Abstract
Mammalian homologues of genes that control oogenesis in other organisms may play similar roles in mammalian ovarian development. In Drosophila melanogaster, GUSTAVUS (GUS) protein physically interacts with and is necessary for the proper posterior localization of VASA protein, and thus is required for specification of germ cells. We identified two mouse genes, SSB-1 and SSB-4 (SPRY domain SOCS box protein), whose protein products share 75% identity and are each approximately 70% identical to Drosophila GUS. Both SSB-1 and SSB-4 mRNA were detectable in mouse ovaries by Northern blotting of total and poly(A) + RNA, but were expressed in few other tissues. SSB-1 was detectable in testes, although the 3′-untranslated region of the mRNA was considerably shorter than the ovarian mRNA. In situ hybridization and RT-PCR analysis of ovaries revealed that both genes were expressed in granulosa cells at all stages of follicular development. In contrast, expression was barely detectable in in oocytes. Immunoblotting analysis revealed that SSB-1 protein was present in follicles at different stages of growth, and immunocytochemistry confirmed that SSB-1 and SSB-4 were detectable in granulosa cells of primary and subsequent stage follicles and that they were present in both mural and cumulus granulosa cells of antral follicles. These results establish that GUS-related proteins, which in Drosophila are restricted to the germ cells, are in the mouse instead expressed in the granulosa cells and are present throughout folliculogenesis. Based on their tissue-restricted pattern of expression and apparent abundance in granulosa cells, we propose that SSB-1 and SSB-4 play key roles in regulating granulosa cell physiology.
Introduction
Development of a mature fertilizable oocyte requires the coordinated expression and interaction of a variety of gene products in the oocyte and the surrounding granulosa cells. Understanding the molecular basis of follicular development is a prerequisite for revealing the genetic basis of differential fertility, the aetiology of some types of infertility, and for improving assisted reproductive technology. To this end, several strategies have been developed to identify oocyte-specific genes whose function can then be experimentally addressed. One approach has used in silico analysis of public databases to identify expressed sequence tags that are enriched in cDNA libraries from oocytes (Rajkovic et al. 2001): this has uncovered a number of key genes required for oocyte growth (Rajkovic et al. 2004). A second approach has been to use subtractive hybridization procedures to identify transcripts enriched in oocytes compared with another cell type: this has also yielded previously unknown oocyte-specific genes (Zeng & Schultz 2003, Vallee et al. 2005).
Another strategy is to search for mammalian homologues of genes already known to play key roles during oogenesis in other organisms. The potential advantage of this approach is that the gene candidates need not be restricted to those expressed mainly or exclusively in the oocyte. Moreover, they can be selected from genes that have already been demonstrated experimentally to play a role in oogenesis. A rich source of these genes is the fly, Drosophila melanogaster, in which the genetic basis of oogenesis has been intensively studied. For example, an apparent mammalian homologue of the Drosophila nanos gene, which is required in the fly to prevent germ-line stem cells from undergoing differentiation (Wang & Lin 2004), is required in mice for the proliferation or migration of primordial germ cells (Tsuda et al. 2003). Genes closely related to Drosophila tudor, which encodes a component of the pole plasm where the primordial germ cells develop, and staufen have also been identified in mammalian male germ cells, although their function in mammals is not yet known (Saunders et al. 2000, Chuma et al. 2003, Smith et al. 2004).
vasa is another gene first uncovered in Drosophila. It encodes an RNA helicase and is a component of the pole plasm that is inherited by and specifies the germ cells. vasa is required to establish localized translation of at least two mRNAs, oskar (no known mammalian homologue) and nanos, within the polar plasm. VASA protein itself accumulates at the pole plasm, although the mRNA is uniformly distributed in the oocyte (reviewed in Johnstone and Lasko (2001). A murine homologue of vasa (mouse vasa homologue, Mvh) has been identified (Fujiwara et al. 1994). Mvh is expressed in embryonic germ cells as well as in small oocytes and in male germ cells. Genetic deletion of Mvh caused arrest of developing spermatocytes around the stage of pachytene but, surprisingly, has no apparent effect on oogenesis (Tanaka et al. 2000).
Recently, a novel protein was identified in the fly, and was termed GUSTAVUS (GUS); this protein interacts physically with VASA and is required for localization of VASA at the pole plasm and thus for specification of the germ cells (Styhler et al. 2002). GUS contains two well-conserved protein domains: a SPRY domain, which was first identified in ryanodine receptors and is thought to mediate protein–protein interactions (Wang et al. 2005); and an SOCS box, which has been implicated in ubiquitination of proteins, thus targeting them for proteasomal degradation. To determine whether a GUS homologue might be expressed and functional in germ cells, we undertook a search for murine genes encoding proteins that were similar to GUS. We report that the protein products of the genes SSB-1 and SSB-4 (SPRY domain SOCS box protein) bear substantial similarity to GUS. Unexpectedly, although SSB-1 and SSB-4 are expressed in the ovary, they are barely detectable in the germ cells. Rather, and in contrast to gus, they are expressed in granulosa cells of the ovarian follicle. We discuss potential roles of these proteins in granulosa cell function.
Materials and Methods
Animals
Experiments were carried out using CD-1 mice (Charles River, St-Constant, Quebec, Canada). All animal procedures followed the guidelines of the Canadian Council on Animal Care and were approved by the Animal Care Committee of the McGill University Health Centre.
Northern blot hybridization
Sequences within the coding sequences of SSB-1 and the coding and part of the 3′-untranslated region (UTR) of SSB-4 were amplified by PCR. Antisense RNA probes were prepared from the PCR products by ligation to a T7 promoter adapter (Lig’nScribe, Roche) followed by incubation with T7 RNA polymerase (Roche) in the presence of digoxygenin-labelled UTP (Roche). Mouse total RNA from different tissues (Ambion, Austin, TX, USA) was stored at −80 °C. RNA (2 μg/lane) was separated on denaturing agarose gels, transferred by downward capillary blotting (Turboblotter, Mandel Scientific, Guelph, Ontario, Canada) to a nylon membrane (Roche) and fixed by exposure to u.v. light. Membranes were hybridized with the RNA probes and bound probe visualized using a commercial detection kit (Roche). Poly(A) + RNA was isolated from total RNA of mouse liver, ovary and kidney (Oligotex, Qiagen) and analysed as for total RNA.
In situ hybridization
Antisense and sense RNA probes were prepared as described above. Ovaries were fixed in freshly prepared 4% para-formaldehyde, dehydrated, embedded in paraffin, sectioned at 7 μm and mounted on glass slides. Following rehydration, hybridization was carried out overnight at 45 °C in hybridization buffer (40% deionized formamide, 10% dextran sulfate, 4 × SSC, 1 × Denhardt’s solution, 10 mM dithiothreitol (DTT), 1 mg/ml yeast DNA, 1 mg/ml denatured and sheared salmon sperm DNA). Probe was present at 0.5–1 ng/μl hybridization buffer. Bound probe was revealed using an anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche). Activity was detected using NBT/BCIP (Roche) as the chromogen following the manufacturer’s directions.
Collection of ovarian oocytes and ovulated eggs
Granulosa–oocyte complexes were collected from mice up to 3 weeks of age by either enzymatic digestion of the ovaries or by puncturing the follicles with a 30G needle, as previously described (McLay & Clarke 1997). To collect ovulated eggs at metaphase II, 7- to 8-week-old female mice were injected with 7.5 IU pregnant mares’ serum gonadotrophin (Sigma) followed by 5 IU human chorionic gonadotrophin (hCG) 44 h later to induce superovulation. Egg masses were recovered at 16 h post-hCG and oocytes were freed from the granulosa cells using hyaluronidase. The granulosa–oocyte complexes or oocytes were transferred using a pipette to storage at −80 °C for up to 4 weeks.
Reverse transcription and PCR
RNA was extracted from oocytes using Trizol (Invitrogen) and reverse transcribed into cDNA using Moloney murine leukemia virus (MMLV) as previously described (Mohamed et al. 2004). cDNA corresponding to 15 oocyte-equivalents was subjected to PCR using a thermal cycler (Biometra UNO Thermoblock, version 3.30) using the following conditions: 95 °C for 5 min and (94 °C for 45 s, 56 °C for 45 s, 72 °C for 1 min) × 35 cycles; the last cycle was followed by a 5-min extension at 72 °C.
Generation of antibodies against SSB-1 and SSB-4
Peptides corresponding to amino acids 15–31 (DPTYR-PLKQELQGLDYC) of SSB-1 and 16–32 (EPALRPAKRELR-GLEPG) of SSB-4 were synthesized and injected into rabbits to generate antibodies (Biosynthesis, TX, USA). Affinity-purified antibodies were prepared by the supplier.
Immunoblotting and immunoprecipitation
Ovaries were rapidly homogenized in 2 × loading buffer (125 mM Tris, pH 6.8, 20% glycerol, 4% SDS, 0.1% bromophenol blue, 10% β-mercaptoethanol), denatured at 95 °C for 5 min, and centrifuged to pellet debris. Granulosa cell, granulosa–oocyte and oocyte extracts were prepared by transferring the cells into an Eppendorf tube, withdrawing excess medium and adding an appropriate volume of sample loading buffer, and denaturing as above. Protein electrophoresis, transfer to PVDF membranes and immunoblotting were carried out as previously described (Allard et al. 2002). Primary antibodies were used at a dilution of 1:1000, secondary antibodies conjugated to horseradish peroxidase (HRP, Promega) at a concentration of 1:5000. HRP activity was revealed using ECL + (Amersham) following the manufacturer’s directions.
For immunoprecipitation, ovaries of 8-week-old CD-1 mice were homogenized in cold lysis buffer (50 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1% NP-40) with one tablet of protease inhibitor cocktail (Roche) added to every 10 ml buffer. The sample was then centrifuged at 14 000 g at 4 °C for 15 min, and the supernatant was collected; 10 μg of antibody were added to the supernatant (containing 5–10 mg total protein) in a final volume of 600 μl and mixed by rotation for 4 h at 4 °C. Protein A-Sepharose beads (150 μl; Upstate, Charlottesville, VA, USA) were washed four times with PBS, then twice with lysis buffer and added to 150 μl of the protein–antibody mixture. This was mixed by rotation overnight at 4 °C. Following a brief centrifugation, the supernatant was removed and the beads washed three times with lysis buffer; 150 μl of 2 × loading buffer were added to the beads and the mixture was boiled for 5 min. Samples were run on 10% SDS-polyacrylamide gels and subjected to immunoblotting.
Immunostaining
Ovaries were excised and fixed overnight at 4 °C in freshly prepared 4% para-formaldehyde, washed in PBS, dehydrated and embedded in paraffin. Five micrometer sections were exposed to 3% H2O2 in absolute methanol for 10 min, then washed three times in H2O. The slides were placed in an antigen retrieval solution (9 ml of 0.1 M citric acid and 41 ml of 0.1 M sodium citrate in 450 ml H2O) and boiled for 10 min. After cooling, the slides were washed three times in PBS containing 0.1% Tween-20 (PBST). They were treated with a blocking solution (PBST, 3% BSA) for 1 h, then incubated overnight at 4 °C with affinity-purified anti-SSB diluted 1:200 in blocking solution. After three 30-min washes in PBST, the sections were incubated with biotinylated secondary antibodies (ABC kit, Vector Laboratories, Burlington, Ontario, Canada), washed and incubated with the ABC complex following the manufacturer’s instructions. The sections were washed and stained using the AEC color substrate (Vector Laboratories), then washed and counterstained with hematoxylin for 30 s. Slides were mounted in glycerol.
Cell culture and transfection conditions
KK-1 and NT-1 cells were generously supplied by Professor I Huhtaniemi (Imperial College London, UK). HeLa and DC3 cells were generously supplied by Professor R Farookhi (McGill University). CHO cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), or DMEM/F12 for the KK-1 and NT-1 cells, supplemented with 10% fetal bovine serum and transfected at 70–90% confluency. The coding sequences of SSB-1 and SSB-4 were inserted into pTriEX-2 and sequenced to confirm their identity. For transfection, 4 μg plasmid and 10 μl lipofectamine (Invitrogen) were incubated separately in serum-free medium for 5 min, combined and allowed to stand for 20 min, then added drop-wise to the plate containing the cells. Culture medium was changed 6 h after transfection. Cells were harvested by scraping or trypsinization 24 and 48 h after transfection.
Primers
Northern analysis
SSB-1 (497 nt): forward, ATAACGACCGTTCGCTCAAC; reverse, AGTCCGTTCAGTAGCGCAT. SSB-4 coding region (823 nt): forward, CGGGATCCATGGGTCAGAAGC; reverse, CCCAAGCTTTCACTGGTACTG. SSB-4 3′-UTR (533 nt): forward, AAAAGCACCTGGCCTTACCT; reverse, CACAAAGATGCCAAATGGTG.
In situ hybridization
SSB-1 (236 nt): forward, GGCGTAACCGTCTCTACCAC; reverse, AGTCCGTTCAAGTAGCGCAT. SSB-4 coding region (280 nt): forward, GCACCCAGTAGCCCAGAGCA; reverse, AAGGAATCTGGCAGAGCAAA. SSB-4 3-UTR (289 nt): forward, AAAAGCACCTGGCCTTACCT; reverse, CTTCACCCTGAATGATGGCT.
RT-PCR (each pair spans an intron)
ZP3 (467 nt): forward, GCACCTTCCTACTCCACGAC; reverse, ATCCACCGTGAACTGGAGAG. FSH-R (401 nt): forward, GAGGCCTTCCAGAATCTTCC; reverse, CTGGCCCTCAACTTCTTCAG. SSB-1 (558 nt): forward, ATAACGACCGTTCGCTCAAC; reverse, GAACGCCGGCA-CAGGTCCAT. SSB-4 (576 nt): forward, CCGGATACAAGTC-GAGAGGA; reverse, TGCTCTAGGCTACTGGGTGC.
Results
Homologues of the Drosophila gus gene are present in the mouse and other vertebrates
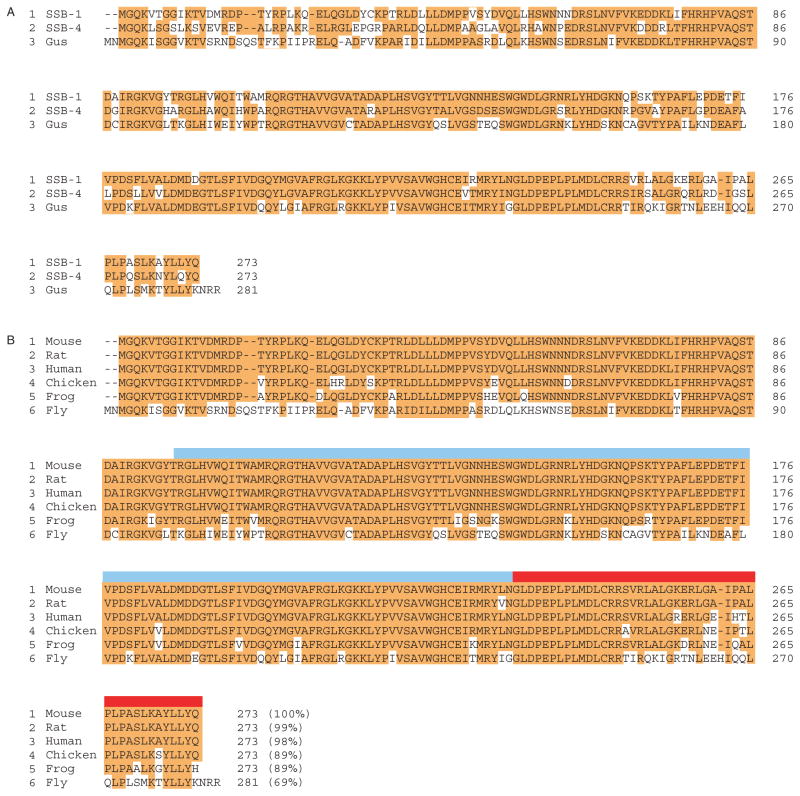
To search for mouse genes related to gus, we screened GenBank using the full-length Drosophila gus sequence (CG2944_RB). This screen identified two closely related genes, SSB-1 (SPRY-domain SOCS box protein 1, NP_083311) and SSB-4 (NP_660116), whose protein products are approximately 75% identical and each approximately 70% identical to Drosophila GUS (Fig. 1A). Further screening using the SSB-1 sequence revealed apparent SSB-1 homologues in both mammalian and non-mammalian species (Fig. 1B). All of the SSB-1-related proteins contain a SPRY domain and a SOCS box, as shown in the figure. Moreover, the protein sequences are remarkably conserved. Mouse SSB-1, for example, is 98% identical to human SSB-1 and 89% identical to Xenopus SSB-1. Thus, SSB-1 and presumably SSB-4 are to be highly conserved. Two other SSB family proteins (SSB-2 and SSB-3), which share less sequence similarity with SSB-1 and SSB-4, have also been identified (Wang et al. 2005).
Figure 1.
(A) Amino acid sequence of mouse SSB-1 (NP_083311) and SSB-4 (NP_660116) and Drosophila GUS (CG2944_RB). Identical residues are highlighted in red. SSB-1 and SSB-4 are about 75% identical and each about 70% identical to GUS. (B) Amino acid sequence of SSB-1 in different organisms. Identical residues are highlighted in red. Note that only four amino acids differ between human and mouse sequences. The blue band indicates the SPRY domain, the tan band the SOCS box. Gus, Gustavus; Fly, Drosophila; Frog, Xenopus.
SSB-1 and SSB-4 mRNAs are expressed in the mouse ovary and testis
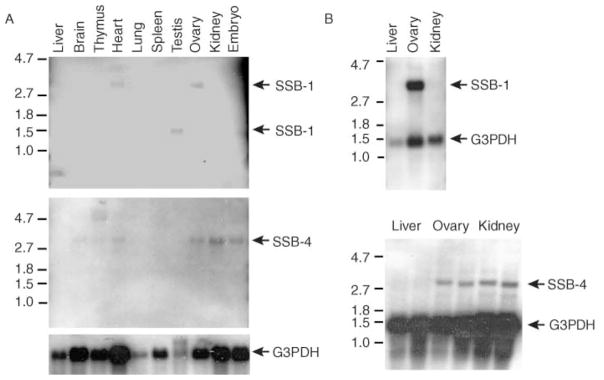
To examine the expression of SSB-1 and SSB-4 in mouse tissues, we screened Northern blots using probes corresponding to the coding regions of SSB-1 and SSB-4 and the 3′-UTR of SSB-4. Among the ten tissues screened, the SSB-1 probe detected a single band of about 3 kb in heart and ovary and of about 1.4 kb in testis (Fig. 2). No expression was detectable under these conditions in liver, brain, thymus, lung, spleen or kidney. To confirm that the RNA detected was potentially translatable, poly(A) + RNA from the mouse liver, ovary and kidney was isolated and probed. SSB-1 transcripts were detected as a prominent 3 kb band in ovary but not in liver and kidney (Fig. 2).
Figure 2.
Analysis of SSB-1 and SSB-4 expression by Northern blotting. Total RNA (A) or poly(A) + RNA (B) were analysed using probes corresponding to SSB-1 or SSB-4. The SSB-1 probe recognizes a species of approximately 3 kb in ovary and heart and a species of approximately 1.4 kb in testis. The SSB-4 probe recognizes a species of about 2.8 kb in heart, ovary, kidney and whole embryo. Glucose-3-phosphate dehydrogenase (G3PDH) was used as a loading control.
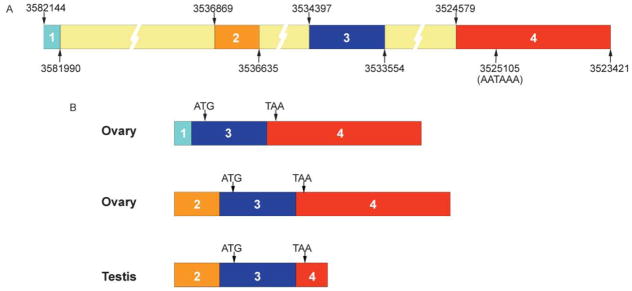
By comparing the published cDNA (BC057563) and genomic (NT_039258.4) sequences of SSB-1, we were able to map a putative exon–intron structure (Fig. 3A). To determine which exons were represented in the SSB-1 expressed in ovary and testis, we designed primers within the different putative exons and used these to PCR-amplify cDNA from these tissues (data not shown). The results revealed that SSB-1 expressed in the ovary consists of either exon 1 or exon 2 spliced to exon 3 and exon 4 (Fig. 3B). Transcripts containing both exon 1 and exon 2 were not detected. SSB-1 expressed in the testis consists of exons 2 and 3 together with a truncated fragment of exon 4. This truncation appears to arise from the use of a polyadenylation signal that is not used in the ovarian transcript. The absence of most of exon 4 from the testicular transcript accounts for its relatively small size. These differences in primary sequence of the transcripts exist solely within the 5′- and 3′-UTRs; the coding sequence of all transcripts is identical.
Figure 3.
(A) Putative exon–intron structure of mouse SSB-1. Published cDNA and genomic sequences of SSB-1 were compared putative exon–intron map. Nucleotide positions are as in NT_039268.4. A putative polyadenylation sequence that may account for the truncation of exon 4 in testis is indicated. (B) Structure of SSB-1 transcripts in ovary and testis as determined by RT-PCR using exon-specific primers and sequencing. Ovarian transcripts contain either exon 1 or exon 2 but not both. The coding sequence, indicated by textured bars, begins within exon 3 and ends within exon 4 and is identical in all transcripts.
Using the SSB-4 coding-region probe, a single 2.8 kb band was detected in ovary and also in brain, heart, ovary, kidney and embryo (Fig. 2). Two transcripts (2.8 and 4.7 kb) were detected in thymus. SSB-4 was not detected in liver, lung, spleen or testis (Fig. 2). Analysis of poly(A) + mRNA confirmed that a 2.8 kb SSB-4 transcript was present in ovary and kidney, but not in liver (Fig. 2). As SSB-4 expression was relatively weak when total RNA was analysed, these analyses were repeated using a probe from the 3′-UTR of SSB-4. The same expression patterns were observed (data not shown). These results indicate that SSB-1 is expressed in mouse gonadal tissue of males and females, and SSB-4 in the ovary, as well as in a small number of other tissues.
SSB-1 and SSB-4 mRNAs are detectable in ovarian granulosa cells but not in oocytes
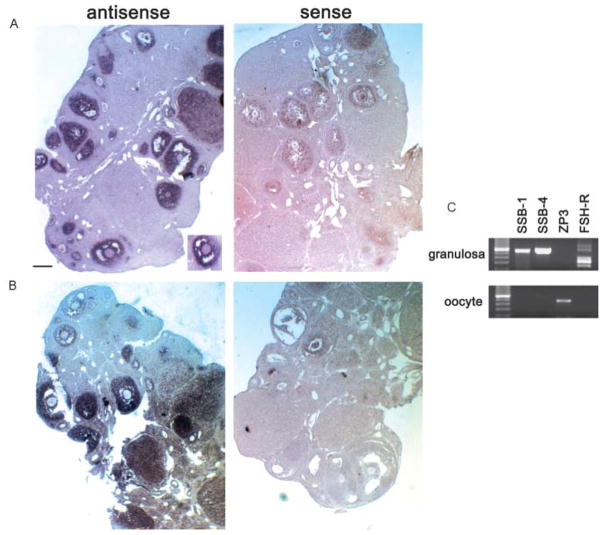
To define the cellular localization of SSB-1 and SSB-4 transcripts in mouse ovary, in situ hybridization using digoxigenin-labeled RNA probes was performed. Hybridization signals corresponding to SSB-1 were highly concentrated in the granulosa cells of follicles at different stages of growth (Fig. 4A), whereas no signals above background were detected in oocytes or the stromal cells. A similar pattern of expression was observed for SSB-4 (Fig. 4B). This result was unexpected in view of the results obtained in Drosophila, where gus expression is restricted to the developing oocyte and nurse cells (Styhler et al. 2002). Therefore, to clarify the sites of SSB-1 and SSB-4 expression in the mouse, fully grown oocytes and their surrounding granulosa cells were collected and analysed separately using RT-PCR (Fig. 4C). PCR products corresponding to SSB-1 and SSB-4 were observed in granulosa cells, as was a product corresponding to the follicle-stimulating hormone (FSH) receptor that is known to be expressed in these cells. Expression of the zona pellucida-3 (zp-3) gene was not detected, confirming that the granulosa cells were not contaminated with oocytes. In contrast, neither SSB-1 nor SSB-4 transcripts were detected at a significant level in oocytes, whereas zp-3 expression was detected. Nonetheless, in the case of SSB-4, the RT-PCR analysis did reveal a weak signal in oocytes (Fig. 4C). These results indicate that the granulosa cells are the major site of expression of SSB-1 and SSB-4 in the ovary.
Figure 4.
(A and B) Analysis of SSB-1 (A) and SSB-4 (B) expression by in situ hybridization. Ovaries were fixed in 4% para-formaldehyde, embedded in paraffin and sectioned at 7 μm thickness. The right side shows results using sense-strand probes. Hybridization signals are concentrated over granulosa cells of primary and more advanced follicles. The inset in panel A shows an absence of signal over the oocyte. (C) Analysis of SSB-1 and SSB-4 expression by RT-PCR. Purified populations of granulosa cells and oocytes were collected and subjected to RT-PCR using primers corresponding to SSB-1, SSB-4, ZP-3 and the FSH receptor. SSB-1 and SSB-4 are expressed in granulosa cells but are not detectable in oocytes. Restriction enzyme digestion confirmed that the SSB-1 primers did not amplify SSB-4 cDNA and vice versa (not shown). The absence of ZP3 signal in granulosa cells confirms the absence of contaminating oocytes, and the absence of FSH-R signal in oocytes confirms the absence of granulosa cells.
SSB-1 and SSB-4 proteins are detectable in granulosa cells but not oocytes
During development of the follicle to the antral stage, the granulosa cells become separated into two morphologically distinct sub-populations – the cumulus granulosa cells that surround the oocyte and the mural granulosa cells that lie inside of the basement membrane – thecal cell layer that constitutes the follicular wall. The mural cells are sometimes further subdivided according to whether they contact the antrum or the follicle wall. Molecular and functional differences exist between the cumulus and mural granulosa cells (Joyce et al. 2001, Eppig et al. 2005). To investigate whether SSB-1 protein was expressed throughout the follicle, we raised a polyclonal antibody against a short peptide corresponding to its N-terminal region that is relatively dissimilar to SSB-4 (amino acids 15–31, see Fig. 1) and affinity-purified it.
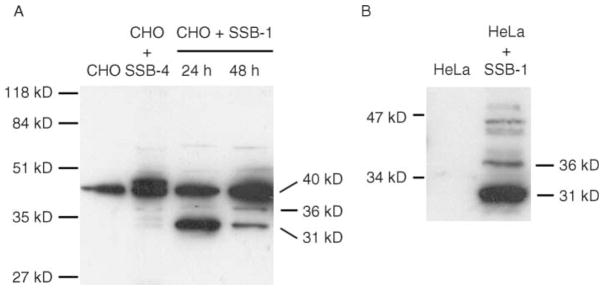
Anti-SSB-1 recognized a protein migrating at about 31 kDa in CHO cells harvested 24 h after transfection with a construct encoding SSB-1, but not in non-transfected cells or in cells transfected with a construct encoding SSB-4 (Fig. 5A). Forty-eight hours after transfection, a 36 kDa protein had appeared. These results imply that SSB-1 can be modified intracellularly to generate a slower-migrating form. This antibody also recognized a species of about 40 kDa in non-transfected CHO cells and in tissues where we did not detect SSB-1 mRNA (data not shown). Similarly, transfected HeLa cells also displayed several slow-migrating species in addition to the predicted 31 kDa protein (Fig. 5B). Taken, together, these results indicate that SSB-1 can be modified within the cell to generate multiple forms that differ in relative molecular mass (Mr).
Figure 5.
Detection of multiple SSB-1-related species in transfected cells. (A) CHO cells that were not transfected, transfected with a plasmid encoding SSB-4 or transfected with a plasmid encoding SSB-1 were harvested 24 or 48 h post-transfection and imunoblotted using affinity-purified anti-SSB-1. The 40 kDa band in all lanes is probably unrelated to SSB-1 (see text). SSB-1-transfected cells display a 31 kDa band 24 h after transfection and a weaker 36 kDa band that is more intense 48 h after transfection. (B) HeLa cells were not transfected or were transfected with the construct encoding SSB-1 and blotted using affinity-purified anti-SSB-1. In addition to the 31 kDa band, several higher molecular mass immunoreactive species are present.
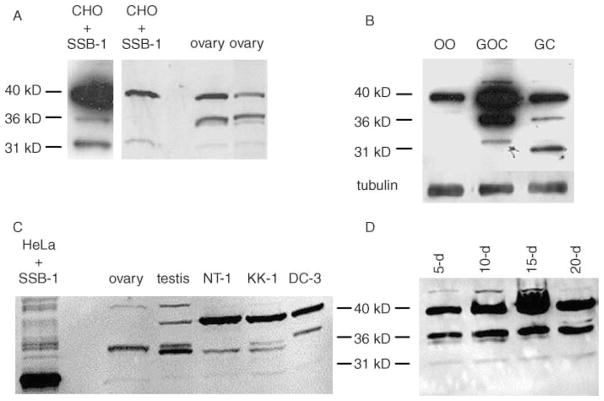
When whole-ovary extracts were probed using anti-SSB-1, both the 31 and 36 kDa bands were detected, as well as the apparently unrelated 40 kDa band (Fig. 6A). To verify the ovarian cell type expressing SSB-1, granulosa cell–oocyte complexes were collected and either used directly for blotting or separated into oocytes and granulosa cells, and analysed separately. As shown in Fig. 6B, the 31 and 36 kDa species were present in the granulosa–oocyte complexes and the purified granulosa cells but not in the purified oocytes. To confirm these results, we also analysed SSB-1 expression in immortalized cell lines derived from granulosa cells (see Havelock et al. (2004) for review). KK-1 and NT-1 cells were derived from granulosa cell tumours of transgenic mice expressing SV40 T-antigen under the control of the inhibin α-subunit promoter (Kananen et al. 1995, Rilianawati et al. 1999). DC3 cells were derived from rat granulosa cells transformed using SV40 in vitro (Fitz et al. 1989). All three lines expressed the 31 kDa species as well as species migrating near 36 kDa. The slight differences in electrophoretic mobility of the 36 kDa species observed in the different lines may reflect post-translational modification. Taken together, these results are consistent with the data obtained by in situ hybridization analysis and RT-PCR, and reinforce the conclusion that SSB-1 is expressed in granulosa cells of growing follicles but not in female germ cells.
Figure 6.
Detection of SSB-1 in ovarian granulosa cells and cell lines. (A) Comparison of transfected CHO cells and whole-ovary extracts blotted using anti-SSB-1. The left panel shows a long exposure of a different CHO cell blot from that shown in the right panel. Both contain the 31, 36 and 40 kDa immunoreactive species. (B) Ovaries were digested using collagenase to yield granulosa–oocyte complexes or using collagenase and trypsin to dissociate the cell types. Oocytes (OO), complexes (GOC) or granulosa cells (GC) were collected and immunoblotted. The 31 and 36 kDa species are present in complexes and purified granulosa cells but not in purified oocytes. The increased mobility of the 31 kDa species in the granulosa cell lane was not reproducible. (C) Extracts were prepared from whole ovaries and testes and from immortalized granulosa cell lines: NT-1 and KK-1 from mouse and DC3 from rat. The 31 and 36 kDa species are present in all groups, with the latter apparently existing as a doublet. (D) Granulosa–oocyte complexes were collected from mice at different ages (shown in days), yielding populations enriched for complexes at successively more advanced stages of folliculogenesis. SSB-1 is present at all stages of follicular growth.
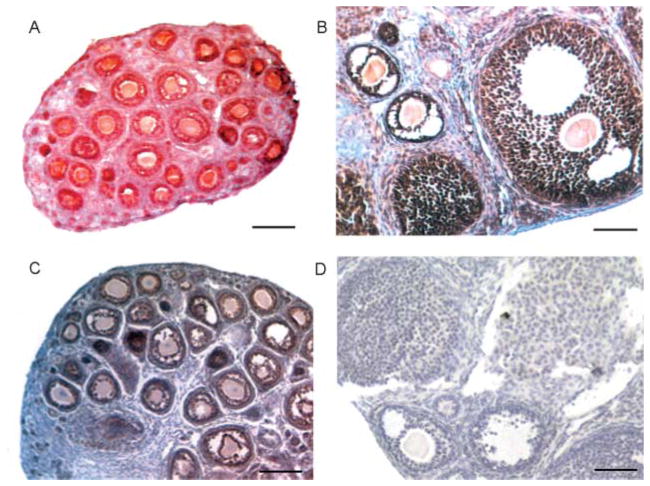
To determine whether expression of SSB-1 was developmentally regulated during follicular growth, we took advantage of the fact that, during the first three weeks ?of life, a relatively large pool of follicles undergoes synchronous growth. Thus, ovaries obtained from mice of increasing age during this time are enriched for follicles at successively more advanced stages of development. We isolated granulosa–oocyte complexes from 5- to 20-day-old mice. SSB-1 was present in complexes at all stages of growth (Fig. 6D). In addition, we stained ovarian histological sections using anti-SSB1. Strong staining was detected in granulosa cells of growing follicles and in both the mural and cumulus granulosa of antral follicles (Fig. 7A and B), but not when the primary antibody was replaced by rabbit immunoglobulin at the same concentration (Fig. 7D). Moreover, the same staining pattern was observed using a different antibody raised against the N-terminal 60 amino acids of SSB-1 (data not shown). Similarly, antibodies raised against SSB-4 that recognized transfected SSB-4 but not SSB-1, confirming their subtype specificity (data not shown), stained the granulosa cells of growing and fully grown follicles (Fig. 7C). Figure 7 also shows a weak staining reaction in the oocytes. This staining was also observed when the primary antibody was omitted (see Fig. 7D), indicating that the detection system produces a certain degree of non-specific staining in the oocyte cytoplasm. Taken together, these results indicate that SSB-1 and SSB-4 are expressed in granulosa cells in primary follicles and all subsequent stages, and in both the mural and antral granulosa cells of antral follicles.
Figure 7.
Detection of SSB-1 and SSB-4 by immunohistochemistry. Ovaries were fixed in 4% para-formaldehyde, embedded in paraffin and sectioned at 5 μm thickness. Sections were subjected to an antigen retrieval technique. (A and B) SSB-1 is detected in granulosa cells, including both mural and cumulus cells of antral follicles, but not in the thecal cells or interstitial tissue. (C) SSB-4 is detected in granulosa cells in follicles at different stages of growth. (D) Sections processed as above but using the same concentration of rabbit IgG as the primary antibody show no specific staining. Non-specific staining of oocytes is sometimes observed using this immunodetection procedure.
Discussion
We have identified two members of the SSB gene family that are very similar in sequence and presumably homologous to the gus gene of Drosophila. All of these proteins carry a SPRY domain and a SOCS box. Sequence similarity is strongest in the SPRY domain and is significant in the SOCS box. Moreover, SSB-1 is highly conserved among mice, humans and Xenopus. In addition, SSB-1 and SSB-4 proteins are about 70% identical to Drosophila GUS. This is considerably higher than the protein sequence identity between MVH, NANOS and TUDOR and their apparent homologues in Drosophila. Taken together, these results suggest that the biochemical function of GUS is conserved in SSB-1 and SSB-4 and that this function is probably important in a variety of organisms.
By Northern analysis, we observed that SSB-1 and SSB-4 were expressed in specific tissues. This result differs slightly from a recent database analysis indicating that these genes are expressed in many tissues (Wang et al. 2005). This suggests that, notwithstanding a widespread low-level expression of these genes, the major sites of SSB-1 and SSB-4 function may be restricted to a few tissues. One of these tissues is likely to be the ovary, where both SSB-1 and SSB-4 mRNA and protein were abundantly expressed in the granulosa cells of growing follicles. Moreover, expression was detected in granulosa cells from follicles of different sizes, suggesting that SSB-1 and SSB-4 are constitutively expressed in these cells, at least from the earliest stages of follicle growth.
Expression of SSB-1 and SSB-4 in ovarian granulosa cells was unexpected, as it contrasts with the expression of gus in Drosophila, which is restricted to the oocyte and nurse cells (Styhler et al. 2002). We could not detect SSB-1 mRNA or protein in mouse oocytes, although a weak expression of SSB-4 mRNA was apparent. This result concords with a recent report of gene expression in oocytes at different stages of development (Pan et al. 2005). The ovarian granulosa cells are thought to originate from the ovarian rete cords, which are composed of cells that invaded the ovary from the overlying mesonephros during embryonic development. In contrast, the germ cells are derived from posterior embryonic ectoderm (reviewed in Yasuhisa Matsui (2005)). Thus, the oocyte and granulosa cells do not share an embryonic lineage as do the oocyte and nurse cells of Drosophila. The expression pattern of SSB-1 and SSB-4 also contrasts with other mammalian homologues of genes regulating oogenesis in flies. In these other instances – including vasa, nanos, tudor and fat facets – the mammalian homologues are also expressed in the germ cells (Paules et al. 1989, Noma et al. 2002, Chuma et al. 2003, Tsuda et al. 2003, Smith et al. 2004). The mechanism responsible for this lineage switch in expression pattern of SSB-1 and SSB-4 is unknown, although our mRNA expression data indicate that it is controlled at the level of transcription.
SSB-1 has a predicted molecular mass of about 31 kDa and histidine-tagged SSB-1 migrates at about this position (Wang et al. 2005). We also observed a 31 kDa protein in HeLa and CHO cells transfected with SSB-1. Unexpectedly, these cells also showed several immunoreactive species of higher Mr that became more prominent with extended culture. Importantly, these species were not observed in non-transfected cells (Fig. 5) or in cells co-transfected with SSB-1 and an siRNA targeting this mRNA (Y Xing & H J Clarke, unpublished observations), indicating that they require expression of SSB-1. Among these species was one of about 36 kDa that co-migrated with an immunoreactive species detected in both primary granulosa cells and immortalized granulosa cell lines. As we have been unable to characterize these immunoreactive species, we cannot formally rule out that they represent proteins unrelated to SSB-1 that are both induced by SSB-1 and also recognized by anti-SSB-1. A hypothesis that seems more conservative, however, is that SSB-1 can be subject to post-translational modifications. Thus, we propose that the immunoreactive species migrating at 36 kDa in granulosa cells represents a post-translationally modified form of SSB-1.
As discussed in the Introduction, GUS is required for appropriate localization of VASA and the generation of polarity in Drosophila eggs. There has been considerable recent interest in the possibility that mammalian eggs may also manifest a developmentally relevant polarity, although this remains highly controversial (Gardner 2001, Hiiragi & Solter 2004). MVH is expressed in both oocytes and spermatocytes, but is required only for spermatogenesis (Tanaka et al. 2000). Our data showing that SSB-1 and SSB-4 are not expressed in oocytes provide further evidence that an SSB-/MVH-mediated polarization mechanism analogous to that of Drosophila does not operate in mammalian oocytes. However, we have not examined SSB expression in primordial germ cells, which express MVH (Toyooka et al. 2000). Thus, it is possible that SSB and MVH function together at this stage of gametogenesis.
The function of SSB-1 and SSB-4 in granulosa cells remains to be identified. Several lines of evidence suggest that SSB family proteins may physically interact with other proteins. GUS interacts with VASA, as revealed by co-immunoprecipitation experiments (Styhler et al. 2002). Although the site of interaction has not been identified, the SPRY domains of other proteins mediate protein–protein interactions. In the case of SSB-1, its SPRY domain mediates its binding to the tyrosine kinase domain of MET receptor (Wang et al. 2005). Moreover, SSB-1 binding enhances transcriptional activation following MET stimulation by hepatocyte growth factor. In this context, it is interesting to note that MET is expressed in granulosa cells (Yang & Park 1995, Parrott & Skinner 1998). It is tempting to speculate that SSB-1 may play a key role in regulating MET-mediated cellular responses in these cells.
Acknowledgments
This work was supported by the Program in Oocyte Health of the Canadian Institutes for Health Research (H J C). We thank Professors Ilpo Huhtaniemi (Imperial College London, UK) and Riaz Farookhi (McGill University) for generously supplying cell lines. The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
References
- Allard P, Champigny MJ, Skoggard S, Erkmann JA, Whitfield ML, Marzluff WF, Clarke HJ. Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. Journal of Cell Science. 2002;115:4577–4586. doi: 10.1242/jcs.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mechanisms of Development. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biology of Reproduction. 2005;73:351–357. doi: 10.1095/biolreprod.105.041798. [DOI] [PubMed] [Google Scholar]
- Fitz T, Wah R, Schmidt W, Winkel C. Physiologic characterization of transformed and cloned rat granulosa cells. Biology of Reproduction. 1989;40:250–258. doi: 10.1095/biolreprod40.2.250. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. PNAS. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. Specification of embryonic axes begins before cleavage in normal mouse development. Development. 2001;128:839–847. doi: 10.1242/dev.128.6.839. [DOI] [PubMed] [Google Scholar]
- Havelock JC, Rainey WE, Carr BR. Ovarian granulosa cell lines. Molecular and Cellular Endocrinology. 2004;228:67–78. doi: 10.1016/j.mce.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Solter D. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. 2004;430:360–364. doi: 10.1038/nature02595. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annual Review of Genetics. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Joyce IM, Pendola FL, O’Brien M, Eppig JJ. Regulation of prostaglandin-endoperoxide synthase 2 messenger ribonucleic acid expression in mouse granulosa cells during ovulation. Endocrinology. 2001;142:3187–3197. doi: 10.1210/endo.142.7.8268. [DOI] [PubMed] [Google Scholar]
- Kananen K, Markkula M, Rainio E, Su J, Hsueh A, Huhtaniemi I. Gonadal tumorigenesis in transgenic mice bearing the mouse inhibin alpha-subunit promoter/simian virus T-antigen fusion gene: characterization of ovarian tumors and establishment of gonadotropin-responsive granulosa cell lines. Molecular Endocrinology. 1995;9:616–627. doi: 10.1210/mend.9.5.7565808. [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. The ability to organize sperm DNA into functional chromatin is acquired during meiotic maturation in murine oocytes. Developmental Biology. 1997;186:73–84. doi: 10.1006/dbio.1997.8581. [DOI] [PubMed] [Google Scholar]
- Mohamed O, Dufort D, Clarke HJ. β-Catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Developmental Dynamics. 2004;231:416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- Noma T, Kanai Y, Kanai-Azuma M, Ishii M, Fujisawa M, Kurohmaru M, Kawakami H, Wood SA, Hayashi Y. Stage- and sex-dependent expressions of Usp9x, an X-linked mouse ortholog of Drosophila Fat facets, during gonadal development and oogenesis in mice. Mechanisms of Development. 2002;119:S91–S95. doi: 10.1016/s0925-4773(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Pan H, O’Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Developmental Biology. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK. Developmental and hormonal regulation of hepatocyte growth factor expression and action in the bovine ovarian follicle. Biology of Reproduction. 1998;59:553–560. doi: 10.1095/biolreprod59.3.553. [DOI] [PubMed] [Google Scholar]
- Paules RS, Buccione R, Moschel RC, Vande Woude GF, Eppig JJ. Mouse Mos protooncogene product is present and functions during oogenesis. PNAS. 1989;86:5395–5399. doi: 10.1073/pnas.86.14.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Yan MSC, Klysik M, Matzuk M. Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertility and Sterility. 2001;76:550–554. doi: 10.1016/s0015-0282(01)01966-5. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- Rilianawati, Rahman NA, Huhtaniemi I. Hormonal regulation of proliferation of granulosa and Leydig cell lines derived from gonadal tumors of transgenic mice expressing the inhibin-[alpha] subunit promoter/simian virus 40 T-antigen fusion gene. Molecular and Cellular Endocrinology. 1999;149:9–17. doi: 10.1016/s0303-7207(99)00004-0. [DOI] [PubMed] [Google Scholar]
- Saunders PTK, Pathirana S, Maguire SM, Doyle M, Wood T, Bownes M. Mouse staufen genes are expressed in germ cells during oogenesis and spermatogenesis. Molecular and Human Reproduction. 2000;6:983–991. doi: 10.1093/molehr/6.11.983. [DOI] [PubMed] [Google Scholar]
- Smith JM, Bowles J, Wilson M, Teasdale RD, Koopman P. Expression of the tudor-related gene Tdrd5 during development of the male germline in mice. Gene Expression Patterns. 2004;4:701–705. doi: 10.1016/j.modgep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-boc containing protein, GUSTAVUS. Developmental Cell. 2002;3:865–876. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes and Development. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mechanisms of Development. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Vallee M, Gravel C, Palin M-F, Reghenas H, Stothard P, Wishart DS, Sirard M-A. Identification of novel and known oocyte-specific genes using complementary DNA subtraction and microarray analysis in three different species. Biology of Reproduction. 2005;73:63–71. doi: 10.1095/biolreprod.104.037069. [DOI] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing EM, Wu G. The SPRY domain-containing SOCS box protein 1 (SSB-1) interacts with MET and enhances the hepatocyte growth factor-induced Erk-Elk-1-serum response element pathway. Journal of Biological Chemistry. 2005;280:16393–16401. doi: 10.1074/jbc.M413897200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Yang XM, Park M. Expression of the hepatocyte growth factor/scatter factor receptor tyrosine kinase is localized to epithelia in the adult mouse. Laboratory Investigation. 1995;73:483–491. [PubMed] [Google Scholar]
- Yasuhisa Matsui DO. Mechanisms of germ-cell specification in mouse embryos. BioEssays. 2005;27:136–143. doi: 10.1002/bies.20178. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biology of Reproduction. 2003;68:31–39. doi: 10.1095/biolreprod.102.007674. [DOI] [PubMed] [Google Scholar]