Inverse and erythrodermic psoriasis are rare subtypes of psoriasis. Whereas the former is characterized by shiny erythematous non-scaly plaques in the body folds, the latter has widespread redness with fine scale, covering over 80% of the body-surface area, and can be life-threatening. Both are considered to be clinical subtypes of chronic plaque psoriasis, and often co-exist or evolve from plaque psoriasis (Boyd and Menter, 1989; Omland and Gniadecki, 2015), but the pathogenic mechanisms involved are unknown, and current treatments are frequently unsatisfactory (Rosenbach et al., 2010).
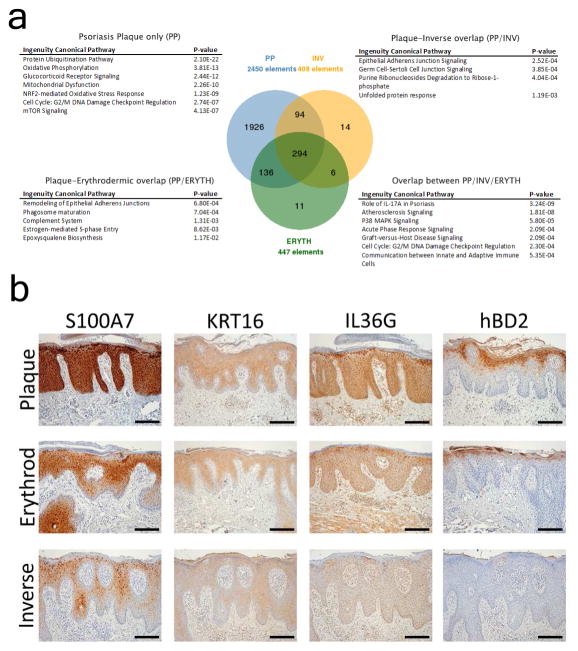
To assess shared and unique processes between chronic plaque, inverse, and erythrodermic psoriasis we analyzed archived formalin-fixed paraffin-embedded biopsies of clinically and histologically confirmed chronic plaque (n=12), inverse (n=40) and erythrodermic psoriasis cases (n=30) and healthy control skin (n=12) using Affymetrix ST 2.1 Arrays. Compared with healthy skin, psoriatic plaque lesions yielded 2450 differentially expressed genes (DEGs) (FDR, p<0.05), inverse psoriasis lesions yielded 408 DEGs (FDR, p<0.05) and erythrodermic psoriasis lesions yielded 447 DEGs (FDR, p<0.05) (Figure 1A). In total 294 genes were found to be shared among the three disease subtypes (FDR, p<0.05). While the overlap only accounted for 12% of the DEGs in chronic plaque psoriasis, it accounted for 66% and 72% of DEGs in erythrodermic and inverse psoriasis respectively.
Figure 1. Unique and shared canonical pathways between chronic plaque, inverse and erythrodermic psoriasis.
Venn diagram of the overlap between chronic plaque (PP), inverse (INV) and erythrodermic (ERYTH) psoriasis, and enriched canonical pathways. The overlap group represented the majority of differentially expressed genes in both inverse and erythrodermic group (>66%) but only about 12% of the DEGs in chronic plaque psoriasis. Dominant canonical pathway in the shared group was IL-17 responses (A). Histologic confirmation of several genes shared between chronic plaque, inverse and erythrodermic psoriasis (B) (scale bar = 100μm).
Disease processes specific to each psoriasis subtype were analyzed using Ingenuity Pathway Analysis. Canonical processes unique to chronic plaque psoriasis included protein ubiquitination pathway (p=2.10E-22), oxidative phosphorylation (p=3.81E-13), glucocorticoid receptor signaling (p=2.44E-12), mitochondrial dysfunction (2.26E-10), and mTOR signaling (p=4.13E-07). Genes unique to chronic plaque psoriasis included NFKBIZ (3.6-fold increased) and DDX58 (2.4-fold), genes recently implicated in the pathogenesis of psoriasis (Johansen et al., 2015; Tsoi et al., 2015). Overlapping canonical processes between plaque and inverse psoriasis included epithelial adherens junction signaling (p=2.52E-4) and unfolded protein responses (p=1.19E-03), while the overlap between plaque and erythrodermic psoriasis included remodeling of epithelial adherence junction (p=6.8E-04), and phagosome maturation (p=7.0E-04). The numbers of genes unique to inverse and erythrodermic psoriasis were too small for analyses of enriched biologic processes (Supplementary File 1). Genes shared among all three clinical presentations included S100A7 (32-fold up in PP, 8.7-fold up in inverse psoriasis, 6-fold up in erythrodermic psoriasis), KRT16 (15-, 5- and 4.8-fold respectively), IL36G (10-, 3.7- and 3.9-fold respectively), and human β-defensin 2 (hBD2/DEFB4, 10-, 4.5-, 3.5-fold respectively). These were confirmed by immunohistochemistry (Figure 1B).
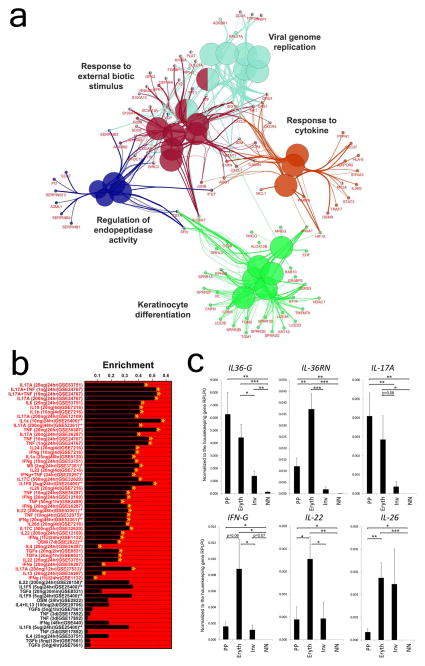
The most enriched canonical pathway from the group of genes shared among plaque, inverse and erythrodermic psoriasis included IL-17A signaling (p=3.24E-09), followed by p38 MAPK signaling (p=5.8E- 05) and communication between innate and adaptive immune cells (p=5.35E-04). The key biologic nodes in the shared gene set included keratinocyte differentiation, regulation of endopeptidase activity, response to external biotic stimulus, viral genome replication, and response to cytokines (Figure 2A). We next screened the overlapping gene set to determine if such genes were disproportionately induced by cytokine treatments applied to cultured keratinocytes using cytokine-response analyses previously described by our group (Swindell et al., 2013a; Swindell et al., 2013b). Consistent with the Ingenuity pathway analysis results, the overlapping gene set amongst the three transcriptomes was most strongly enriched with respect to genes induced by IL-17A in cultured KCs, with strong enrichment also observed with respect to genes induced by IL-17A+TNF and TNF (Figure 2B). QRT-PCR confirmed elevated mRNA expression of IL-17A in all three clinical presentations, with the highest expression in chronic plaque psoriasis, but lowest in inverse psoriasis (Figure 2C). IL-17A mRNA expression was about 10-fold higher in plaque compared to inverse psoriasis (p<0.01). The reason for this difference is not clear and will need to be addressed in future studies. Similarly, IL36G mRNA expression was elevated in all three but highest in plaque psoriasis. In contrast IL36RN, the receptor antagonist for the IL-36 family of cytokines, had the highest expression in erythrodermic psoriasis (p<0.01). Interestingly, IFNG and IL22 mRNA expression was the highest in erythrodermic psoriasis compared to both plaque and inverse psoriasis (about 5-fold higher compared to both plaque and inverse psoriasis, p<0.05), whereas, in stark contrast, IL26 mRNA expression, a cytokine recently implicated in the pathogenesis of psoriasis (Meller et al., 2015), was higher in both erythrodermic and inverse psoriasis compared to plaque psoriasis (i.e. erythrodermic vs plaque psoriasis 6-fold higher, p<0.01) (Figure 2C).
Figure 2. Overlapping and unique biologic and cytokine responses in chronic plaque, inverse and erythrodermic psoriasis.
Using biologic process analyses by Cytoscape and ClueGo the shared genes between chronic plaque, inverse, and erythrodermic psoriasis belonged to five major groups, including keratinocyte differentiation and response to cytokine (A). Using gene-set enrichment analyses IL-17A signatures were observed to dominate the shared gene set along with IL-17A+TNF and TNF responses (B). QRT-PCR of chronic plaque (n=12), inverse (n=40) and erythrodermic psoriasis (n=30) revealed differences in the mRNA expression of key cytokines among the three clinical presentations (C). (data shown with S.E.M, two-tailed t-test, * p<0.05, ** p<0.01, *** p<0.001).
These results demonstrate that IL-17A is the major shared pathway for chronic plaque, inverse and erythrodermic psoriasis. This is consistent with recently published case series showing six out of eight erythrodermic patients achieving complete clearance with anti-IL-17 treatment (Saeki et al., 2015), but similar data on flexural psoriasis does not yet exist. In addition, our data demonstrates pronounced differences in the expression of several key pro-inflammatory cytokines between each clinical subtype, suggesting that while IL-17 may provide a common inflammatory pathway, other inflammatory signals may modify the clinical presentation and contribute to unique manifestations of each subtype. This includes the non-pustular presentation of erythrodermic psoriasis, and its higher risk of septicemia (Green et al., 1996), particularly from skin pathogens including Staphylococcus aureus (Prystowsky and Cohen, 1995), although the extent of skin involvement is likely to be a contributing factor as well. Similarly, the clinical appearance of inverse psoriasis, with its decreased scale and thinner shinier plaques, may therefore be less due to its localization in moist, warm milieu of flexural body sites (Omland and Gniadecki, 2015), but more due to differences in the composition of the inflammatory network in skin, and variation in growth factors such as epiregulin, heparin-binding EGF, amphiregulin, and vascular endothelial growth factor, which in our study was found to be increased in chronic plaque but not in erythrodermic or inverse psoriasis (data not shown).
In summary, the findings presented here provide information that may change the existing therapeutic approach to treating patients with inverse and erythrodermic psoriasis, such as the use of anti-IL-17 treatments. This provides direction for the design of future clinical trials, which will be needed to validate our conclusions. Furthermore, our data provide to our knowledge previously unreported insights into how variations in the cytokine network in psoriatic skin may shape different clinical manifestations of psoriasis.
Acknowledgments
The work was in part supported by a research grant from Novartis, the University of Michigan Babcock Endowment Fund (A.J., J.J.V, J.E.G.), NIH awards K08-AR060802 and R01-AR069071 (J.E.G.), K01-AR064765 (A.J.), K08-AR063668 (J.M.K.) the A. Alfred Taubman Medical Research Institute Kenneth and Frances Eisenberg Emerging Scholar Award (J.E.G.), and Doris Duke Charitable Foundation Grant #2013106 (J.E.G).
Footnotes
SUPPLEMENTARY
Supplementary Methods and Supplementary Files are in the Supplementary Material.
CONFLICTS OF INTEREST
Dr. Johann E. Gudjonsson has received research support from Amgen, AbbVie and Novartis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyd AS, Menter A. Erythrodermic psoriasis. Precipitating factors, course, and prognosis in 50 patients. J Am Acad Dermatol. 1989;21:985–91. [PubMed] [Google Scholar]
- Green MS, Prystowsky JH, Cohen SR, Cohen JI, Lebwohl MG. Infectious complications of erythrodermic psoriasis. J Am Acad Dermatol. 1996;34:911–4. doi: 10.1016/s0190-9622(96)90078-x. [DOI] [PubMed] [Google Scholar]
- Johansen C, Mose M, Ommen P, Bertelsen T, Vinter H, Hailfinger S, et al. IkappaBzeta is a key driver in the development of psoriasis. Proc Natl Acad Sci U S A. 2015;112:E5825–33. doi: 10.1073/pnas.1509971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller S, Di Domizio J, Voo KS, Friedrich HC, Chamilos G, Ganguly D, et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16:970–9. doi: 10.1038/ni.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omland SH, Gniadecki R. Psoriasis inversa: A separate identity or a variant of psoriasis vulgaris? Clin Dermatol. 2015;33:456–61. doi: 10.1016/j.clindermatol.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Prystowsky JH, Cohen PR. Pustular and erythrodermic psoriasis. Dermatol Clin. 1995;13:757–70. [PubMed] [Google Scholar]
- Rosenbach M, Hsu S, Korman NJ, Lebwohl MG, Young M, Bebo BF, Jr, et al. Treatment of erythrodermic psoriasis: from the medical board of the National Psoriasis Foundation. J Am Acad Dermatol. 2010;62:655–62. doi: 10.1016/j.jaad.2009.05.048. [DOI] [PubMed] [Google Scholar]
- Saeki H, Nakagawa H, Ishii T, Morisaki Y, Aoki T, Berclaz PY, et al. Efficacy and safety of open-label ixekizumab treatment in Japanese patients with moderate-to-severe plaque psoriasis, erythrodermic psoriasis and generalized pustular psoriasis. J Eur Acad Dermatol Venereol. 2015;29:1148–55. doi: 10.1111/jdv.12773. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC genomics. 2013a;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Xing X, Voorhees JJ, Elder JT, Gudjonsson JE. Modulation of epidermal transcription circuits in psoriasis: new links between inflammation and hyperproliferation. PloS one. 2013b;8:e79253. doi: 10.1371/journal.pone.0079253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015;6:7001. doi: 10.1038/ncomms8001. [DOI] [PMC free article] [PubMed] [Google Scholar]