Abstract
Histone deacetylase (HDAC) regulation is an essential process in myogenic differentiation. Inhibitors targeting the activity of specific HDAC family members have been shown to enhance the cardiogenic differentiation capacity of discrete progenitor cell types; a key property of donor cell populations contributing to their afforded benefits in cardiac cell therapy applications. The influence of HDAC inhibition on cardiac-derived mesenchymal stromal cell (CMC) transdifferentiation or the role of specific HDAC family members in dictating cardiovascular cell lineage specification has not been investigated. In the current study, the consequences of HDAC inhibition on patient-derived CMC proliferation, cardiogenic program activation, and cardiovascular differentiation/cell lineage specification were investigated using pharmacologic and genetic targeting approaches. Here, CMCs exposed to the pan-HDAC inhibitor sodium butyrate (NaBu) exhibited induction of a cardiogenic transcriptional program and heightened expression of myocyte and endothelial lineage-specific markers when coaxed to differentiate in vitro. Further, shRNA knockdown screens revealed CMCs depleted of HDAC1 to promote the induction of a cardiogenic transcriptional program characterized by enhanced expression of cardiomyogenic- and vasculogenic-specific markers, a finding which depended upon and correlated with enhanced acetylation and stabilization of p53. Cardiogenic gene activation and elevated p53 expression levels observed in HDAC1-depleted CMCs were associated with improved aptitude to assume a cardiomyogenic/vasculogenic cell-like fate in vitro. These results suggest that HDAC1 depletion-induced p53 expression alters CMC cell fate decisions and identify HDAC1 as a potential exploitable target to facilitate CMC-mediated myocardial repair in ischemic cardiomyopathy.
Keywords: Cardiac mesenchymal stromal cells, histone deacetylases, cardiogenic gene expression, histone deacetylase inhibitors, cardiogenic differentiation, vasculogenic differentiation, p53
Graphical abstract
HDAC1 Regulates the Expression of a Cardiogenic Program in Human Cardiac Mesenchymal Stromal Cells (CMCs) through a p53-Dependent Mechanism. CMCs depleted of HDAC1 exhibited induction of a cardiogenic transcriptional program characterized by enhanced expression of cardiomyogenic (MEF2C, NKX2.5, and TBX5)- and vasculogenic (CD31) factors. This phenomenon required and correlated with enhanced acetylation and stabilization of the tumor suppressor protein p53. Cardiogenic gene activation and elevated p53 expression levels observed in HDAC1-depleted CMCs were associated with heightened proclivity to assume a cardiomyogenic/vasculogenic cell-like fate in vitro. These results indicate HDAC1 depletion-induced p53 expression alters CMC cell fate decisions and identify HDAC1 as a potential exploitable target to facilitate CMC-mediated myocardial repair in ischemic cardiomyopathy.
Introduction
While cell therapy has demonstrated salutary effects on cardiac function in heart failure patients [1-5], the efficacy of this therapeutic modality is significantly limited by the poor engraftment, survival, and differentiation of transplanted cells [6, 7]. Understanding the precise mechanisms underlying stem cell-mediated cardiac repair will be vital for developing effective cell therapy strategies. Although the mechanism by which transplanted stem cells mitigate detrimental cardiac remodeling and improve ventricular performance remains unclear, both donor cell cardiogenic differentiation [8-10] and paracrine signaling [11] have been implicated as contributing factors.
Many investigators have focused on cardiogenic differentiation when developing novel strategies for cell therapy. For instance, cytokine-mediated guided cardiopoiesis was shown to enhance the cardiac reparative aptitude of transplanted bone marrow-derived mesenchymal stem cells (BM-MSCs) in a murine model of ischemic cardiomyopathy [10]. The reparative cytotype of these “cardiopoietically” primed BM-MSCs was characterized by elevated expression of cardiogenic transcription factors (i.e. Nkx2.5, Tbx5, and Mef2c) that function cooperatively to regulate genetic programs involved in cardiomyogenic differentiation. Further, cell types more oriented towards a cardiovascular phenotype, such as those derived from the heart, possess greater therapeutic potential than those derived from extra-cardiac sources (e.g. BM-MSCs). For example, cardiac-derived mesenchymal stromal cells (CMCs), which more efficiently express cardiogenic markers vis-à-vis those isolated from bone marrow (BMCs), exhibit superior tissue retention, infarct infiltration, and proclivity to differentiate into adult myocytes when injected into infarcted rat hearts [12]. In addition to their purported cardiomyogenic differentiation capabilities, in vivo genetic fate mapping studies of endogenous CMCs in murine infarct models have shown these cells to directly contribute to cardiac neovascularization following cardiac injury [13]. These studies suggest that the cardiac stromal compartment is an important cell reservoir for more effective cardiac repair. Currently, the developmental potential of CMCs is poorly understood and little is known regarding the molecular pathways dictating their differentiation towards mature cardiovascular cell types.
Chromatin remodeling plays a central role in determining mesenchymal stem cell lineage-specific differentiation [14]. Chromatin modifying agents, such as histone deacetylase (HDAC) inhibitors, have been shown to promote myogenic differentiation of various adult progenitor cell types (i.e. cardiac side population (SP) cells [15] and BM-MSCs [16]), presumably via the induction of cardiogenic transcription factors (e.g. Gata-4, Mef2c, and Tbx5). Additional studies have identified specific HDAC family members to be associated with the regulation of myogenic differentiation in distinct progenitor cell types, including HDAC4 in murine c-kitpos cardiac stem cells (CSCs) [17], HDAC7 in C2C12 myoblasts [18], and HDAC1 in rat BM-MSCs [19] – suggesting cell context specific effects for particular HDAC family members in directing cell fate decisions. To date, eighteen mammalian HDACs have been identified; these are stratified into four distinct families based on sequence homology to yeast original enzymes which include the zinc-dependent (class I [HDACs 1, 2, 3, and 8], class II [HDACs 4, 5, 6, 7, 9, and 10], and class IV [HDAC 11]) and nicotinamide adenine dinucleotide (NAD+) requiring enzymes (class III [Sirt 1-7]), referred to as sirtuins [20]. Contrary to what their name may suggest, there also exists a host of nonhistone protein substrates that are targeted and regulated by HDAC activity. Thus, in addition to their role in the epigenetic regulation of gene expression, chiefly through the deacetylation of nucleosomal histone proteins, alterations in the acetylation status of nonhistone substrates (e.g. transcription factors or other proteins) constitute an additional mechanism by which HDACs can modulate various cellular processes. Consistent with their epigenetic regulatory activity, HDAC inhibitors have been shown to function as chromatin modifying agents that can temporally regulate the control of gene expression in mammalian cells [20]. Furthermore, as the epigenetic landscape is central in modulating cellular events that direct progenitor cell differentiation, as well as somatic cell reprogramming [21], significant attention has been placed on the ability of HDAC inhibitors to alter or direct the differentiation of both stem and adult progenitor cell types (reviewed in [22]).
The influence of HDAC inhibition on CMC differentiation is unknown. Furthermore, the role of specific HDAC family members in dictating CMC cardiovascular cell lineage specification has not been investigated. Accordingly, we employed both pharmacologic and genetic approaches to investigate the consequences of HDAC inhibition on CMC biology and, specifically, on CMC cardiogenic gene transcription and its impact on cardiovascular cell lineage commitment. Our results demonstrate that the epigenetic regulator HDAC1 modulates transcription of a core cardiogenic program in human CMCs through a mechanism involving the induced acetylation and elevated expression of the tumor suppressor protein p53. CMCs depleted of HDAC1 exhibited induction of a cardiogenic transcriptional program characterized by enhanced expression of cardiomyogenic, vasculogenic, and stemness-related transcripts, an effect that was diminished by p53 inhibition. Augmentation of lineage-specific markers in HDAC1-depleted CMCs was associated with heightened competency to adopt a cardiomyogenic/vasculogenic cell-like fate in vitro. Overall, these data suggest the inhibition of HDAC1 can indirectly alter CMC cell fate decisions via enhancing the expression of p53.
Materials and Methods
Cells, cell culture, and treatment
Primary normal human dermal fibroblasts (HDMFs; ATCC® PCS-201-010™) were maintained in DMEM medium (GIBCO) containing 20% FBS (Seradigm), 0.2 mM L-Glutamine (Gibco), and 100 U/ml penicillin/streptomycin (Gibco). Murine myoblasts (C2C12; ATCC® CRL-1772) were cultured in DMEM medium + GlutaMAX™ (Gibco) containing 10% FBS (Seradigm) and 100 U/ml penicillin/streptomycin (Gibco). Primary patient-derived CMCs were propagated in Ham’s F12 medium (Gibco) supplemented with 10% FBS (Seradigm), 20 ng/ml Recombinant Human bFGF (PeproTech), 0.2 mM L-Glutamine (Gibco), 0.005 U/ml human Erythropoietin (Sigma), and 100 U/ml penicillin/streptomycin (Gibco). Human HEK293FT cells (Life Technologies) used in lentiviral production were grown in DMEM medium (Gibco) containing 10% FBS (Seradigm) and 0.2 mM L-Glutamine (Gibco). All cell lines were propagated under standard incubation conditions at 37°C with 5% atmospheric CO2 and, further, were enzymatically passaged using TrypLE™ (Life Technologies) when approaching ≈70% confluence.
Human cardiac stromal cell (CMC) isolation
Human CMCs were isolated from discarded atrial appendage specimens collected from patients during routine coronary artery bypass surgery at Jewish Hospital (University of Louisville, Louisville, KY). De-identified right atrial appendage specimens were acquired via written consent agreement according to the approved protocol by the Institutional Review Boards on human subject research (IRB number: 03.052J) at the University of Louisville. Right atrial auricles were transferred to 35mm dishes and washed three times with ice cold PBS. Tissues were then manually minced to obtain fragments <1mm3 in dimension and resuspended in serum free Ham’s F12. Auricle fragments were next incubated at 37°C in a shaking incubator and enzymatically digested using Collagenase type II solution (≈2 mg/ml; Worthington Biochemical Corporation). Following enzymatic dissociation, released cells were pelleted at 400×g for 5 min and washed in complete medium (Ham’s F12 (Gibco), 10% FBS (Seradigm), 20 ng/ml Recombinant Human bFGF (PeproTech), 0.2 mM L-Glutamine (Gibco), 0.005 U/ml human Erythropoietin (Sigma), and 100 U/ml penicillin/streptomycin (Gibco). Cells were resuspended in complete medium and plated in 6-well tissue culture grade plates for passage 0 initial expansions. Medium was replaced at 24 h after the initial plating step and subsequently replaced every 48 h thereafter. Cells were expanded until 70% confluence at which time they were trypsinized (TrypLE™, Life Technologies) and transferred to T75 tissue culture flasks for further expansion. CMCs utilized in experiments were not used beyond passage 8.
Flow cytometry
Cells were immunophenotyped using PE-conjugated antibodies raised against human mesenchymal (CD73, CD90, CD105), hematopoietic (CD45), and stem cell (CD117) markers, as directed by the manufacturer’s instructions. Live stained and unstained control cells were analyzed using a BD LSR II Flow Cytometer (BD Biosciences). 10,000 events were collected per sample and analyzed using Flowing Software version 2.5.0 (Perttu Terho, Turku Centre for Biotechnology). Live cells were gated according to light scatter characteristics and resultant population PE fluorescence intensity plotted on dot plots. The percentage of PE-positive cells corresponding to their respective cell surface antigens were determined by gating against unstained (autofluorescence controls) negative controls according to previously described guidelines [23, 24]. For intracellular protein staining, cells were fixed with 4% formaldehyde for 10 min at room temperature, washed in 1X PBS, and permeabilized with 90% methanol on ice for 30 min. Cells were incubated with primary antibodies or isotype controls diluted in incubation buffer (1% BSA, 1X PBS) for 1 h at room temperature, washed by centrifugation (600 × g) in incubation buffer, and resuspended in fluorochrome-conjugated secondary antibodies for 30 min at room temperature. Flow cytometry was performed as described above and the percentage of positive cells determined by gating against corresponding isotype controls. A complete list of utilized antibodies is listed in Supporting Information Table S1.
Statistical analysis
Statistical analyses were performed using the descriptive statistics data analysis toolpak available in the Microsoft Office Excel program. The Student’s two-tailed t-test, after ascertaining parametric distribution, was used to analyze differences between groups. Data are reported as means ± SEM.
Additional materials and procedures are detailed in Supporting Information Supplementary Methods.
Results
CMCs exhibit a mesenchymal immunophenotype
For the experiments described in the current study, cells exhibiting plastic adherence in tissue-culture grade dishes were utilized following their propagation in culture for no less than 24 h. CMCs displayed a distinctive mesenchymal immunophenotype characterized by positivity for CD73, CD90, and CD105 that closely resembled the phenotype of their dermal-derived stromal counterparts, HDMFs (human dermal fibroblasts) (Supporting Information Fig. S1). Both CMCs and HDMFs exhibited low or no expression of the hematopoietic marker CD45 and the putative cardiac stem cell (CSC) marker c-kit [25] (Supporting Information Fig. S1). Although they shared many similarities with HDMFs, CMCs were notably heterogeneous in CD90 expression, consistent with previous investigations of CMCs [12] and mesenchymal cells derived from tissues other than bone marrow [26]. CMCs exhibiting plastic adherence at 24 h were immunophenotypically indistinguishable from those propagated in long-term culture (passage 8; data not shown).
Sodium butyrate reduces CMC proliferation, increases histone acetylation, and alters cardiac-specific gene transcription
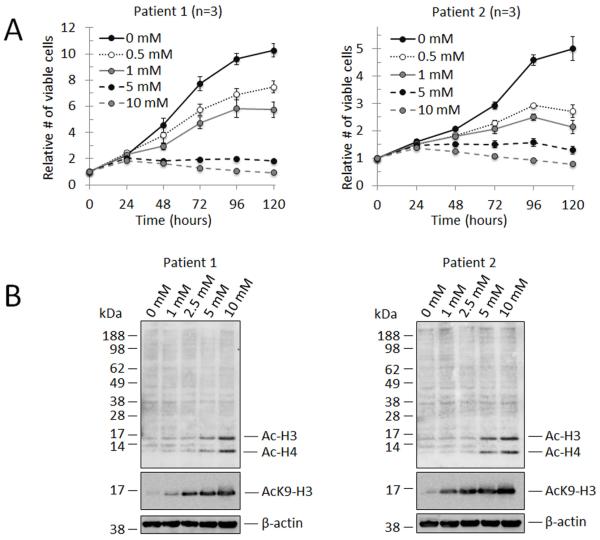
The pan-HDAC inhibitor, sodium butyrate (NaBu), exerts both concentration- and cell type-specific effects on proliferation and gene expression, and also influences tissue-specific stem cell differentiation dynamics [22, 27, 28]. The consequences of NaBu-mediated HDAC inhibition on CMC gene transcription and differentiation have not been previously described. We performed preliminary NaBu dose-response experiments on patient-derived samples to empirically investigate the effect of escalating doses of NaBu on CMC proliferation rates and their correlation with the extent of HDAC inhibition (Fig. 1). In Fig. 1A, the growth of CMCs from two different patients was assessed at 24 h intervals for 5 days with NaBu concentrations ranging from 0 to 10 mM. Although growth kinetics were different, CMCs from both patients revealed a similar dose-dependent decrease in cell proliferation with increasing concentrations of NaBu exposure. Notably, CMCs subjected to NaBu concentrations of 5 mM or greater exhibited near complete inhibition of growth (Fig. 1A). Next, the level of HDAC inhibition within this concentration range was assessed following 72 h of NaBu exposure by immunoblotting for acetylated-lysine (AcK) or acetylated-lysine 9 of histone-H3 (AcK9-H3) (Fig. 1B). Immunoblots revealed a dose-dependent increase in the quantity of acetylated histones (H3 and H4) with increasing concentrations of NaBu, which inversely correlated with declining CMC proliferation rates.
Figure 1.
Sodium butyrate (NaBu) exerts dose-dependent effects on CMC proliferation and histone acetylation. (A): Growth curves corresponding to two independent patient-derived CMC samples exposed to increasing concentrations of NaBu. Relative cell viability was calculated at 24 h intervals over a period of 5 ds (n=3 for each). Values are mean ± SEM. (B): Representative immunoblots (n=2 for each) detecting acetylated nucleosomal histone proteins in cell lysates from CMCs treated with NaBu for 72 h; employed antibodies include anti-AcK (top panel; anti-acetylated lysine), anti-AcK9-H3 (middle panel; anti-acetylated lysine 9 of histone H3), and anti-β-actin (bottom panel).
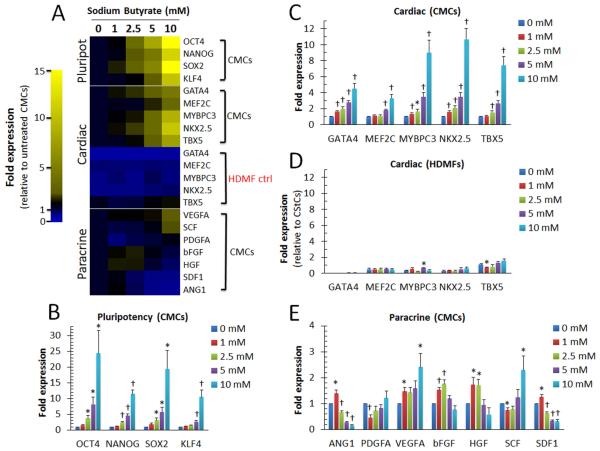
Previous studies have identified multiple genetic targets affected by HDAC inhibition in various mammalian cell types, including those associated with pluripotency [29], lineage-specific differentiation [15, 16], and cytokine production [30]. In Fig. 2A, quantitative PCR (qPCR)-based gene expression assays were used to assess the concentration-dependent effects of NaBu on CMC gene transcription. Human CMCs exposed to NaBu for 72 h exhibited a dose-dependent increase in stemness-related transcripts (Fig. 2 A and B), as well as both early (homeobox transcription factor NKX2.5, T-box transcription factor TBX5, and GATA binding protein 4 GATA4) and late (myocyte enhancer factor MEF2C and myosin-binding protein MYBPC3) cardiac lineage-specific markers (Fig. 2A and C); conversely, NaBu exposure failed to augment the expression of several investigated non-cardiac lineage markers (including lymphoid (CD8), primitive endoderm (AFP), or neuronal (TUBB3)), suggesting selectivity for cardiogenic gene transcription in CMCs (Supporting Information Fig. S2). The selective effects of NaBu on the induction of CMC cardiac-gene transcription also demonstrated cell context specificity as such observations were not recapitulated in HDMFs under the same conditions (Fig. 2A and D). NaBu exerted less pronounced effects on the expression of transcripts encoding soluble cytokines (i.e. ANG1, PDGFA, VEGFA, bFGF, HGF, SCF, and SDF1) (Fig. 2A and E) – paracrine factors implicated in stem cell-mediated cardiac repair [31]. Elevated doses of NaBu (>2.5 mM) resulted in a significant reduction in ANG1 and SDF1 in CMCs (Fig. 2A and E). While many of these factors remained largely unchanged across all concentrations (e.g. PDGFA, bFGF, and HGF), NaBu concentrations exceeding 5 mM resulted in a significant increase in pro-angiogenic VEGFA and the chemotactic ligand/pro-proliferative stem cell factor SDF1 (Fig. 2A and E).
Figure 2.
Concentration-dependent effects of NaBu exposure on CMC gene transcription. (A): Representative heat map summarizing qPCR-based gene expression assays employing gene specific primers targeting (B): stemness- (CMCs, n=6), (C-D): cardiac- (CMCs, n=8; HDMFs, n=3), and (E): paracrine- (CMCs, n=8) associated transcripts in cells treated with increasing concentrations of NaBu for 72 h. Yellow indicates an increase in fold change (>1) and blue a decrease in fold change (<1). Values are mean ± SEM. * P <0.05 (Relative to 0 mM) and † P <0.005 (Relative to 0 mM).
Sodium butyrate-treated CMCs are more oriented towards a cardiomyogenic/vasculogenic cell fate following their differentiation in vitro
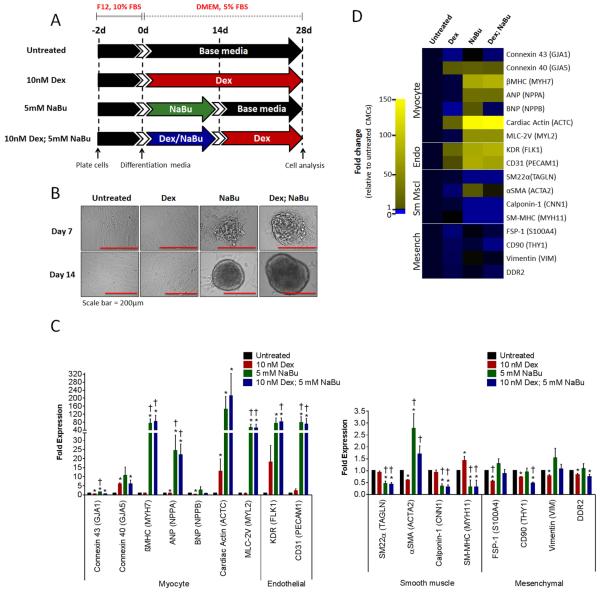
We next sought to investigate the consequences of NaBu-mediated cardiogenic program activation on CMC differentiation in vitro. Using a method routinely employed to promote the cardiomyogenic differentiation of adult cardiac progenitors [12, 25, 32, 33], CMCs were coaxed to differentiate in dexamethasone (Dex)-based medium under reduced serum conditions for 4 weeks (Fig. 3A). Relative to untreated and 10 nM Dex-treated controls, CMCs pretreated with 5 mM NaBu or in combination with dexamethasone (10 nM Dex; 5 mM NaBu) showed evidence of cell aggregation by day 7 in culture, which became more complex and prominent by day 14 (Fig. 3B). On day 28, qPCR-based gene expression assays were performed using transcript-specific primers targeting various cardiac cell lineage markers (i.e. myocyte, endothelial, smooth muscle, and mesenchymal/fibroblast) (Fig. 3C and D). qPCR results summarized in the accompanying heat map reveal a marked increase in both myocyte (GJA5, βMHC, ANP, ACTC, MLC-2V) and endothelial (KDR and CD31)-specific markers with a concomitant reduction in a number of smooth muscle (SM22α, CNN1, and SM-MHC) transcripts in CMC samples pretreated with 5 mM NaBu or combination, relative to untreated controls (Fig. 3C). Dex alone showed minimal influence on CMC cardiogenic differentiation compared with samples containing NaBu and exhibited modest increases in only a fraction of the detected myocyte (GJA5 and ACTC, P<0.05 vs. untreated) and endothelial (KDR and CD31, P>0.05 vs. untreated)-specific transcripts relative to untreated CMCs (Fig. 3D) – demonstrating Dex to be an ineffective CMC cardiomyogenic differentiation agent. The combination of NaBu and Dex did not have a synergistic effect on cardiogenic differentiation. Although NaBu alone and combination treatments yielded markedly similar results regarding the identity and magnitude of activated myocyte (GJA5, βMHC, ANP, ACTC, MLC-2V) and endothelial lineage-specific genes (KDR and CD31), NaBu alone showed a marginal advantage over combinatorial-treatments in promoting CMC cardiomyocyte gene transcription (Fig. 3D); NaBu alone demonstrated a mean increase in all evaluated myocyte associated transcripts including GJA1, GJA5, and BNP, which were either significantly decreased in combination treated CMCs (GJA1 and BNP) relative to untreated or their mean expression was similar to Dex-treated controls (GJA5) (Fig. 3D). Although NaBu alone and combination exposed CMCs showed analogous effects in terms of decreased smooth muscle gene transcription, CMCs treated with combination exhibited a reduction in CD90 and DDR2 mesenchymal markers (this was not observed in CMCs treated with NaBu alone) (Fig. 3D). In summary, NaBu exposure resulted in the activation of a cardiogenic transcriptional program in CMCs whose expression pattern suggests heightened commitment towards a myocyte and/or vasculogenic cell fate following their cardiogenic differentiation in vitro.
Figure 3.
NaBu-treated CMCs assume a cardiomyogenic/vasculogenic cell-like fate in vitro. (A): In vitro cardiogenic differentiation schematic and timeline. Untreated, dexamethasone (Dex), sodium butyrate (NaBu), or combination (Dex; NaBu) treated CMCs were cultured under reduced serum conditions for 28 ds to induce cardiogenic differentiation. (B): Representative phase contrast images of untreated, Dex, NaBu, or combination treated CMCs at 7 and 14 ds post induction of differentiation. (C): Real-time qPCR assays assessing the expression of various cardiovascular cell lineages (myocyte, endothelial, smooth muscle and mesenchymal/fibroblast) in differentiated CMCs. Values are mean (n=8) ± SEM. * P <0.05 (Relative to untreated) and † P <0.05 (Relative to Dex). (D): Heat map summarizing resultant qPCR-based gene expression assays. Yellow indicates an increase in fold change (>1) and blue a decrease in fold change (<1).
Targeted inhibition of HDAC1 enhances the expression of pluripotent, cardiac, and endothelial lineage-specific markers in human CMCs
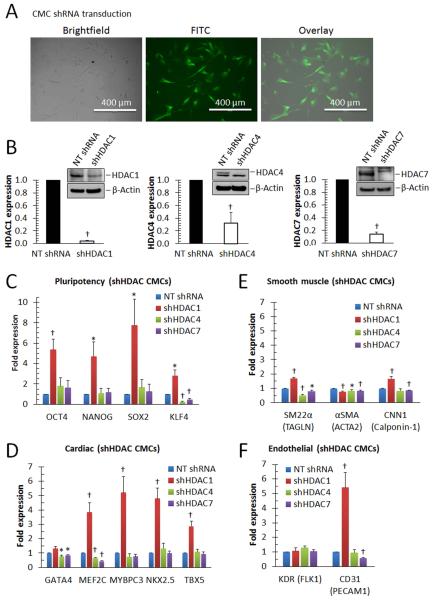
The pan-HDAC inhibitor NaBu targets both class I (HDAC1, 2, 3, and 8) and class IIa (HDAC4, 5, 7, and 9) HDAC family members [34]. Of these, HDAC1, HDAC4, and HDAC7 have been individually implicated in the induction of cardiac lineage differentiation in distinct progenitor cell populations (e.g. BM-MSCs [19], c-kit+ cardiac progenitors [17], and C2C12 myoblasts [18], respectively) following their targeted inhibition. For this reason, short hairpin RNA-interference (shRNAi) constructs, stably targeting HDAC1, 4, or 7 (Fig. 4), were employed to i) investigate the effects of these HDAC family members on the activation of CMC cardiogenic gene transcription, ii) identify what lineage-specific transcriptional networks they modulate, and iii) ascertain which of these may underlie resultant cardiogenic gene expression alterations observed with NaBu-mediated HDAC inhibition. With lentiviral transduction efficiencies exceeding 85% (CMCs infected with GFP control vector [Fig. 4A]), CMCs harboring shRNAi constructs targeting HDAC1 (shHDAC1), 4 (shHDAC4), or 7 (shHDAC7) exhibited significant reductions in cognate mRNA (4%, 32%, and 14%, respectively) and corresponding protein (<40%, <1%, and <1%, respectively) expression levels relative to non-target shRNA controls (NT shRNA) (Fig. 4B). Next, gene expression assays were performed (72 h after transduction) to assess early HDAC-knockdown-mediated transcriptional changes in stemness-related, cardiac, smooth muscle, and endothelial-specific transcripts (Fig. 4C-F). shHDAC1-transduced CMCs exhibited a discernible increase in stemness-related (OCT4, NANOG, SOX2, and KLF4) (Fig. 4C) and cardiac-specific (MEF2C, MYBPC3, NKX2.5, and TBX5) (Fig. 4C) transcripts relative to NT, shHDAC4, and shHDAC7 transduced CMCs. Such changes were accompanied by minor, yet statistically significant, increases in two of the three assessed smooth muscle markers (SM22α and CNN-1) in shHDAC1-transduced CMCs compared with NT, shHDAC4, and shHDAC7 (Fig. 4E). Further, although shHDAC1, 4, and 7 produced no changes in the expression of the endothelial marker KDR (Fig. 4F), there was a robust increase in CD31 expression in shHDAC1 relative to NT shRNA (Fig. 4F); this was not observed with either shHDAC4 or shHDAC7.
Figure 4.
CMCs depleted of HDAC1 exhibit enhanced expression of stemness-, cardiac-, and endothelial lineage-specific markers. (A): Epifluorescence microscopy images depicting efficiency of CMC lentiviral transduction (PGK-GFP control vector). (B): The efficiency of HDAC knockdown in CMCs transduced with shHDAC1, shHDAC4, shHDAC7, or non-targeting shRNA control (NT shRNA) was evaluated via real-time qPCR (bar graphs; n=7) and Western analysis (insets; n=3). Values are mean (n=7) ± SEM. † P <0.005 (Relative to NT shRNA). (C-F): The influence of HDAC knockdown on stemness-, cardiac-, smooth muscle- or endothelial-lineage specific gene transcription, respectively, was assessed via real-time qPCR 72 h after lentiviral transduction. Values are mean (n=7) ± SEM. * P <0.05 (Relative to NT shRNA) and † P <0.005 (Relative to NT shRNA).
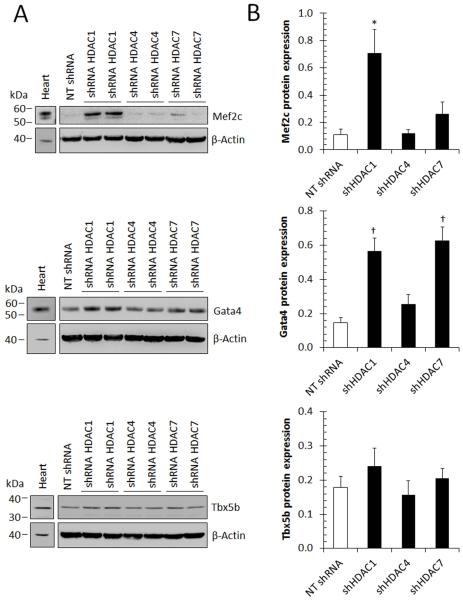
Congruent with qPCR-based gene expression assays, immunoblots (Fig. 5) confirmed enhanced expression of MEF2C, GATA4, and TBX5b (the predominant isoform expressed in more differentiated cells [35]) proteins in shHDAC1-transduced CMCs relative to NT controls. Contrary to shHDAC1, the targeted inhibition of HDAC4 or HDAC7 did not result in the induction of MEF2C or TBX5b proteins; however, a marginal increase in GATA4 protein was observed with HDAC7 knockdown compared to NT – but not to the magnitude detected in shHDAC1 CMCs (Fig. 5). Further, although HDAC1 knockdown was associated with enhanced NKX2.5 and CD31 gene transcription, neither of these proteins could be detected in total protein extracts isolated from shHDAC1 transduced CMCs or in other samples harboring shHDAC4, 7, or NT control constructs (data not shown). Taken together, these results show that depletion of HDAC1, but not HDAC4 or HDAC7, in CMCs results in the simultaneous activation of a cardiomyogenic and vasculogenic transcriptional program. This is accompanied by an early increase in the expression of cardiomyogenic transcription factor proteins (i.e. MEF2C, GATA4, and TBX5b), suggesting increase commitment towards cardiac parenchyma.
Figure 5.
HDAC1 knockdown augments cardiogenic protein expression in human CMCs. (A): Immunoblots evaluating the expression of cardiogenic-specific proteins (Mef2C; n=5, Gata4; n=5, and Tbx5b; n=4) 72 h after cell transduction with HDAC1, HDAC4, HDAC7, or non-target control (NT) shRNA constructs. (B): Densitometric quantification of accompanying Mef2c (n=5), Gata4 (n=5), or Tbx5 (n=4) immunoblots. Values represent mean protein expression (relative to β-actin) ± SEM. * P <0.05 (Relative to NT shRNA) and † P <0.005 (Relative to NT shRNA).
CMCs depleted of HDAC1 adopt a cardiomyocyte-/endothelial-cell-like fate following their differentiation in vitro
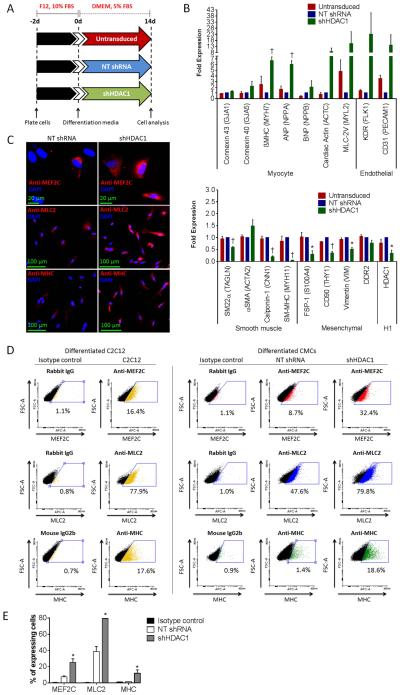
Next, the effects of HDAC1 depletion on CMC in vitro cardiogenic differentiation were examined. Untransduced, shHDAC1-transduced, or NT control CMCs were cultured in DMEM base medium under reduced serum conditions for a period of two weeks to promote cardiogenic differentiation (Fig. 6A). At the conclusion of differentiation, gene expression assays were performed using transcript-specific primers targeting various cardiovascular cell markers to assess resultant lineage commitment. As illustrated in Fig. 6B, HDAC1-depleted CMCs exhibited a pronounced increase in the mean expression of both myocyte and endothelial-specific transcripts relative to NT shRNA and untransduced controls. Those showing the greatest changes (i.e. βMHC, ANP, ACTC, MLC-2V, KDR, and CD31) exhibited expression increases that ranged from 6 to 23 fold relative to NT shRNA-transduced CMCs (Fig. 6B); the increase in MLC-2V, KDR, and CD31 expression was not statistically significant, possibly because of the variability among patient samples. In stark contrast to the heightened expression of myocyte and endothelial lineage markers, HDAC1-depleted CMCs displayed a substantial decrease in all smooth muscle and mesenchymal-specific transcripts – with the exception of αSMA, which showed a minor 0.5-fold increase in expression. In agreement with these observations, immunocytochemical (Fig. 6C) and flow cytometric (Fig. 6D-E) analyses revealed a significant increase in the total number of cells that express myogenic transcription factor Mef2c (25.3±4.5% vs. 7.5±1.2%; shHDAC1 vs. NT shRNA; p<0.05) and contractile machinery component proteins, MHC (11.9±4.3% vs. 1.6±0.1%; shHDAC1 vs. NT; p<0.05) and MLC2 (79.4±0.4% vs. 38.7±6.2%; shHDAC1 vs. NT; p<0.05), in differentiated HDAC1-depleted CMCs relative to NT controls. Despite showing greater expression of these myogenic proteins, spontaneous contractions were not observed in post differentiated HDAC1-depleted CMCs. Thus, overall, CMCs depleted of HDAC1 appear to lose expression of archetypal markers of mesenchymal and smooth muscle cell lineages and assume an expression pattern more characteristic of a cardiomyocyte-/endothelial-cell-like fate in vitro.
Figure 6.
HDAC1-depleted CMCs exhibit heightened commitment towards a myocyte- and endothelial-cell-like fate following their differentiation in vitro. (A): In vitro cardiogenic differentiation schematic and timeline. Untransduced, NT shRNA control, or shHDAC1 transduced CMCs were cultured under reduced serum conditions for 14 d to induce cardiogenic differentiation. (B): Real-time qPCR assays assessing the expression of cardiovascular cell lineage markers (myocyte, endothelial, smooth muscle and mesenchymal/fibroblast) in differentiated CMCs. Values are mean (n=3) ± SEM. * P <0.05 (Relative to NT shRNA) and † P <0.005 (Relative to NT shRNA). (C): Immunocytochemical (n=3) and (D): flow cytometric (n=3) detection of myocyte lineage-specific proteins [Mef2C (myocyte-specific enhancer factor 2C), MLC2 (cardiac myosin light chain 2), and MHC (cardiac myosin heavy chain)] in differentiated CMCs. Differentiated C2C12 myoblasts (cultured under reduced serum conditions for 14 d) served as a positive control in FACS. (E): Graph depicting flow cytometric-based quantification of the percentage of differentiated CMCs expressing Mef2C, MLC2, or MHC proteins. Values are mean (n=3) ± SEM. * P <0.05 (shHDAC1 vs. NT shRNA).
HDAC1 inhibition-mediated activation of cardiac- and endothelial-specific gene transcription is regulated by p53
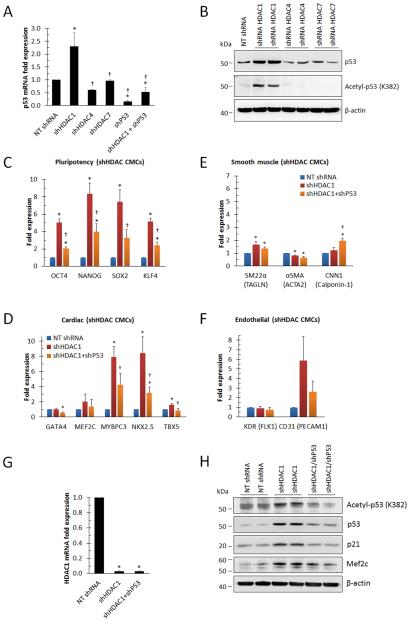
Mechanisms underlying the induction of myocyte- and endothelial-specific gene transcription in HDAC1-depleted CMCs may include modification of chromatin structure and/or changes in protein function through alterations in nucleosomal-histone or non-histone protein acetylation, respectively. In light of the fact that p53 is known to be negatively regulated via HDAC1-mediated deacetylation [36] and that p53 can promote a shift in cardiac fibroblast cell fate via direct regulation of lineage-specific differentiation programs [13], we sought to investigate the effects of HDAC1 depletion on p53 expression and whether it has a role in mediating cardiogenic gene transcription in CMCs. qPCR was performed to evaluate the influence of HDAC depletion on p53 gene transcription (Fig. 7A). Compared with NT shRNA controls, HDAC7-depleted CMCs showed no change in P53 expression whereas those depleted of HDAC4 exhibited a decrease (≈40%). In contrast to HDAC4 and HDAC7, HDAC1-depleted CMCs displayed a statistically significant increase in P53 mRNA (≈130%) relative to NT shRNA controls (Fig. 7A). These results were confirmed in p53 immunoblots containing total protein lysates isolated from NT, HDAC1, HDAC4, or HDAC7-transduced CMCs (Fig. 7B). Here, HDAC1 depleted CMCs showed a robust increase in p53 protein levels compared with HDAC4 or HDAC7-transduced cells which showed little to no change relative to NT controls.
Figure 7.
p53 regulates cardiogenic gene activation in HDAC1-depleted CMCs. (A): Real-time qPCR evaluating the expression of p53 in CMCs 72 h after transduction with NT control, HDAC1, HDAC4, HDAC7, p53, or HDAC1 and p53 shRNA constructs. Values are mean (n=4 patient samples) ± SEM. * P <0.05 (Relative to NT shRNA) and † P <0.05 (Relative to shHDAC1). (B): Representative immunoblots detecting p53 and acetylated p53 (Acetyl-p53; acetylated lysine 382) in total protein extracts derived from CMCs 72 h after infection with NT control, HDAC1, HDAC4, or HDAC7 shRNA lentiviral vectors. (C-F): Real-time qPCR assessing the expression of stemness-, cardiac-, smooth muscle- or endothelial-lineage specific transcripts, respectively, in CMCs transduced with NT control, HDAC1, or p53 and HDAC1 shRNA constructs. Values are mean (n=4) ± SEM. * P <0.05 (Relative to NT-shRNA) and † P <0.05 (Relative to shHDAC1). (G): Real-time qPCR quantifying the expression of HDAC1 in CMCs transduced with NT control, HDAC1, or p53 and HDAC1 shRNA constructs. Values are mean (n=4) ± SEM. * P <0.05 (Relative to NT shRNA). (H): Representative immunoblots detecting p53 (n=3), p21 (n=2), acetylated p53 (n=3), and Mef2C (n=3) in protein extracts isolated from NT control, HDAC1, or HDAC1 and p53 shRNA transduced CMCs.
Previous studies have shown ectopically expressed dominant negative HDAC1 isoforms to promote p53 acetylation (resulting in enhanced p53 stabilization/expression in murine fibroblasts) [36] and HDAC inhibitor-induced p53 acetylation at K373/K382 to extend p53 half-life (by blocking p53 ubiquitination), enhance p53 recruitment to promoter regions, and increase the transcriptional activity of downstream p53-responsive genes (i.e. p21) [37]. Thus, Western analysis was performed to evaluate whether HDAC1 depletion effectively induced p53 acetylation in CMCs (Fig. 7B). Immunoblots employing antibodies raised against acetylated lysine 382 of p53 (Acetyl-p53 [K382]) revealed a pronounced increased in the total amount of acetylated p53 in protein lysates isolated from HDAC1 depleted CMCs , whereas the depletion of either HDAC4 or HDAC7 in CMCs had no effect on p53 protein acetylation (Fig. 7B). Thus, the targeted depletion of HDAC1, but not HDAC4 or HDAC7, resulted in increased p53 protein acetylation and enhanced p53 expression.
Next, we sought to investigate whether p53 functions to regulate cardiogenic gene transcription in HDAC1-depleted CMCs. If p53 is responsible for the activation of the cardiogenic transcriptional program observed in HDAC1-depleted CMCs, then inhibiting p53 expression should effectively mitigate this induction. To this end, the gene expression patterns of CMCs infected with either shHDAC1 alone or co-infected with shHDAC1 and shP53 were compared (Fig. 7C-F). Of note, the shP53 construct utilized here was shown to efficiently reduce P53 message levels by approximately 84% in shP53 alone-transduced CMCs, whereas co-transduction of CMCs with shP53 and shHDAC1 sufficiently reduced the enhanced expression of P53 message in response to HDAC1 depletion to a level equal to or below that of NT controls (Fig. 7A). qPCR-based gene expression assays showed that the induction of stemness- (OCT4, NANOG, SOX2, and KLF4), myocyte- (MEF2C, MYBPC3, NKX2.5, and TBX5), and endothelial (CD31)-related transcripts in HDAC1-depleted CMCs were attenuated in CMCs co-infected with shHDAC1 and shP53 (Fig. 7C, D, and F, respectively) relative shHDAC1 alone. Although smooth muscle markers were again shown to be largely unaffected by HDAC1 inhibition, CNN1 was enhanced in shHDAC1/shP53 transduced CMCs, suggesting that P53 may negatively regulate its expression (Fig. 7E). The ability of shP53 to reduce the activation of stemness- , myocyte-, and endothelial-associated transcripts in HDAC1-depleted CMCs was not associated with transcriptional de-repression of HDAC1, as both shHDAC1 and shHDAC1/shP53-transduced CMCs exhibited similar levels of HDAC1 expression (Fig. 7G). qPCR assay results implicating p53 as a regulator of cardiogenic factor expression in HDAC1-depleted CMCs were further corroborated in complementary immunoblots comparing shHDAC1 and shHDAC1/shP53 transduced CMCs (Fig. 7H). Here the elevated levels of acetylated p53, total p53, p21, and myogenic transcription factor Mef2c in HDAC1-depleted CMCs were effectively reduced via the targeted inhibition of p53. Taken together, these results suggest that HDAC1 inhibition activates a cardiogenic transcriptional program and that this program is, at least in part, regulated by the tumor suppressor p53.
Discussion
HDAC inhibitors have distinct effects on the differentiation of tissue-specific adult progenitors towards mature parenchymal cell types that correspond to their tissue of origin. For example, HDAC inhibitors have been shown to promote the neuronal differentiation of adult neural progenitors [38], ductal differentiation of pancreatic cells [39], and cardiomyocytic differentiation of cardiac side populations cells [15]. Such findings have significant therapeutic implications for the use of HDAC inhibitors in tissue regeneration. Deciphering the precise mechanisms of action of HDAC inhibitors and understanding the role of specific HDAC isoforms in regulating cell type-specific gene expression programs, differentiation dynamics, and cell lineage specification is important to enable their exploitation for tissue repair. In the current study we sought to investigate the impact of HDAC inhibition on CMC cardiogenic gene regulation and lineage-specific differentiation in vitro using both pharmacologic and gene targeting approaches.
Our results demonstrate that NaBu produces distinct effects on CMC cardiogenic gene transcription compared with their dermal-derived equivalents, HDMFs; specifically, NaBu resulted in a dose-dependent induction of early (NKX2.5, GATA4, and TBX5) and late (MEF2C) cardiac lineage-specific transcription factors in patient-derived CMCs, which may be interpreted as initiation of cardiogenic differentiation or at least priming for such a process. Consistent with the early upregulation of these cardiogenic factors, a comprehensive analysis of lineage-specific gene transcription, corresponding to various cardiovascular cell lineages, revealed that CMCs pretreated with NaBu displayed expression profiles reminiscent of cardiomyocytes and endothelium when stimulated to differentiate in vitro. This enhanced expression of both myocytic (connexin43, βMHC, ANP, cardiac actin, and MLC-2V) and endothelial markers (KDR and CD31) was accompanied by a concomitant reduction in a number of smooth muscle (i.e. SM22α, calponin-1, and SM-MHC) and mesenchymal (i.e. CD90 and DDR2) transcripts. The effects elicited by NaBu exposure on CMC cardiogenic gene transcription and lineage-specific differentiation were closely recapitulated with shRNA-mediated, isoform-selective depletion of HDAC1, but not HDAC4 or HDAC7 (both of which have been previously implicated in the induction of myogenic differentiation of other progenitor cell types [17, 18]). Specifically, we found that shRNA-mediated HDAC1 depletion also induced the early upregulation of cardiogenic (MEF2C, MYBPC3, NKX2.5, and TBX5) and endothelial (CD31) associated markers. These early cardiogenic expression alterations were associated with elevated transcription of myocytic- and endothelial-lineage specific transcripts, and decreased expression of smooth muscle- (e.g. SM22α, calponin-1, and SM-MHC) and mesenchymal-associated markers (e.g. FSP1, CD90, and vimentin), when coaxed to differentiate in vitro. Taken together, our results indicate HDAC1 inhibition promotes a shift towards a developmental gene expression program that favors myocyte and endothelial commitment at the expense of the smooth muscle and mesenchymal cell fate in human CMCs – revealing a hitherto unknown role of HDAC1 in influencing CMC cell fate decisions.
Investigation into the mechanisms underlying cardiogenic gene activation in HDAC1-depleted CMCs revealed enhanced p53 expression as a contributor to this induction. The p53 transcription factor has been identified as a direct target of HDAC1 activity; the acetylation status of this protein is known to have important consequences on its stability, expression, and function [36, 37]. In illustrating this notion, Ito and colleagues demonstrated the overexpression of a dominant-negative HDAC1 mutant isoform in murine fibroblasts to enhance DNA damage-induced p53 acetylation [36]. This modification prevented MDM2-dependent ubiquitination and resulted in enhanced p53 stability, elevated p53 expression, and more robust induction of its respective downstream transcriptional targets (i.e. p21 and MDM2) [36]. Whether p53 protein acetylation is required to enhance its transcriptional activity has been a long standing debate, however, Zhao and colleagues demonstrated HDAC inhibition-mediated acetylation of p53 at K373/382 to enhance its binding to the promoter region of p21 and, in a related fashion, to increase p21 expression [37] – suggesting that acetylation of p53 influences both its stability and transcriptional activity. Whether p53 acetylation alone is required for the induction of HDAC1 inhibition-induced cardiac gene transcription in CMCs is unknown. Consistent with such observations, HDAC1-depleted CMCs exhibited enhanced p53 protein acetylation at K382 and heightened expression levels of itself and p53-responsive genes (p21). This phenomenon was restricted to HDAC1 depletion, as HDAC4 or HDAC7 knockdown had no effect on either p53 acetylation or expression. Furthermore, we were able to demonstrate that shRNA-mediated knockdown of p53 mitigated the induction of stemness (OCT4, NANOG, SOX2, and KLF4)-, cardiac- (MEF2C, MYBPC3, NKX2.5, and TBX5), and endothelial (CD31)-associated transcripts in CMCs depleted of HDAC1. These data implicate enhanced p53 acetylation and elevated p53 protein expression levels as a potential mechanism facilitating cardiogenic gene activation in HDAC1-depleted CMCs. Thus, the results of our study provide a direct biological link between p53 signaling and lineage-specific transcriptional program activation in human CMCs.
While p53 has a well characterized role in the modulation of cell-cycle arrest, apoptosis, and DNA repair, a number of studies have identified additional functions for this protein in the regulation of cellular differentiation programs and developmental pathways [40]. Early studies have shown that p53 expression and its transcriptional activity are important for myogenic differentiation of embryonic stem cells (via regulating the expression of mesodermal master genes Brachyury and Mesp1) [41] and skeletal muscle precursors [42-44]. Our finding that HDAC1 inhibition stimulates the induction of a cardiogenic developmental program in CMCs through a p53-dependent mechanism is consistent with these prior observations. While the function of p53 in directing CMC cell fate decisions has not been well characterized, a recent study by Ubil and colleagues revealed that p53 signaling constitutes an endogenous mechanism by which native CMCs can undergo mesenchymal-endothelial transition and contribute to cardiac neovascularization in the infarcted murine heart [13], an effect that was attributed to the ability of p53 to directly bind and activate the expression of endothelial-specific genes and transcription factors which coordinate endothelial differentiation [13]. Such data corroborate the findings of this study in which the artificial induction of p53 in CMCs by means of HDAC1 depletion was demonstrated to be associated with heightened expression of endothelial markers in vitro. It should be emphasized, however, that in addition to the upregulation of endothelial markers, we also observed a simultaneous activation of myocyte-specific transcripts, suggesting that p53 may also play a role in CMC cardiomyocyte commitment – a phenomenon that was not directly assessed in the aforementioned lineage tracing studies that followed the fate of endogenous cardiac fibroblasts in infarcted murine hearts [13]. Although endogenous CMCs have not been shown to directly contribute to the formation of new cardiomyocytes in vivo, Rossini and colleagues have recently shown that transplanted human CMCs can give rise to both cardiomyocytes and endothelium in a rat infarct model, which support cardiac repair [12] and suggests a degree of intrinsic cellular plasticity. In view of the inherent aptitude of CMCs for cardiomyocyte and endothelial differentiation, and given the role of p53 in driving myogenic and endothelial differentiation programs, our study suggests that HDAC1 inhibition-mediated activation of p53 in CMCs results in increased commitment towards such cardiovascular parenchymal cell types. While our study highlights p53 as a premier target responsible for cardiogenic gene activation in CMCs depleted of HDAC1, the existence of a panoply of other known and unidentified substrates (both histones and non-histone proteins) that are likely subject to histone deacetylase activity makes it arduous to decipher the precise mechanisms responsible for this effect.
Summary
Our study demonstrates that human CMCs depleted of HDAC1 adopt a cardiomyocyte-/endothelial-cell-like fate following their differentiation in vitro, and that cardiac- and endothelial-specific gene transcription induced by HDAC1 inhibition is regulated, in part, by the tumor suppressor p53. Taken together, our results implicate HDAC1 as a potential modulator of CMC cell fate decisions. Our findings have significant implications for the use of HDAC inhibitors in cardiac regenerative therapies. Specifically, this work suggests that isoform-selective inhibitors of HDAC1 may be a useful strategy to promote the transdifferentiation of endogenous or exogenously sourced CMCs and to enhance the effects of CMC-based cell therapy on cardiac function in the setting of heart failure.
Supplementary Material
Acknowledgments
This work was supported by NIH grants P01 HL-78825 and 1 UM1 HL-113530 (CCTRN). Additional funding was provided by the University of Louisville’s School of Medicine Basic Grant Program (awarded to JBM).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
Author contributions:
Joseph Moore: Conception and design, financial support, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
John Zhao: Conception and design, collection and assembly of data, data analysis and interpretation
Matthew Keith: Collection and assembly of data, data analysis, final approval of manuscript
Alok Amraotkar: Provision of study materials, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Marcin Wysoczynski: Concept and design, provision of study materials, data analysis and interpretation, final approval of manuscript
Kyung Hong: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval
Roberto Bolli: Conception and design, financial support, provision of study materials, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Bolli R, Chugh AR, D'Amario D, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Chugh AR, Beache GM, Loughran JH, et al. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation. 2012;126:S54–64. doi: 10.1161/CIRCULATIONAHA.112.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrtovec B, Poglajen G, Lezaic L, et al. Comparison of Transendocardial and Intracoronary CD34(+) Cell Transplantation in Patients With Nonischemic Dilated Cardiomyopathy. Circulation. 2013;128:S42–S49. doi: 10.1161/CIRCULATIONAHA.112.000230. [DOI] [PubMed] [Google Scholar]
- 4.Vrtovec B, Poglajen G, Lezaic L, et al. Effects of Intracoronary CD34(+) Stem Cell Transplantation in Nonischemic Dilated Cardiomyopathy Patients 5-Year Follow-Up. Circulation Research. 2013;112:165–173. doi: 10.1161/CIRCRESAHA.112.276519. [DOI] [PubMed] [Google Scholar]
- 5.Sanganalmath SK, Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res. 2013;113:810–834. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong KU, Li QH, Guo YR, et al. A highly sensitive and accurate method to quantify absolute numbers of c-kit plus cardiac stem cells following transplantation in mice. Basic Research in Cardiology. 2013;108 doi: 10.1007/s00395-013-0346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith MCL, Bolli R. "String Theory" of c-kit(pos) Cardiac Cells A New Paradigm Regarding the Nature of These Cells That May Reconcile Apparently Discrepant Results. Circulation Research. 2015;116:1216–1230. doi: 10.1161/CIRCRESAHA.116.305557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon CH, Koyanagi M, Iekushi K, et al. Mechanism of improved cardiac function after bone marrow mononuclear cell therapy: role of cardiovascular lineage commitment. Circulation. 2010;121:2001–2011. doi: 10.1161/CIRCULATIONAHA.109.909291. [DOI] [PubMed] [Google Scholar]
- 9.Orlic D, Kajstura J, Chimenti S, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 10.Behfar A, Yamada S, Crespo-Diaz R, et al. Guided cardiopoiesis enhances therapeutic benefit of bone marrow human mesenchymal stem cells in chronic myocardial infarction. J Am Coll Cardiol. 2010;56:721–734. doi: 10.1016/j.jacc.2010.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnecchi M, Zhang ZP, Ni AG, et al. Paracrine Mechanisms in Adult Stem Cell Signaling and Therapy. Circulation Research. 2008;103:1204–1219. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossini A, Frati C, Lagrasta C, et al. Human cardiac and bone marrow stromal cells exhibit distinctive properties related to their origin. Cardiovascular Research. 2011;89:650–660. doi: 10.1093/cvr/cvq290. [DOI] [PubMed] [Google Scholar]
- 13.Ubil E, Duan J, Pillai IC, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514:585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leu YW, Huang TH, Hsiao SH. Epigenetic reprogramming of mesenchymal stem cells. Adv Exp Med Biol. 2013;754:195–211. doi: 10.1007/978-1-4419-9967-2_10. [DOI] [PubMed] [Google Scholar]
- 15.Oyama T, Nagai T, Wada H, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. Journal of Cell Biology. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang G, Tian J, Feng C, et al. Trichostatin A Promotes Cardiomyocyte Differentiation of Rat Mesenchymal Stem Cells After 5-Azacytidine Induction or During Coculture With Neonatal Cardiomyocytes Via a Mechanism Independent of Histone Deacetylase Inhibition. Cell Transplantation. 2012;21:985–996. doi: 10.3727/096368911X593145. [DOI] [PubMed] [Google Scholar]
- 17.Zhang LX, DeNicola M, Qin X, et al. Specific inhibition of HDAC4 in cardiac progenitor cells enhances myocardial repairs. American Journal of Physiology-Cell Physiology. 2014;307 doi: 10.1152/ajpcell.00187.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao CZ, Liu Y, Lam M, et al. Histone deacetylase 7 (HDAC7) regulates myocyte migration and differentiation. Biochimica Et Biophysica Acta-Molecular Cell Research. 2010;1803:1186–1197. doi: 10.1016/j.bbamcr.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu DF, Yao Y, Su ZZ, et al. Downregulation of HDAC1 Is Involved in the Cardiomyocyte Differentiation from Mesenchymal Stem Cells in a Myocardial Microenvironment. Plos One. 2014;9 doi: 10.1371/journal.pone.0093222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu WS, Parmigiani RB. Marks PA Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 21.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kretsovali A, Hadjimichael C, Charmpilas N. Histone Deacetylase Inhibitors in Cell Pluripotency, Differentiation, and Reprogramming. Stem Cells International 2012. doi: 10.1155/2012/184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney M, Gratama JW, Chin-Yee IH, et al. Isotype controls in the analysis of lymphocytes and CD34+ stem and progenitor cells by flow cytometry - Time to let go! Cytometry. 1998;34:280–283. [PubMed] [Google Scholar]
- 24.O'Gorman MRG, Thomas J. Isotype controls - time to let go? Cytometry. 1999;38:78–80. [PubMed] [Google Scholar]
- 25.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 26.Montesinos JJ, Flores-Figueroa E, Castillo-Medina S, et al. Human mesenchymal stromal cells from adult and neonatal sources: comparative analysis of their morphology, immunophenotype, differentiation patterns and neural protein expression. Cytotherapy. 2009;11:163–176. doi: 10.1080/14653240802582075. [DOI] [PubMed] [Google Scholar]
- 27.Kruh J. Effects of Sodium-Butyrate, a New Pharmacological Agent, on Cells in Culture. Molecular and Cellular Biochemistry. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 28.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruau D, Ensenat-Waser R, Dinger TC, et al. Pluripotency associated genes are reactivated by chromatin-modifying agents in neurosphere cells. Stem Cells. 2008;26:920–926. doi: 10.1634/stemcells.2007-0649. [DOI] [PubMed] [Google Scholar]
- 30.Yamanegi K, Kawabe M, Futani H, et al. Sodium valproate, a histone deacetylase inhibitor, modulates the vascular endothelial growth inhibitor-mediated cell death in human osteosarcoma and vascular endothelial cells. Int J Oncol. 2015;46:1994–2002. doi: 10.3892/ijo.2015.2924. [DOI] [PubMed] [Google Scholar]
- 31.Duran JM, Makarewich CA, Sharp TE, et al. Bone-Derived Stem Cells Repair the Heart After Myocardial Infarction Through Transdifferentiation and Paracrine Signaling Mechanisms. Circulation Research. 2013;113:539–552. doi: 10.1161/CIRCRESAHA.113.301202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Amario D, Fiorini C, Campbell PM, et al. Functionally competent cardiac stem cells can be isolated from endomyocardial biopsies of patients with advanced cardiomyopathies. Circ Res. 2011;108:857–861. doi: 10.1161/CIRCRESAHA.111.241380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu ED, Dul E, Sung CM, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. Journal of Pharmacology and Experimental Therapeutics. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 35.Georges R, Nemer G, Morin M, et al. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Mol Cell Biol. 2008;28:4052–4067. doi: 10.1128/MCB.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito A, Kawaguchi Y, Lai CH, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Lu S, Wu L, et al. Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1) Mol Cell Biol. 2006;26:2782–2790. doi: 10.1128/MCB.26.7.2782-2790.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh J, Nakashima K, Kuwabara T, et al. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28:6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molchadsky A, Rivlin N, Brosh R, et al. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31:1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 41.Hadjal Y, Hadadeh O, Yazidi CE, et al. A p38MAPK-p53 cascade regulates mesodermal differentiation and neurogenesis of embryonic stem cells. Cell Death Dis. 2013;4:e737. doi: 10.1038/cddis.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soddu S, Blandino G, Scardigli R, et al. Interference with p53 protein inhibits hematopoietic and muscle differentiation. Journal of Cell Biology. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazzaro G, Bossi G, Coen S, et al. The role of wild-type p53 in the differentiation of primary hemopoietic and muscle cells. Oncogene. 1999;18:5831–5835. doi: 10.1038/sj.onc.1202962. [DOI] [PubMed] [Google Scholar]
- 44.Halevy O. P53 Gene Is up-Regulated during Skeletal-Muscle Cell-Differentiation. Biochemical and Biophysical Research Communications. 1993;192:714–719. doi: 10.1006/bbrc.1993.1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.