Summary
The Kruppel-like transcription factor zinc finger protein 217 (ZNF217) (mouse homolog ZFP217) contributes to tumorigenesis by dysregulating gene expression programs. The newly discovered molecular function of ZFP217 in controlling N6-methyladenosine (m6A) deposition in embryonic stem cells (ESCs) sheds new light on the role of this transcription factor in tumor development.
Keywords: ZNF217, cancer, stem cell, N6-methyladenosine, RNA methylation
The zinc finger protein 217 is a versatile pro-oncogenic factor
ZNF217 belongs to the Kruppel-like family of transcription factors and contains eight C2H2 zinc finger motifs and a proline-rich transactivation domain. The ZNF217 gene is located at the 20q13 chromosomal region that is commonly amplified in various human cancers, and the expression of ZNF217 is strongly associated with poor clinical prognosis [1, 2]. Mammary epithelial tumor cells ectopically expressing ZFP217 and transplanted into mammary fad pads of immune compromised mice resulted in increased tumor burden [3]. These observations suggested ZNF217/ZFP217 to be pro-oncogenic. Importantly to the understanding of ZNF217/ZFP217-mediated pathogenesis, ZNF217/Zfp217 expression levels are not only associated with gene amplification but also influenced by promoter methylation and miRNA-mediated targeting [1].
ZNF217/ZFP217 controls a variety of networks and intracellular pathways by orchestrating several mechanisms of action. First, ZNF217/ZFP217 has been suggested to regulate oncogenic gene expression by acting as a transcriptional repressor of tumor suppressor genes [2]. Second, ZNF217/ZFP217 may also promote cancer progression and pluripotency by activating the expression of pro-oncogenic genes and core stem cell transcripts, respectively [3, 4]. Third, ZFP217 was found to restrict m6A deposition at pluripotency RNAs, protecting such transcripts from rapid degradation [5]. This mechanism of action, together with ZFP217-mediated transcriptional activation of the stem cell signature gene expression, would maintain embryonic stem cell (ESC) self-renewal and somatic cell reprogramming [5]. Given that ESCs share several hallmarks with cancer cells, it is reasonable to propose that altered m6A homeostasis is associated with tumor development [6, 7]. In this Forum article, we will focus on ZNF217/ZFP217-associated pathogenesis and discuss the novel role of ZFP217 in m6A deposition.
ZNF217 as a Transcriptional Repressor
ZNF217 binds DNA in a sequence-specific manner through the sixth and seventh zinc fingers [1]. Furthermore, ZNF217 cooperates in transcriptional silencing programs by recruiting chromatin modifiers such as Jarid1b/Plu-1, G9a, and EZH2, all of which are histone methyltransferases that transfer methyl groups to specific lysine residues in histones to mediate gene silencing (Figure 1A) [8]. Other repressor proteins associated with ZNF217 include the C-terminal binding protein 1/2 (CtBP1/2), histone deacetylase 2 (HDAC2), lysine demethylase 1 (LSD1), and the corepressor of REST (CoREST) [9]. ZNF217 functionally represses gene expression by direct and indirect mechanisms. For instance, E-Cadherin is silenced by direct binding of ZNF217 at the proximal promoter and by recruitment of the CtBP co-repressor complex, resulting in stimulation of cancer cell migration, invasion and anchorage-independent growth.
Figure 1.
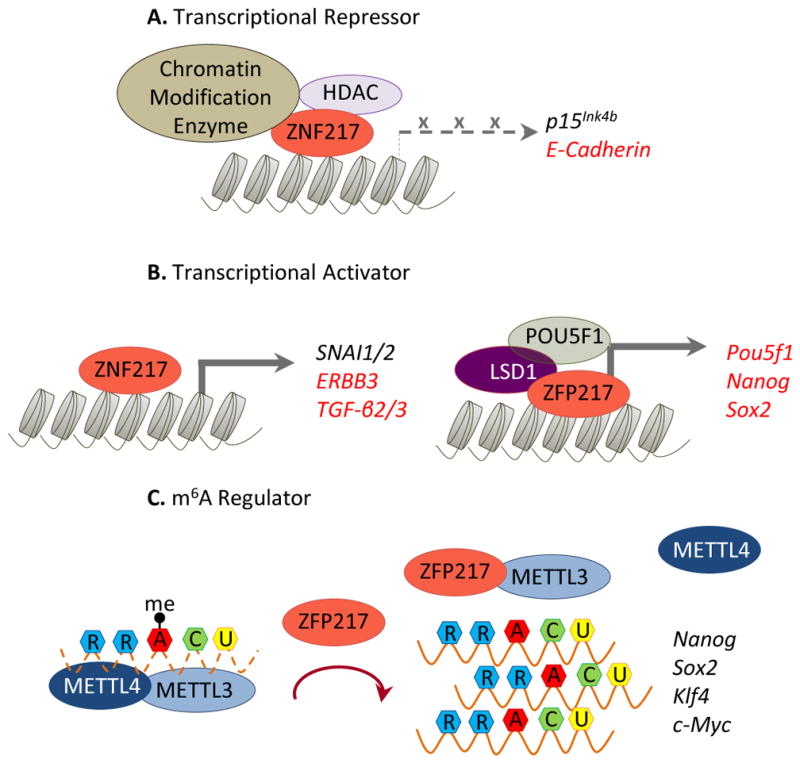
The multifaceted control imposed by ZNF217/ZFP217. ZNF217/ZFP217 regulates a variety of molecular programs by direct binding at the promoter of target genes (red) or by indirect regulation (black), either acting as a transcriptional activator (A) or repressor (B), and by controlling m6A deposition at RNA (C). RRACU indicates the m6A consensus motif.
An alternative mechanism of gene repression involves impairment of active demethylation of the p15Ink4b tumor suppressor gene by the ZNF217/CoREST complex. The anti-proliferative effects of TGF-β at early stages of tumorigenesis are in part mediated by SMAD2/3 and thymine DNA-glycosylase (TDG), which demethylate the p15Ink4b gene prior to its activation. ZNF217/CoREST prevented the recruitment of SMAD2/3 and TDG, and recruited the de novo methyltransferase DNA (cytosine-5)-methyltransferase 3 alpha (DNMT3A) at the p15Ink4b promoter [10], promoting its methylation and thus silencing p15Ink4b expression.
ZNF217 as a Transcriptional Activator
Although ZNF217 was first described as a transcriptional repressor [2], it has been shown that ZNF217 exerts many biological functions through activation of specific gene expression programs [1, 4] (Figure 1B), thus acting as a complex double-faceted transcriptional regulator. In ESCs, murine ZFP217 directly binds to promoter and enhancer regions of the core pluripotency factors Pou5f1, Nanog, and Sox2, and activates their expression to maintain the ESC-identity signature [5]. In mammary stem cells (CD24MedCD49fHigh), where the expression of ZFP217 is enriched [3], ZFP217 prompts cancer stem cell (CSC) function through upregulation of SNAI1 and SNAI2, which modulate epithelial-mesenchymal transition (EMT) and metastasis. ZNF217 also promotes EMT in human mammary epithelial cells, through direct transcriptional activation of TGF-β2 and/or TGF-β3, resulting in the activation of the TGF-β-mediated SMAD signaling pathway [1]. In breast cancer, ZNF217 directly upregulates ERBB3 expression facilitating the formation of the ERBB2/ERBB3 heterodimer [4], which results in the activation of the oncogenic mitogen-activated protein kinases (MAPK) and phosphoinositide 3-kinase (PI3K)/AKT, promoting tumor survival, proliferation and invasion [1].
ZFP217 regulates N6-Methyladenosine mRNA deposition in mouse ESC
N6-methyladenosine (m6A) is the most abundant post-transcriptional modification in RNA [11]. The core mammalian m6A methyltransferase complex includes the methyltransferase-like 3 (METTL3, also known as MT-A70) and the methyltransferase-like 14 (METTL14). The complex associates with Wilm’s tumor 1 associating protein (WTAP), which is required for catalytic activity of the m6A methyltransferase in vivo [11]. Enrichment of m6A in RNA effectively influences all aspects of RNA metabolism, including mRNA stability, alternative splicing, mRNA translation efficiency, 5′ untranslated region cap-independent translation, RNA-protein interactions, and microRNA processing, resulting in alterations in a cascade of cellular processes [11].
ZFP217 also has crucial regulatory functions in m6A deposition, adding an additional layer of complexity to ZFP217 functions [5] (Figure 1C). ZFP217 interacts with METTL3, hindering METTL3 binding to RNAs. Moreover, METTL4, which is required for METTL3 methyltransferase activity, does not interact with ZFP217, strongly suggesting that METTL3-ZFP217 is held in an inactive complex. In pluripotent ESCs, the high level of ZFP217 strongly suppresses METTL3 methyltransferase activity, preventing core ESC transcripts from aberrant methylation. During cell differentiation, expression of ZFP217 and its target genes rapidly decreases, and METTL3 is released and able to catalyze m6A methylation at the remaining pluripotency transcripts, triggering ESC differentiation.
Concluding remarks
An increasing body of research indicates that ZNF217 modulates both physiological and pathological cellular functions through coordination of complex distinct biological activities. ZNF217 cooperates with several integrated circuits governing hallmark capabilities within ESCs and cancer cells, promoting the expansion of a pool of progenitor-like cells [3]. ZNF217-mediated molecular functions involve ZNF217 direct binding at the promoter of target genes, recruitment of chromatin modifiers, and impairment of DNA methylation. Recently, ZNF217 has been identified as a negative regulator of m6A RNA methylation in ESCs [5]. This novel ZFP217-mediated mechanism may potentially function in tumor initiation and progression. Interestingly, the m6A modification has also been detected in prokaryotic and unicellular eukaryotic DNA, which is usually referred to as 6mA. In C. elegans 6mA increases trans-generationally in worms mutant for spr-5 [12], an ortholog of the mammalian LSD1. Given that LSD1 is intimately associated with ZNF217 [8], and the involvement of ZNF217 in transcriptional and post-transcriptional regulation, epigenetic mechanisms may regulate m6A modification of target transcripts not just in development, but also in diverse pathological processes, including cancer. Elucidating the role of ZNF217 in coordinating epigenetic with epitranscriptomic networks could predict cancer risk, achieve early diagnosis, track the prognosis of tumor fate, and ultimately provide valuable targets for novel therapeutic approaches.
Acknowledgments
We sincerely apologize to authors whose work could not be included due to space limitations. M.J.W. is supported by the Senior Scholar Award in Aging AG-SS-2482-10 and NIH grant CA154903. D.-F.L. is the CPRIT scholar in Cancer Research and supported by NIH Pathway to Independence Award R00 CA181496 and CPRIT grant RR160019. We thank Yifei Sun for critically reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cohen PA, et al. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015;6(39):41566–81. doi: 10.18632/oncotarget.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinlan KG, et al. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta. 2007;1775(2):333–40. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Littlepage LE, et al. The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2012;2(7):638–51. doi: 10.1158/2159-8290.CD-12-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krig SR, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282(13):9703–12. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilo F, et al. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell. 2015;17(6):689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62(3):335–45. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banck MS, et al. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009;4(2):100–6. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thillainadesan G, et al. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28(19):6066–77. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thillainadesan G, et al. TGF-beta-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell. 2012;46(5):636–49. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–26. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161(4):868–78. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


