein disulphide bridges anchor the protamines.
The replacement of histones by protamines is a multistep process during the final stages of spermatogenesis (reviewed in Wouters-Tyrou et al., 1998). DNA is complexed with histones and organized in nucleosomes throughout meiosis to the haploid spermatid stage. As a prelude to removal of the histones, the stable nucleosome structure is relaxed by processes linked with acetylation of histone H4 (Meistrich et al., 1992) and ubiquitination of H2B in mice and H3 in rats (reviewed in Jason et al., 2002). During spermatid elongation and nuclear condensation, histones are replaced by transition proteins TP1–4, which in turn are replaced by pro-protamines, one of which is cleaved to produce mature P2 (Yelick et al., 1987). Haplo-insufficiency of the genes encoding P1 or P2 results in aberrant sperm chromatin condensation and structure, and decreased fertility (Cho et al., 2001). Although protamines P1 and P2 are the major nucleoproteins of mature spermatozoa, human sperm chromatin may contain up to 15% histones (Gatewood et al., 1987). Moreover, by fluorescent in situ hybridization, histone H2B co-localizes with telomeric DNA in spermatozoa (Zalensky et al., 1997), and the telomere-binding protein complex isolated from human spermatozoa contains a variant of H2B (Gineitis et al., 2000). This evidence indicates that even in mature spermatozoa, some chromatin remains arranged into nucleosomes. Nonetheless, chromatin of mature spermatozoa is transcriptionally inert, presumably as a result of its unique structure and composition.
Morphological and molecular remodelling of the sperm chromatin at fertilization
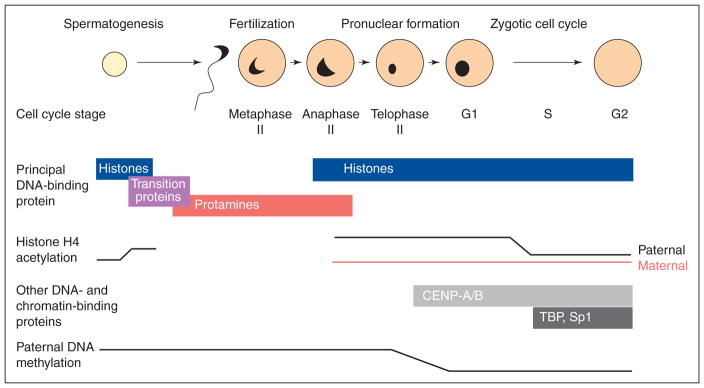
At fertilization, the oocyte faces the challenge of remodelling the condensed sperm chromatin into an accessible, transcriptionally competent form. This process is described below and in Fig. 1. Upon entry into the oocyte, the sperm nucleus undergoes marked morphological changes. This remodelling follows a characteristic tri-phasic pattern (Wright and Longo, 1988; Adenot et al., 1991). In the first phase, during the period that the oocyte is completing anaphase II, the paternal chromatin disperses. Although the change in volume is difficult to calculate, the area occupied by the chromatin as seen using the light microscope increases about threefold. While the oocyte chromosomes are completing telophase II, the chromatin transiently recondenses into a small mass, the area it occupies decreasing by about one-half. In the final phase, the chromatin extensively decondenses within the male pronucleus and the area it occupies increases about 10-fold, in synchrony with the oocyte chromatin that decondenses in the separate female pronucleus.
Fig. 1.
Summary of sperm chromatin remodelling in mammals. During spermatogenesis, sperm histones become acetylated and are replaced by transition proteins, which are then replaced by protamines. After fertilization, the protamines are replaced by oocyte-supplied histones. This replacement occurs as the oocyte completes the second meiotic division and coincides with a transient recondensation of the paternal chromatin. After the protamine–histone exchange, 5-methylcytosine becomes undetectable on the paternal DNA. The paternal and maternal chromatin then decondense extensively within separate pronuclei, and the histones on the paternal chromatin become deacetylated. Other oocyte proteins (centromeric proteins CENP-A/B, and transcription factors TBP and Sp1) imported into the pronuclei further modify the chromatin and DNA replication begins.
Morphological remodelling is accompanied by the replacement of protamines by histones. Inactivation of sulphydryl-dependent enzymes by iodoacetamide or depletion of oocyte glutathione prevents sperm decondensation, implying that the reduction of inter-protamine sulphydryl bonds is a necessary step for their removal (reviewed by Perreault, 1992). A variety of techniques, including autoradiographic detection of tritiated chromatin-associated proteins and immunostaining using antibodies against sperm basic proteins, have established that protamines are removed from sperm chromatin by the end of oocyte anaphase II (Ecklund and Levine, 1975; Kopecny and Pavlok, 1975; Rodman et al., 1981). In a similar way, histones become immunologically detectable before the first round of embryonic DNA replication begins (Nonchev and Tsanev, 1990; Adenot et al., 1997). Nonetheless, the precise timing of histone assembly on to the paternal chromatin at fertilization has not been established. However, in an experimental study in which oocytes were inseminated at the beginnning of maturation, histone assembly was closely correlated with the transient recondensation observed morphologically (McLay and Clarke, 1997). It seems likely that this same relationship holds during normal fertilization and that the morphological recondensation reflects histone assembly onto the chromatin.
After histone assembly and its organization into nucleosomes, the paternal chromatin becomes decondensed within the male pronucleus. During this time, additional proteins must be imported from the ooplasm into the pronucleus (as indicated by its continued enlargement), where they become directly or indirectly associated with the chromatin. For example, kinetochore proteins are assembled on to male chromatin during pronuclear development. Using the CREST antibody, which recognizes centromeric proteins CENP-A and CENP-B, Schatten et al. (1988) showed that mature mouse spermatozoa did not contain immunodetectable kinetochores. Shortly after fertilization, while the male pronucleus was still growing, CENP-A–B appeared on the paternal chromatin. Likewise, the proteins required for DNA replication must also be assembled onto the paternal chromatin. Priming the genome for DNA replication has been shown to involve several steps in somatic cells, for example, assembly during late telophase of minichromosome maintenance proteins MCM2–7 on DNA to form pre-replication complexes (Dimitrova et al., 2002). It seems reasonable to suppose that during chromatin remodelling at fertilization, MCM proteins would also enter the male pronucleus to prepare the paternal chromatin for the S-phase.
Surprisingly, although the male and female pronuclei reside in the same ooplasm, several differences become established between them during the first cell cycle. During the early portion of the first cell cycle, histone H4 in the paternal pronucleus is hyperacetylated compared with the female pronucleus. However, this difference disappears before DNA replication begins (Adenot et al., 1997). This may reflect the fact that histones H3 and H4 are acetylated when assembled on to DNA and subsequently become deacetylated (Verreault, 2000).
A second difference is that, although transcriptional activity in the one-cell embryo is quite low compared with subsequent stages, transcription of both endogenous and microinjected genes is higher in the paternal than maternal pronucleus (Aoki et al., 1997; Rastelli et al., 2001). The mechanism underlying this difference is not entirely clear. One clue may come from evidence that plasmid DNA injected into the paternal pronucleus may not become assembled into chromatin. First, expression of the genes injected into the maternal pronucleus, but not the paternal pronucleus, is increased in embryos cultured in the presence of histone deacetylase inhibitors (Wiekowski et al., 1993; Majumder et al., 1997). Second, injection of histones into the paternal pronucleus both reduces injected-gene expression and produces an upregulation in response to the deacetylase inhibitors (Rastelli et al., 2001). However, this explanation seems insufficient, as the paternal DNA appears to be efficiently assembled into chromatin. It is also known that the transcription factors TBP and Sp1 preferentially accumulate in the paternal pronucleus (Worrad et al., 1994). Hence, the higher transcriptional activity in the paternal genome may also reflect the higher abundance of transacting factors. The differences described above are not known to persist after the genomes unite in the same nucleus after first mitosis, although factors could associate preferentially with maternally or paternally derived chromatin within the same nucleus.
An unexpected difference between the pronuclei that appears within the first few hours after fertilization and persists during the early embryonic cell cycles is an apparent demethylation of the paternal DNA. In contrast, the maternal DNA remains methylated during this time. Thus, an anti-5-methyl-cytosine antibody (5 mC) stains both male and female chromatin of mouse embryos for the first few hours after fertilization but only the female pronucleus by about 6 h after fertilization (Mayer et al., 2000; Santos et al., 2002). In addition, sequencing of DNA following treatment with sodium bisulphite (which converts unmodified cytosine to uracil but leaves 5-methyl-cytosine unchanged) and PCR amplification indicates that, at the sites analysed, the paternal DNA frequently becomes demethylated whereas the maternal DNA at the same sites does not (Oswald et al., 2000). Demethylation follows histone assembly on the paternal DNA but precedes DNA replication (Santos et al., 2002). These results imply that an ooplasmic activity rapidly demethylates the remodelled paternal DNA after fertilization, but does not affect the maternal DNA residing in the same ooplasm. The molecular nature of this activity is entirely unknown. Moreover, the function of selective paternal demethylation, which has also been observed in embryos of cows, pigs and rats (Dean et al., 2001) but not sheep (Wilmut et al., 2002), remains to be elucidated.
Chromatin remodelling activity resides in the oocyte
What is the source of the activity that remodels the sperm chromatin at fertilization? Current evidence supports the intuitively attractive idea that it is the oocyte. First, the extent of remodelling of the sperm chromatin depends on the stage of the oocyte at insemination. When oocytes arrested at prophase I are inseminated, the sperm chromatin remains highly condensed, manifesting no morphological changes (although demembranated spermatozoa injected into the oocyte nucleus become dispersed) (Usui and Yanagimachi, 1976; Clarke and Masui, 1986; Maeda et al., 1998). When oocytes undergoing meiotic maturation are inseminated, the sperm chromatin becomes dispersed and then recondenses, often forming individual chromosomes (Clarke and Masui, 1986), and acquires histones (McLay and Clarke, 1997). These changes are similar to those occurring after normal fertilization at metaphase II except that, as maturing oocytes are not activated by sperm penetration, no pronuclei are formed. However, when oocytes are parthenogenetically activated and inseminated several hours later, the sperm chromatin fails to progress beyond an initial dispersion (Usui and Yanigimachi, 1976; Szollosi et al., 1994). Second, when protein synthesis is inhibited during meiotic maturation, sperm chromatin does not progress beyond the stage of initial dispersion (McLay and Clarke, 1997). Third, as assayed by the morphological changes, only a limited number of sperm nuclei can be remodelled within one oocyte (Witkowska, 1981; Clarke and Masui, 1986). Fourth, when prophase I oocytes are enucleated and allowed to mature, they are unable morphologically to remodel sperm chromatin (Balakier and Tarkowski, 1980).
These results indicate that the activity responsible for the early events of remodelling, namely, the initial dispersion and recondensation and the assembly of histones, becomes functional during meiotic maturation. It requires a factor(s) present in the oocyte nucleus and protein synthesis during maturation, and some component of the activity is present in a limited amount. The activity disappears within a few hours after activation of the oocyte. One-cell embryos during the first mitotic division can induce the early events of sperm chromatin remodelling (Maleszewski et al., 1999). Hence, it may be that M-phase cytoplasm is generally permissive for sperm chromatin dispersion and recondensation and assembly of histones. The later events of remodelling, including the extensive decondensation of the chromatin within the male pronucleus and the import of nuclear proteins, however, require transition from M-phase to interphase. This cell-cycle transition is triggered by Ca2+ oscillations (Ducibella et al., 2002) normally induced in the metaphase II oocyte by a phospholipase C in the fertilizing spermatozoa (Saunders et al., 2002) that initiate destruction of cyclin B (Nixon et al., 2002) and consequent loss of activity of cyclin-dependent kinase 1.
Oocyte factors implicated in sperm chromatin remodelling
Although remodelling comprises many changes in the structure and composition of the paternal chromatin, the best-understood changes pertain to the replacement of protamines by histones. Protamine removal requires reduction of the intermolecular disulphide bonds and, as the nucleoprotein exchange is sensitive to phosphorylation inhibitors, may also require their phosphorylation (reviewed in Perreault, 1992). The histones are likely supplied by the oocyte, which accumulates a pool during oogenesis (Wassarman and Mrozak, 1981). Histone assembly into chromatin is an energy-requiring process, and antimycin A, which blocks ATP hydrolysis, prevents paternal chromatin remodelling (Wright and Longo, 1988). In oocytes treated with antimycin A, the paternal chromatin undergoes the initial dispersion but fails to recondense or decondense within a pronucleus. The same result is seen in oocytes penetrated by more than three spermatozoa (Clarke and Masui, 1986). These results taken together reinforce the model that ATP-dependent assembly of oocyte-supplied histones on to the paternal chromatin is a prerequisite for proper remodelling.
In somatic cells, histone assembly onto chromatin requires the activity of chaperone proteins. Results obtained using Xenopus indicate that chaperones also mediate histone assembly onto the paternal chromatin at fertilization. Xenopus spermatozoa contain histones H3 and H4, but lack histones H2A and H2B, which have been replaced by the sperm-specific proteins, X and Y (Philpott and Leno, 1992). Xenopus eggs contain two abundant acidic proteins: nucleoplasmin, which interacts with H2A–H2B dimers and possibly H3–H4 tetramers (Philpott and Leno, 1992; Philpott et al., 2000); and N1, which interacts with H3–H4 tetramers (Dilworth et al., 1987). Nucleoplasmin is essential for histone assembly on to paternal chromatin at fertilization. Purified nucleoplasmin removes proteins X and Y from, and assembles histones H2A and H2B on to, paternal chromatin tetramers in vitro. More importantly, immunodepletion of nucleoplasmin from cell-free egg preparations prevents both this nucleoprotein exchange and pronuclear formation. Nucleoplasmin is present in immature oocytes and becomes heavily phosphorylated during meiotic maturation. Phosphorylation substantially increases its histone-assembly activity (Leno et al., 1996). As the paternal chromatin already possesses H3 and H4, N1 is not required for its remodelling at fertilization.
Subsequent studies have identified other proteins from various organisms that can remodel sperm chromatin, as assayed by different criteria (Table 1). However, no proteins with these activities have been described in mammalian oocytes. Such a protein must possess at least two characteristics. First, it must interact directly or indirectly with histones, and potentially with protamines. Second, as remodelling at fertilization occurs on non-replicating DNA, it must be able to assemble histones on to DNA in the absence of DNA replication. One approach to identify mammalian remodelling factors is to search for genes that share sequence similarity with Xenopus nucleoplasmin. Two potential homologues have been identified to date: nucleoplasmin 2 (Npm-2) and Npm-3. Npm-2 appears not to play a role in paternal chromatin remodelling at fertilization (M. Matzuk et al., personal communication). Npm-3 protein (MacArthur and Shackleford, 1997) is 42% identical (67% similar) to Xenopus nucleoplasmin and its encoding mRNA is expressed in growing and fully grown oocytes (Fig. 2). A second candidate has emerged, somewhat paradoxically, from DNA replication studies. After passage of the replication fork, chromatin assembly occurs in two stages: replication-dependent assembly of the H3–H4 tetramer by chromatin-assembly factor 1, followed by addition of H2A–H2B dimers by nucleosome-assembly protein 1 (NAP-1). First isolated from human extracts and later identified in diverse species, NAP-1 is able to assemble nucleosomes onto unreplicated DNA in vitro, as judged by nuclease digestion or supercoiling assays. NAP-1 mRNA is also expressed in oocytes (Fig. 2) and its ability to assemble nucleosomes in the absence of DNA replication may indicate a function at fertilization.
Table 1.
Oocyte factors implicated in sperm chromatin remodelling
| Factor | Source | Activity | Reference |
|---|---|---|---|
| Nucleoplasmin | Xenopus | Removal of X and Y from, and assembly of H2A and H2B onto, sperm chromatin | Philpott et al. (2000) |
| N1 | Xenopus | Assembly of H3 and H4 onto new chromatin | Dilworth et al. (1987) |
| HIRA | Xenopus | Replication-independent nucleosome assembly onto plasmids | Ray-Gallet et al. (2002) |
| Template-activating factor Iβ | Xenopus | Dispersion of sperm chromatin, removal of sperm-specific basic proteins | Matsumoto et al. (1999) |
| Nucleosome-assembly protein-1 | Drosophila | Dispersion of Xenopus sperm chromatin in vitro | Ito et al. (1996) |
| p22 | Drosophila | Dispersion of Xenopus sperm chromatin in fly oocyte extract; release of sperm-specific proteins | Kawasaki et al. (1994) |
| DF 31 | Drosophila | Dispersion of Xenopus sperm chromatin in vitro; release of sperm-specific proteins | Crevel and Cotterill (1995) |
| Sésame gene product | Drosophila | Formation of male pronucleus | Loppin et al. (2001) |
| – | Mytilus (surf clam) | Decondenses spermatozoa in egg extracts | Rice et al. (1995) |
| – | Spisula solidissima (mussel) | Protein characteristics similar to nucleoplasmin | Herlands and Maul (1994) |
Fig. 2.
Expression of mRNA encoding nucleoplasmin 3 (Npm3) and nucleosome-assembly protein 1 (NAP-1) in mammalian oocytes at different stages of development. RT–PCR was performed using cDNA prepared from 30 oocytes. No RT: reverse transcriptase was omitted from the sample; M: 100-nt molecular size markers; GI: growing, incompetent; GC: growing, competent; FG: fully grown; MII: metaphase II.
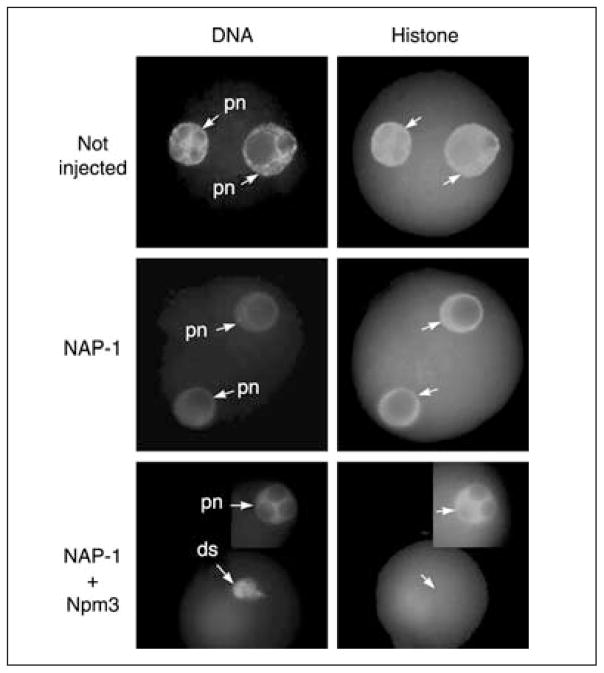
By microinjecting antisense oligonucleotides, the present authors substantially reduced the concentrations of Npm-3 and NAP-1 mRNAs in mature oocytes. Oocytes were then inseminated to test their ability to assemble histones onto paternal DNA. NAP-1 antisense oligonucleotides had no effect on sperm chromatin remodelling. In contrast, Npm3 antisense oligonuclotides alone, or Npm3 and mNAP antisense oligonucleotides together, prevented histone assembly and formation of the male pronucleus in a significant number of oocytes (Fig. 3; Table 2). These experiments indicate that Npm-3 may serve a function similar to Xenopus nucleoplasmin during paternal chromatin remodelling at fertilization. In addition, a histone-binding protein that is closely related to N1 (which interacts with H3–H4 tetramers) is encoded by the mammalian NASP gene (Richardson et al., 2000). A partial cDNA that matches NASP is present in a mouse oocyte cDNA library (http://lgsun.grc.nia.nih.gov/cDNA), indicating that it may participate in assembling histones H3 and H4 on to the paternal chromatin.
Fig. 3.
Effect of microinjection of antisense oligonucleotides on sperm chromatin morphology and histone assembly after fertilization in mammals. Oocytes arrested at prophase I were injected with oligonucleotides targeting nucleosome-assembly protein 1 (NAP-1) or nucleoplasmin 3 (Npm3), allowed to mature to metaphase II, and then fertilized. The zygotes were fixed and stained 4 h later using 4,6-diamino-2-phenylindole (DAPI) to assess maternal and paternal chromatin morphology (left panels) and an anti-histone antibody to assess histone content (right panels). Top: control uninjected oocyte. Two pronuclei (pn) containing immunodetectable histones are visible. Middle: oocyte injected with NAP-1 antisense oligonucleotides. Two pronuclei containing immunodetectable histone are visible. Bottom: oocyte injected with NAP-1 and Npm3 antisense oligonucleotides. Female pronucleus (inset) is fully developed and contains histones. Paternal chromatin has not progressed beyond the dispersed (ds) state and does not contain detectable histones.
Table 2.
Histone assembly onto paternal chromatin in mammalian oocytes injected with oligonucleotides targeting nucleoplasmin 3 (Npm3) or nucleosome-assembly protein 1 (NAP-1)
| Oocyte treatment | Number of experiments | Number of oocytes | Histones on paternal chromatin
|
|
|---|---|---|---|---|
| Detected | Not detected | |||
| Not injected | 3 | 23 | 22 | 1 |
| Npm3 oligonucleotides | 3 | 29 | 27 | 2 |
| NAP-1 oligonucleotides | 3 | 24 | 16 | 8 |
| Npm3 + NAP-1 oligonucleotides | 3 | 20 | 11 | 9 |
As described earlier, changes in acetylation of histone also occur during the first cell cycle after fertilization. This indicates that the oocyte contains histone acetyl-transferases (HATs) and histone deacetylases (HDACs). Xenopus has been useful in isolating these proteins, because of the large size of the oocytes, and the large maternal stores of histones in comparison with mammalian oocytes. HAT1 has been isolated from frog oocyte extracts in complexes with histone-binding RbAp48 and 14-3-3 proteins (Imhof and Wolffe, 1999). This complex acetylates free histone H4, but not nucleosomal H4, and is localized in the oocyte nucleus, where acetylated free histones are stored. Regarding deacetylases, an oocyte-specific form of HDAC is synthesized during oogenesis and persists until the mid-blastula stage (Ladomery et al., 1997). Maternal HDAC forms complexes with RbAp28–46 and possesses deacetylase activity (Ryan et al., 1999). Similar proteins will likely be found in mammalian oocytes, although pools as large as those found in Xenopus may be unnecessary, given that the embryonic genome activates much earlier in mammals than in frogs.
The assembly of histones is only one component of the global remodelling of the sperm chromatin at fertilization. Subsequent chromatin maturation may involve the positioning of nucleosomes as well as the addition of other DNA-binding proteins. For example, ACF, which contains the chromatin-remodelling engine ISWI, was identified from Drosophila by its ability, in concert with NAP-1, to assemble and also position nucleosomes (Ito et al., 1999). Members of SWI–SNF are involved in chromatin remodelling and could function at fertilization. Two mammalian homologues, BRM and BRG, are present in mouse eggs and embryos. Deletion of BRM permits viable offspring, possibly due to BRG replacing BRM (Reyes et al., 1998).
Chromatin remodelling activity develops during oogenesis
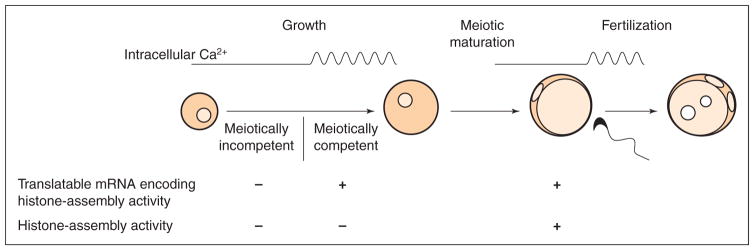
As the ability to remodel sperm chromatin resides in the oocyte, it must develop during oogenesis. McLay and Clarke (1997) and McLay et al. (2002) used a functional assay to test the ability of growing and maturing oocytes to assemble histones on to paternal chromatin, which they termed a histone-assembly activity. Fully grown oocytes were inseminated, incubated for different periods, then fixed and stained with DAPI to observe chromatin morphology, and with specific antibodies to monitor histone assembly. With this assay, it was found that when maturing oocytes were inseminated shortly after germinal vesicle breakdown (GVBD), histones were assembled onto the paternal chromatin. However, histone assembly failed to occur when protein synthesis was inhibited during maturation; moreover, this failure could not be rescued by microinjecting histones into the oocyte. These results indicate that histone-assembly activity develops during meiotic maturation through a process requiring new protein synthesis.
This assay was applied to oocytes at different stages of growth (McLay et al., 2002). The results revealed that oocytes acquired histone-assembly activity at the same time that they acquired meiotic competence (defined as the ability to undergo GVBD in vitro). However, these activities were separable, as when incompetent oocytes were cultured in vitro for several days to allow them to acquire meiotic competence, they did not necessarily also acquire histone-assembly activity. As competent, but not incompetent, oocytes manifest spontaneous Ca2+ oscillations, incompetent oocytes were treated with SrCl2. This triggered Ca2+ oscillations in a fraction of the oocytes, and a similar fraction was now able to assemble histones onto paternal chromatin after insemination. This finding indicates that Ca2+ may promote development of the histone-assembly activity, although the nature of the link is obscure.
These results may be incorporated into the following model (Fig. 4). Growing oocytes accumulate mRNAs encoding factors such as NAP-1 or Npm3 that are required for the development of histone-assembly activity. These mRNAs reach a threshold amount or become translatable at the time of meiotic competence, possibly under the influence of Ca2+-regulated events occurring during growth. Upon entry of the oocyte into metaphase of meiotic maturation, the mRNAs are translated to produce functional histone-assembly activity. In this context, it may be noted that the 3′-untranslated region of the mRNA encoding Npm3 contains a putative cytoplasmic polyadenylation element, a sequence in the 3′-untranslated region of other mRNA species that controls their translational activation during maturation (reviewed by de Moor and Richter, 2001). Thus, at the time of fertilization, histone-assembly activity is functional and assembles histones onto the paternal chromatin, completing an early, critical step in the production of a transcriptionally competent paternal genome.
Fig. 4.
A model for the development during oogenesis of the activity that assembles histones on to paternal chromatin at fertilization in mammals. During growth, oocytes accumulate mRNAs encoding proteins that mediate DNA replication-independent assembly of histones on to chromatin. These mRNAs become translatable or reach a threshold level at the time that oocytes acquire meiotic competence, and through a process promoted by Ca2+ oscillations. During meiotic maturation the mRNAs are translated to generate histone-assembly activity. Although the activity is functional in maturing oocytes, it normally is not manifested until fertilization activates fully mature metaphase II oocytes.
Paternal chromatin remodelling and somatic nuclear remodelling
Chromatin remodelling by oocytes has attracted particular interest of late because of its potential role in reprogramming somatic nuclei during cloning. Somatic nuclei transferred into an oocyte undergo marked changes in biochemical composition, likely including the import of chromatin remodelling factors (reviewed in Kikyo and Wolffe, 2000). The identification of the molecular players that regulate remodelling of the paternal chromatin at fertilization may lead to better insight into this process. In support of this, nucleoplasmin plays a role in removing linker histones H1 and H1* from the chromatin of erythrocyte nuclei in Xenopus egg extracts and replacing them with the oocyte-specific variant, H1M, and HMG1 (Dimitrov and Wolffe, 1996). This change produces chromatin similar to that found in early embryos, and also an increase in gene transcription. Perhaps functionally or structurally similar proteins play analogous roles in reprogramming somatic nuclei in mammalian oocytes after nuclear transplantation.
Acknowledgments
The authors thank the members of their laboratory for critical review of the manuscript. Owing to a restriction on the number of references that may be listed, recent reports have been cited, which will lead readers to previous relevant studies. Work in the authors’ laboratory is supported by the Canadian Institutes of Health Research.
References
Key reference are identified by asterisks.
- *.Adenot PG, Szollosi MS, Geze M, Renard JP, Debey P. Dynamics of paternal chromatin changes in live one-cell mouse embryo after natural fertilization. Molecular Reproduction and Development. 1991;28:23–34. doi: 10.1002/mrd.1080280105. [DOI] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Ao A, Erickson RP, Winston RM, Handyside AH. Transcription of paternal Y-linked genes in the human zygote as early as the pronucleate stage. Zygote. 1994;2:281–287. doi: 10.1017/s0967199400002100. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Balakier H, Tarkowski AK. The role of germinal vesicle karyoplasm in the development of male pronucleus in the mouse. Experimental Cell Research. 1980;128:79–85. doi: 10.1016/0014-4827(80)90389-4. [DOI] [PubMed] [Google Scholar]
- *.Balhorn R. A model for the structure of chromatin in mammalian sperm. Journal of Cell Biology. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Experimental Cell Research. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nature Genetics. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- Clarke HJ, Masui Y. Transformation of sperm nuclei to metaphase chromosomes in the cytoplasm of maturing oocytes of the mouse. Journal of Cell Biology. 1986;102:1039–1046. doi: 10.1083/jcb.102.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. DF 31, a sperm decondensation factor from Drosophila melanogaster: purification and characterization. EMBO Journal. 1995;14:1711–1717. doi: 10.1002/j.1460-2075.1995.tb07160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proceedings National Academy of Sciences USA. 2001;98:13, 734–13, 738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, Richter JD. Translational control in vertebrate development. International Review of Cytology. 2001;203:567–608. doi: 10.1016/s0074-7696(01)03017-0. [DOI] [PubMed] [Google Scholar]
- *.Dilworth SM, Black SJ, Laskey RA. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO Journal. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, Gilbert DM. Mammalian nuclei become licensed for DNA replication during late telophase. Journal of Cell Science. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducibella T, Huneau D, Angelichio E, Xu Z, Schultz RM, Kopf GS, Fissore R, Madoux S, Ozil JP. Egg-to-embryo transition is driven by differential responses to Ca2+ oscillation number. Developmental Biology. 2002;250:280–291. [PubMed] [Google Scholar]
- Ecklund PS, Levine L. Mouse sperm basic nuclear protein –electrophoretic characterization and fate after fertilization. Journal of Cell Biology. 1975;66:251–262. doi: 10.1083/jcb.66.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO Journal. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. Journal of Cell Biology. 2000;151:1591–1597. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlands L, Maul GG. Characterization of a major nucleoplasmin-like germinal vesicle protein which is rapidly phosphorylated before germinal vesicle breakdown in Spisula solidissima. Developmental Biology. 1994;161:530–537. doi: 10.1006/dbio.1994.1051. [DOI] [PubMed] [Google Scholar]
- Imhof A, Wolffe AP. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry. 1999;38:13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Molecular Cellular Biology. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes and Development. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LJM, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: a tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Philpott A, Avilion AA, Berrios M, Fisher PA. Chromatin decondensation in Drosophila embryo extracts. Journal of Biological Chemistry. 1994;269:10 169–10, 176. [PubMed] [Google Scholar]
- Kikyo N, Wolffe AP. Reprogramming nuclei: insights from cloning, nuclear transfer and heterokaryons. Journal of Cell Science. 2000;113:11–20. doi: 10.1242/jcs.113.1.11. [DOI] [PubMed] [Google Scholar]
- Kopecny V, Pavlok A. Autoradiographic study of mouse spermatozoan arginine-rich nuclear protein in fertilization. Journal of Experimental Zoology. 1975;191:85–96. doi: 10.1002/jez.1401910109. [DOI] [PubMed] [Google Scholar]
- Ladomery M, Lyons S, Sommerville J. Xenopus HDm, a maternally expressed histone deacetylase, belongs to an ancient family of acetyl-metabolising enzymes. Gene. 1997;198:275–280. doi: 10.1016/s0378-1119(97)00325-9. [DOI] [PubMed] [Google Scholar]
- Leno GH, Mills AD, Philpott A, Laskey RA. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. Journal of Biological Chemistry. 1996;271:7253–7256. doi: 10.1074/jbc.271.13.7253. [DOI] [PubMed] [Google Scholar]
- Loppin B, Berger F, Couble P. The Drosophila maternal gene sesame is required for sperm chromatin remodeling at fertilization. Chromosoma. 2001;110:430–440. doi: 10.1007/s004120100161. [DOI] [PubMed] [Google Scholar]
- MacArthur CA, Shackleford GM. Npm3: a novel, widely expressed gene encoding a protein related to the molecular chaperones nucleoplasmin and nucleophosmin. Genomics. 1997;42:137–140. doi: 10.1006/geno.1997.4353. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. The ability to organize sperm DNA into functional chromatin is acquired during meiotic maturation in murine oocytes. Developmental Biology. 1997;186:73–84. doi: 10.1006/dbio.1997.8581. [DOI] [PubMed] [Google Scholar]
- *.McLay DW, Carroll J, Clarke HJ. The ability to develop an activity that transfers histones onto sperm chromatin is acquired with meiotic competence during oocyte growth. Developmental Biology. 2002;241:195–206. doi: 10.1006/dbio.2001.0499. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Yanagimachi H, Tateno H, Usui N, Yanagimachi R. Decondensation of the mouse sperm nucleus within the interphase nucleus. Zygote. 1998;6:39–45. doi: 10.1017/s0967199400005062. [DOI] [PubMed] [Google Scholar]
- Majumder S, Zhao Z, Kaneko K, DePamphilis ML. Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO Journal. 1997;16:1721–1731. doi: 10.1093/emboj/16.7.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszewski M, Borsuk E, Koziak K, Maluchnik D, Tarkowski AK. Delayed sperm incorporation into parthenogenetic mouse eggs: sperm nucleus transformation and development of resulting embryos. Molecular Reproduction and Development. 1999;54:303–310. doi: 10.1002/(SICI)1098-2795(199911)54:3<303::AID-MRD11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Nagata K, Miyaji-Yamaguchi M, Kikuchi A, Tsujimoto M. Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins. Molecular Cellular Biology. 1999;19:6940–6952. doi: 10.1128/mcb.19.10.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Embryogenesis – demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Molecular Reproduction and Development. 1992;31:170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- Nixon VL, Levasseur M, McDougall A, Jones KT. Ca2+ oscillations promote APC/C-dependent cyclin B degradation during metaphase arrest and completion of meiosis in fertilizing mouse eggs. Current Biology. 2002;12:746–750. doi: 10.1016/s0960-9822(02)00811-4. [DOI] [PubMed] [Google Scholar]
- Nonchev S, Tsanev R. Protamine-histone replacement and DNA replication in the male mouse pronucleus. Molecular Reproduction and Development. 1990;25:72–76. doi: 10.1002/mrd.1080250113. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Current Biology. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Perreault SD. Chromatin remodeling in mammalian zygotes. Mutation Research. 1992;296:43–55. doi: 10.1016/0165-1110(92)90031-4. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- Philpott A, Krude T, Laskey RA. Nuclear chaperones. Seminars in Cell and Developmental Biology. 2000;11:7–14. doi: 10.1006/scdb.1999.0346. [DOI] [PubMed] [Google Scholar]
- *.Rastelli L, Robinson K, Xu YB, Majumder S. Reconstitution of enhancer function in paternal pronuclei of one-cell mouse embryos. Molecular Cellular Biology. 2001;21:5531–5540. doi: 10.1128/MCB.21.16.5531-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Molecular Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO Journal. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Garduno R, Itoh T, Katagiri C, Ausio J. Nucleoplasmin-mediated decondensation of Mytilus sperm chromatin. Identification and partial characterization of a nucleoplasmin-like protein with sperm-nuclei decondensing activity in Mytilus californianus. Biochemistry. 1995;34:7563–7568. doi: 10.1021/bi00023a001. [DOI] [PubMed] [Google Scholar]
- Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O’Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. Journal of Biological Chemistry. 2000;275:30 378–30 386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- Rodman TC, Pruslin FH, Hoffmann HP, Allfrey VG. Turnover of basic chromosomal-proteins in fertilized-eggs – a cytoimmunochemical study of events in vivo. Journal of Cell Biology. 1981;90:351–361. doi: 10.1083/jcb.90.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J, Llinas AJ, White DA, Turner BM, Sommerville J. Maternal histone deacetylase is accumulated in the nuclei of Xenopus oocytes as protein complexes with potential enzyme activity. Journal of Cell Science. 1999;112:2441–2452. doi: 10.1242/jcs.112.14.2441. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Developmental Biology. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC zeta: a sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. Development. 2002;129:3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- Schatten G, Simerly C, Palmer DK, Margolis RL, Maul G, Andrews BS, Schatten H. Kinetochore appearance during meiosis, fertilization and mitosis in mouse oocytes and zygotes. Chromosoma. 1988;96:341–352. doi: 10.1007/BF00330700. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Szollosi MS, Borsuk E, Szollosi D. Relationship between sperm nucleus remodelling and cell cycle progression of fragments of mouse parthenogenotes. Molecular Reproduction and Development. 1994;37:146–156. doi: 10.1002/mrd.1080370205. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Ray PF, Ao A, Winston RM, Handyside AH. Paternal transcripts for glucose-6-phosphate dehydrogenase and adenosine deaminase are first detectable in the human preimplantation embryo at the three- to four-cell stage. Molecular Reproduction and Development. 1997;48:442–448. doi: 10.1002/(SICI)1098-2795(199712)48:4<442::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Usui N, Yanagimachi R. Behavior of hamster sperm nuclei incorporated into eggs at various stages of maturation, fertilization, and early development. The appearance and disappearance of factors involved in sperm chromatin decondensation in egg cytoplasm. Journal of Ultrastructure Research. 1976;57:276–288. doi: 10.1016/s0022-5320(76)80117-7. [DOI] [PubMed] [Google Scholar]
- Verreault A. De novo nucleosome assembly: new pieces in an old puzzle. Genes and Development. 2000;14:1430–1438. [PubMed] [Google Scholar]
- Wassarman PM, Mrozak SC. Program of early development in the mammal: synthesis and intracellular migration of histone H4 during oogenesis in the mouse. Developmental Biology. 1981;84:364–371. doi: 10.1016/0012-1606(81)90405-x. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Developmental Biology. 1993;159:366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Beaujean N, de Sousa PA, Dinnyes A, King TJ, Paterson LA, Wells DN, Young LE. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- Witkowska A. Pronuclear development and the 1st cleavage division in polyspermic mouse eggs. Journal of Reproduction and Fertility. 1981;62:493–498. doi: 10.1530/jrf.0.0620493. [DOI] [PubMed] [Google Scholar]
- Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo – developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120:2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- Wouters-Tyrou D, Martinage A, Chevaillier P, Sautiere P. Nuclear basic proteins in spermiogenesis. Biochimie. 1998;80:117–128. doi: 10.1016/s0300-9084(98)80018-7. [DOI] [PubMed] [Google Scholar]
- Wright SJ, Longo FJ. Sperm nuclear enlargement in fertilized hamster eggs is related to meiotic maturation of the maternal chromatin. Journal of Experimental Zoology. 1988;247:155–165. doi: 10.1002/jez.1402470207. [DOI] [PubMed] [Google Scholar]
- Yelick PC, Balhorn R, Johnson PA, Corzett M, Mazrimas JA, Kleene KC, Hecht NB. Mouse protamine 2 is synthesized as a precursor whereas mouse protamine 1 is not. Molecular Cell Biology. 1987;7:2173–2179. doi: 10.1128/mcb.7.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalensky AO, Tomilin NV, Zalenskaya IA, Teplitz RL, Bradbury EM. Telomere–telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells. Experimental Cell Research. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]