Abstract
Fishes (i.e., teleost fishes) are the largest group of vertebrates. Although their immune system is based on the fundamental receptors, pathways, and cell types found in all groups of vertebrates, fishes show a diversity of particular features that challenge some classical concepts of immunology. In this chapter, we discuss the particularities of fish immune repertoires from a comparative perspective. We examine how allelic exclusion can be achieved when multiple Ig loci are present, how isotypic diversity and functional specificity impact clonal complexity, how loss of the MHC class II molecules affects the cooperation between T and B cells, and how deep sequencing technologies bring new insights about somatic hypermutation in the absence of germinal centers. The unique coexistence of two distinct B-cell lineages respectively specialized in systemic and mucosal responses is also discussed. Finally, we try to show that the diverse adaptations of immune repertoires in teleosts can help in understanding how somatic adaptive mechanisms of immunity evolved in parallel in different lineages across vertebrates.
1 Introduction
An immune system is characterized by two linked properties: a somatic learning process to make a self–nonself discrimination and a mechanism for determining the class of the response that optimally rids the target
Melvin Cohn (Cohn 1994)
When a pathogen confronts a host organism, it stimulates defense mechanisms leading to its elimination, reduction, or containment. Besides innate pathways based on factors encoded in the genome as “ready to use” units, systems that undergo somatic modifications offer opportunities of focused “adaptive” responses to pathogens. Specific recognition of antigens by lymphocyte receptors diversified through VDJ somatic rearrangements is the archetype of such systems of adaptive response.
During their differentiation, immunoglobulin (Ig)—respectively, T-cell receptor (TR)—loci are subjected to random genomic rearrangements of V, D, and J gene segments, leading to the expression of a unique antigen receptor by each lymphocyte. The universe of antigenic motifs is matched to a large population of lymphocytes through the specific recognition of an epitope by a given receptor unique to a lymphocyte clone. During the differentiation of lymphocytes, epitope-specific, selective processes lead to deletion of most autoreactive cells and expansion of mature T and B cells. These populations are dramatically affected during antigen-driven responses, for example, during pathogen infections, as clones specific of pathogen epitopes are expanded.
The concept of immune repertoire was created to describe the diversity of lymphocyte receptors involved in this network of interactions. Niels Jerne referred to the immune repertoire as a dual concept integrating both the potential diversity allowed by the genetic resources of the genome and the available set of receptors expressed in a given tissue at a given moment (Jerne 1971). This notion of “immune repertoires” represents a useful tool to describe lymphocyte and receptor populations, their development, and their modifications by responses to infections.
In teleost fish, three immunoglobulin classes have been described: IgM, IgD, and IgT. While IgM constitutes the main systemic immunoglobulin, IgT plays the prevalent role in mucosal surfaces. The role of IgD in fish immunity remains to be elucidated. Both IgM and IgD are co-expressed in B cells found both in systemic and mucosal lymphoid areas, whereas IgT is uniquely expressed by a B-cell subset devoid of IgM and IgD expression. The three isotypes share the same genomic repertoire of VH gene segments; IgM and IgD heavy chains are generally produced by alternative splicing of constant exons from a common long transcript, while mRNAs for IgT heavy chains are transcribed from another genomic region. Importantly, B lymphocytes express either IgM and IgD or IgT, which distinguishes two fundamental B-cell subsets. However, it has been shown that a third B-cell subset uniquely expressing IgD exists both in catfish and rainbow trout. TCR isotypes are much more conserved across vertebrates compared to immunoglobulins, and for the time being two main T-cell subsets, which, respectively, express αβ and γδ T-cell receptors, have been described in teleosts.
In this review, we examine a number of distinctive features of B- and T-cell immunity in fishes and show how repertoire studies shed light on particular somatic adaptations found in these animals. We first review the great diversity of teleost fishes and the canonical features of adaptive immunity they share with other (jaw) vertebrates. We then discuss selected mechanisms or features that represent distinctive adaptations of the fish immune system. Finally, we consider how these adaptations can help in understanding how the somatic adaptive mechanisms of immunity evolved in parallel in different lineages.
2 Common Conserved Features of Immune Repertoires Across the Great Diversity of Teleost Fishes
2.1 The Diversity of Fishes
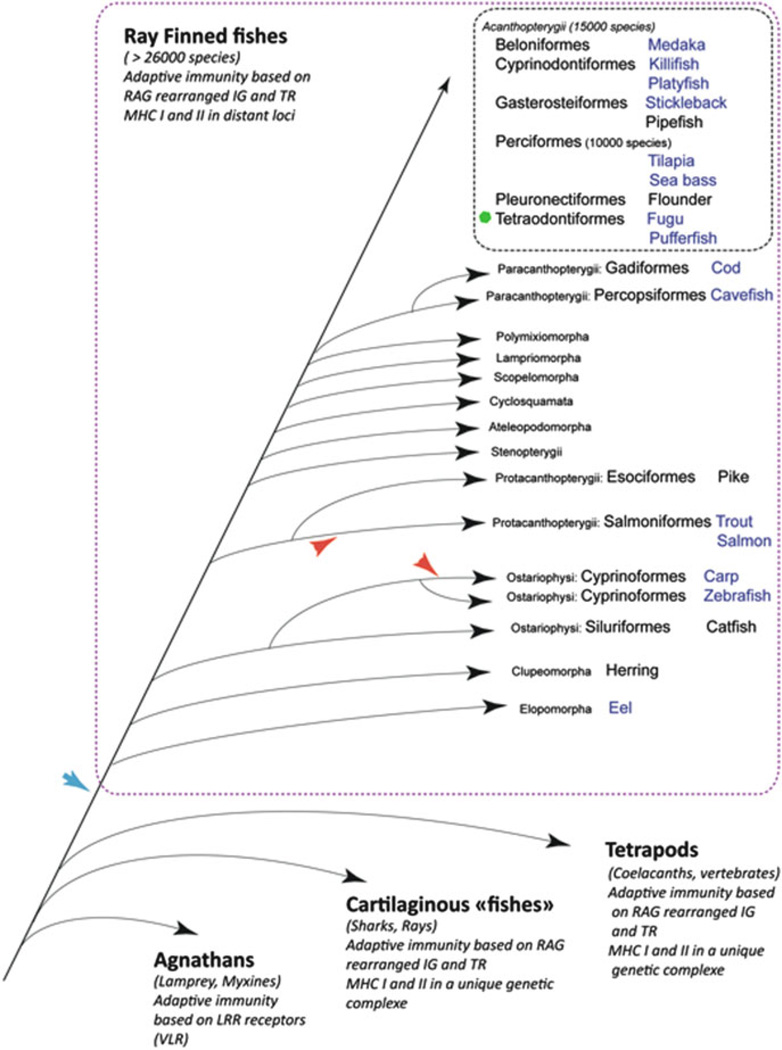
Following Nelson (Nelson 2006), a fish is “a poikilothermic vertebrate with gills and with limbs in the shape of fins.” In this chapter we focus on bony fishes which are by far the largest group of vertebrates with more than 26,000 species (Helfman et al. 2010), while there are about 10,000 species of birds and 5000 species of mammals. The diversity of shape, size (from 8 to 10 mm gobies and Danionella to very large sunfishes, swordfishes, and tunas), life span, and adaptations is spectacular. Most species are marine (about 60 %), with the remainder primarily living in freshwater and about 1 % moving between salt- and freshwater in their life cycle. Fishes have colonized almost all aquatic environments and evolve special adaptations to extreme habitats such as deep sea, polar regions, strong currents, caves, and seasonal water bodies in arid regions. Some species are warm-blooded, while other species living in cold environments have antifreeze peptides in the blood. Fish physiological adaptations to physical parameters such as pressure, temperature, alkalinity and salinity, light, high-energy water zones, etc., have been extensively studied, but the impact of these adaptations on immunity remains poorly known. It is certainly significant, however, as such adaptations lead to changes at the anatomical level (e.g., deep sea fish have lost the swim bladder) as well as at cellular level and blood composition or even at molecular scale (with adaptations of proteins including enzymes to different temperature and pressure ranges). Importantly, adaptation to multiple environments brought fishes in contact to diverse types of pathogen exposure, which likely represents the most important selection pressure on the defense system.
Fishes share the basic components of their immune system with all other jaw vertebrates (Gnathostomes) (Flajnik and Du Pasquier 2013, Table 1), of which the oldest fossils have been found in Ordovician sediments. The jaw acquisition likely has been pivotal for the later evolution of vertebrates, as it made possible diverse adaptations to a great number of ecological niches and food resources (Romer 1962). This shift from the microphage diet of agnathans should have modified significantly the interactions of the fish ancestors with their pathogens, as well as with the commensal bacterial flora in their gut (Matsunaga and Rahman 1998). It is precisely at this step of vertebrate evolution—in early Gnathostomes—that a new adaptive immunity emerged, in contrast to the VLR-based specific antigen recognition found in Agnathans (Herrin and Cooper 2010). The novel adaptive immune system was based on antigen receptors made of Ig domains and diversified by genomic rearrangements mediated by RAG in specialized cells, the lymphocytes. As for VLR, the expression of a unique receptor per clone allowed clonal somatic selection of lymphocytes by their cognate antigen. Fishes and other jaw vertebrates also inherited from these early ancestors a common array of innate immune pathways and receptors, which were later amplified, reduced, or lost in the different lineages.
Table 1.
Characteristics of the adaptive immune system in teleosts and mammals
| Teleosts | Mammals | |
|---|---|---|
| Primary organs | Thymus/head kidney | Thymus/bone marrow |
| Secondary organs | Spleen | Spleen/lymph nodes |
| Mucosa-associated lymphoid tissues |
+ (no Peyer’s Patch) |
+ |
| Germinal centers/FDCs | −a | + |
| B cells | ||
| Immunoglobulins | ||
| Heavy chain (locus configuration) | IgD, IgM, IgT/Zb (translocon)c |
IgD, IgM, IgA, IgG, IgE (translocon) |
| Light chain (locus configuration) | Kappa, lambda, sigma, sigma cart (multi-cluster) |
Kappa/lambda (translocon/multi cluster) |
| Somatic hypermutation | + | + |
| Affinity maturation | Low efficiency | High efficiency |
| Class switch | − | + |
| T cells | ||
| CD4/CD8 subsets | +d | + |
| Th1/Th2/Th17 | + | + |
| TCR αβ/γδ | + | + |
FDCs follicular dendritic cell
Putative primordial germinal centers have been characterized
In medaka and catfish the IgT/Z has not been identified
More than one functional IgH locus in most of studied teleost species
Absence of the cd4 and mhc II functional genes in some species (e.g. cod and pipefish)
Bony fishes appear in late Ordovician (≈440 My) with Acanthodians, but the oldest fossils of teleosts were found only in Triassic deposits (≈200 My). The major radiation of teleosts occurred during Cretaceous, leading to the main groups of modern ray-finned fishes. The number of species and the diversity of adaptations make it one of the great successes of vertebrates. While fish inherited the basic components of the jaw vertebrate immune system, an open question is how such a spectacular expansion did affect immunity and somatic selection of lymphocyte populations. In fact, the immune system has been extensively studied in only a few key fish species: mainly aquaculture fishes, like carp, catfish, trout, and salmon, and among the model species, essentially the zebrafish. Importantly, complete genome sequences are now available for many species belonging to a number of fish families, revealing several particular features—including traits important for immunity—of the evolution of the group. As shown in Fig. 1, species in which a complete genome sequence is available cover the main fish lineages. These genomes still represent a minute fraction of the whole fish diversity, but their analysis clearly showed that a whole genome duplication occurred during the early evolution of ray-finned fishes. Pairs of duplicated genes from this early event provided a large resource for subfunctionalization and likely favored the diversity of defense mechanisms among other adaptations. Fish genome diversity was further increased by lineage-specific events of genome duplication and/or contraction (Fig. 1). Specific expansions of gene families were also frequent. A striking example in the immune system is the fish-specific trim family (fintrims) (van der Aa et al. 2009); this multigenic family shows various degrees of expansion across fishes and diversified through distinct mechanisms. For example, local duplications occurred in zebrafish, leading to multiple clusters, while retropositions were the key mechanisms in the medaka, leading to many intronless genes. With respect to the adaptive immune system, duplicated loci may create significant complications of the finely tuned clonal selection pathways. For example, the expression of multiple major histocompatibility complex (MHC) molecules in an Ag-presenting cell might perturbate lymphocyte selection or lead to low density of Ag-presenting molecules. In fact, only one or a few copies of MHC gene duplicates are generally conserved. The fish MHC is unique among vertebrates since genes encoding MHC class I (MHC I) and MHC class II (MHC II) molecules are not encoded in the same genomic region but in two distant loci, which may each correspond to old duplicated regions (Kelley et al. 2005). Another important question is the maintenance of allelic exclusion with multiple loci of Ig and/or TCR loci. Efficient mechanisms certainly have been developed for this, as several fish species have maintained multiple Ig loci; however, many duplicated Ig segments have become pseudogenes. Finally, the presence of multiple families of transposable elements in fish genomes likely has been an additional driver for the evolution of fish genomes and multigenic families.
Fig. 1.
Schematic phylogenetic tree of fishes. Taxonomy as in Helfman et al. (2010). The blue arrow indicates the whole genome duplication (WGD) which occurred during the early evolution of ray-finned fishes and led to a tetraploidization followed by a rediploidization. Red arrows indicate examples of posterior WGD in specific branches. The green dot denotes an example of genome contraction in tetraodontiformes. A genome sequence is available for all species in blue
3 Selected Distinctive Features of Fish Immune Repertoires
3.1 How Allelic Exclusion Can Be Achieved with Multiple, Complex, and Large Loci Found in Some Fish Species?
The adaptive immune system can recognize millions of different antigens/antigenic determinants in a highly specific way. This fundamental property was modeled by Jerne (1955) and Burnet (1957) in a Darwinian perspective: the world of antigens was matched to a large population of lymphocytes through a specific recognition of an epitope by a given receptor unique to each lymphocyte clone. The genetic basis of this unique specificity of the Ag receptor expressed by each lymphocyte was not obvious even after the discovery of Ig gene rearrangements explained the generation of the molecular diversity of antibodies (Abs). The mechanisms ensuring an allelic exclusion to allow the expression of a unique receptor at the surface of B and T cells were progressively discovered mainly in the mouse.
In humans and mice, the IgH or TCR beta locus is activated in proB (T) cells and a D–J recombination occurs. This is followed by a locus contraction allowing the V to DJ rearrangement, which only occurs in one allele. Finally, the VDJ rearrangement is expressed on the surface of immature B or T cells as a pre-B-cell receptor (pre-BCR) or pre-T-cell receptor (pre-TCR) that allows to test its “functionality.” If the expression of the pre-receptor leads to the right signal, loci of IgH (TCR beta) are closed and rearrangements of the light chain (TCR alpha) locus start. If the first VDJ joining is nonproductive or encodes a non-pairing chain, the second IgH/TCR beta allele is rearranged. Although the exact mechanisms are not fully understood, the current models suggest the presence of controls at multiple levels including the accessibility of the RAG recombinase to the different alleles within chromatin, the locus topology, and repositioning of loci to pericentromeric heterochromatin (Oltz 2001; Jhunjhunwala et al. 2009). Importantly, the allelic exclusion at the genomic level is not absolute especially for the second rearrangement (i.e., for IgL or TCR alpha/gamma). Regulation at the expression level is therefore also important to ensure that a unique receptor is mainly expressed by a given lymphocyte.
The enzymatic complex RAG is well conserved in jaw vertebrates. However, the accessibility of the antigen receptor genes to RAG, as well as the modalities of the VDJ rearrangements, has evolved with the organization of the Ig and TCR loci, which differ significantly between fishes and tetrapods (Hirano et al. 2011; Das et al. 2012). Studies in different vertebrate species including rabbit, chicken, and shark indicate that the VDJ rearrangement does not seem to always follow an ordered model (Hsu 2009). In chicken, for example, there is no sequential rearrangement of IgH and IgL genes; however, most of chicken B cells present only one functionally rearranged Ig allele, the other remaining in DJ configuration (Weill et al. 2002; Ratcliffe 2006). It was suggested that the V rearrangement happens in one allele randomly and that this is an event of such low efficiency that the probability to occur in both alleles is very low (stochastic/asynchronous model) (Weill et al. 2002). In such a model of simultaneous rearrangement of IgH and IgL genes, there would likely be no pre-BCR.
In non-tetrapod vertebrates, this question has been studied mainly in cartilaginous fishes as the presence of multiple VDJ recombination units in the genome represents an obvious issue for allelic exclusion (Hsu et al. 2006). In Chondrichthyans, the IgH locus is arranged in multiple independent clusters consisting in a few gene segments (V–D–D–J–C) and representing recombination units, as there is no evidence for intercluster recombination. These clusters can even be situated on different chromosomes (Rast and Litman 1998). In this context, a locus contraction would not have the same regulatory potential as in a translocon configuration. In fact, the close proximity of the gene segments makes them recombine all at once and to completion (Rast et al. 1998). How different recombination units mutually exclude each other remains unknown.
Eason et al. (2004) identified different productive IgH gene transcripts in isolated single peripheral blood lymphocytes from the clearnose skate (Raja eglanteria), suggesting a simultaneous expression from multiple IgH rearrangements, potentially from different loci. However, most of the IgH loci are not characterized in this species, and some of them have germline-joined VHDJH, which makes these results difficult to evaluate. In the nurse shark (Ginglymostoma cirratum), there are no germline-joined VHDJH and the multiple IgH genes can rearrange autonomously (Zhu et al. 2011). Only one or a few IgH genes were found to be completely rearranged in each B cell, leading to only one functional transcript; the other loci remain in germline configuration. Interestingly, most IgH loci were partly rearranged in thymocytes, suggesting that a differential permissive state of chromatin in this region controls the recombination events in B-cell versus T-cell precursors (Malecek et al. 2008).
In fish, mechanisms of allelic exclusion remain largely unknown. The structure of TCR and Ig loci resembles the one found in mammals, rather than the one of Chondrichthyans: TCR beta and alpha/delta as well as IgH loci are organized as translocons while IgL and TCR gamma are made of multiple clusters. An important feature of fish Ig loci is the very large number of VH gene segments found in many species, especially in salmonids (Yasuike et al. 2010). Additionally, as bony fish underwent one or several additional genome-wide duplications (Petit et al. 2004) compared to most tetrapod species, the number of loci has increased. Thus, Salmo salar (Yasuike et al. 2010), Oncorhynchus mykiss (Hansen et al. 2005), Oryzias latipes (Magadán-Mompó et al. 2011), and Gasterosteus aculeatus (Bao et al. 2010; Gambón-Deza et al. 2010) present more than one functional IgH locus per haplotype. Thus, as Chondrichthyans, polyploid Xenopus, and genetically manipulated models with multiple IgH loci such as mice triallelic for IgH, the clonality of the system in bony fish requires not only allelic exclusion but also locus exclusion (Du Pasquier and Hsu 1983; Barreto et al. 2001).
Regarding IgL loci, the genomic organization of IgL was found to harbor clusters of V segments in opposite transcriptional polarity to J and C segments in trout and zebrafish. This configuration implies that primary gene rearrangements would be generated by inversion, which maximizes secondary rearrangements at IgL loci and serves as an important mechanism for receptor editing (Hsu and Criscitiello 2006). The impact of this structure on allelic/locus exclusion is not understood.
Importantly, the presence of a pre-BCR in fish has not been confirmed. In human and in the mouse, the pre-BCR consists of a homodimer of μ heavy chains associated with surrogate light chains (VpreB, homologous to a Vλ, and lambda 5, homologous to a Jλ-Cλ (Mårtensson et al. 2007)) and with the transmembrane signal molecules (Igα and Igβ). To date, homologues of the VpreB and λ5 genes have not been found in teleost genomes. In several fish species including zebrafish (Haire et al. 2000), Atlantic cod (Daggfeldt et al. 1993; Hsu and Criscitiello 2006), and medaka (Magadán-Mompó et al. 2013), IgL transcripts without a V segment have been reported. However, these JCL transcripts may produce functional surrogate JCκ proteins, as reported in humans for germline Vκ and JCκ transcripts encoding proteins that can functionally replace VpreB and λ5 (Francés et al. 1994; Rangel et al. 2005). Alternatively, it would be interesting to study if any of the many teleost light chain subtypes/isotypes could play the role of a surrogate light chain. Regarding the pre-TCR, of which the surrogate chain pTalpha that associates to TR beta has been described only recently in birds (Smelty et al. 2010), nothing is known in fish.
Altogether, an allelic (and interlocus) exclusion seems to be required in fish as in other vertebrates to ensure the clonal responses that have been observed, but the mechanisms remain to be discovered. In fact, a better knowledge of the early stages of differentiation of B and T lymphocytes, especially good molecular markers, would be critical to uncover such mechanisms. It is not even excluded to date that genic conversion might play a role in the diversity generation of the primary Ig repertoire in fish. Another important aspect is the diversity of fishes, as multiple solutions might have been selected in different fish groups to ensure lymphocyte clonality.
3.2 What Does Clonal Complexity of Responses to Pathogens Reveal About Roles of Fish Ig Isotypes and the Structure of T-/B-Cell Responses?
In fish, particularities of immune responses to Ag—for example, in Atlantic cod—might suggest they do not always follow the paradigm of the clonal selection theory. However, a number of features shared between the immune system of fish and mammals (or other tetrapods) advocate analogous sets of lymphocytes and response mechanisms. For example, the typical counterparts of coreceptors (CD4, CD8, CD28, etc.), as well as Th1 and Th17 cytokines, suggest the presence of the similar types of T-cell subsets in fish and mammals. With respect to fish B lymphocytes, cells expressing, respectively, IgT or IgM are specialized in mucosal versus systemic immunity, reminding the roles of IgA and IgM/IgG in mammals, respectively. The analysis of the immune complexity of fish naive repertoires, as well as of responses to pathogens, has just started to decipher to what extent fish and mammalian adaptive immunity may be similar.
Spectratyping of TCR beta CDR3 after a systemic viral infection revealed a strong polyclonal secondary response in the spleen, implicating most of the rainbow trout TCR beta V families (Boudinot et al. 2001, 2002). As observed in humans and in mice, there was a public response consisting of a number of slightly varying CDR3 sequences differing by conservative amino acid substitutions. This was observed in all tested individuals of a trout clone sharing the same genetic background. Also, the same CDR3 protein sequence could be encoded by different nucleotide sequences produced by distinct rearrangements, which was a good evidence of Ag-driven clonal selection. These observations showed that salmonids have an available TCR beta repertoire diverse enough to allow public responses, besides a large diversity of private responses that were observed only in some individuals. After DNA vaccination by intramuscular injection of an expression plasmid for a viral G protein, the complexity of the response dropped, and only the public component could be detected by CDR3 spectratyping (Boudinot et al. 2004). This response was apparently specific for the Ag, as it was never observed after injection of an expression plasmid for another viral protein, the nucleocapsid. These analyses are not highly sensitive as they are performed in the absence of in vitro restimulation because the necessary reagents are not available in the trout model. However, they indicate that DNA vaccination elicits a TCR beta response mainly restricted to an epitope located on the target of the neutralizing Ab response (the viral G protein), which is absolutely essential for the protection in this disease model. This observation evokes the idea of a T-/B-cell cooperation for the anti-G response, although it does not provide a direct evidence for it. With the recent production of MoAbs against CD8, CD4, and potentially other T-cell markers, it would be important to determine if this public response is mediated either by CD8+, presumably cytotoxic, T cells (which have been detected close to the site of DNA vaccine injection (Utke et al. 2008) or by CD4+ helper T cells. Furthermore, it would be interesting to investigate if responses similar to the public component observed in double haploid trout would also be found in genetically mixed farmed or wild trout populations, as suggested by preliminary results (Boudinot et al. 2004).
Such CDR3 spectratyping using V-, C-, and J-specific primers can be used in an “immunoscope” strategy to identify and target particular strong components of the adaptive response or to provide an overview of the degree of oligoclonality of a repertoire. However, it does not allow to describe the whole clonal composition and complexity of a lymphocyte population. Such comprehensive analyses have been made possible by the development of deep sequencing technologies, which provide a complete or quasi complete description of sequence distribution within a V/C or VJ PCR product (Six et al. 2013). These technologies have been used to analyze the B-cell repertoire in two fish species: zebrafish and rainbow trout.
A first seminal study used high-throughput sequencing to perform a global analysis of the μ IgH repertoire in 14 adult zebrafish (Weinstein et al. 2009). This work showed that a large part of the potential combinations of V, D, and J genes are indeed used in the available repertoire, with VDJ frequency patterns often common to different individuals, as observed in mouse or in human. Different methods of the number of different μ IgH chains estimated that 1200–3500 are expressed per individual zebrafish. This large dataset was subjected to several in-depth analyses that revealed additional features of the zebrafish IgH repertoire. Using methods of statistical physics, Mora et al. investigated pairwise correlations between residues in sequences comprising the end of the VH gene segment and the CDR3 region (Mora et al. 2010). Their analysis confirms that each individual fish repertoire covers only a small part of the potential repertoire and that the VDJ combinations expressed are correlated between fishes. In contrast, each fish has its own CDR3 diversity and 13 individuals are not enough to express the whole potential diversity of this region. Overall, these analyses conclude that (1) the antibody diversity is not directly limited by the number of VDJ sequences present in the genome and that (2) about half of the repertoire diversity of μ IgH CDR3 is unique to each individuals, while the other half is shared by many (or all) fishes. Importantly, this represent a large potential for public responses, still keeping each fish able to mount individual specific components of responses to pathogens. Importantly, these proportions (50 % shared/50 % individual) might be different in large species or in fishes exposed to a vast diversity of pathogens in their ecological niche: intuitively, one would expect that the “minimal shared component” selected by the pathogenic/antigenic landscape would represent a smaller proportion of the repertoire of bigger species having much higher numbers of B cells. However, this view is likely oversimplistic, and experimental studies will be necessary to understand the repertoire dynamics and statistical properties in diverse fish species. Interestingly, an independent network analysis of the same dataset identified two distinct groups of fishes: one group with a uniform use of VJ combinations and network of clonotypes and another group with some VJ combinations present at much higher frequencies and highly connected to each other. While these fishes had not been intentionally immunized, the clonal structure of the second group was interpreted as a set of clonotypes participating to an adaptive (pathogen-driven?) response (Ben-Hamo and Efroni 2011).
When studying the μ IgH repertoire of a 2-week-old zebrafish, a high stereotypy, i.e., a preferential use of a small number of VDJ combinations among the potential diversity, was observed (Jiang et al. 2011). Such similar primary μ IgH repertoires expressed in different individuals—which remind observations published in mice at the level of expression of VH families (Huetz et al. 1993)—had apparently disappeared by 1 month in young fishes: many dominant VDJ combinations are not observed anymore at this stage. To differentiate (1) the rate of rearrangement for a given V(D)J combination and (2) its promotion by clonal selection during the formation of the repertoire, the authors distinguished the total number of reads and the number of lineages (identified by the junction sequence) for a given V(D)J. Interestingly, while the number of reads per VDJ was highly correlated between different fishes only at the 2-week time point, the “VDJ lineage diversity” (i.e., the diversity of clonotypes per V(D)J combination) appeared highly stereotyped in older animals and even highly correlated between fishes of different ages.
In rainbow trout, a similar technology was used to investigate the clonal structure of the B-cell response against a fish rhabdovirus, the viral hemorrhagic septicemia virus (VHSV). The response was analyzed in spleen by a combined approach of CDR3 spectratyping and deep sequencing of the IgH transcripts (Castro et al. 2013). Double haploid fish were vaccinated using an attenuated strain of VHSV, and challenged 3 weeks later. In naive animals, bell-shaped CDR3 spectratypes suggested a polyclonal repertoire for the three isotypes, while deep sequencing revealed the presence of a few IgM- and IgT-secreting cells in the spleen. The analysis of the modifications of IgM, IgD, and IgT repertoires after the viral challenge and secondary response revealed complex and highly diverse IgM responses involving all VH subgroups and dominated by a few large public and private clones. No public IgT response was identified, but a fair number of amplified clonotypes were detected, showing that this Ig class specialized in mucosal protection also participates to some degree to the response against a systemic viral infection in the spleen. In contrast, the IgD response to the infection appeared to be negligible in this context.
The clonal complexity of this response and its public IgM component were consistent with the general rules of B-cell response and clonal selection known in mammals, suggesting that general properties were already in place in the common ancestors of fish and tetrapods and were conserved in both lineages. However, the implication of all VH subgroups is intriguing and might be explained partly by polyclonal bystander activation during the secondary responses to an acute infection. It will be important to determine how the primary response is structured and to what extent it is stable over the following months in order to better understand the mechanisms of B-cell memory.
Remarkably, fish have been among the first models in which global repertoire studies were conducted using deep sequencing, and their repertoires are currently among the best described of all vertebrates. With new sequencing techniques coming such as IgL/IgH-coupled sequencing and molecular bar-coded reads, we anticipate that rejuvenated repertoire studies will help in understanding the structure of responses, bases of protection, and mechanisms of memory both in fish and Vertebrates in general.
3.3 Independent Loss of MHC Class II Molecules Occurred in Two Different Evolutionary Branches of Fish; Is T Cell Help Dispensable for Good Ig Response to Pathogens?
In men and mice, B-cell responses to T-dependent (TD) antigens require CD4+ helper T cells to cooperate with B cells expressing an Ag-specific Ig and presenting the antigen. This cooperation leads to the synthesis of Ag-specific Abs and finally to the induction of immune memory. In contrast, T-independent (TI) antigens, which often have repeated structure such as viral capsids, can trigger production of IgM in the absence of T cells.
It has been known for a long time that responses to murine TD and TI antigens required cooperation between different cell types in fish. Using an in vitro PFC test and catfish leukocyte culture models, Miller and Clem (Miller and Clem 1984; Miller et al. 1985) studied hapten-carrier effect and primary responses to both TI (TNP-LPS adsorbed on bentonite particles) and TD (DNP-BSA; TNP10-KLH; TNP10-HorseSA). They could demonstrate that mixing lymphocytes expressing Ig (surf-Ig+) and macrophages allowed responses to a TI antigen, while responses to TD antigens required the presence of surf-Ig+ lymphocytes, macrophages, and surf-Ig− lymphocytes (comprising helper T cells). This remarkable work was the first evidence that fish B cells cooperate with T cells to build responses to TD antigens, as in mammals. Many other indications confirmed this view in other fish species, such as the reduction of the secondary Ab response to human gamma globulin in young trout when fry had been thymectomized at 1 month post hatch (Tatner 1986).
Furthermore, orthologues of CD4 and CD8 coreceptors have been found in fish, and they are specific for T-cell subsets that generally have similar functions to their mammalian counterparts: while the CD8+ population contains cytotoxic cells (Somamoto et al. 2009; Takizawa et al. 2011; Nakanishi et al. 2011), the CD4/CD4rel+ populations appear to be able to help B cells (Toda et al. 2011; Somamoto et al. 2014). Typical polymorphic mhc I and II and the key genes for antigen processing and presentation molecules also have been found in many fish species; the structure and expression pattern of mhc genes suggest that they present the antigen to CD8+ and CD4+ T cells, respectively, as in other vertebrates. Collectively, these observations suggest that the antigen processing, presentation, and B- and T-cell antigen recognition are conserved in jaw vertebrates including fish, setting up a general mode of lymphocyte selection and cooperation during responses.
However, the antibody response of Atlantic cod (Gadus morhua, Gadidae) did not follow this paradigm. Despite extensive efforts to immunize cod using different antigens and protocols, it remained for a long time impossible to induce a typical specific antibody response (reviewed in Pilstrom et al. (2005)). However, the level of serum “natural” antibodies was very high in non-immunized cod when compared to other fish species (Israelsson et al. 1991; Pilström and Petersson 1991; Magnadottir et al. 1999). The search for a genetic explanation in cod Ig sequences and loci was unsuccessful, leading to the idea that a deficiency in MHC class II likely was the reason for the lack of specific antibody response in this species (Pilstrom et al. 2005). This hypothesis was confirmed when the complete genome sequence of cod was deciphered (Star et al. 2011): the mhc II genes were absent, as well as the invariant chain (Ii21) which is involved in the assembly of the MHC II/peptide complex. Also, the gene encoding the coreceptor CD4 was represented only by a truncated pseudogene, indicating that the Atlantic cod had lost the classical pathway of antigen presentation by mhc II to helper T cells.
Lack of mhc II sequence also has been observed in the transcriptome of another gadoid, the burbot (Lota lota) (Star et al. 2011), suggesting that the deficiency is probably shared within the gadoid lineage. Strikingly, the absence of mhc II and cd4 genes and a nonfunctional li21 was reported in a phylogenetically distant species, the pipefish Syngnathus typhle (Syngnathidae) (Table 2). This observation indicates that the loss of the key genes of the MHC II pathway occurred at least twice independently during the evolution of teleosts.
Table 2.
Absence of functional genes from the MHCII pathway in cod and pipefish
| Gene | Cod | Pipefish |
|---|---|---|
| MHCI | + | + |
| MHCIIa | − | − |
| MHCIIb | − | − |
| Invariant chain | − | Nonfunctional |
| CD3e | + | + |
| CIITA | + | − |
| RFX5 | + | + |
| RFX7 | + | + |
| CD4 | Truncated | − |
| CD8a | + | + |
| CD8b | + | − |
These particular situations in cod and pipefish raise several fundamental questions about somatic selection of lymphocytes and the importance of cooperation between T and B cells in the fish defense system:
How mhc II-deficient immune systems, which would lead to a strong decrease of resistance to pathogens in mammals, could be adopted by very successful species as Atlantic cod?
Can such species mount efficient Ig responses without MHC II pathway?
Do they have compensatory mechanisms that make their defense system well adapted to their ecological niche?
Several evolutionary pathways explaining the absence of mhc II genes in cod have been discussed by Star and Jentoft (Star and Jentoft 2012). A first scenario involves genetic drift and hypothesizes that the MHC II pathway is not crucial for defense against parasites. Other scenarios are based on directional selection: for example, the MHC II pathway could have metabolic costs that would not be compensated by its contribution to the protection of the individual against pathogens or other defense mechanisms could have compensated for the deficiency. Such scenarios may be conditioned by the environmental conditions, and it has been proposed that the cold marine environment may have favored the mhc II loss in the cod lineage. However, the mhc II deficiency of the pipefish and the conservation of mhc II in a large number of fishes living in cold sea do not strongly support this hypothesis.
Following Star et al. (2011), it is tempting to link the loss of the mhc II pathway in cod to the particularities of its repertoire of immune genes, especially to the high number of mhc I genes (Persson et al. 1999). Interestingly, the amphibian axolotl has a notoriously poor humoral response that was associated with the limited presentation capacity of its non-polymorphic MHC II molecules (Tournefier et al. 1998); as the Axolotl possesses multiple polymorphic mhc I genes (Sammut et al. 1999), mhc I-based compensatory mechanisms may represent an interesting case of convergence with cod. In fact, a detailed analysis of the large repertoire of cod mhc I sequences (Malmstrøm et al. 2013) identified a particular gene subset with a novel combination of two endosomal sorting motifs in the cytoplasmic tail: a tyrosine-based motif associated with exogenous peptide presentation by cross-presenting MHC I molecules and a dileucine-based motif associated with normal MHC II functionality in mammals. Although these observations evoke MHC II function and a specific adaptation to antigen cross-presentation by MHC I molecules, it does not explain how this mechanisms could actually compensate for the loss of the MHC II pathway. Would this cross-presentation be restricted to cytotoxic T cells (providing Th1-like responses)? Or might specialized MHC I molecules expressed on B cells cross-present extracellular pathogen-derived peptides to CD8+ T cells with TH2-like functions? While initial immunization trials did not elicit specific Ab production in cod, it was recently reported that the administration of emulsions of bacterin antigen in mineral oil leads to robust and specific antibody responses mainly targeting the LPS and A-layer components (Schrøder et al. 2009; Arnesen et al. 2010; Mikkelsen et al. 2011). The mechanism by which such immunizations succeed in eliciting good Ab responses may rely on Ag stabilization in oil deposits or on the induction of local inflammatory and co-stimulatory signals. Hence, in the absence of MHC II presentation and T-cell help, Ab responses to pathogen TI epitopes could be pivotal for the fish defense and may account for the protection afforded by oil-based vaccines against Vibrios in challenges performed 7 weeks post vaccination. In fact, Ab secondary responses to TD, but also to TI antigens, are significantly prolonged as compared to the primary responses in rainbow trout (Ma et al. 2013); such particular features of fish B-cell responses could explain increased protection long after vaccination even if Ab responses are fully thymus independent.
Thus, when a fish species is able to mount efficient anamnestic responses to TI antigens, the loss of TD response due to genomic inactivation of a key gene of the MHC II pathway may not affect significantly the fitness, at least in some ecological niches where pathogens can be controlled by TI responses. Of note, it seems that the fish MHC II pathway is less sophisticated than the one of mammals—as it lacks, for example, the DM chains (Dijkstra et al. 2013)—hence it could be less critical for the overall resistance to pathogens. The remarkable plasticity of fish genomes and the complexity of their chromosome evolution across a vast adaptive radiation certainly increase the probability of independent events of mhc II loss in several fish lineages. Furthermore, the location of mhc I and II genes in distinct regions of the genome, a feature unique to fish, may have facilitated the deletion of mhc II genes without impact on the mhc I region. The other elements of the MHC II pathway could have been lost by genetic drift after the first inactivating event, in the absence of “protective” selection pressure. It is difficult to decide if compensatory mechanisms (e.g., via MHC I-based cross-presentation) represent adaptations posterior to the initial event or a preexisting context favorable to the loss of the MHC II pathway. In this respect, it will be interesting to characterize the diversity of the mhc I genes in the pipefish. Another question is what factor(s) led to the success of individuals deficient in mhc II within their own species or population: lucky neutral evolution or selection of advantageous genomic event(s) parallel to the mhc II inactivation? Many other pathways remain possible.
3.4 Are Fish Ig Sequences Diversified in the Absence of Germinal Centers? New Insights from Deep Sequencing Datasets
The mammalian immune system can respond specifically to antigens and allows Ag-specific memory after a first encounter: the secondary response is faster and based on antibodies with higher affinity compared to the primary response. In mice and humans, this strong increase of affinity is based on the ability of immune system to modify the immunoglobulin diversity engaged in the response through mutation of Ig genes in responding B cells within germinal centers (GC), followed by clonal selection of B cells expressing high-affinity BCRs (Shlomchik and Weisel 2012). In a typical primary TD immune response, Ag-stimulated B cells rapidly move to the T-/B-cell interface of lymphoid tissues where they interact with CD4+ helper T cells and cytokines. T cells then induce B-cell proliferation and differentiation into short-lived antibody-producing plasma cells, or B-cell translocation to the germinal centers where the variable region of rearranged Ig genes may undergo somatic hypermutation. B cells finally differentiate into long-lived plasma cells or memory B cells (Zielinski et al. 2011).
A number of studies showed that in fishes as in mammals, the B-cell repertoire diversity is not only due to V(D)J recombination in adult individuals but also based on the hypermutation of the variable region. In fact, fishes express the enzyme activation-induced cytidine-deaminase (AID), which is responsible for Ig somatic hypermutation and class switch recombination in mammals and other tetrapods (Barreto et al. 2005). Although fishes lack conventional class switch (Stavnezer and Amemiya 2004), fish AID has similar biochemical functions and deaminates cytosine, thus inducing point mutations; it even mediates class switch in mouse B cells (Barreto et al. 2005). The mutational pattern observed in Ig sequences from catfish (Yang et al. 2006), zebrafish (Marianes et al. 2011), and also in nurse shark (Diaz et al. 1999) reminds actually the one described in mammals, with a preference for transitions but no major bias for mutation of G:C or A:T pairs (Diaz et al. 2001).
However, the capacity to perform somatic hypermutation in Ig genes does not imply that efficient antigen-driven selection and affinity maturation of immune response occur. It is generally accepted that the affinity maturation in fishes and other ectotherms is much less efficient than in mammals (Wilson et al. 1992; Yang et al. 2006), based on two different types of data: (1) measurements of the affinity of Ag-specific serum antibodies using analytical tools and (2) computing of the dS/dN ratio of synonymous (S) and non-synonymous (N) mutations in variable regions of immunoglobulin genes.
The classical analytical tools, such as equilibrium dialysis or fluorescence quenching, only provide an estimation of the average affinity for all Ag-specific antibodies within a serum sample, which is moderately sensitive. The use of new analytical techniques, as solid-phase ELISA (Shapiro et al. 1996), has allowed the fractionation of an Ab response into affinity subpopulations (Kaattari et al. 2002) and more sensitive analyses. Such experiments revealed a consistent increase of affinity for the Ag during the antibody immune response in teleosts (Ye et al. 2011): after immunization of rainbow trout with the TNP-KLH in Freund’s complete adjuvant, a significant proportion of the lower affinity subpopulations appeared during the first 5 weeks and decline after a few months, giving way to higher affinity subpopulations with an increase of affinity reaching 100-fold, which is very low but reminds that observed in mammals in secondary responses.
A typical pattern of synonymous (S) and non-synonymous (N) mutations within Ig variable region is observed after affinity maturation, with low dS/dN ratio specifically in CDRs. In fish, very few works have addressed precisely this process. Somatic mutation was detected within VH regions of channel catfish Ig sequences. However, dS/dN was not significantly different in FR and CDR (Yang et al. 2006), suggesting that the process potentially induced a repertoire diversification that was not followed by efficient clonal selection of high-affinity B cells. In contrast, the distribution of mutations focused on CDRs in immunoglobulin kappa-like light chain in medaka suggested selection and affinity maturation; additionally, the dS/dN ratio was consistent with an antigen-driven selection in 5 out of 13 mutated sequences (Magadán-Mompó et al. 2013). In zebrafish, the expansion of VJC clonal lineages with an increased frequency of mutation might also suggest selection of B cells expressing hypermutated Ig (Marianes et al. 2011).
A comprehensive assessment of the hypermutation changes of Ig sequences has become theoretically feasible with massive parallel sequencing methods (Weinstein et al. 2009). As mentioned above, simultaneous sequencing of Ig transcripts from millions of B cells recently gave access to a whole-organism IgH repertoire of small organisms such as zebrafish (Jiang et al. 2011). To measure the effect of hypermutation and clonal selection during zebrafish development, junctional diversity and VH sequence mutations were compared as a function of sequence abundance among different age groups of fish. Importantly, it appeared that mutated sequences dominate highly expressed VDJ combinations in older fish, but not in young animals, which was consistent with an accumulation of B-cell clones expressing hypermutated IgH with age.
In these studies, fishes were naturally (and progressively) immunized (i.e., not intentionally), and further studies using targeted immunizations or vaccines will have to address directly the question of affinity maturation: isolation and Ig gene analysis of activated Ag-specific B cells will have to be done and the Ig affinity evaluated as in the work of Dooley et al. in nurse shark new antigen receptor (IgNAR) (Dooley et al. 2006). In nurse shark hyperimmunized with hen egg white lysozyme (HEL), HEL-binding clones expressed IgNAR with hypermutated V regions, and an increase in affinity was confirmed. Admittedly, the presence of light chains in teleostean immunoglobulins complicates such analyses. However, advances in high-throughput sequencing of VH:VL pairs (DeKosky et al. 2013; Tan et al. 2014) likely will allow a comprehensive analysis of VH:VL gene usage and somatic hypermutation in FACS-sorted Ag-specific B-cell populations. Such methods will certainly be more powerful than the current complex and expensive methods based on single sorting and RT-PCR (Tanaka et al. 2010).
It is still generally believed that the reason for the poor affinity maturation of Ab responses in ectothermic vertebrates is an inefficient selection of high-affinity clones. This is consistent with the absence of true GC in these organisms, although lymphotoxin-β KO mice succeed to perform good affinity maturation of Ab responses in the absence of GC (Matsumoto et al. 1996). The affinity maturation pathways have not been characterized in these mice, but other studies have identified hypermutation and/or selection outside of conventional microenvironments (Schröder et al. 1996). On the other hand, cell clusters were observed in the spleen and kidney of channel catfish and goldfish, which were put forward as putative primordial GC (Saunders et al. 2010; Barreto and Magor 2011). These cell clusters contain melanomacrophages that express CSFR1, B cells, and CD4+ T cells, as well as AID+ cells. In the model proposed, CD4+ cells would play a similar role as their counterparts in mammals, providing survival and differentiation signals to B cells; melanomacrophages would act as follicular dendritic cells (FDCs) in the GC of mammals, trapping and presenting Ag or Ag-antibody complexes on their surface. Thus, Ag-specific B cells with hypermutated Ig genes would be selected on their capacity to bind the (limiting) antigen on melanomacrophages.
Another question is the contribution of long-lived plasma cells and true memory B cells to the maintenance of an effective humoral adaptive immunity in teleosts. Results obtained in several models indicate that serum Ag-specific antibodies are not detectable 1–2 years after exposition and that Ag-specific plasma cells are not any more visualized by ELISPOT in head kidney if no challenge is performed. This has been observed in channel catfish infected with the ciliate Ichthyophthirius multifiliis (Findly et al. 2013) and in rainbow trout immunized with an expression plasmid for the G of IHNV (Kurath et al. 2006) or with Streptococcus iniae bacterin (Costa et al. 2012). Taken together, these data may suggest that IgM+ memory B cells, not long-lived plasma cells, likely are responsible for the long-term protective immunity in teleosts. In fact, a population of true memory B cell remains to be identified and characterized in fish.
Memory and long-term maintenance of Ag-specific B-cell immunity in species where mhc II and/or cd4 genes have been lost, such as Atlantic cod or pipefish, is another interesting issue. Cod-specific Ab responses seem to be mainly directed to prototypic TI antigen as LPS and A-layer components (Espelid et al. 1991; Gudmundsdóttir et al. 2009). In mice and humans, such antigens that bypass T-cell help indeed induce specific memory B cells, whose secondary activation is inhibited by the presence of Ag-specific antibodies (Brodeur and Wortis 1980; Obukhanych and Nussenzweig 2006). The quality and life span of these cellular subsets likely depend on the strength of BCR-derived signals and complementary effects of toll-like receptor (TLR) stimulation and inflammatory cytokines (Alugupalli et al. 2007; Defrance et al. 2011). Their existence in cod and pipefish remains unproven.
A better understanding of mechanisms involved in induction and maintenance of long-lived and memory B cells in teleosts will be critical to develop efficient vaccines affording a durable protective immunity, especially against pathogens that evolve subversion strategies. In that sense, there is still a very long way to go.
3.5 What Are the Particular Features of the Mucosal Repertoires in Fish?
Fishes have the most extensive and complex mucosal surfaces of interaction with environment among vertebrates: not only gut and respiratory mucosa (gills) but also all the skin surface is a bona fide mucosa. Even in the best studied models, the immunity of these tissues remains poorly understood, although significant advances have recently been made in the understanding of mucosal B-cell responses in teleosts.
3.5.1 Fishes Possess a Mucosal Specialized Ig Isotype, But IgM (and IgD?) May Also Participate in the Protection of Mucosa
In 2005 a new teleost fish immunoglobulin (Ig) isotype was identified. In rainbow trout this Ig was named IgT (Hansen et al. 2005), whereas in zebrafish it was called IgZ (Danilova et al. 2005). This Ig has been identified in all analyzed teleost fish, except in medaka and in channel catfish. However, the complete genome of catfish is not available thus far, and it remains possible that the catfish orthologue for IgT has not yet been found. Rainbow trout IgT and IgM use the same VH segments, which rearrange to either to Dτ or Dμ segments. Analogous to the structure of the TCRα/δ locus, Igτ Dτ, Jτ, Cτ genes are located downstream of the VH and upstream of the IgM Dμ, Jμ, Cμ (Hansen et al. 2005), suggesting that VH–DμJμ precludes further VHDτJτ rearrangement. Combined with the lack of obvious switch regions in the IgT locus, commitment to IgT or IgM appears to involve alternative V(D)J recombination rather than isotype switching.
It was not until 2010 that IgT was biochemically characterized and its function in mucosal immunity revealed (Zhang et al. 2010). It was shown that while plasma IgT is a monomeric Ig (~180 kDa), gut mucus IgT is polymeric (4–5 monomers). Significantly a previously unknown IgT+ B-cell lineage was identified. This B-cell subset expresses surface IgT, but not IgM or IgD, thus constituting the first vertebrate B-cell lineage devoid of surface IgD expression. IgT+ B cells represented the predominant B-cell subset in the gut. Moreover, plasma-like cells uniquely producing IgT could also be detected. The same study identified a polymeric Ig receptor in rainbow trout (tpIgR) whose putative secretory component (sC) was found associated with gut mucus but not serum IgT and IgM. This result strongly suggested that like in mammals, pIgR in fish is involved in the transport of polymeric IgT and IgM from the mucosal epithelium into the gut lumen. The IgT/IgM ratio was found much higher in the gut mucus when compared to that in serum, thus suggesting that IgT could play a role in gut mucosal immunity. Confirming this hypothesis it was shown that trout infected with Ceratomyxa shasta (a gut parasite) induced parasite-specific IgT responses only in the gut mucus, but not in serum. Conversely, the serum of surviving fish contained significant parasite-specific IgM, but not IgT titers. Moreover, fish that survived parasite infection contained large accumulations of IgT+ B cells in the gut, whereas the number of IgM+ B cells did not change with respect to control fish. Overall, this model provided the first evidence in teleost fish or any other non-tetrapod species for a compartmentalization of immunoglobulin isotypes into mucosal (IgT) and systemic (IgM) areas in response to pathogenic challenge. Supporting further the idea that IgT is involved in mucosal homeostasis, it was found that IgT, similar to mammalian IgA, was the prevalent Ig found coating the gut microbiota. This finding represented the first example of coating of microbiota by a nonmammalian mucosal Ig and pointed to a conserved role of mucosal Igs in the control of the bacterial microbiota at mucosal surfaces. Moreover, this finding provided further evidence for the specialization of IgT in mucosal homeostasis. More recently, very similar responses were reported in the skin mucosa of rainbow trout (Xu et al. 2013). Similar to what was found in the gut mucus, IgT in the skin mucus was mainly detected in polymeric form. In addition, IgT+ B cells also constituted the main B-cell subset of the trout skin-associated lymphoid tissue (SALT). Fish that survived infection with a skin pathogen (Ichthyophthirius multifiliis) showed large accumulations of IgT+, but not IgM+ B cells, in the SALT. Along with these substantial increases in IgT+ B cells, large increases of IgT protein (~10-fold) mucus were detected, while IgM levels remained unchanged. Moreover, survivor fish had significant titers of parasite-specific IgM in the serum, but not in the mucus of most fish. Conversely, significant titers of parasite-specific IgT were detected in the skin mucus but not in serum. Supporting further the prevalent role of IgT in the SALT, it was found that like in the gut, a majority of SALT microbiota were coated with IgT. Essentially very similar IgT responses to those described in the trout GALT and SALT were also found in the trout gill-associated lymphoid tissue (Dr. Sunyer, personal communication). Moreover, a mucosal-associated lymphoid tissue has recently been described in the nose of rainbow trout (NALT) (Tacchi et al. 2014). In this tissue, IgT+ B cells are the predominant B-cell subset, thus suggesting also that IgT plays a major role in NALT immunity. Overall, these findings support further the initial discovery in the gut, indicating that IgT is an immunoglobulin specialized in mucosal immunity. Thus, it seems reasonable to suggest that IgT responses in all main fish mucosal areas operate under the guidance of primordial common principles. While it is likely that IgT is the major responder in mucosal surfaces, IgM responses have also been detected in the gut and skin mucosa upon vaccination or infection (Salinas et al. 2011). Whether fish IgD plays a role in systemic or mucosal immunity remains unknown, although IgD responses were not detected in the gill of rainbow trout upon infection with I. multifiliis (Dr. Sunyer, personal communication), nor in the spleen of trout infected upon systemic viral infection (Castro et al. 2013).
Future repertoire studies of mucosal B-cell populations in healthy or infected fish will provide more insights about the clonal structure of B-cell response in these tissues. It is tempting to speculate that IgT+ B cells, like IgA+ B cells in mammals, could have a particular repertoire with a few very large clones amplified in mucosal tissues (Stoel et al. 2005). The compartmentalization of IgT+ B-cell repertoires in different mucosae of the individual will also be an important issue to clarify for a better understanding of mucosal fish defenses.
Indeed, the absence of switch in fish likely introduces important differences in the selection constraints exerted on mucosal B cells between fish and mammals: IgT+ B cells constitute a separate lineage of B cells and express IgT from their early differentiation stages, while mammalian B cells are all first selected on the basis of a membrane-bound IgM. One might imagine that a fully independent differentiation pathway of mucosal B cells in fish might bring more freedom to select specialized resident B-cell subsets for each mucosal territory.
3.5.2 Fish Anatomical Structures Represent Particular Microenvironments for B and T Cells, which Constraint Available Repertoires and Initiation of Adaptive Responses
It is generally accepted from observations made in men and mice that the adaptive immune response is initiated in special microenvironments within secondary lymphoid organs. This view is challenged by the anatomical differences between fishes and mammals: while fishes are able to mount protective immune responses, they lack lymph nodes and germinal centers. In the model proposed by Kaattari and colleagues, B-cell differentiation occurs in the fish kidney (mainly in the anterior kidney), from which mature B lymphocytes migrate to the blood and immune tissues including spleen and likely mucosal territories. It is generally believed that upon encounter with the Ag, B cells then differentiate into plasmablasts, proliferate, and migrate to the kidney where they become (long-lived) plasma cells. It is not clear where the interactions between B cells and T cells occur. Interestingly, a number of observations realized in mammals support the notion that T cells are able to respond to the Ag outside of a lymph node environment (Hofmann et al. 2010); for example, LTα−/− mice are devoided of lymph nodes but have a vigorous cell-mediated immunity (De Togni et al. 1994). In fish, adaptive response and T-/B-cell cooperation likely take place in the major secondary peripheral lymphoid organ, the spleen; although not clearly regionalized, this organ contains ellipsoidal blood vessels with a layer of macrophages specialized in antigen trapping as macrophages of the marginal zone in mammals and melanomacrophage aggregate.
The mechanisms of the initiation of adaptive responses in fish mucosa remain even more elusive. Like all jaw vertebrates, fish possess lymphoid tissues associated to mucosa. Macrophages, B and T lymphocytes as well as granulocytes are found in the gut and constitute the GALT, which is not made of encapsulated structures as Peyer’s patches (Rombout et al. 2011). Specialized structures are also present in gills, with aggregations of lymphocytes including a large proportion of T cells in the interbranchial lymphoid tissue (ILT) (Haugarvoll et al. 2008; Koppang et al. 2010). Moreover, fish skin constitutes a true mucosa as it is not keratinized and is coated by mucus (Xu et al. 2013).
The absence of lymph nodes and delimitated structures—especially in the skin—rises the issue of the selection of a relevant available local repertoire, well adapted to and actually shaped by the interactions with the microbiota present at mucosal surfaces. Such a repertoire would ideally reflect the selection of lymphocyte populations by the microbe populations (which can be more or less stable depending on the tissue and on the environment) and would express a diversity of receptors sufficient locally to mount efficient responses to a large set of potential pathogens. Thus, the structure of the B- and T-cell repertoires in naive and infected animals would provide many insights into the spatial dynamics of adaptive responses in the absence of integrated secondary structures as lymph nodes.
Although IgM+ and IgT+ B cells are present in fish gut, gills, and skin, the corresponding repertoires remain almost unknown. In mammals, CDR3 spectratyping analysis showed that the gut IgA (and IgM) repertoire contained highly expanded peaks suggesting oligoclonal proliferations (Holtmeier et al. 2000; Stoel et al. 2005). However, a recent study using deep sequencing found that the gut IgA available repertoire comprises a large diversity of low-frequency clones in addition to the highly expanded ones (Lindner et al. 2012). In fact, this likely reflects an equilibrium between the impact of gut bacteria on the B-cell repertoire and the influence of secreted Ig on the composition of the microbiota (Wei et al. 2011). It will be interesting to determine if a similar available repertoire is found in the gut of naive fish, with a limited number of highly expanded clones expressing IgM or IgT. It is interesting to note that in mice the selection of the IgA repertoire depends on T cells, microbiota, and RORgt but not on Peyer’s patches; as fishes possess a true RORgt orthologue, IgT repertoire might be determined by a similar pathway (Lindner et al. 2012).
While it remains unknown if such a structure of B-cell repertoire is shared by fish and mouse or human, CDR3 spectratyping of TCRβ in rainbow trout has revealed important differences in T-cell diversity. In men and mice, early studies of intraepithelial T lymphocytes (IEL) found oligoclonal repertoires of dominant clones in adult individuals (Gross et al. 1994; Regnault et al. 1994). In contrast, youngster IEL repertoires were highly diverse and polyclonal, with bell-shaped CDR3 length distributions in mammals and birds (Dunon et al. 1994; Williams et al. 2004). In contrast, rainbow trout IEL repertoire did not show restricted diversity in the young adult, suggesting very different selective constraints in fish compared to chicken, rodent, and human (Bernard et al. 2006). However, more restricted repertoires were found in older fish from farms, suggesting that IEL diversity might be reduced—or at least that large clones might be positively selected—in older fishes living in natural conditions (data not shown). While it is increasingly clear that commensal bacteria of the gut microbiota play a critical role in the peripheral selection of T-cell subsets such as Treg (Lathrop et al. 2011), it must be noted that these observations in trout integrated all TCRβ-expressing T cells, which might have masked even strong effects of selection on minor cell subsets. More detailed studies on sorted T-cell populations at different ages would be necessary to clarify the mechanisms of IEL selection in fish and differences in men and mice.
Mucosal responses also are particular. As previously stated, large accumulation of IgT+ (but not IgM+) B cells was found in the gut of rainbow trout surviving infection with the parasite Ceratomyxa shasta (myxosporidian) and in skin epidermis of fish infected by the ciliated Ichthyophthirius (Zhang et al. 2010; Xu et al. 2013). As fish B cells express either IgM or IgT, these observations suggest that mucosal adaptive response triggers activation and proliferation of specialized B cells expressing the mucosal isotype, while IgM+ B cells responding to the pathogens would migrate and lead to a systemic response with circulating secreted Ab. The relative importance of local proliferation at the infection site versus infiltration and concentration of Ag-specific IgT+ B cells from other territories remains unknown. However, it seems unlikely that the IgT+ B cells locally present in the skin would be diverse enough to match any possible pathogen; hence, IgT+ B cells likely would have to gather at inflammatory sites and the Ag-specific cells sampled and locally amplified. In interbranchial lymphoid tissue (ILT), a transcriptome analysis found a small delayed increase in IgT transcripts after infection with infectious salmon anemia virus by immersion, suggesting an expansion of IgT-expressing B cells (Austbø et al. 2014).
Regarding mucosal T cells, comparison of repertoires of naive and VHSV-infected trout showed that IEL were responsive to the infection; the most significant alterations of CDR3 length profiles were found for the same Vβ segments in the gut and in the spleen. Amplified clonotypes were also found in both tissues of infected fish, suggesting that T-cell clonotypes selected by a systemic viral infection were shared between gut mucosa and other peripheral (non-mucosal) lymphoid tissues. However, repertoire analysis of the response to the bacterium Yersinia ruckeri administered per anal indicated that gut and spleen contained different compartments at least during the first phase of the response (unpublished data). A first infection was performed using a vaccinal strain and was followed by a second infection with a virulent strain 3 weeks later; the TCRβ repertoire was characterized 15 and 35 days after the second infection. Bacterium was not detected in the gut, but modifications of the IEL TCRβ repertoire were already very clear, while in contrast, spleen and head kidney repertoires were not significantly altered. Three weeks later, the TCRβ repertoire has been drastically modified in both tissues: the CDR3 length profiles of IEL are overall normalized (bell shaped as in naive animals), while those in spleen were highly biased. These observations showed that there is at least some level of compartmentalization between spleen and gut T-cell populations in rainbow trout, regarding responses to local infections. However, as for B-cell responses, the respective contributions of local T cells expanded in the mucosa versus T cells recruited from other tissues (mucosal or not) remain unclear.
Understanding the impact of the environment and local microbiota on the lymphocyte repertoires of fish mucosa is critical to improve natural resistance and to validate better vaccines against many pathogens that are notoriously difficult to fight; also, it provides an interesting model for comparative study with human mucosal immunity and may identify novel general mechanisms.
4 Conclusions
Somatic diversification of fish Ag receptors generally reflects the unity of RAG-based immunity across vertebrates, with shared recombination mechanisms and common patterns of clonal selection. However, unique configurations of adaptive immune system have been discovered in fish, such as the lack of mhc II and CD4+ T cells in cod and pipefish, which questions the flexibility of the regulation of adaptive immunity.
The fish-specific Ig isotypes reflect the evolutionary plasticity of Ig classes across vertebrates. Interestingly, fish IgT, amphibian IgX, and mammalian IgA appear as convergent-specific adaptations to protect mucosa. In contrast, the four TcR types (α, β, γ, δ) are ubiquitously conserved from Chondrichthyans to mammals. This is certainly linked to the locked structure of the MHC/Ag/TcR tri-complex; however, this may not fully explain the conservation of TcR classes, especially for TcR γδ. A better understanding of fish TcR γδ functions might shed light on these aspects.
With very different lymphoid anatomical structures and microenvironments, fish and mammals provide a very good subject for comparative approaches to distinguish fundamental conserved properties of B or T cells and convergent adaptations driven by the necessity to fight pathogens in critical tissues or at entry points. Thus, the similarity of fish pronephros and mammalian bone marrow is striking as they both host B-cell differentiation and constitute the survival niche for memory B cells. In the same line, the lack of lymph nodes in fish raises issues about the sites of encounters between lymphocytes and antigens and about modalities of T-/B-cell cooperation.
Beyond the interest of the particularities of fish adaptive immunity, recent studies of fish immune repertoires have reactivated the debate on determinism and contingency in the generation of the expressed Ag-receptor diversity. One easily conceives that the unique immunological history of each individual would allege that contingency plays a key role in selecting somatically diversified immune repertoires. In keeping with this, the high complexity of the regulation of VDJ recombination (epigenetic status of VDJ segments, RSS efficiency, etc.) has impeded a full understanding of the establishment of primary repertoires. There are clear biases in the recombination of given VDJ segments, partly due to the RSS sequences, but the choice of recombining segments appears intuitively largely contingent. Interestingly, however, recent studies of zebrafish primary repertoire provided evidences in favor of determinism in this respect (Jiang et al. 2011). In fact, it remains difficult to evaluate the respective contributions of determinism and contingency in building immune repertoires; quoting Darwin: “I have hitherto sometimes spoken as if the variations so common and multiform in organic beings {…} had been due to chance. This, of course, is a wholly incorrect expression, but it serves to acknowledge plainly our ignorance of the cause of each particular variation.”1 Comparative studies will certainly provide important new insights about the importance of determinism in somatic adaptation of immune repertoires in the future: to this respect, the large diversity of fishes provides a rich resource to assess the impact of size, temperature regulation, and other parameters. This issue has even direct and important practical implications in aquaculture, as the determined arm of repertoire composition is accessible to genetic selection, and the contingent arm due to the immunological history of the fish is the target of vaccinations.
Acknowledgments
This article is dedicated to the memory of Steve Kaattari, who pioneered the study of fish B cells and will be missed by fish immunologists after passing away in November 2014.
This work was supported by Institut National de la Recherche Agronomique, by the European Commission under the Work Programme 2012 of the 7th Framework Programme for Research and Technological Development of the European Union (Grant Agreement 311993 TARGETFISH), and by the National Institutes of Health Grant R01GM085207 (to J.O.S.). We acknowledge S. Fillatreau, T. Mora, A. Six, and Dr. G. Wiegertjes for helpful discussions.
Footnotes
The origin of the species, Chap. 5, Laws of Variation
Contributor Information
Susana Magadan, Email: Susana.Magadan@jouy.inra.fr, Virologie et Immunologie Moléculaires, Institut National de la Recherche Agronomique, Jouy-en-Josas, France.
Oriol J. Sunyer, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA, USA
Pierre Boudinot, Email: Pierre.Boudinot@jouy.inra.fr, Virologie et Immunologie Moléculaires, Institut National de la Recherche Agronomique, Jouy-en-Josas, France.
References
- Alugupalli KR, Akira S, Lien E, Leong JM. MyD88- and Bruton’s tyrosine kinase-mediated signals are essential for T cell-independent pathogen-specific IgM responses. J Immunol. 2007;178:3740–3749. doi: 10.4049/jimmunol.178.6.3740. [DOI] [PubMed] [Google Scholar]
- Arnesen KR, Mikkelsen H, Schrøder MB, Lund V. Impact of reattaching various Aeromonas salmonicida A-layer proteins on vaccine efficacy in Atlantic cod (Gadus morhua) Vaccine. 2010;28:4703–4708. doi: 10.1016/j.vaccine.2010.04.100. [DOI] [PubMed] [Google Scholar]
- Austbø L, Bergva Aas I, König M, et al. Transcriptional response of immune genes in gills and the interbranchial lymphoid tissue of Atlantic salmon challenged with infectious salmon anaemia virus. Dev Comp Immunol. 2014;45:107–114. doi: 10.1016/j.dci.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Bao Y, Wang T, Guo Y, et al. The immunoglobulin gene loci in the teleost Gasterosteus aculeatus. Fish Shellfish Immunol. 2010;28:40–48. doi: 10.1016/j.fsi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Barreto VM, Magor BG. Activation-induced cytidine deaminase structure and functions: a species comparative view. Dev Comp Immunol. 2011;35:991–1007. doi: 10.1016/j.dci.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Barreto V, Meo T, Cumano A. Mice triallelic for the Ig heavy chain locus: implications for VHDJH recombination. J Immunol. 2001;166:5638–5645. doi: 10.4049/jimmunol.166.9.5638. [DOI] [PubMed] [Google Scholar]
- Barreto VM, Pan-Hammarstrom Q, Zhao Y, et al. AID from bony fish catalyses call switch recombination. J Exp Med. 2005;202:733. doi: 10.1084/jem.20051378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hamo R, Efroni S. The whole-organism heavy chain B cell repertoire from Zebrafish self-organizes into distinct network features. BMC Syst Biol. 2011;5:27. doi: 10.1186/1752-0509-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Six A, Rigottier-Gois L, et al. Phenotypic and functional similarity of gut intraepithelial and systemic T cells in a teleost fish. J Immunol. 2006;176:3942–3949. doi: 10.4049/jimmunol.176.7.3942. [DOI] [PubMed] [Google Scholar]
- Boudinot P, Boubekeur S, Benmansour A. Rhabdovirus infection induces public and private T cell responses in teleost fish. J Immunol. 2001;167:6202–6209. doi: 10.4049/jimmunol.167.11.6202. [DOI] [PubMed] [Google Scholar]
- Boudinot P, Boubekeur S, Benmansour A. Primary structure and complementarity-determining region (CDR) 3 spectratyping of rainbow trout TCRbeta transcripts identify ten Vbeta families with Vbeta6 displaying unusual CDR2 and differently spliced forms. J Immunol. 2002;169:6244–6252. doi: 10.4049/jimmunol.169.11.6244. [DOI] [PubMed] [Google Scholar]
- Boudinot P, Bernard D, Boubekeur S, et al. The glycoprotein of a fish rhabdovirus profiles the virus-specific T-cell repertoire in rainbow trout. J Gen Virol. 2004;85:3099–3108. doi: 10.1099/vir.0.80135-0. [DOI] [PubMed] [Google Scholar]
- Brodeur PH, Wortis HH. Regulation of thymus-independent responses: unresponsiveness to a second challenge of TNP-Ficoll is mediated by hapten-specific antibodies. J Immunol. 1980;125:1499–1505. [PubMed] [Google Scholar]
- Burnet FM. A modification of Jerne’s theory of antibody production using the concept of clonal selection. Aust J Sci. 1957;20:67–69. doi: 10.3322/canjclin.26.2.119. [DOI] [PubMed] [Google Scholar]
- Castro R, Jouneau L, Pham H-P, et al. Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013;9:e1003098. doi: 10.1371/journal.ppat.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. The wisdom of hindsight. Annu Rev Immunol. 1994;12:1–62. doi: 10.1146/annurev.iy.12.040194.000245. [DOI] [PubMed] [Google Scholar]
- Costa G, Danz H, Kataria P, Bromage E. A holistic view of the dynamisms of teleost IgM: a case study of Streptococcus iniae vaccinated rainbow trout (Oncorhynchus mykiss) Dev Comp Immunol. 2012;36:298–305. doi: 10.1016/j.dci.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Daggfeldt A, Bengtén E, Pilström L. A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics. 1993;38:199–209. doi: 10.1007/BF00211520. [DOI] [PubMed] [Google Scholar]
- Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- Das S, Hirano M, Tako R, et al. Evolutionary genomics of immunoglobulin-encoding Loci in vertebrates. Curr Genomics. 2012;13:95–102. doi: 10.2174/138920212799860652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Togni P, Goellner J, Ruddle NH, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 1994;264:703–707. doi: 10.1126/science.8171322. [DOI] [PubMed] [Google Scholar]
- Defrance T, Taillardet M, Genestier L. T cell-independent B cell memory. Curr Opin Immunol. 2011;23:330–336. doi: 10.1016/j.coi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Ippolito GC, Deschner RP, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Velez J, Singh M, et al. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- Diaz M, Flajnik MF, Klinman N. Evolution and the molecular basis of somatic hypermutation of antigen receptor genes. Philos Trans R Soc Lond B Biol Sci. 2001;356:67–72. doi: 10.1098/rstb.2000.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Grimholt U, Leong J, et al. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol Biol. 2013;13:260. doi: 10.1186/1471-2148-13-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H, Stanfield RL, Brady RA, Flajnik MF. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci USA. 2006;103:1846–1851. doi: 10.1073/pnas.0508341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier L, Hsu E. Immunoglobulin expression in diploid and polyploid interspecies hybrid of Xenopus: evidence for allelic exclusion. Eur J Immunol. 1983;13:585–590. doi: 10.1002/eji.1830130714. [DOI] [PubMed] [Google Scholar]
- Dunon D, Schwager J, Dangy JP, et al. T cell migration during development: homing is not related to TCR V beta 1 repertoire selection. EMBO J. 1994;13:808–815. doi: 10.1002/j.1460-2075.1994.tb06323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eason DD, Litman RT, Luer CA, et al. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- Espelid S, Rødseth O, Jørgensen T, et al. Vaccination experiments and studies of the humoral immune responses in cod, Gadus morhua L., to four strains of monoclonal-defined Vibrio anguillarum. J Fish Dis. 1991;14:185–197. [Google Scholar]
- Findly RC, Zhao X, Noe J, et al. B cell memory following infection and challenge of channel catfish with Ichthyophthirius multifiliis. Dev Comp Immunol. 2013;39:302–311. doi: 10.1016/j.dci.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Flajnik M, Du Pasquier L. Evolution of the immune system. In: Paul W, editor. Fundamental Immunology. 7th. New York: Wolters Kluwer & Lippincott Williams & Wilkins; 2013. pp. 67–128. [Google Scholar]
- Francés V, Pandrau-Garcia D, Guret C, et al. A surrogate 15 kDa JC kappa protein is expressed in combination with mu heavy chain by human B cell precursors. EMBO J. 1994;13:5937–5943. doi: 10.1002/j.1460-2075.1994.tb06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambón-Deza F, Sánchez-Espinel C, Magadán-Mompó S. Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev Comp Immunol. 2010;34:114–122. doi: 10.1016/j.dci.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Gross GG, Schwartz VL, Stevens C, et al. Distribution of dominant T cell receptor beta chains in human intestinal mucosa. J Exp Med. 1994;180:1337–1344. doi: 10.1084/jem.180.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdóttir S, Magnadóttir B, Björnsdóttir B, et al. Specific and natural antibody response of cod juveniles vaccinated against Vibrio anguillarum. Fish Shellfish Immunol. 2009;26:619–624. doi: 10.1016/j.fsi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Haire RN, Rast JP, Litman RT, Litman GW. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogenetics. 2000;51:915–923. doi: 10.1007/s002510000229. [DOI] [PubMed] [Google Scholar]
- Hansen J, Landis E, Phillips R. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugarvoll E, Bjerkås I, Nowak BF, et al. Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J Anat. 2008;213(2):202–209. doi: 10.1111/j.1469-7580.2008.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman G, Collette B, Facey D, Bowen B. The diversity of fishes. 2nd. Hoboken: Wiley Blackwell; 2010. [Google Scholar]
- Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185:1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–157. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- Hofmann J, Greter M, Du Pasquier L, Becher B. B-cells need a proper house, whereas T-cells are happy in a cave: the dependence of lymphocytes on secondary lymphoid tissues during evolution. Trends Immunol. 2010;31:144–153. doi: 10.1016/j.it.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Holtmeier W, Hennemann A, Caspary WF. IgA and IgM V(H) repertoires in human colon: evidence for clonally expanded B cells that are widely disseminated. Gastroenterology. 2000;119:1253–1266. doi: 10.1053/gast.2000.20219. [DOI] [PubMed] [Google Scholar]
- Hsu E. V(D)J recombination: of mice and sharks. Adv Exp Med Biol. 2009;650:166–179. doi: 10.1007/978-1-4419-0296-2_14. [DOI] [PubMed] [Google Scholar]
- Hsu E, Criscitiello MF. Diverse Immunoglobulin Light Chain Organizations in Fish Retain Potential to Revise B Cell Receptor Specificities. J Immunol. 2006;177(4):2452–2462. doi: 10.4049/jimmunol.177.4.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E, Pulham N, Rumfelt LL, Flajnik MF. The plasticity of immunoglobulin gene systems in evolution. Immunol Rev. 2006;210:8–26. doi: 10.1111/j.0105-2896.2006.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huetz F, Carlsson L, Tornberg UC, Holmberg D. V-region directed selection in differentiating B lymphocytes. EMBO J. 1993;12:1819–1826. doi: 10.1002/j.1460-2075.1993.tb05830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson O, Petersson A, Bengtén E, et al. Immunoglobulin concentration in Atlantic cod, Gadus morhua L., serum and -reactivity between anti-cod-antibodies and immunoglobulins from other species. J Fish Biol. 1991;39:265–278. [Google Scholar]
- Jerne NK. The natural selection theory of antibody formation. Proc Natl Acad Sci USA. 1955;41:849. doi: 10.1073/pnas.41.11.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerne N. Proceedings of CIBA Foundation Symposium, CIBA. Amsterdam; 1971. What precedes clonal selection? Ontogeny of acquired immunity; pp. 1–15. [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Weinstein JA, Penland L, et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci USA. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaattari SL, Zhang HL, Khor IW, et al. Affinity maturation in trout: clonal dominance of high affinity antibodies late in the immune response. Dev Comp Immunol. 2002;26:191–200. doi: 10.1016/s0145-305x(01)00064-7. [DOI] [PubMed] [Google Scholar]
- Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- Koppang EO, Fischer U, Moore L, et al. Salmonid T cells assemble in the thymus, spleen and in novel interbranchial lymphoid tissue. J Anat. 2010;217:728–739. doi: 10.1111/j.1469-7580.2010.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath G, Garver KA, Corbeil S, et al. Protective immunity and lack of histopathological damage two years after DNA vaccination against infectious hematopoietic necrosis virus in trout. Vaccine. 2006;24:345–354. doi: 10.1016/j.vaccine.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner C, Wahl B, Föhse L, et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209:365–377. doi: 10.1084/jem.20111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Ye J, Kaattari SL. Differential compartmentalization of memory B cells versus plasma cells in salmonid fish. Eur J Immunol. 2013;43:360–370. doi: 10.1002/eji.201242570. [DOI] [PubMed] [Google Scholar]
- Magadán-Mompó S, Sánchez-Espinel C, Gambón-Deza F. Immunoglobulin heavy chains in medaka (Oryzias latipes) BMC Evol Biol. 2011;11:165. doi: 10.1186/1471-2148-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadán-Mompó S, Zimmerman AM, Sánchez-Espinel C, Gambón-Deza F. Immunoglobulin light chains in medaka (Oryzias latipes) Immunogenetics. 2013;65:387–396. doi: 10.1007/s00251-013-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnadottir B, Jonsdottir H, Helgason S, et al. Humoral immune parameters in Atlantic cod (Gadus morhua L.), II: the effects of size and gender under different environmental conditions. Comp Biochem Physiol. 1999;122:181–188. doi: 10.1016/s0305-0491(98)10157-8. [DOI] [PubMed] [Google Scholar]
- Malecek K, Lee V, Feng W, et al. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008;6:e157. doi: 10.1371/journal.pbio.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Jentoft S, Gregers TF, Jakobsen KS. Unraveling the evolution of the Atlantic cod’s (Gadus morhua L.) alternative immune strategy. PLoS One. 2013;8:e74004. doi: 10.1371/journal.pone.0074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianes AE, Zimmerman M, Zimmerman AM. Targets of somatic hypermutation within immunoglobulin light chain genes in zebrafish. Immunology. 2011;132:240–255. doi: 10.1111/j.1365-2567.2010.03358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Lo SFF, Carruthers CJJ, et al. Affinity maturation without germinal centres in lymphotoxin-alpha-deficient mice. Nature. 1996;382:462–466. doi: 10.1038/382462a0. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Rahman A. What brought the adaptive immune system to vertebrates? The jaw hypothesis and the seahorse. Immunol Rev. 1998;166:177–186. doi: 10.1111/j.1600-065x.1998.tb01262.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen H, Lund V, Larsen R, Seppola M. Vibriosis vaccines based on various serosubgroups of Vibrio anguillarum O2 induce specific protection in Atlantic cod (Gadus morhua L.) juveniles. Fish Shellfish Immunol. 2011;30:330–339. doi: 10.1016/j.fsi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Miller NW, Clem LW. Microsystem for in vitro primary and secondary immunization of channel catfish (Ictalurus punctatus) leukocytes with hapten-carrier conjugates. J Immunol Methods. 1984;72:367–379. doi: 10.1016/0022-1759(84)90006-1. [DOI] [PubMed] [Google Scholar]
- Miller NW, Sizemore RC, Clem LW. Phylogeny of lymphocyte heterogeneity: the cellular requirements for in vitro antibody responses of channel catfish leukocytes. J Immunol. 1985;134:2884–2888. [PubMed] [Google Scholar]
- Mora T, Walczak AM, Bialek W, Callan CG. Maximum entropy models for antibody diversity. Proc Natl Acad Sci USA. 2010;107:5405–5410. doi: 10.1073/pnas.1001705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Toda H, Shibasaki Y, Somamoto T. Cytotoxic T cells in teleost fish. Dev Comp Immunol. 2011;35:1317–1323. doi: 10.1016/j.dci.2011.03.033. [DOI] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the world. 4th. Hoboken: Wiley; 2006. [Google Scholar]
- Obukhanych TV, Nussenzweig MC. T-independent type II immune responses generate memory B cells. J Exp Med. 2006;203:305–310. doi: 10.1084/jem.20052036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltz EM. Regulation of antigen receptor gene assembly in lymphocytes. Immunol Res. 2001;23:121–133. doi: 10.1385/IR:23:2-3:121. [DOI] [PubMed] [Google Scholar]
- Persson AC, Stet RJ, Pilström L. Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics. 1999;50:49–59. doi: 10.1007/s002510050685. [DOI] [PubMed] [Google Scholar]
- Petit J, Stange-Thomann N, Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Pilström L, Petersson A. Isolation and partial characterization of immunoglobulin from cod (Gadus morhua L.) Dev Comp Immunol. 1991;15:143–152. doi: 10.1016/0145-305x(91)90005-j. [DOI] [PubMed] [Google Scholar]
- Pilstrom L, Warr G, Stromberg S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fish Sci. 2005;71:961–971. [Google Scholar]
- Rangel R, McKeller MR, Sims-Mourtada JC, et al. Assembly of the kappa preB receptor requires a V kappa-like protein encoded by a germline transcript. J Biol Chem. 2005;280:17807–17814. doi: 10.1074/jbc.M409479200. [DOI] [PubMed] [Google Scholar]
- Rast JP, Litman GW. Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev. 1998;166:79–86. doi: 10.1111/j.1600-065x.1998.tb01254.x. [DOI] [PubMed] [Google Scholar]
- Rast JP, Amemiya CT, Litman RT, et al. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- Ratcliffe MJ. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev Comp Immunol. 2006;30:101–118. doi: 10.1016/j.dci.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Regnault A, Cumano A, Vassalli P, et al. Oligoclonal repertoire of the CD8 alpha alpha and the CD8 alpha beta TCR-alpha/beta murine intestinal intraepithelial T lymphocytes: evidence for the random emergence of T cells. J Exp Med. 1994;180:1345–1358. doi: 10.1084/jem.180.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombout JH, Abelli L, Picchietti S, et al. Teleost intestinal immunology. Fish Shellfish Immunol. 2011;31:616–626. doi: 10.1016/j.fsi.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Romer A. The vertebrate body. 3rd. Philadelphia: Saunders WB Co.; 1962. [Google Scholar]
- Salinas I, Zhang Y-A, Sunyer JO. Mucosal immunoglobulins and B cells of teleost fish. Dev Comp Immunol. 2011;35:1346–1365. doi: 10.1016/j.dci.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut B, Du Pasquier L, Ducoroy P, et al. Axolotl MHC architecture and polymorphism. Eur J Immunol. 1999;29:2897–2907. doi: 10.1002/(SICI)1521-4141(199909)29:09<2897::AID-IMMU2897>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Saunders HL, Oko ALAL, Scott ANAN, et al. The cellular context of AID expressing cells in fish lymphoid tissues. Dev Comp Immunol. 2010;34:669. doi: 10.1016/j.dci.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Schröder AE, Greiner A, Seyfert C, Berek C. Differentiation of B cells in the nonlymphoid tissue of the synovial membrane of patients with rheumatoid arthritis. Proc Natl Acad Sci USA. 1996;93:221–225. doi: 10.1073/pnas.93.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrøder MB, Ellingsen T, Mikkelsen H, et al. Comparison of antibody responses in Atlantic cod (Gadus morhua L.) to Vibrio anguillarum, Aeromonas salmonicida and Francisella sp. Fish Shellfish Immunol. 2009;27:112–119. doi: 10.1016/j.fsi.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Shapiro D, Adkison M, Kaattari S. Antibody affinity analysis using the ELISA. In: Lefkovitz I Jr, editor. Immunology methods manual: comprehensive sourcebook of techniques. New York: Academic; 1996. p. 2353e65. [Google Scholar]
- Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- Six A, Mariotti-Ferrandiz ME, Chaara W, et al. The past, present, and future of immune repertoire biology – the rise of next-generation repertoire analysis. Front Immunol. 2013;4:413. doi: 10.3389/fimmu.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelty P, Marchal C, Renard R, et al. Identification of the pre-T-cell receptor alpha chain in nonmammalian vertebrates challenges the structure-function of the molecule. Proc Natl Acad Sci USA. 2010;107:19991–19996. doi: 10.1073/pnas.1010166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somamoto T, Okamoto N, Nakanishi T, et al. In vitro generation of viral-antigen dependent cytotoxic T-cells from ginbuna crucian carp, Carassius auratus langsdorfii. Virology. 2009;389:26–33. doi: 10.1016/j.virol.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Somamoto T, Kondo M, Nakanishi T, Nakao M. Helper function of CD4+ lymphocytes in antiviral immunity in ginbuna crucian carp, Carassius auratus langsdorfii. Dev Comp Immunol. 2014;44:111–115. doi: 10.1016/j.dci.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Star B, Jentoft S. Why does the immune system of Atlantic cod lack MHC II? Bioessays. 2012;34:648–651. doi: 10.1002/bies.201200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Stoel M, Jiang H, van Diemen CC, et al. Restricted IgA repertoire in both B-1 and B-2 cell-derived gut plasmablasts. J Immunol. 2005;174:1046–1054. doi: 10.4049/jimmunol.174.2.1046. [DOI] [PubMed] [Google Scholar]
- Tacchi L, Musharrafieh R, Larragoite ET, et al. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun. 2014;5:5205. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa F, Dijkstra JM, Kotterba P, et al. The expression of CD8α discriminates distinct T cell subsets in teleost fish. Dev Comp Immunol. 2011;35:752–763. doi: 10.1016/j.dci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Tan Y-C, Blum LK, Kongpachith S, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nakasone H, Yamazaki R, et al. Single-cell analysis of T-cell receptor repertoire of HTLV-1 Tax-specific cytotoxic T cells in allogeneic transplant recipients with adult T-cell leukemia/lymphoma. Cancer Res. 2010;70:6181–6192. doi: 10.1158/0008-5472.CAN-10-0678. [DOI] [PubMed] [Google Scholar]
- Tatner MF. The ontogeny of humoral immunity in rainbow trout, Salmo gairdneri. Vet Immunol Immunopathol. 1986;12:93–105. doi: 10.1016/0165-2427(86)90114-5. [DOI] [PubMed] [Google Scholar]
- Toda H, Saito Y, Koike T, et al. Conservation of characteristics and functions of CD4 positive lymphocytes in a teleost fish. Dev Comp Immunol. 2011;35:650–660. doi: 10.1016/j.dci.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Tournefier A, Laurens V, Chapusot C, et al. Structure of MHC class I and class II cDNAs and possible immunodeficiency linked to class II expression in the Mexican axolotl. Immunol Rev. 1998;166:259–277. doi: 10.1111/j.1600-065x.1998.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Utke K, Kock H, Schuetze H, et al. Cell-mediated immune responses in rainbow trout after DNA immunization against the viral hemorrhagic septicemia virus. Dev Comp Immunol. 2008;32:239–252. doi: 10.1016/j.dci.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Van Der Aa LM, Levraud J, Yahmi M, et al. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;23:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Weill JC, Cocea L, Reynaud C-A. Allelic exclusion: lesson from GALT species. Semin Immunol. 2002;14:213–215. doi: 10.1016/s1044-5323(02)00045-3. discussion 227–228. [DOI] [PubMed] [Google Scholar]
- Weinstein JA, Jiang N, White RA, et al. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–811. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AM, Bland PW, Phillips AC, et al. Intestinal alpha beta T cells differentiate and rearrange antigen receptor genes in situ in the human infant. J Immunol. 2004;173:7190–7199. doi: 10.4049/jimmunol.173.12.7190. [DOI] [PubMed] [Google Scholar]
- Wilson M, Hsu E, Marcuz A, et al. What limits affinity maturation of antibodies in Xenopus the rate of somatic mutation or the ability to select mutants? EMBO J. 1992;11:4337–4347. doi: 10.1002/j.1460-2075.1992.tb05533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Parra D, Gómez D, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci USA. 2013;110:13097–13102. doi: 10.1073/pnas.1304319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Waldbieser GC, Lobb CJ. The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J Immunol. 2006;176:1655. doi: 10.4049/jimmunol.176.3.1655. [DOI] [PubMed] [Google Scholar]
- Yasuike M, De Boer J, Von Schalburg KR, et al. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genomics. 2010;11:486. doi: 10.1186/1471-2164-11-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kaattari IM, Kaattari SL. The differential dynamics of antibody subpopulation expression during affinity maturation in a teleost. Fish Shellfish Immunol. 2011;30:372–377. doi: 10.1016/j.fsi.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Salinas I, Li J, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Feng W, Weedon J, et al. The multiple shark Ig chain genes rearrange and hypermutate autonomously. J Immunol. 2011;187:2492. doi: 10.4049/jimmunol.1101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski CE, Corti D, Mele F, et al. Dissecting the human immunologic memory for pathogens. Immunol Rev. 2011;240:40–51. doi: 10.1111/j.1600-065X.2010.01000.x. [DOI] [PubMed] [Google Scholar]