Summary
Eukaryotic messenger RNA (mRNA) undergoes chemical modification both at the 5′cap [1, 2] and internally [3–14]. Among internal modifications, m6A, by far the most abundant, is present in all eukaryotes examined, including mammals [3–6], flies [15], plants [16, 17] and yeast [18, 19]. m6A modification plays an essential role in diverse biological processes. Over the past few years, our knowledge relevant to establishment and function of this modification has grown rapidly. This review focuses on technologies that have facilitated m6A detection in mRNAs, identification of m6A methylation enzymes and binding proteins, and potential functions of the modification at the molecular level. Regarding m6A function at cellular or organismal levels or in disease, please refer to other recent reviews [20–23].
Methods to detect m6A
Historically, methods used to detect and quantify overall m6A levels on mRNA have included chromatography [3–6], two-dimensional cellulose thin-layer chromatography (2D-TLC) [17, 24], dot-blotting [25], and liquid chromatography-tandem mass spectrometry (LC-MS/MS) [25] (Table 1). Based on these studies, it is now accepted that m6A frequency is 3–5 residues per mRNA [1–5, 26]. Methods employed until recently, however, could not reveal m6A location, a task that has proven challenging. Although the m6A methyl group is found at the Watson-Crick base-pairing site and perturbs Adenosine and Uridine (A/U) or A/T (thymidine) pairing, it does not completely block reverse transcriptase, as does m1A RNA modification [27], and there is no chemical treatment analogous to bisulfite conversion of 5mC available to convert m6A to a different and detectable nucleotide [28]. Therefore, for a long time, it remained unclear which mRNAs even exhibit m6A.
Table 1.
Methods used to detect and map m6A modification of polyadenylated RNA.
| Method | Purpose | Pros | Cons |
|---|---|---|---|
| 2D-TLC [17, 24] | Measure overall m6A levels | Well-established protocol; Quantitative; High sensitivity. | Requires radioactivity; Does not cover all A sites due to specificity of ribonucleases used for 5′ end labeling. |
| LC-MS/MS [25] | Quantitative; Covers all A sites; Very high sensitivity. | Requires special equipment and expertise; High cost. | |
| Dot blot [25] | Low cost; No radioactivity required. | Nonspecific antibody binding; Low sensitivity. | |
| Ligation assay [55] | Measure m6A levels at a specific site | Easy to set up. | Not applicable to endogenous m6A sites; Requires radioactivity. |
| Primer extension [53] | Easy to set up. | Relies on specific RTase; Cannot detect low abundant m6A sites. | |
| SMRT [54] | Single-molecule, real time detection. | Requires special equipment; Quantification not reported. | |
| SCARLET [56] | Precise locations mapped; Highly quantitative. | Requires radioactivity and sophisticated methodology. | |
| m6A- meRIP-Seq [29, 30] | Map m6A locations genome-wide | Well established protocol; High sensitivity. | IP/antibody can yield false- positives; Low resolution. |
| PA-m6A- RIP-Seq [36] | Higher resolution than meRIP-seq. | Need to use living cells to incorporate 4-SU; thus cannot be applied to tissue or clinical samples. IP/antibody can yield false- positives. |
|
| miCLIP-Seq [37, 38] | Single-nucleotide resolution. | High reliance on specific antibody to generate a signature at m6A sites. | |
| m6A-LAIC- seq [39] | Only method to quantify percentage of methylated vs. unmethylated RNAs. | Cannot detect specific m6A sites. | |
| Two-color microarray [47] | Antibody not required. | Low sensitivity. |
In 2012, however, two groups independently developed technology that coupled RNA immunoprecipitation using an m6A -specific antibody to next generation high throughput sequencing (m6A meRIP-Seq) to map m6A sites in the mammalian transcriptome [29, 30] (Table 1). Initially, m6A was mapped to over 7000 coding and non-coding mammalian polyadenylated (polyA) RNAs [29, 30]. Since then, over 10,000 m6A -methylated polyA RNAs have been reported from various organisms and cell types, ranging from yeast to mammalian reprogrammed pluripotent stem cells [29–39]. A consensus m6A methylation motif, RRACH (R = G or A; H = A, C or U), was identified from high throughput data [29, 30], in agreement with biochemical studies [40–42]. Recently, the consensus motif was redefined as DRACH (D = A, G or U), based on a study reporting that the nucleotide at the -2 position relative to m6A can also be U [38]. Many m6A sites are highly conserved between species, suggesting evolutionary importance of the modification [29, 30]. There is general agreement that m6A is highly enriched at 3′-UTRs (untranslated regions) [29, 30, 36–38], and early meRIP-seq studies suggested that m6A is located near stop codons [29, 30, 32]. However, a later study with improved detection resolution suggested that m6A sites are present in 3′-UTR but there is no preference for m6A to locate around stop codons [37]. Some m6A modifications have also been found flanking 5′- and 3′-splice sites of exons, spatially overlapping with binding sites for mRNA splicing factors [43, 44], suggestive of a splicing function. Since N6, 2 -O-dimethyladenosine (m6Am), a modification that occurs exclusively on first nucleotide of mRNAs [45], can also be recognized by anti-m6A antibody, m6A abundance at 5′-UTRs remained unclear until a recent study distinguished these modifications using improved technology. This study showed m6Am enrichment at transcription start sites [38]. In contrast, much lower m6A levels were detected at 5′-UTRs [38]. Nevertheless, the same group later reported that m6A but not m6Am at the 5′-UTR regulates cap-independent mRNA translation [46].
One limitation of the original meRIP-seq method is its relatively low resolution: m6A can be mapped within a 100–200 nt transcript region but precise positions cannot be identified [29, 30]. Efforts from multiple laboratories have improved meRIP-seq resolution. Using yeast as a system, one study (2013) employed an improved computational algorithm to predict m6A at almost single-nucleotide resolution [31] (Table 1). Additionally, (2014) a photo-crosslinking-assisted m6A sequencing strategy (PA-m6A -seq) has been used to improve resolution [36] (Table 1). In 2015, two groups adapted ultraviolet (UV) CLIP (cross-linking immunoprecipitation) to accurately locate tens of thousands of m6A residues in mammalian mRNAs with single-nucleotide resolution [37, 38] (Table 1). Both studies screened different m6A antibodies and found that some can induce specific mutational signatures around m6A residues after UV light-induced antibody/RNA cross-linking and reverse transcription. This approach can map m6A at single-nucleotide resolution.
Another limitation of meRIP-seq m6A detection methods is their reliance on antibody-based IP procedures, which are often associated with false positives [31]. To circumvent this problem, alternative technologies have been developed. These include m6A detection by 1) two-color tiling microarray [47] based on m6A interference with A/T pairing [48–52] (Table 1); 2) reverse transcription based methods, based on changes in kinetics of specific reverse transcriptases by m6A base modifications [53, 54] (Table 1); and 3) ligation-based assays such as site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography (SCARLET) [55, 56] (Table 1). Although these methods cannot yet be carried out in a high-throughput manner comparable to meRIP-seq and some are applicable to only specific transcripts, they provide a complementary approach to confirm specific m6A sites identified by meRIP-seq.
An important question is, among sites modified by m6A, what fraction of transcripts are m6A -tagged versus untagged. For example, for the long noncoding RNA (lncRNA) MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), four precise m6A sites were mapped using SCARLET technology [56]. The proportion of MALAT1 transcripts modified at these sites varies between 11% and 77% in HeLa cells, suggesting variation in modification of a potential m6A site could have functional consequences for methylated versus unmethylated RNAs. Recently, one group developed a technology termed m6A-LAIC-seq (m6A-level and isoform-characterization-sequencing) to detect m6A methylated vs. unmethylated mRNA transcripts [39] (Table 1). m6A-LAIC-seq method modifies the standard m6A-meRIP-seq protocol by using excess m6A antibody, including RNA spike-in controls to improve quantification, and sequencing full-length rather than fragmented PolyA RNAs. Use of excess m6A antibody and full-length transcripts ensures that all m6A -containing PolyA RNAs are pulled down in the IP’ed fraction and not in the flow-through. Therefore, the proportion of PolyA RNAs containing m6A is calculated as the ratio of transcripts detected in IP’d vs. flow-through fractions. The authors reported that, for most genes, less than 50% of transcripts contained m6A methylation and proportions differed between cell types. This method for the first time quantified the proportion of m6A methylated vs. unmethylated transcripts on a genome-wide scale. However, since full-length mRNAs were used, m6A locations were not defined; thus the resolution of this method is at the mRNA rather than the m6A site level. Furthermore, this method cannot distinguish m6Am-from m6A-containing mRNAs. Hence, novel methods are required to map a fraction of specific m6A sites.
m6A methyltransferases and demethylases
m6A is a reversible modification. An effort to purify enzymes that synthesize m6A began in the 1990s [57, 58]. Methyltransferase-like 3 or METTL3 (also known as MTA-70) was reported as a putative m6A methyltransferase in 1997 [59]. Not until 2014 did four studies [33, 35, 44, 60] report significant interaction between METTL3 and the previously uncharacterized protein METTL14, which also harbors an MTA domain [33, 35, 44, 60, 61]. Two of them reported that a combination of METTL3 and METTL14 showed remarkably greater in vitro methyltransferase activity than did METTL3 or METTL14 alone, suggesting that they functioned synergistically [35, 60] (Fig. 1). This prediction was confirmed by recent reports of the crystal structure of a METTL3/METTL14 heterodimer [62–64]. Those studies focused on the METTL3 or METTL14 methyltransferase domain and adjacent motifs and were based on ligand-free, methyl group donor S-adenosyl methionine (SAM)-bound states [62–64]. Interestingly, previous studies [35, 60] reported that METTL14 displayed higher methyltransferase activity than did METTL3 in in vitro methylation assays, suggesting METTL14 as the predominant catalytic subunit. In contrast, structural analysis supports a model in which METTL3 serves as the catalytic subunit, which binds SAM, while METTL14 plays a structural role and potentially functions in RNA substrate binding via the positively charged groove formed between METTL3 and METTL14 [62–64]. One particular structural study suggested that while both METTL3 and METTL14 display a predicted catalytic motif, the METTL14 SAM binding domain is blocked while that of METTL3 is hollow, allowing binding [63]. The authors of that study suggest that high METTL14 activity in a methylation assay could be due to METTL3 contamination [63], explaining conflicting conclusions emerging from biochemical versus structural studies. As yet, the structure of a METTL3/METTL14 RNA complex has not been solved, an achievement that would provide important information relevant to substrate sequence specificity.
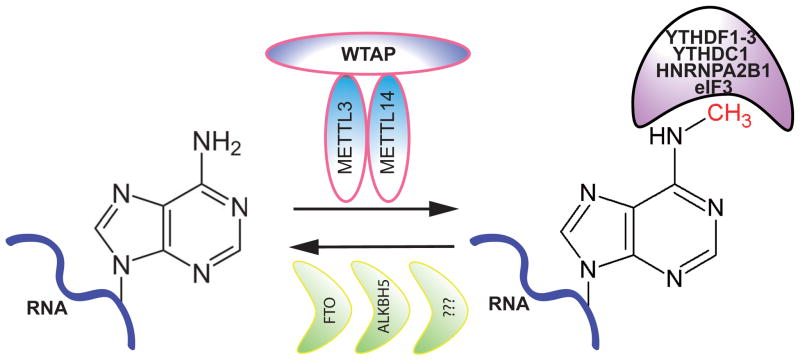
Figure 1. Formation, removal, and recognition of m6A.
METTL3/METTL14 were identified as core components of an N6-methyladenosine methyltransferase complex. Both form a heterodimer catalyzing m6A formation. WTAP has been identified as a METTL3- and METTL14-interacting protein. The presence of WTAP does not alter METTL3/METTL14 methyltransferase activity in vitro, but WTAP has a critical role in m6A formation in vivo through an unknown mechanism. Other METTL3/METTL14-interacting proteins have been identified, but their activities remain to be determined. Two Alkb family members, FTO and ALKBH5, reportedly serve as m6A demethylases and remove m6A in an oxidative manner, although additional unknown m6A demethylases may also serve this function. Several m6A binding proteins are reported, including multiple YTH family members (YTHDF1–3 and YTHDC1), heterogeneous ribonucleoprotein HNRNPA2B1, and eIF3.
The RRACH motif has been identified as enriched at m6A sites; however, only a small fraction of RRACH motifs exhibit m6A [29, 30]. How METTL3/METTL14 is recruited to a specific transcript and why some RRACH motifs become modified and others do not remains poorly understood. It is hypothesized that RNA binding proteins (RBPs) interacting with METTL3/METTL14 may recruit these proteins. Several METTL3/METTL14-interacting proteins have been identified. The most well-established is Wilms tumor 1 associated protein (WTAP), which is an RBP that displayed high affinity to METTL3/METTL14 [33, 44] (Fig. 1). METTL3/WTAP interactions are conserved in yeast [65]. Although WTAP does not alter METTL3/METTL14 methyltransferase activity in vitro, its loss promotes transcriptome-wide m6A depletion in cells [33, 44], demonstrating that it is required for m6A modification and suggesting it may direct METTL3/METTL14 onto targets via RNA-binding activity. Indeed, WTAP PAR-CLIP analysis reported direct WTAP binding to RNA and m6A enrichment at WTAP/RNA binding sites [44]. Nonetheless, how WTAP recognizes RRACH motifs and facilitates methylation of adenosine within them is unknown. In addition to WTAP, thirteen other proteins have been identified in a METTL3-interacting protein network [33]. Knockdown of one, KIAA1429, decreased global m6A levels [33]. Functions of other proteins identified in the network remain unknown.
m6A methyl groups are removed by m6A demethylase (Fig. 1). Two members of the alpha-ketoglutarate-dependent dioxygenase Alkb family, Fat mass and obesity-associated protein FTO and Alkylation Repair Homolog 5 (ALKBH5), reportedly remove m6A in an oxidative manner [25, 66]. FTO was first shown to demethylate 3-methylthymine on single-stranded DNA (ssDNA) [67]. Later, a group showed that FTO demethylates 3-methyluridine in ssRNA [68] in vitro. In 2011, the same group reported that m6A -modified RNA was the primary FTO substrate [25]. Overexpression of FTO or ALKBH5 in cells decreases global m6A levels, but knockdown or knockout of either only mildly increases m6A levels [25, 66], suggesting the existence of other demethylases or perhaps a synergy between ALKBH5 and FTO that has not been studied. The crystal structures of both FTO and ALKBH5 have been reported, and small molecule inhibitors targeting their demethylase activities have been developed based on these structures [69–71]. For example, the natural product rhein, derived from herbs, is among the most effective FTO m6A demethylase inhibitors [72]. As yet, it is unclear whether FTO or ALKBH5 target the same or different methylated mRNAs.
m6A binding proteins
Like methylated DNA and histone protein tails, m6A-modified RNA is recognized by specific proteins, or readers, that transmit the code to downstream effectors. In 2012, using methylated vs. non-methylated RNA probes as baits, several m6A-interacting proteins in mammalian cells, including members of the YTH domain-containing family, such as YTHDF2 and YTHDF3, were pulled down [30] (Fig. 1). Later, a study demonstrated direct binding of YTHDF2 to m6A RNA [34]. Since then, additional YTH family proteins were identified as m6A binders, including YTHDF1 and YTHDC1 [73–75] (Fig. 1). In agreement, while biochemical and structural analysis revealed YTH as a general RNA binding domain [76], kinetic analysis demonstrated that the binding affinity between YTH domains to m6A-modified RNA is 10 times higher than that to non-m6A RNA [77, 78]. Furthermore, PAR-CLIP analysis of YTHDF1, YTHDF2 and YTHDC1 identified genome-wide YTH protein binding sites overlapping with the RRACH motif [34, 73, 74]. Together, these studies strongly support direct interaction of YTH proteins and m6A -modified RNA. However, it is noteworthy that YTH domain affinity for m6A is moderate and much lower than that of DNA methylation binding proteins such as Methyl-CpG Binding Domain Protein 1 (MBD1) and Methyl-CpG-binding protein 2 (Mecp2) for 5mC [79]. In addition, YTHDF1/2 CLIP-seq data clearly showed that YTH proteins also bind to RNA sites that lack m6A in vivo [34, 74]. Therefore, observations derived from these studies, which have greatly advanced our knowledge of how m6A exerts its function, underscore the importance of using cells engineered to lack m6A as controls in analyzing YTH domain protein function in m6A modification to ensure specificity of effects. While YTHDF1/2 proteins are generally defined as cytoplasmic m6A “readers”, YTHDC1 is a nuclear reader. A different nuclear m6A binding protein was recently reported, the heterogeneous ribonucleoprotein HNRNPA2B1 [80] (Fig. 1). m6A also reportedly binds eukaryotic initiation factor 3 (eIF3), a critical component of translation initiation complex [46] (Fig. 1). The proposed functions of these binding are discussed below.
m6A regulates mRNA activity through diverse mechanisms
m6A destabilizes RNA
Early studies in 1970s hinted that RNA methylation functions in regulating mRNA stability [26, 81]. Multiple recent studies report that loss of m6A methyltransferase activity accompanied by decreased m6A modification increases transcript stability [32–35, 82], suggesting that m6A modification destabilizes RNA (Key Figure, Fig. 2). Multiple underlying mechanisms have been proposed.
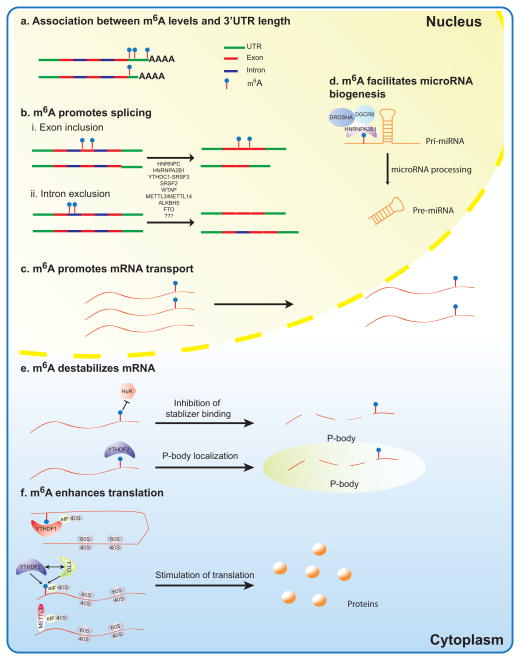
Key Figure Figure 2.
Diverse molecular mechanisms of m6A.
(a) Association between m6A levels and 3′UTR length, (b) m6A promotes splicing, (c) m6A promotes mRNA transport, (d) m6A facilitates microRNA biogenesis, (e) m6A destabilizes mRNA, and (f) m6A enhances translation. As indicated, these activities occur in the nucleus, cytoplasm, or both.
One study reports that YTHDF2 binds to m6A, which then translocates mRNA from the translation machinery to processing bodies (P-bodies), where it is degraded [34]. However, a later study challenged this model by showing that YTHDF2 does not interact with core components of the P-body and that, instead, YTHDF2 directly recruits the CCR4-NOT deadenylase complex to destabilize RNAs that contain m6A [83]. These discrepant findings may reflect direct and indirect mechanisms on YTHDF2 regulated m6A-mRNA stability. In addition to YTHDF2, two groups reported that all three YTHDF proteins (YTHDF1–3) regulate HIV-1 RNA expression; one study reported that all three promoted HIV-1 RNA expression [84], while the other reported that they repress HIV-1 RNA expression [85]. One thing that both studies agree on is that all three YTHDF proteins regulate gene expression in the same manner, in contrast to previous studies reporting that YTHDF2 destabilizes mRNA [34], while YTHDF1 promotes protein synthesis [74]. These differences may reflect genome-wide versus gene-specific effects but warrant future investigation.
Another study showed that m6A blocks mRNA binding to the mRNA stabilizer human antigen R (HuR or ELAVL1) [35]. In that study, the authors observed that whether m6A blocks or facilitates mRNA and HuR interaction depends on the distance between m6A and HuR binding sites [35]. When sites are in close proximity, m6A promotes HuR binding, consistent with previous work that identified HuR as a m6A binding protein by employing an m6A site probe next to a HuR-binding U track [30]; while when m6A and HuR binding sites were far apart, the presence of m6A decreased HuR binding [35]. Since predicted RNA motifs favoring m6A modification or HuR binding differ substantially in sequence, endogenous m6A and HuR sites may not always co-localize. Thus, it was proposed that m6A is more likely to block HuR-RNA binding, destabilizing mRNA in vivo. Since this work was not performed on a transcriptome-wide scale, future studies should address the scope of this interaction.
m6A alters RNA structure to modulate RNA/protein interaction
It is well-established that secondary and tertiary structure governs RNA function [86, 87]. Since m6A destabilizes A/U pairing [47–52], it is reasonable to predict that m6A can alter thermostability of an RNA duplex to change RNA secondary structure and function. Indeed, using a technology known as in vivo click selective 2′-hydroxyl acylation and profiling experiment (icSHAPE), which can determine endogenous RNA secondary structure, one group compared RNA base-pairing status of the m6A consensus motif GGACU in wildtype vs. METTL3 knockout mESCs [88]. They reported that the GGACU motif is less structured in wildtype ESCs than in ESCs lacking m6A, suggesting that m6A may help transit paired RNA to unpaired [88]. Another study further demonstrated that m6A -mediated RNA structural changes alter RNA/protein interactions [89]. These authors reported that m6A on a stem-loop region of the lncRNA MALAT1 altered local RNA structure to enhance MALAT1 binding to the RBP HNRNPC (Heterogeneous Nuclear Ribonucleoprotein C). They named this type of m6A-containing region an “m6A-switch” and identified thousands of potential RNA sequences that could function in a “switch” using sequential HNRNPC-PAR-CLIP followed by m6A-RIP-Seq in wildtype vs METTL3/METTL14 knockdown cells. Most switches were located in introns of coding and non-coding RNAs and potentially regulate alternative splicing [89].
m6A enhances mRNA translation
Several mechanisms have been proposed relevant to m6A effects on translation (Fig. 2). In 2015, a study reported that the m6A binding protein YTHDF1 interacts with eIF3 to promote efficient translation of m6A -modified mRNAs [74]. Later, two studies reported that cellular stress, such as heat-shock, increases m6A modification at mRNA 5′-UTRs and promotes mRNA translation [46, 90]. One study showed that m6A promoted cap-independent mRNA translation in the absence of the cap-binding factor eIF4E, since m6A directly binds eIF3 to recruit the 43S complex, initiating translation [46]. The other study showed that in the nucleus, heat-shock induced 5′-m6A is protected from FTO-mediated demethylation by nuclear-translocated YTHDF2 [90]. This model is supported by observations that the affinity of m6A RNA for the YTH domain is greater than that of m6A RNA for FTO [77, 78, 91]. Yet another study reported that METTL3 directly interacts with the translation initiation factor eIF3 to promote translation of a subset of mRNAs, independent of METTL3 methyltransferase activity or YTHDF1 or YTHDF2 binding [92]. It remains unclear whether and how these mechanisms co-exist in cells. It would now be informative to identify RNA substrates for each of these mechanisms in order to understand the significance of each in normal or conditioned, such as heat-shocked, cells.
About half of mammalian m6A sites are located in coding sequence [29]. One study employed biochemical, structural and single-molecule methods to address the function of m6A modification in mRNA/tRNA interactions using E. coli ribosomes as a system [93]. The authors showed that, although X-ray crystallographic analyses indicate that m6A base-pairs with uridine during the decoding process, m6A modification can act as a barrier to tRNA accommodation and translation elongation in a manner that depends on position and context of m6A within codons [93]. These authors propose that such dynamic changes could modulate coupled co-translational processes such as protein folding, suggesting that m6A may allow a single gene to encode proteins of different functional forms.
m6A promotes exon inclusion and enhances mRNA splicing
m6A-related proteins, including METTL3/METTL14/WTAP of the methyltransferase complex, FTO and ALKBH5 demethylases, or m6A binding proteins YTHDF2 and YTHDC1, all reportedly localize in nuclear organelles known as speckles, which are enriched in pre-mRNA splicing factors [25, 34, 44, 59, 66, 75], suggesting a role for m6A RNA modification in mRNA splicing (Fig. 2). Within the METTL3/METTL14/WTAP complex, WTAP is required for METTL3 and METTL14 accumulation in nuclear speckles and most mRNA species bound by WTAP and METTL14 were transcribed from genes known to give rise to mRNAs with multiple splicing variants [44]. Loss of function studies further show that depletion of either METTL3 or WTAP results in transcriptome-wide changes in RNA splicing [44, 80]. Together, these data suggest that m6A methyltransferase activity regulates mRNA splicing.
One study reports that FTO depletion enhances m6A levels in regions flanking 5′- and 3′-splice sites and promotes binding of the splicing factor SRSF2, increasing exon inclusion [94]. ALKBH5 knockdown cells show loss of phosphorylated SC35, a marker of nuclear speckles, suggesting that ALKBH5 regulates speckle formation, an effect dependent on ALKBH5 demethylase activity [66].
The m6A nuclear reader YTHDC1 reportedly binds the pre-mRNA splicing factors SRSF3 and SRSF10 competitively, and promotes exon inclusion by facilitating SRSF3 but repressing SRSF10 in their nuclear speckle localization and RNA binding [75]. Another nuclear m6A reader HNRNPA2B1 reportedly directly binds a set of m6A-tagged nuclear transcripts and modulates their splicing in a manner comparable to METTL3, as evidenced by a strong positive correlation between global changes in alternative splicing and depletion of either HNRNPA2B1 or METTL3 [80].
m6A promotes mRNA transport into the cytoplasm
ALKBH5 knockout mice show moderate increases in m6A levels and accelerated mRNA export to the cytoplasm [66] (Fig. 2). As a mechanism, the authors of that study focused on the splicing factor ASF/SF2 (Alternative Splicing Factor), as it co-localizes with ALBKH5 in nuclear speckles [66]. It is also well-established that ASF/SF2 hypophosphorylation switches its function from that of a splicing factor to an adaptor protein functioning in mRNA nuclear export [95, 96]. Interestingly, ALKBH5-deficient cells not only show ASF/SF2 hypophosphorylation and loss of ASF localization to nuclear speckles, but relocalization of the ASF/SF2 kinase SRPK1 (Serine/threonine-protein kinase 1) from nucleus to cytoplasm. Thus the authors propose that SRPK1 relocation underlies ASF/SF2 hypophosphorylation, enhancing mRNA transport to the cytoplasm. Importantly, the observed phenotypes in ALKBH5 knockout cells can only be rescued by the overexpression of wild-type but not mutant ALKBH5 lacking demethylase activity, suggesting that m6A modification regulates mRNA transport. The exact mechanism remains unclear.
m6A levels are associated with usage of alternative polyA (APA) sites
A UV CLIP study that mapped m6A sites in the mammalian transcriptome at single-nucleotide resolution reported a positive correlation between m6A density and the length of the last exon [37] (Fig. 2). The authors then simultaneously knocked down METTL3, METTL14, and WTAP and examined APA usage in a subset of mRNAs. They found that upon global reduction of m6A levels, a greater number of genes showed proximal APA usage, raising the possibility that some m6A residues may inhibit proximal polyadenylation. In agreement, another study measured the fraction of m6A -methylated vs. nonmethylated RNAs and reported that m6A levels are positively correlated with 3′-UTR length [39]. Mechanisms underlying these activities remain undetermined.
An interaction between m6A modification and microRNA pathway (Fig. 2)
It was reported that METTL3-mediated m6A methylation of primary microRNAs facilitates primary microRNA processing by the DGCR8 microprocessor complex [97] (Fig. 2). This group further identified HNRNPA2B1 as a nuclear m6A reader mediating this process [80]. Interestingly, another group reported that m6A levels are regulated by the microRNA machinery and by microRNAs [98]. In that study, the authors proposed that microRNA regulates m6A formation by modulating METTL3/mRNA binding, presumably in the cytoplasm. However, it is unclear how microRNA-modulated METTL3/mRNA binding affects m6A methylation, since METTL14 is a nuclear protein [92] and m6A methylation likely occurs in the nucleus. Nevertheless, these studies suggest cellular interaction of two major RNA regulatory mechanisms: m6A mRNA modification and microRNAs. Detailed mechanisms remain to be investigated.
Concluding Remarks and Future Perspective
There has been an enormous expansion in our knowledge of m6A modification over the last few years. Nonetheless, fundamental questions relevant to regulation and activity of this modification remain (See Outstanding Questions Box). For example, we do not yet know why m6A methyltransferases methylate some but not all mRNAs. We also do not yet comprehend what factors control the extent of m6A modification of a particular mRNA. Mechanisms that maintain the balance between formation and removal of m6A are not yet well defined, nor is it understood how m6A binding proteins compete with the demethylases. While numerous functions of m6A modification are proposed, based largely on genome-wide data, the ultimate test of mutating endogenous m6A sites followed by phenotypic analysis is lacking. In addition, although m6A is present on rRNA, tRNA, snRNA (small nuclear RNA), snoRNA (small nucleolar RNA), and lncRNAs [38, 99], it is not known whether characterized m6A readers recognize modified RNAs outside the context of mRNAs. In short, our journey down the road to understand m6A mechanism and function has just begun!
Outstanding Questions.
Why m6A methyltransferases methylate some but not all mRNAs? What factors control the extent of m6A modification of a particular mRNA?
What maintains the balance between formation and removal of m6A? How are m6A levels regulated in cells?
Would characterized m6A mRNA reader recognize m6A modified tRNA, rRNA, or snoRNA? Is there any crosstalk between different types of RNAs through m6A modification?
m6A regulates mRNA activities through diverse mechanisms. How do these mechanisms co-exist in cells?
Trends.
N6-methyladensone or m6A is the most abundant internal messenger RNA (mRNA) modification that tags tens of thousands eukaryotic transcripts. Technologies to detect m6A have improved rapidly. Now we can map m6A methylome at a single nucleotide resolution and determine the proportion of methylated vs. unmethylated transcripts in a high throughput manner.
The “writer”, “eraser”, and “reader” of m6A modification have been reported. These discoveries have greatly facilitated our understanding towards the functional significance of m6A.
Emerging evidence suggests that m6A plays critical roles in regulating diverse mRNA activities, from processing to localization and translation. Therefore, reversible m6A modification represents a new and crucial layer of gene expression regulation in eukaryotes.
Acknowledgments
We thank the NIH for funding P30 CA023168 and R01 GM110090 to J.C.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Furuichi Y, et al. 5′-Terminal m-7G(5′)ppp(5′)G-m-p in vivo: identification in reovirus genome RNA. Proc Natl Acad Sci U S A. 1975;72(2):742–5. doi: 10.1073/pnas.72.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei CM, et al. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–86. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 3.Desrosiers R, et al. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(10):3971–5. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255(5503):28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 5.Perry RP, et al. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell. 1975;4(4):387–94. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- 6.Wei CM, et al. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15(2):397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- 7.Melcher T, et al. Editing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J Biol Chem. 1995;270(15):8566–70. doi: 10.1074/jbc.270.15.8566. [DOI] [PubMed] [Google Scholar]
- 8.Polson AG, et al. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380(6573):454–6. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 9.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delatte B, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282–5. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 13.Dominissini D, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–6. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311–6. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 15.Levis R, Penman S. 5′-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. Journal of molecular biology. 1978;120(4):487–515. doi: 10.1016/0022-2836(78)90350-9. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy TD, Lane BG. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5′-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Canadian journal of biochemistry. 1979;57(6):927–31. doi: 10.1139/o79-112. [DOI] [PubMed] [Google Scholar]
- 17.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant cell. 2008;20(5):1278–88. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clancy MJ, et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic acids research. 2002;30(20):4509–18. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodi Z, et al. Yeast targets for mRNA methylation. Nucleic acids research. 2010;38(16):5327–35. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue Y, et al. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–55. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 22.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–26. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klungland A, Dahl JA. Dynamic RNA modifications in disease. Curr Opin Genet Dev. 2014;26:47–52. doi: 10.1016/j.gde.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77(1–2):142–4. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 25.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7(12):885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. 2005;12:141–177. [Google Scholar]
- 27.Motorin Y, et al. Identification of modified residues in RNAs by reverse transcription-based methods. Methods in enzymology. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 28.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(5):1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155(6):1409–21. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell. 2014;15(6):707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell reports. 2014;8(1):284–96. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology. 2014;16(2):191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen K, et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angewandte Chemie. 2015;54(5):1587–90. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes & development. 2015;29(19):2037–53. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature methods. 2015;12(8):767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molinie B, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13(8):692–8. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Csepany T, et al. Sequence specificity of mRNA N6-adenosine methyltransferase. The Journal of biological chemistry. 1990;265(33):20117–22. [PubMed] [Google Scholar]
- 41.Harper JE, et al. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic acids research. 1990;18(19):5735–41. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rottman FM, et al. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76(12):1109–14. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 43.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24(12):1403–19. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014 doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schibler U, Perry RP. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res. 1977;4(12):4133–49. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer KD, et al. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, et al. Genome-wide detection of high abundance N6-methyladenosine sites by microarray. RNA. 2015;21(8):1511–8. doi: 10.1261/rna.051474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engel JD, von Hippel PH. Effects of methylation on the stability of nucleic acid conformations: studies at the monomer level. Biochemistry. 1974;13(20):4143–58. doi: 10.1021/bi00717a013. [DOI] [PubMed] [Google Scholar]
- 49.Engel JD, von Hippel PH. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. The Journal of biological chemistry. 1978;253(3):927–34. [PubMed] [Google Scholar]
- 50.Micura R, et al. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic acids research. 2001;29(19):3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kierzek E. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Research. 2003;31(15):4472–4480. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137(5):2107–15. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harcourt EM, et al. Identification of a selective polymerase enables detection of N(6)-methyladenosine in RNA. J Am Chem Soc. 2013;135(51):19079–82. doi: 10.1021/ja4105792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilfan ID, et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J Nanobiotechnology. 2013;11:8. doi: 10.1186/1477-3155-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai Q, et al. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic acids research. 2007;35(18):6322–9. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19(12):1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. The Biochemical journal. 1992;288( Pt 1):233–40. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bokar JA, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. The Journal of biological chemistry. 1994;269(26):17697–704. [PubMed] [Google Scholar]
- 59.Bokar JA, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3(11):1233–47. [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10(2):93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bujnicki JM, et al. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. Journal of molecular evolution. 2002;55(4):431–44. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- 62.Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016:5. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P, et al. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63(2):306–17. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, et al. Structural basis of N-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016 doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 65.Agarwala SD, et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS genetics. 2012;8(6):e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia G, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS letters. 2008;582(23–24):3313–9. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng C, et al. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289(17):11571–83. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W, et al. Crystal structure of the RNA demethylase ALKBH5 from zebrafish. FEBS letters. 2014;588(6):892–8. doi: 10.1016/j.febslet.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han Z, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–9. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 72.Chen B, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134(43):17963–71. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 73.Xu C, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nature chemical biology. 2014;10(11):927–9. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao W, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Molecular cell. 2016;61(4):507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Z, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285(19):14701–10. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Theler D, et al. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic acids research. 2014;42(22):13911–9. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu T, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24(12):1493–6. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hashimoto H, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40(11):4841–9. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alarcon CR, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299–308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bachellerie JP, et al. Biosynthesis and utilization of extensively undermethylated poly(A)+ RNA in CHO cells during a cycloleucine treatment. Nucleic Acids Res. 1978;5(8):2927–43. doi: 10.1093/nar/5.8.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–6. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 83.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kennedy EM, et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell host & microbe. 2016;19(5):675–85. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tirumuru N, et al. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016:5. doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rouskin S, et al. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485):701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 88.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544):486–90. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou S, et al. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep. 2016;6:25677. doi: 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin S, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular cell. 2016;62(3):335–45. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choi J, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nature structural & molecular biology. 2016;23(2):110–5. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ben-Haim MS, et al. FTO: linking m6A demethylation to adipogenesis. Cell research. 2015;25(1):3–4. doi: 10.1038/cr.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y, et al. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci U S A. 2004;101(26):9666–70. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279(30):31745–9. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 97.Alarcon CR, et al. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–5. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen T, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell stem cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 99.Cantara WA, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic acids research. 2011;39(Database issue):D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]