Abstract
RNA molecules such as long non-coding RNAs play critical roles in regulating gene expression, chromosome architecture, and the modification states of chromatin. Recent developments suggest that RNA may also influence gene expression and chromatin patterns through the interaction of nascent transcripts with their DNA template via the formation of co-transcriptional R-loop structures. R-loop formation over specific, conserved, hotspots occurs at thousands of genes in mammalian genomes and represents an important and dynamic feature of mammalian chromatin. Here, focusing primarily on mammalian systems, I describe the accumulating connections and possible mechanisms linking R-loop formation and chromatin patterning. The possible contribution of aberrant R-loops to pathological conditions is also discussed.
Keywords: R-loop, transcription, chromatin, RNA:DNA hybrids, genomics, gene regulation
Basic determinants of co-transcriptional R-loop formation
During transcription, one strand of the DNA double helix is copied into a complementary RNA transcript. RNA synthesis occurs within the context of a moving transcription bubble where the two strands of DNA are physically separated and the nascent transcript is held through a transient 8 bp RNA:DNA hybrid. The transcription elongation complex architecture ensures that the outgoing nascent RNA does not become entangled with the DNA helix as the RNA polymerase (RNAP) moves forward [1] (Figure 1). For this reason, R-loop structures (See Glossary) that can form upon reannealing of the nascent RNA to the template DNA strand have long been considered rare and accidental byproducts of transcription.
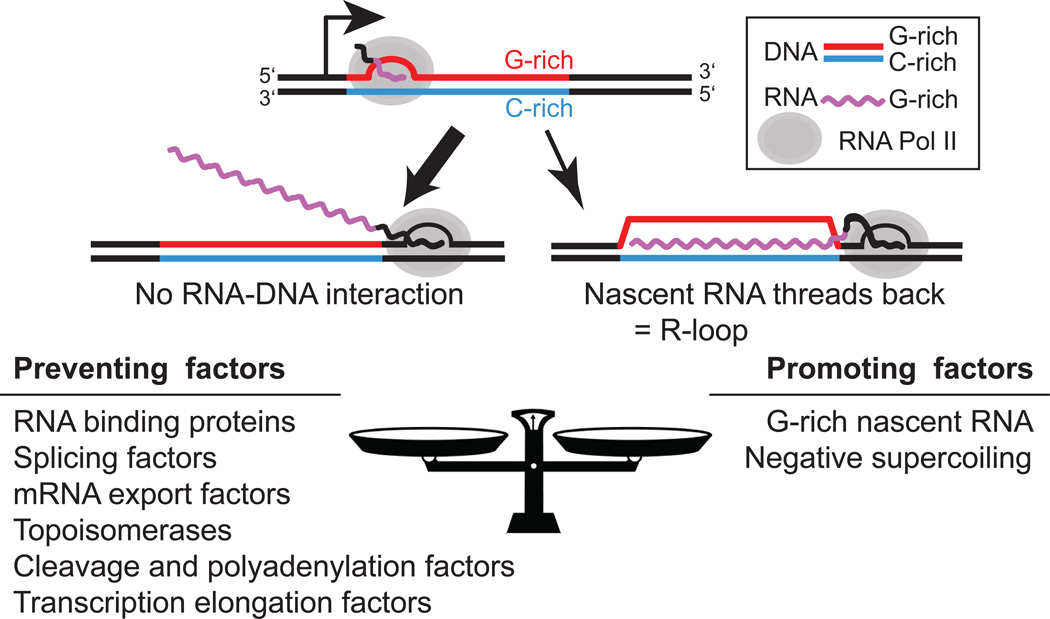
Figure 1.
(Top) schematic description of co-transcriptional R-loop formation. In most cases, transcription proceeds without R-loop formation (left; thick arrow). In some cases, however, in particular over DNA regions characterized by GC strand asymmetries, the nascent RNA can reanneal and lead to R-loop formation. (Bottom) A brief list of factors know to prevent or facilitate R-loop formation.
In vitro transcription studies of mammalian switch regions first revealed that R-loop formation could be highly efficient [2, 3]. Additional studies demonstrated that the efficiency of R-loop formation related directly to the sequence of the transcribed region. Repetitive switch regions characterized by strong G clustering in close proximity to the transcription start site (TSS) efficiently form R-loops in vitro when transcription results in a G-rich RNA [4, 5]. Similarly, in vitro transcription through unique, non-repetitive, human and mouse CpG island (CGI) promoter sequences leads to efficient R-loop formation when the transcribed regions are GCrich, possess strong GC skew, and transcription results in a G-rich nascent transcript [6]. These observations are consistent with the fact that RNA:DNA hybrids made from G-rich RNA strands have a higher thermodynamic stability than DNA-DNA duplexes [7]. Thus, R-loop formation can be understood as a competition between the nascent RNA and the non-template DNA strand for re-annealing with the template DNA strand behind the advancing RNA polymerase [5, 8]. Factors that are conducive to reannealing of the RNA strand, including favorable RNA:DNA base-pair energetics and negative supercoiling, facilitate R-loop formation [9–11] (Figure 1).
Prevalent and conserved R-loop formation in mammalian genomes
R-loops can now be effectively mapped at endogenous loci in genomic DNA using a variety of methods including low throughput, single molecule approaches based on non-denaturing sodium bisulfite footprinting [12] and high throughput, population average techniques based on the S9.6 anti-RNA:DNA hybrid antibody [13, 14] (Box 1). Initial evidence for endogenous Rloop formation came from footprinting analysis of murine switch regions, where R-loops were detected upon induction of the corresponding upstream promoter in primary mouse B cells [12]. Consistent with thermodynamic predictions, these R-loops often initiated in proximity to G clusters and terminated over regions of lower G density [15–17]. R-loop initiation and termination sites, however, were heterogeneous, resulting in structures of variable lengths ranging from a few hundred base-pairs up to over 1 kilobase. Similar results were obtained upon footprinting R-loops at three different CGI promoters in the human and mouse genomes [6]. R-loops therefore form in mammalian chromosomes and individual R-loops can be long, covering stretches of DNA corresponding to multiple nucleosomes.
Box 1. Methods for R-loop mapping.
Non-denaturing (native) sodium bisulfite treatment allows R-loop footprinting by triggering the deamination of intrinsically unpaired cytosines, such as those on the looped-out strand of an R-loop, to uracil. After PCR amplification and sequencing of individual DNA molecules, R-loops can be detected by the presence of long, strand-specific, C to T conversion “footprints”. Importantly, these footprints are sensitive to pre-treatment with Ribonuclease H, an enzyme that degrades RNA strands specifically in the context of RNA:DNA hybrids [84]. The S9.6 antibody and its sequence-independent sub-nanomolar affinity for RNA:DNA hybrids [13, 14] enabled R-loop immunoprecipitation from genomic material [30, 92]. This reagent enabled Rloop profiling at genome scale through DNA:RNA ImmunoPrecipitation coupled to high-throughput DNA sequencing, DRIP-seq [6]. Variations on the DRIP-seq method have now enabled the profiling of R-loops at higher resolution and in a strand-specific manner either through sequencing of RNA strands involved in R-loop formation after cDNA synthesis (DRIP-RNA-seq [22] or DRIPc-seq [19]), or through sequencing of the template DNA strand (RDIP-seq [18] and S1-DRIP-seq [93]).
High-throughput R-loop mapping studies revealed that R-loop formation in mammalian genomes is much more prevalent than initially thought, occurring over tens of thousands of genomic loci [6, 18–21] and covering up to 150 megabases of sequence space or nearly 5% of the human genome [19]. Consistent with results from in vitro transcription and R-loop footprinting, CGI promoters are major R-loop hotspots in the human genome [6, 18, 20, 22]. Rloops formed at CGI promoters were preceded by significant GC skew. The observation that GC skew is a conserved characteristic of mammalian CGI promoters suggest that these loci are prone to R-loop formation in a range of organisms [23]. It is worth noting that promoter Rloops tend to peak 1–2 kb downstream of the TSS and are distinct from the short (40–60 bp) nascent transcripts held within the transcription complex at sites of promoter-proximal pausing [24]. The 3’-end of genes surrounding the polyadenylation site (PAS) is another major hotspot of R-loop formation [19, 20, 22]. Terminal R-loops often do not associate with positive GC skew [19] and terminal GC skew, contrary to promoter GC skew, is conserved only in a small subset of vertebrate genes [23]. Thus, terminal R-loops may not be driven by the energetic favorability of the RNA:DNA hybrid. In addition, the ends of genes, in particular convergent genes that are notable R-loop hotspots [19], are thought to associate with positive supercoiling during transcription elongation [25]. The lack of favorable sequence and topological characteristics at gene ends raises questions regarding the mechanism(s) underlying terminal R-loop formation. Interestingly, these structures were only observed for transcripts undergoing PAS-dependent cleavage and polyadenylation [19]. In budding yeast, mutations in cleavage and polyadenylation factors as well as other 3’-end processing factors lead to R-loop-mediated genomic instability [26–28]. R-loop structures were further proposed to play a key step in the transcription termination process [19, 29–31]. It is possible that terminal R-loops are formed as an intermediate in transcription termination and 3’-processing and that their formation involves the activity of as yet unknown specialized complexes. While gene ends are clearly R-loop hotspots, the majority of genic R-loops form within gene bodies [18, 19]. Gene body R-loops typically occupy only a small portion of a gene’s sequence space. This indicates that R-loop formation is not just a simple consequence of transcription but results from interplay between transcription, DNA sequence, topology, and possibly other characteristics of the chromatin template.
R-loops have now been profiled in multiple human and mouse cell lines [18, 19, 32] including primary human fibroblasts [21] and murine embryonic stem cells [19, 22]. The promoter and terminator regions of genes are conserved R-loop formation hotspots both within cell lines [18, 19, 32] and across species [22], including over orthologous gene pairs [19]. Gene body R-loop signal is also often conserved, albeit slightly less so than the signal at gene extremities. In addition to genic R-loop formation, repetitive regions of mammalian genomes are also capable of structure formation (Box 2). Collectively, these studies highlight that R-loop formation occurs at specific, conserved loci associated with transcription initiation, elongation, and termination.
Box 2. R-loop formation in intergenic / repetitive regions.
While genic R-loops have been the best studied so far, evidence suggest that these structures can also form over intergenic and/or repetitive regions. Ribosomal DNA (rDNA) arrays for instance, are known R-loop hotspots in Bacteria [83], yeast [92], and mammals [18]. Bright S9.6 staining over mammalian nucleoli [6, 87] supports the notion that nucleoli are a strong Rloop hotspots although the extent to which this signal is solely representative of RNA:DNA hybrids or also caused by abundant double-stranded ribosomal RNAs remains unclear. Recent mapping data suggest that RNA:DNA hybrids form over exons of mammalian rDNA units and over the intergenic spacer region [18]. In addition, evidence exists that R-loop can form over telomeric regions in yeast [94] and mammals. In this latter case, transcription initiation upstream of the G-rich telomeric repeats (5’-TTAGGG-3’) through the CGI-like TERRA promoter, gives rise to R-loop formation [95]. RNA:DNA hybrids were also suggested to form over specific human pericentromeric regions [18] and have been long described to form over mitochondrial DNA from yeast to mouse, where they are involved in priming DNA replication [96–98].
Dynamic turnover of R-loop structures
While genomic studies provide a wealth of information on the range of sequences that can form R-loops, they provide no information on two key parameters: the frequency at which Rloops form and the turnover rate of an R-loop once formed. DRIP-qPCR approaches suggest that R-loop formation frequencies range from 1–10% of input genomic DNA depending on the locus [19, 29, 32]. By contrast, negative loci (untranscribed and/or intergenic regions) range from 0.01–0.1% of input. Thus, while R-loops can form over a surprisingly large fraction of the genome, the frequencies at which they are formed remain relatively modest. Given that the large majority of genic R-loops form co-transcriptionally (see above), R-loop turnover rates can be measured after blocking de novo R-loop formation by inhibiting transcription. Treatment with transcription-blocking drugs combined with a kinetic analysis of R-loop frequencies showed that R-loops are dynamic structures. At promoters, which are easiest to analyze, Rloops are rapidly turned over, showing a half-life of 10 min [19]. This implies that R-loops are continuously formed and resolved and that the retention of nascent transcripts at their site of transcription is a dynamic feature of mammalian chromatin.
Heightened accessibility and RNAP stalling are common features of R-loop chromatin
The formation of long, stable R-loops likely alters local chromatin and numerous studies have focused on defining these effects. Consistent with observations that RNA:DNA hybrids adopt a rigid A form-like conformation [33] and prevent nucleosome wrapping in vitro [34], R-loops associate with DNase I hyper-accessibility at all genic positions, as per DRIPc-seq [19]. This association could be explained by interference with nucleosome re-deposition behind the advancing RNAP [35]. In support of this, levels of H3.3, a histone variant that dynamically replaces H3 lost owing to nucleosome disruption [36], are elevated over both promoter and terminal R-loop regions [19]. R-loops further correlate with chromatin decondensation and lower nucleosome occupancy [37] and, conversely, R-loop destabilization causes chromatin compaction [38]. Finally, loss of the FACT histone chaperone causes R-loop accumulation, suggesting that proper transcription-coupled nucleosome re-deposition prevents R-loop formation [39]. The association of R-loops with hyper-accessible chromatin is generally consistent with long-standing observations that nuclear retained RNAs associate with the maintenance of an open, active, chromatin state and that treatment of cells with ribonucleases or transcription inhibitors leads to the collapse of chromatin architecture [40, 41]. Altogether, these observations suggest that R-loops locally open the chromatin structure possibly by regulating nucleosome occupancy, positioning, and/or turnover. The proposed association between “aberrant” R-loops and chromatin condensation is specifically discussed below.
Another strong hallmark of R-loop forming (R-loop(+)) regions is a local increase in the density of RNAP [19]. This suggests that R-loop formation causes transient RNAP stalling, as observed in vitro [11]. The alternative possibility that RNAP stalling promotes R-loop formation cannot be ruled out, although a recent study of elongation-defective yeast RNAP mutants did not reveal an increase in R-loop formation [42]. Interestingly, promoter distal RNAP pausing sites were described at CGI promoters over R-loop-prone regions characterized by high GC skew [43]. These distal pause sites mediate contacts with long-range enhancers, suggesting that R-loop-associated pausing may participate in the control of gene expression [43]. Interestingly, switch regions, one of the best documented R-loop regions (see above), are also characterized by chromatin accessibility and RNAP stalling [44, 45], suggesting that these two features are consistent hallmarks of R-loop chromatin.
Defining histone modification signatures of R-loop chromatin
In addition to chromatin accessibility and RNAP stalling, high-resolution R-loop maps have enabled the identification of a set of histone modifications that associate with R-loop chromatin under normal conditions [19, 22]. Histone modifications that characterize “aberrant” R-loops formed under altered conditions are described separately below. The existence of such signatures suggests that the transient formation of RNA:DNA hybrids is sensed by chromatin modifying enzymes and translated into specific modification states. Thus, R-loop formation, which occurs at thousands of loci of varied DNA sequence in a transient and reversible manner, may represent an additional layer of epigenetic information.
At promoter regions, R-loops associate with higher levels of histone marks characteristic of active transcription (H3K4me1, H3K4me3, H3 acetylation) and transcription elongation (H3K36me3) (Figure 2) [19, 22]. Analysis of the chromatin of activated switch regions in B cells showed that H3K4me3 and H3 acetylation are recruited upon transcriptional induction [46–48]. Furthermore, genomic off-targets of the activated B cell-specific AID cytidine deaminase, an enzyme that acts on ssDNA including that found at R-loop structures [49], are characterized by high-transcription, RNAP stalling, and the combined R-loop-enriched marks H3K4me1, H3K27Ac, and H3K36me3 [50]. Terminal R-loops were predominantly marked by increased H3K4me1 levels [19] suggesting that H3K4me1 is a common mark of R-loop regions. Terminal R-loops also showed increased recruitment of the p300 acetyltransferase and of the CTCF-cohesin complex, conferring them an enhancer-like and insulator-like chromatin state [19]. Interestingly, these observations may account for properties often linked to efficient transcription terminators including the production of antisense non-coding RNAs and the formation of gene loops [29, 51, 52]. Indeed, evidence suggests that terminal R-loops represent a broad class of efficient transcription terminators [19], as previously suggested [30].
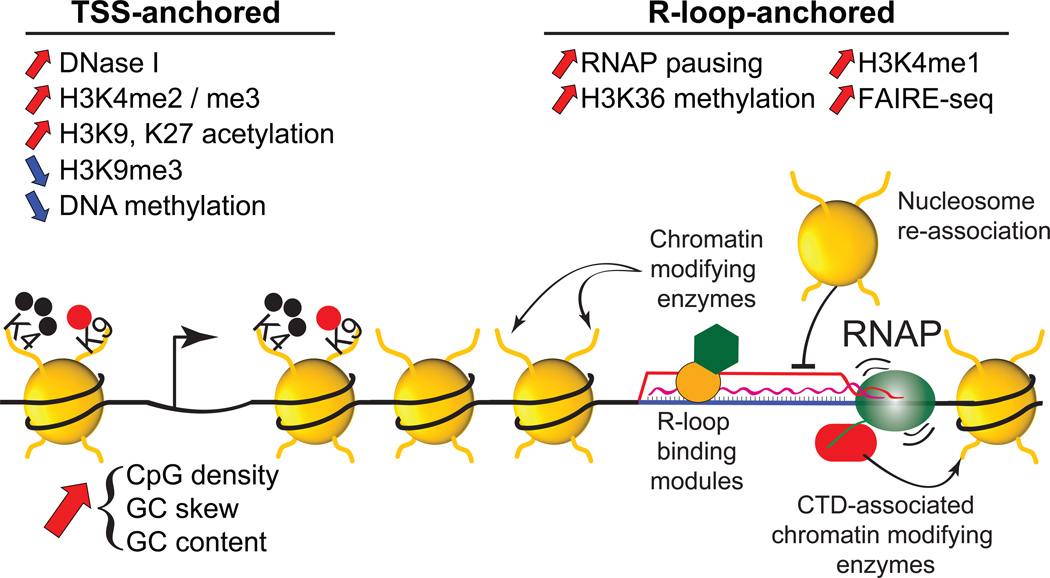
Figure 2.
Summary of chromatin features associated with R-loop formation at promoter regions. The features were broken down depending on their location. TSS-anchored features typically occur in the immediate surrounding of the promoter region, often at CGI promoters marked by increased CpG density, GC skew and GC content. By contrast, R-loop-anchored features were found to physically coincide with the location of R-loops, typically 1-2 kb downstream of the TSS. Multiple mechanisms are envisioned for coupling R-loop formation to chromatin patterning including the recruitment of chromatin modifying complexes through Rloop binding modules, the prevention of nucleosome re-deposition behind the RNAP machinery, and the co-transcriptional recruitment of chromatin modifying enzymes to transiently stalled RNAP complexes.
Mechanisms of R-loop-mediated chromatin patterning
Now that a coherent set of R-loop chromatin signatures is emerging, the task is to decipher which of these marks can be causally linked to R-loop formation and to provide a mechanistic understanding for these relationships. One way that R-loops may affect the chromatin landscape is by favoring the recruitment of chromatin modifying complexes that are normally recruited co-transcriptionally. Trimethylation of histone H3 at lysine 36 (H3K36me3) is mediated by the SETD2 histone methyltransferase upon interaction with the C-terminal domain (CTD) of RNAP [53, 54]. Genes with R-loop(+) promoters show heightened H3K36me3 recruitment precisely over the R-loop formation peak and the associated increased RNAP density [19] (Figure 2). This suggests that R-loops may facilitate recruitment of SETD2 to RNAP complexes by transiently stalling the transcription machinery. Additional histone modifications that are deposited co-transcriptionally, most significantly H3K4 methylation [55, 56], are also enriched over R-loop(+) promoters. In this case, however, it is worth distinguishing between H3K4 trimethylation, which shows a TSS-anchored distribution, from H3K4 monomethylation, which shows an R-loop-anchored distribution 1-2 kilobase downstream of TSS (Figure 2) [19]. In the first case, it is possible that the higher CpG densities and GC contents of R-loop(+) promoters [19] are driving increased H3K4me3. Indeed, CpG density favors the recruitment of the unmethylated CpG binding protein CFP1 [57] and its associated H3K4 methyltransferase SET1/COMPASS complex [55, 58]. In the latter case, it is possible that R-loops play a more direct role in recruiting H3K4me1. While the exact mechanism linking R-loop formation and H3K4me1 deposition remains to be elucidated, we note that several SET1/MLL H3K4 methyltransferase family members were shown to bind ssDNA [59] and that the PAF1 complex, which facilitates both H3K4 and H3K36 methylation [60], is enriched over R-loop regions [19] and may itself be able to sense ssDNA [61]. Interestingly, increased PAF1 recruitment over terminal R-loops may also account for H3K4me1 deposition over these regions and for efficient transcription termination given the ability of the PAF1 complex to recruit termination factors [60]. R-loops may also favor the recruitment of the XRN2 “torpedo” ribonuclease at terminal pause sites, triggering the release of the RNAP machinery upon degradation of the residual transcript downstream of the PAS [30, 31, 62].
Histone acetylation, as discussed above, is a TSS-anchored mark of R-loop(+) promoters (Figure 2). Recent data shows that recruitment of the Tip60-p400 chromatin remodeling and acetylase complex is substantially reduced in murine embryonic stem cells (mESCs) expressing RNASEH1 [22], an enzyme that resolves R-loops. This, together with evidence that many Tip60-p400 targets form R-loops in mESCs, suggests that R-loops contribute to recruiting the Tip60-p400 complex to chromatin [22] through an as-yet-unidentified R-loop binding subunit. Interestingly, RNASEH1 over-expression also led to increased recruitment of the Polycomb Responsive Complex 2 (PRC2) and increased deposition of the PRC2-catalyzed H3K27me3 silencing mark to a subset of genomic targets in mESCs [22]. This suggests that R-loops may prevent PRC2 chromatin loading. Notably, R-loop(+) loci in human carcinoma cells showed significantly higher H3K27me3 levels compared to matched R-loop(-) loci, especially for poorly-expressed genes [19]. One possibility to account for this discrepancy is that pluripotent ESCs possess distinct regulatory mechanisms controlling H3K27me3 deposition compared to differentiated cells. It is worth noting, however, that successfully preventing the recruitment of a complex to chromatin requires an efficient and long-lived inhibitory signal when R-loops form transiently and at modest frequencies.
Altogether, evidence suggests that R-loops may influence chromatin modification states by affecting nucleosome density, through co-transcriptional mechanisms, or even directly via recognition by chromatin regulators. Another speculative mechanism by which R-loops could contribute to chromatin patterning might involve the targeting of lncRNAs (Box 3) [63].
Box 3. Cis or trans R-loops as a mechanism for targeting non-coding RNAs?
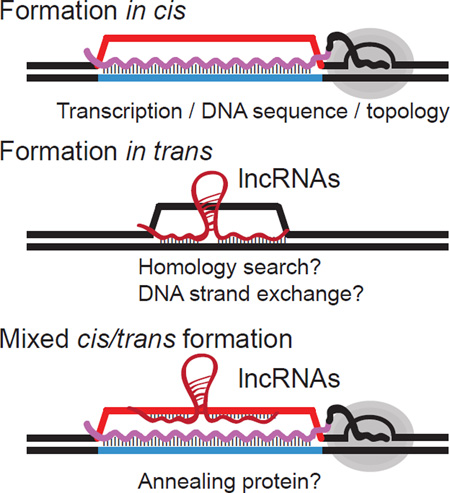
The study of genic R-loops provides strong support for a co-transcriptional mode of formation in cis: (i) in 90% of cases, R-loop are formed co-directionally with transcription [19]; (ii) R-loop levels correlate with expression levels [18, 19, 74]; (iii) R-loop levels respond to transcriptional changes [19, 74]; (iv) DNA-based (DRIP-seq) and RNA-based (DRIPc-seq) R-loop maps agree well with each other [19]; and genes showing allele-specific expression show allele-specific R-loops [6]. It remains possible, however, that R-loops also form in trans. In this case, an RNA strand transcribed from a locus could conceivably hybridize onto a distant locus through regions of RNA-DNA complementarity (Figure I). This mechanism likely requires proteins to catalyze strand invasion and assist in homology searching as is the case for the CRISPR-Cas9 complex [99, 100]. Evidence for a trans R-loop mechanism dependent on the homologous recombination machinery was obtained in budding yeast [101, 102]. Similarly, trans R-loops dependent on the RecA DNA strand exchange protein have been described in E. coli and reconstituted biochemically [103–105]. Trans R-loop formation provides an attractive mechanism for directing long non-coding RNAs (lncRNAs) to potential targets. Recent evidence suggests that the GAL lncRNAs in budding yeast regulate transcriptional induction via trans R-loop-mediated gene looping [106]. In addition to these two mechanisms, a mixed cis/trans model is also possible (Figure I). In this case, co-transcriptional R-loops might provide a ready-made single-stranded DNA (ssDNA) landing pad on the non-template strand for lncRNAs to interact with in trans. Such trans RNA:DNA hybrid formation might be easier to achieve mechanistically and only require the activity of a protein with strand annealing capacity. The conserved Rad52 protein, which mediates strand annealing upon binding to ssDNA and displacement of the ssDNA binding protein RPA [107], is a good candidate for such an activity. Recent evidence shows that Rad52 can catalyze the annealing of an RNA strand to a DNA strand, creating RNA:DNA hybrids [108]. Interestingly, this mixed cis/trans model predicts that transcription itself, through the formation of cis R-loops, might regulate the potential repertoire of some lncRNAs trans targets, as observed [109].
Figure I: possible mechanisms for R-loop formation in cis and in trans, including a mixed cis/trans mechanism. Factors that may contribute to these mechanisms are listed below.
Aberrant R-loops and chromatin condensation
Defects in the THO mRNA export complex in yeast triggers marked genomic instability and increased R-loop formation [64]. This increased R-loop load is accompanied by higher levels of H3S10P [65], a mark typically associated with condensed chromosomes during mitosis [66]. Increased H3S10P and condensation were observed outside of mitosis, and while most visible over pericentromeric regions, they were also observed over coding genic regions. Since H3S10P levels could be suppressed by RNASEH1 expression [65], a link with R-loops was suggested. Accumulation of R-loops and H3S10P in THO-defective conditions was demonstrated in C. elegans and human cells, further arguing that this link is conserved [65]. Thus, under conditions associated with “aberrant” R-loop formation (see below), R-loops appear to associate with H3S10P and chromatin condensation, a startling conclusion given the significant evidence linking R-loops and active, hyper-accessible chromatin under normal conditions. Interestingly, prior studies of the chromatin state of R-loop-prone murine switch regions showed that H3S10P is increased upon transcriptional induction [48]. However, the mark was found in the context of active, acetylated and H3K4 methylated chromatin [48]. Indeed, H3S10P in interphase cells has been associated with transcription and can co-exist with active histone modifications [67–69]. In Drosophila, H3S10 phosphorylation was suggested to permit transcription by counteracting H3K9 dimethylation, a mark of gene silencing [70]. Thus, while evidence suggests that H3S10P associates with R-loop formation, additional work is required to delineate the mechanism underlying this association and to clarify the relationship between H3S10P, R-loops, and the physical state of chromatin.
Recently, R-loop formation has been linked to increased deposition of H3K9me2/me3 and heterochromatin formation in the context of triplet expansion diseases such as Friedrich’s ataxia and Fragile X syndrome [71–73]. This association between R-loops and a silencing histone modification could reflect the fact that R-loops formed over expanded repeat arrays are unusually complex, including formation of hairpins on the displaced ssDNA strand [74], and therefore hard to resolve. Complex, persistent, R-loops might in turn be more prone to double-strand break (DSB) formation, a hallmark of R-loops [75]. Given that DSB repair involves the transient repression of chromatin via H3K9me2/3 deposition [76, 77], the association of Rloops with H3K9 methylated chromatin observed at specific loci may reflect R-loop-induced fragility. Recently, H3K9me2 deposition was also observed broadly over R-loop(+) terminator regions of murine genes and proposed to participate in the transcription termination process [29]. However, re-examination of global datasets did not support this broad association [19]; instead, terminators were shown to adopt an open, insulator-like state. It remains possible, however, that specific terminator regions such as those of highly expressed genes may carry high loads of persistent R-loops that predispose them to H3K9me2/3 deposition via a mechanism similar to that delineated above for triplet repeat expansion loci.
Defining aberrant R-loop formation under pathological conditions
R-loops are well known for their association with genomic instability, a topic extensively covered by several excellent recent reviews [75, 78–82]. This association is clearest in the context of defects in a variety of factors involved in co-transcriptional processes such as splicing, mRNA export, 3’ RNA processing, and transcription elongation, among others. Defects in other factors such as topoisomerases that are thought to prevent R-loop formation [83], or in enzymes such as Ribonucleases H [84] and RNA/DNA helicases that are thought to mediate R-loop resolution, also associate with DSBs and genomic instability [62, 85–89]. Not surprisingly, dysfunctional R-loop metabolism has been implicated in multiple human disorders, particularly cancers and neurological diseases (a topic reviewed in [71, 90, 91]). Despite these clear links, the genomic profiles of R-loops generated under pathological conditions have seldom been examined, raising questions as to what “aberrant”, “increased” or “unscheduled” R-loop formation truly entails. In a recent study, estrogen signaling in breast cells was shown to lead to a burst of R-loop formation at thousands of loci, including estrogen-responsive genes, and to R-loop- and replication-dependent DSBs [32]. Thus, up-regulation of transcriptional programs can lead to elevated genic R-loop loads and fuel cancer-relevant instability and rearrangements. R-loop profiling in Aicardi-Goutières patient cells harboring partial RNASEH2 loss of function, however, did not reveal any significant changes in genic R-loop distribution [21]. Instead RNA:DNA hybrids were unexpectedly found to accumulate over late-replicating, DNA hypomethylated, intergenic regions characterized by facultative heterochromatin [21]. Thus, assumptions about increased genic R-loop levels in pathological situations may not necessarily be correct. Instead, more subtle alterations of R-loop turnover rates caused by mutations in specific factors or by sequence changes at specific R-loop forming loci may be sufficient to lead to persistent R-loops and threaten genome stability. One could even envision that a loss of R-loop formation either globally or locally could lead to detrimental consequences by preventing R-loops from participating in their physiological functions. For instance, R-loop loss could compromise transcription termination mechanisms, thus increasing transcriptional interference and the likelihood of runaway transcription-replication encounters. Additional studies are required to carefully define the distribution, molecular nature, and turnover rates of the aberrant R-loops caused upon alteration of the many factors to which they have been tied.
Concluding remarks
The field of R-loop biology has made rapid progress in recent years thanks in part to technological advances in the detection of endogenous R-loops. While we now understand that R-loops are much more abundant than previously thought, many questions remain (see Outstanding Questions). Efforts should focus in particular on identifying the protein complexes that maintain R-loop homeostasis in normal cells and how breakdown of this equilibrium is linked to disease. These studies will undoubtedly provide new insights into the mechanisms by which nascent RNAs regulate important aspects of chromosome dynamics such as transcription termination, chromatin patterning, replication licensing, and gene looping.
Outstanding questions.
What enzymes and proteins complexes are involved in forming, stabilizing, sensing, and resolving R-loop structures? Our understanding of the metabolism of these structures is still extremely limited. Filling this gap is critical to revealing the eventual cellular functions of R-loops.
What is the fate of the RNA transcripts involved in R-loop formation? Do they represent terminal intermediates that will be targeted for degradation or are they normal intermediates in the life of a transcript?
What are the distinguishing features of pathological and physiological R-loops? Methodologies to measure global R-loop distribution at high resolution as well as dynamic R-loop turnover and individual R-loop footprints will need to be leveraged to address this key question.
How do pathological R-loops trigger genomic instability? An abundant literature shows that conflicts between co-transcriptional R-loops and replication play a key role in driving instability. However, mechanistic insights into the process are still lacking despite recent progress.
Trends.
-
-
R-loop structures are a type of non-B DNA structure which can form during transcription upon reannealing of the nascent RNA to the template DNA template. R-loops can now be profiled globally at high resolution in any genome.
-
-
R-loops in mammalian systems are abundant, covering over 100 megabases of DNA sequence, and form dynamically over conserved regions.
-
-
R-loops play important roles in chromosome dynamics, in particular as it concerns transcription termination and chromatin patterning.
-
-
Deregulation of R-loop metabolism is linked to an increasing number of human diseases, including cancers and several neurodegenerative disorders.
Acknowledgments
Research in my laboratory is supported by NIH grants GM094299, GM113929, and GM120607.
Glossary
- R-loop structure
a type of three-stranded non-B DNA structure in which the non-template DNA strand exists in a looped out single-stranded state owing to the presence of the RNA:DNA hybrid (Figure 1).
- GC skew
a sequence property that describes the strand asymmetry in the distribution of guanine versus cytosine residues.
- Supercoiling
refers to the torsional stress that is exerted on the DNA double helix. In negatively supercoiled DNA, the two DNA strands are slightly underwound compared to B DNA.
- CpG island
a stretch of DNA sequence characterized by high GC content and high CG dinucleotide density compared to the rest of the human genome. CpG islands often map to the beginning of mammalian genes where they serve as promoters. Nearly 60% of human genes possess a CpG island promoter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303(5660):1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 2.Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23(24):5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reaban ME, Lebowitz J, Griffin JA. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem. 1994;269(34):21850–21857. [PubMed] [Google Scholar]
- 4.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29(11):3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy D, et al. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol. 2010;30(1):146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginno PA, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratmeyer L, et al. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry. 1994;33(17):5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 8.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28(1):50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drolet M, Bi X, Liu LF. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem. 1994;269(3):2068–2074. [PubMed] [Google Scholar]
- 10.Phoenix P, et al. Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J Biol Chem. 1997;272(3):1473–1479. doi: 10.1074/jbc.272.3.1473. [DOI] [PubMed] [Google Scholar]
- 11.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A. 2010;107(29):12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4(5):442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 13.Boguslawski SJ, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89(1):123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 14.Phillips DD, et al. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit. 2013;26(8):376–381. doi: 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang FT, et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27(16):5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang FT, et al. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A. 2006;103(13):5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao YP, et al. Detection and characterization of R-loops at the murine immunoglobulin Salpha region. Mol Immunol. 2013;54(2):208–216. doi: 10.1016/j.molimm.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Nadel J, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanz LA, et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell. 2016;63(1):167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginno PA, et al. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23(10):1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim YW, et al. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife. 2015;4 doi: 10.7554/eLife.08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen PB, et al. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol. 2015;22(12):999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartono SR, Korf IF, Chedin F. GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, et al. Proteomic patterns for classification of ovarian cancer and CTCL serum samples utilizing peak pairs indicative of post-translational modifications. Proteomics. 2007;7(22):4045–4052. doi: 10.1002/pmic.200601044. [DOI] [PubMed] [Google Scholar]
- 26.Stirling PC, et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26(2):163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Gonzalez B, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30(15):3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahba L, et al. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44(6):978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014 doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352(6291):aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stork CT, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife. 2016;5 doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noy A, et al. Structure, recognition properties, and flexibility of the DNA.RNA hybrid. J Am Chem Soc. 2005;127(13):4910–4920. doi: 10.1021/ja043293v. [DOI] [PubMed] [Google Scholar]
- 34.Dunn K, Griffith JD. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980;8(3):555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 36.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28(7):672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell WT, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci U S A. 2013;110(34):13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boque-Sastre R, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A. 2015;112(18):5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera-Moyano E, et al. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014;28(7):735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickerson JA, et al. Chromatin architecture and nuclear RNA. Proc Natl Acad Sci U S A. 1989;86(1):177–181. doi: 10.1073/pnas.86.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. 2012;22(2):179–187. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Felipe-Abrio I, et al. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J. 2015;34(2):236–250. doi: 10.15252/embj.201488544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellner WA, Bell JS, Vertino PM. GC skew defines distinct RNA polymerase pause sites in CpG island promoters. Genome Res. 2015;25(11):1600–1609. doi: 10.1101/gr.189068.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, et al. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206(8):1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balter BB, et al. Mice lacking Smu tandem repeats maintain RNA polymerase patterns but exhibit histone modification pattern shifts linked to class switch site locations. Mol Immunol. 2012;52(1):1–8. doi: 10.1016/j.molimm.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22(15):2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li G, et al. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 2013;5(3):702–714. doi: 10.1016/j.celrep.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu K, et al. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol. 2005;25(5):1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang IX, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6(5):906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 52.Grzechnik P, Tan-Wong SM, Proudfoot NJ. Terminate and make a loop: regulation of transcriptional directionality. Trends Biochem Sci. 2014;39(7):319–327. doi: 10.1016/j.tibs.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strahl BD, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22(5):1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25(8):3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 57.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464(7291):1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280(50):41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 59.Krajewski WA, et al. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25(5):1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013;1829(1):116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Jong RN, et al. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure. 2008;16(1):149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 62.Morales JC, et al. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016;12(7):e1006107. doi: 10.1371/journal.pgen.1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12(3):711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52(4):583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102(3):279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 67.Sharma AK, et al. Dynamic alteration in H3 serine 10 phosphorylation is G1-phase specific during ionization radiation induced DNA damage response in human cells. Mutat Res. 2015;773:83–91. doi: 10.1016/j.mrfmmm.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 68.Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007;21(21):2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 70.Cai W, et al. Genome-wide analysis of regulation of gene expression and H3K9me2 distribution by JIL-1 kinase mediated histone H3S10 phosphorylation in Drosophila. Nucleic Acids Res. 2014;42(9):5456–5467. doi: 10.1093/nar/gku173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Groh M, Gromak N. Out of Balance: R-loops in Human Disease. PLoS Genet. 2014;10(9):e1004630. doi: 10.1371/journal.pgen.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groh M, et al. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014;10(5):e1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colak D, et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343(6174):1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loomis EW, et al. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014;10(4):e1004294. doi: 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ayrapetov MK, et al. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A. 2014;111(25):9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gursoy-Yuzugullu O, House N, Price BD. Patching Broken DNA: Nucleosome Dynamics and the Repair of DNA Breaks. J Mol Biol. 2016;428(9 Pt B):1846–1860. doi: 10.1016/j.jmb.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 80.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015 doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 81.Costantino L, Koshland D. The Yin and Yang of R-loop biology. Curr Opin Cell Biol. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan YA, Hieter P, Stirling PC. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014;30(6):245–253. doi: 10.1016/j.tig.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masse E, Phoenix P, Drolet M. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J Biol Chem. 1997;272(19):12816–12823. doi: 10.1074/jbc.272.19.12816. [DOI] [PubMed] [Google Scholar]
- 84.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. Febs J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11(11):1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y, et al. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol Cell. 2014;53(3):484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56(6):777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33(2):406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41(1):21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richard P, Manley JL. R Loops and Links to Human Disease. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salvi JS, Mekhail K. R-loops highlight the nucleus in ALS. Nucleus. 2015;6(1):23–29. doi: 10.1080/19491034.2015.1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.El Hage A, et al. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24(14):1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wahba L, et al. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30(11):1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pfeiffer V, et al. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32(21):2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arora R, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El Hage A, et al. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10(10):e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J Biol Chem. 2008;283(52):36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu B, Clayton DA. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cell Biol. 1995;15(1):580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang F, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351(6275):867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111(27):9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aronica L, et al. The spliceosome-associated protein Nrl1 suppresses homologous recombination-dependent R-loop formation in fission yeast. Nucleic Acids Res. 2016;44(4):1703–1717. doi: 10.1093/nar/gkv1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61(2):212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kasahara M, et al. RecA protein-dependent R-loop formation in vitro. Genes Dev. 2000;14(3):360–365. [PMC free article] [PubMed] [Google Scholar]
- 105.Zaitsev EN, Kowalczykowski SC. A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 2000;14(6):740–749. [PMC free article] [PubMed] [Google Scholar]
- 106.Cloutier SC, et al. Regulated Formation of lncRNA-DNA Hybrids Enables Faster Transcriptional Induction and Environmental Adaptation. Mol Cell. 2016;61(3):393–404. doi: 10.1016/j.molcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci U S A. 1998;95(11):6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keskin H, et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515(7527):436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]