Abstract
Various factors may impact the processes of diversification of a clade. In the marine realm, it has been shown that coral reef environments have promoted diversification in various fish groups. With the exception of requiem sharks, all the groups showing a higher level of diversity in reefs than in non-reef habitats have diets based predominantly on plankton, algae or benthic invertebrates. Here we explore the pattern of diversification of carangoid fishes, a clade that includes numerous piscivorous species (e.g. trevallies, jacks and dolphinfishes), using time-calibrated phylogenies as well as ecological and morphological data from both extant and fossil species. The study of carangoid morphospace suggests that reef environments played a role in their early radiation during the Eocene. However, contrary to the hypothesis of a reef-association-promoting effect, we show that habitat shifts to non-reef environments have increased the rates of morphological diversification (i.e. size and body shape) in extant carangoids. Piscivory did not have a major impact on the tempo of diversification of this group. Through the ecological radiation of carangoid fishes, we demonstrate that non-reef environments may sustain and promote processes of diversification of different marine fish groups, at least those including a large proportion of piscivorous species.
Keywords: carangoid fishes, trait evolution, morphospace, disparity, habitat shift, HiSSE
1. Introduction
Many factors may influence diversification processes and it is particularly challenging to identify the major driving forces explaining the diversity of a clade. Intrinsic factors such as body plan complexity [1] or genetic variance [2] can drive the morphological diversification in a lineage. Additionally, various intrinsic lineage characteristics may constitute novelties that shape the process of diversification. For example, trophic specialization [3,4], locomotor strategy [5,6] and anti-predatory defences [7] may influence lineage and phenotypic diversification rates. Extrinsic factors such as the invasion of a competitor-free region may also provide opportunities for a lineage to radiate into a variety of untapped niches [8], whereas competition may constrain divergence in resource use and thus limit diversification [9].
In the marine realm, tropical coral reefs have been shown to have promoted diversification in several groups of teleost fishes [10–12] as well as requiem sharks [13]. The ecological radiation of many fish families is clearly associated with this highly productive and structurally complex habitat [14,15]. With the exception of requiem sharks, however, all the groups that were shown to possess higher diversity in reef than non-reef environments have diets based mostly or exclusively on plankton, algae or small benthic invertebrates. Little work has so far been devoted to test the hypothesis that reef association may increase rates of diversification in groups that include a large proportion of piscivorous species. Furthermore, relatively few studies of marine ray-finned fishes have tested the hypothesis that other aquatic environments besides coral reefs may also provide ecological opportunities and lead to increased diversification rates. For example, various lineages of marine fishes have colonized freshwater environments during their evolutionary history, and several recent studies have shown that freshwaters may provide open niche space sustaining their diversification, even though the relationship between invasion of the new habitat and the tempo of lineage diversification can be a complex one [16,17]. Open seas have provided ecological opportunity for multiple groups such as cephalopods [18] and cetaceans [19]. Recently, it has been suggested that open seas may have facilitated the radiation of some pelagic fishes [20], even though no tests were performed to support such a conclusion.
Trophic specializations may also promote diversification. For example, a switch to low-quality food sources such as algae, detritus, sponges or corals was associated with higher rates of lineage diversification in some reef fish families [21]. In parrotfishes (Scarinae), the evolution of an intramandibular joint on the lower jaw allows efficient scraping of hard corals promoting morphological diversity in some genera [4]. With regard to the diversity of fish forms, various ecomorphological studies have revealed that the body shape is linked to swimming performance, habitat preference or behaviour [22]. Body shape is an ecologically relevant character and is therefore suited for studying diversification patterns associated with habitat and diet shifts. Recently, body shape analysis has become a powerful tool to characterize changes in morphospace occupation during the evolution of teleosts [23,24], and the quantitative analysis of morphospace can shed light on how ecofunctional diversity arises or changes over evolutionary time [25].
Carangoid fishes, including Carangidae (trevallies, pompanos and jacks), Echeneidae (remoras), Coryphaenidae (dolphinfishes) and Rachycentridae (the cobia), represent a successful group of about 159 species [26]. This marine fish group, which originated during the Late Cretaceous and experienced a major radiation during the Eocene [27], includes both species living in and around reef ecosystems (i.e. reef-associated species), as well as species found in open water habitats or coastal non-reef environments (i.e. non-reef-associated species). Most carangoids are powerful swimmers and active predators constituting a major clade of piscivorous species, whereas some species that feed on zooplankton and remoras are opportunist feeders, eating ectoparasites, shedding skin and scraps of food from the meals of their hosts such as sharks, turtles or large teleosts [28]. A recently published phylogenetic hypothesis suggests multiple transitions from one habitat type to another and dietary shifts across the evolutionary history of carangoids [27]. Even though their evolutionary history has so far received scant attention, carangoids provide an important opportunity to test various hypotheses about both habitat and diet shifts as promoter of taxonomic and morphological diversification (i.e. body size and shape) in marine fishes.
On the basis of the current diversity of carangoid fishes, and according to the hypothesis of ecological opportunity, we anticipated that the rates of lineage and morphological diversification would vary between non-reef and reef habitats. We also expected dietary specialization, such as piscivory, could induce variation in the tempo or mode of diversification, as has been observed in other fish groups [29]. Furthermore, carangoid fishes have a rich fossil record dating to the earliest Eocene [30], which allows us the opportunity to compare the pattern of evolution within both extant and extinct lineages.
Here, we examine the pattern of diversification in carangoid fishes. Using a morphological dataset that includes fossil and extant species, we explore the variation in morphospace occupation through time. We test the hypothesis that carangoid lineages occupying non-reef environments have different diversification rates compared with those occupying reef habitats. We also test whether piscivorous lineages have different diversification rates compared with non-piscivorous lineages.
2. Material and methods
(a). Morphological and ecological data
For the morphological analyses, we studied accessioned museum specimens with additional photographs obtained from online databases (electronic supplementary material, table S1). The dataset contains 384 specimens from 178 carangoid species, including 24 fossil species. Sample sizes within species ranged between one and four individuals (median = two individuals; electronic supplementary material, table S1). We did not include in this study the Eocene percomorph †Ductor, which has been associated with Echeneoidei [31] and used by Friedman et al. in their study on the evolutionary history of remoras [32]. In this last study, the placement of †Ductor as a stem rachycentrid was supported by characters that are broadly homoplastic within percomorphs [32]. For this reason, we prefer to avoid including this taxon until its carangoid affinities have been properly demonstrated with a comparative morphological study. The majority of Miocene and Oligocene carangoid taxa were erected on the basis of incomplete or fragmentary specimens. These species were not included in our study. Only species represented by complete articulated specimens were included in our morphometric analysis.
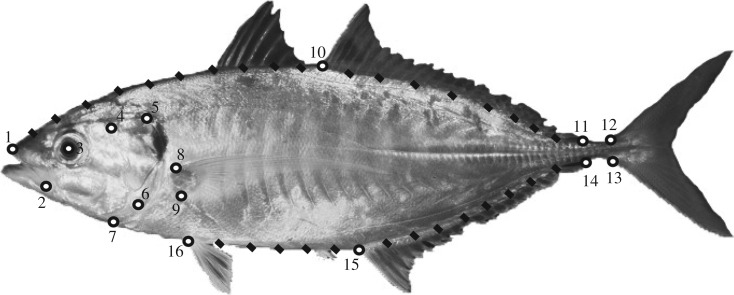
We used landmark-based geometric morphometrics [33] to quantify fish body shape. The x, y coordinates of 16 homologous landmarks (LMs) and 31 semi-landmarks (semi-LMs; figure 1) were recorded using TpsDig [34]. The chosen LMs allow the capture of variation related to body elongation (the ratio between body length and body depth), which are well-known morphological traits directly related to swimming performances, such as manoeuvrability, acceleration rate and sustained swimming [35,36]. Semi-LMs help to capture the curvature of the fish body, especially for the cephalic region where extensive morphological variation occurs in carangoids. Details about the generation of shape data are provided in Frédérich et al. [37]. We used species scores along a principal component axis for exploring phenotypic diversification. In order to compare rates of shape evolution among groups using the time-calibrated phylogenies (see details hereafter), we recalculated PCA using a superimposition based upon species included in the molecular sampling, thus excluding the fossil species. In addition to the study of shape variation, we used maximum body size (total length, TL) as a second morphological trait. We gathered size and ecological data from various sources, including FishBase [28] and the primary literature [38]. For every species in our study, we scored two discrete variables with two states each: (i) habitat type—reef (1) and non-reef (0); (ii) feeding preference—piscivory (1) and non-piscivory (0). Concerning the fossil species, it was not possible to define their diet, whereas their habitat was identified using the sedimentological features of the fossiliferous deposits and their associated fauna.
Figure 1.
The homologous landmarks (LMs) and semi-landmarks (semi-LMs) used in the analysis of the fish body variation illustrated in Alepes djedaba (image modified under Creative Commons licence from original photographs by J. E. Randall, retrieved from http://pbs.bishopmuseum.org/images/JER/). Semi-LMs are represented by solid rectangles, LMs by circles with crescent numeration starting from snout: (1) anterior tip of the snout (premaxilla), (2) posterior end of the maxilla, (3) centre of the orbit, (4) anterodorsal origin of the opercle, (5) posterodorsal edge of the opercle, (6) ventral tip of the subopercle, (7) base of the isthmus, (8) and (9) superior and inferior insertion of the pectoral fin, (10) anterior origin of the posterior lobe of the dorsal fin, (11) posterior insertion of the dorsal fin, (12) insertion of first ray of dorsal lobe of the caudal fin, (13) insertion of first ray of ventral lobe of the caudal fin, (14) posterior insertion of the anal fin, (15) anterior origin of the anal fin and (16) insertion of the pelvic fin.
(b). Phylogeny, ancestral state reconstruction and stochastic mapping
We used the time-calibrated phylogeny described by Santini & Carnevale [27] (electronic supplementary material, figure S1). From the Bayesian posterior distribution generated by BEAST v. 1.8 [35], we randomly sampled 100 trees that we used throughout the study as a way of including uncertainty in tree topology and branch length into our phylogenetic comparative analyses.
We used stochastic character mapping [39] to infer possible ecological histories. The stochastic mapping and the ancestral state reconstruction was produced using the function make.simmap in the phytools package (v. 0.5.0) [40] for R [41]. We then sampled 100 character histories allowing the incorporation of the uncertainty associated with the timing of the transitions between ecological states. For the parametrization of make.simmap, we used the estimated ancestral state and the best model for the transition matrix from our empirical data. To assess the best model for the transition matrix, we fitted a model with equal rate of transition between states and a model with all rates different using the function ace in the R package ape [42]. The likelihood of these two models was then compared using a likelihood ratio test, which suggested the use of unequal rates for the diet preference and equal rates for the habitat preference (see Results).
(c). Lineage diversification
As for the majority of marine teleost groups investigated to date [14], we predicted that the tempo of lineage diversification should be higher for reef-associated carangoid lineages. Similarly, if specialization on piscivory mainly explains the success of carangoid fishes, we predicted that the tempo of lineage diversification should vary between trophic groups. We combined binary state speciation and extinction (BiSSE) [43] and hidden state speciation and extinction (HiSSE) [44] methods to test whether the ecological shift to ‘piscivory’ and ‘reef-association’ triggers an elevated rate of speciation during the carangoid evolution. BiSSE provides a likelihood-based test of whether a discrete character influences the rate of lineage diversification. HiSSE extends the BiSSE framework to account for the presence of unmeasured factors (i.e. hidden states) that could impact diversification rates estimated for the states of observed traits [44]. Thus, for example, state 1 (e.g. piscivory) can take on two states: 1A when the hidden state is absent and 1B when the hidden state is present.
To assess the ecological factors acting on the tempo of lineage diversification in carangoids, we compared the fit of 12 different evolutionary models based on our empirical data (timetree and character states at the tips). The extinction fraction (ɛ) was forced to be equal between states in every model, because cautions are needed for the estimation of extinction from molecular phylogenies [45], and we also aimed to reduce the number of parameters. All models are described in table 1 and differ by net turnover (τ = speciation λ + extinction μ), transition rates (q), and presence or absence of hidden states. As for the parametrization of stochastic mapping, we used the best model for the transition matrix from our empirical data. We used Akaike's information criterion (AIC) scores and weights to compare the fit of the models. A ΔAICc value of four or more was taken as an indication of support for one model over the other following Burnham & Anderson [46]. We performed analyses using functions from the R package HiSSE [44], correcting for incompletely resolved phylogeny. Here, we assumed that the missing species are randomly distributed on the phylogenetic tree.
Table 1.
Results from fitting lineage diversification models. The models are compared with AICc (small-sample corrected AIC) scores and Akaike weights (wtAICc). ΔAICc scores indicate the difference between the candidate model and the best-fitting model. Parameters of the twelve models of lineage diversification in the HiSSE and BiSSE frameworks are described. Habitat (0, non-reef; 1, reef) and diet (0, non-piscivory; 1, piscivory) preferences were coded by binary variables. Tau (τ), epsilon (ɛ) and q refer to ‘net turnover’ rate, extinction fraction and transition rates, respectively.
| trait | model | hidden states | τ | ɛ | q | AICc | ΔAICc | wtAICc |
|---|---|---|---|---|---|---|---|---|
| habitat | BiSSE 1 | no hidden states | equal | equal | equal | 1202.87 | 1.85 | 0.12 |
| BiSSE 2 | no hidden states | vary | equal | equal | 1202.43 | 1.41 | 0.16 | |
| HiSSE 1 | hidden state present for 0 and 1 | all vary | equal | equal | 1203.14 | 2.12 | 0.11 | |
| HiSSE 2 | hidden state present for 0 | all vary | equal | equal | 1201.97 | 0.95 | 0.20 | |
| HiSSE 3 | hidden state present for 1 | all vary | equal | equal | 1203.33 | 2.31 | 0.10 | |
| HiSSE 4 | hidden state present for 0 and 1 | only A and B vary | equal | equal | 1201.02 | 0.00 | 0.31 | |
| diet | BiSSE I | no hidden states | vary | equal | vary | 1168.44 | 3.30 | 0.10 |
| BiSSE II | no hidden states | equal | equal | vary | 1166.71 | 1.57 | 0.24 | |
| HiSSE I | hidden state present for 0 and 1 | all vary | equal | vary | 1165.14 | 0.00 | 0.54 | |
| HiSSE II | hidden state present for 0 | all vary | equal | vary | 1172.04 | 6.90 | 0.02 | |
| HiSSE III | hidden state present for 1 | all vary | equal | vary | 1177.36 | 12.21 | 0.00 | |
| HiSSE IV | hidden state present for 0 and 1 | only A and B vary | equal | vary | 1168.54 | 3.40 | 0.10 |
(d). Morphological diversification
To test whether ecological changes (habitat and diet) induced shifts in the tempo of morphological evolution (i.e. including size and shape) throughout the evolutionary history of jacks and allies, we fitted three different evolutionary models to the data: a single-rate Brownian motion (BM) model (BM1), a BM model with different rate parameters for species living in reef environment and non-reef species (BMS_habitat), and a BM model with different rate parameters for piscivorous and non-piscivorous species (BMS_diet). We fitted these models of continuous trait evolution using the OUwie package [47] for R [41], and compared their fit using AIC scores and weights [46]. For these models fitting in OUwie, we ran the analyses using the simmap trees.
In addition to the visual exploration of morphospace defined by principal components of shape variables, we explored the level of shape disparity and compared it among periods. Additionally, we compared the levels of disparity between habitat groups (reef versus non-reef). This analysis of morphospace allowed a deeper interpretation of the evolution of morphological diversity in carangoid fishes through time. We calculated the level of shape disparity based on Procrustes variance [33] and performed pairwise comparisons between groups (i.e. permutation test, 9999 iterations) using the function morphol.disparity in the R package geomorph [48]. We also used convex hulls to compare the level of shape disparity among periods and between environments in the Euclidean plane made by the first two PCs. Convex hull areas were calculated through nearest-neighbour analysis in PAST v. 3.08 [49].
3. Results
(a). Ancestral state reconstruction and stochastic mapping
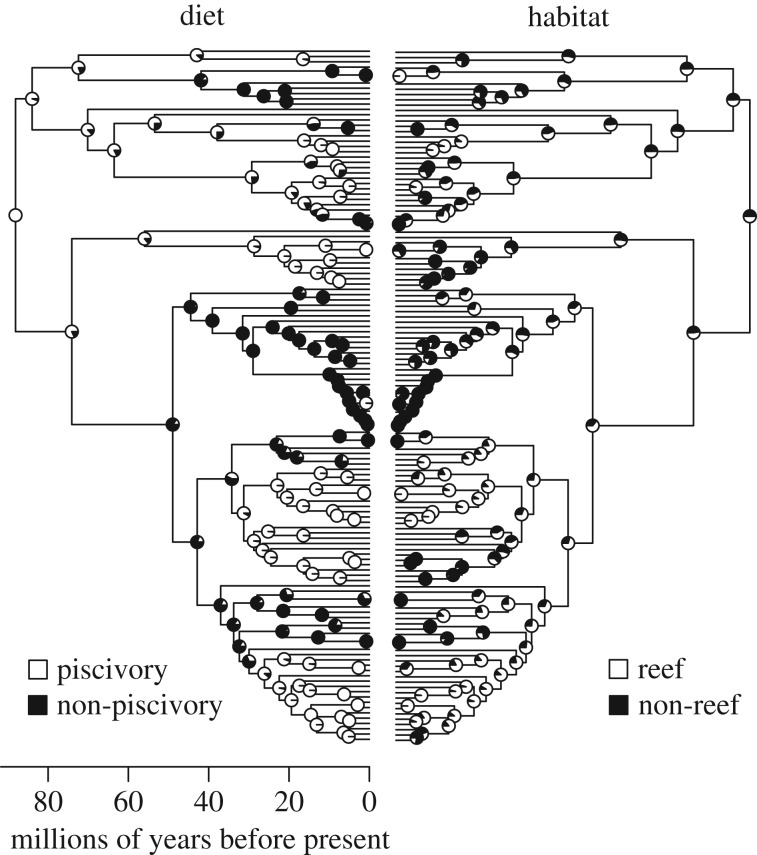
The likelihood ratio tests asked for the use of equal rates for habitat transition (q01 = q10 = 0.035; χ2 = 0.4, p = 0.536) and a matrix of unequal rates for diet preference for stochastic mapping (χ2 = 7.9, p = 0.005). The transition rate from non-piscivory to piscivory (q01 = 0.019) was two times higher than the transition rate from piscivory to non-piscivory (q10 = 0.009). The results of ecological reconstructions suggested that the last common ancestor of all living carangoids was piscivorous (68% of likelihood), whereas the habitat that this taxon inhabited is unknown (50% likelihood for both reef and non-reef environment). Stochastic mapping revealed two major transitions to non-reef environments, one in the genus Trachurus and one in Selene, and at least six shifts to a non-piscivorous diet (figure 2; electronic supplementary material, figure S1).
Figure 2.
Summary of the ecological (diet and habitat) histories on the consensus timetree of carangoid fishes using stochastic mapping. Shading keys refer to ecological categories.
(b). Lineage diversification
The best-fitting models of lineage diversification did not support the presence of hidden states acting on the rate of speciation (table 1). Indeed, no HiSSE model fitted better on the empirical data than BiSSE models. The comparison of the fit of the diversification models failed to find strong support for one model over the others (table 1). Consequently, we were unable to validate an effect of diet or habitat preference on the tempo of lineage diversification.
(c). Morphological diversification
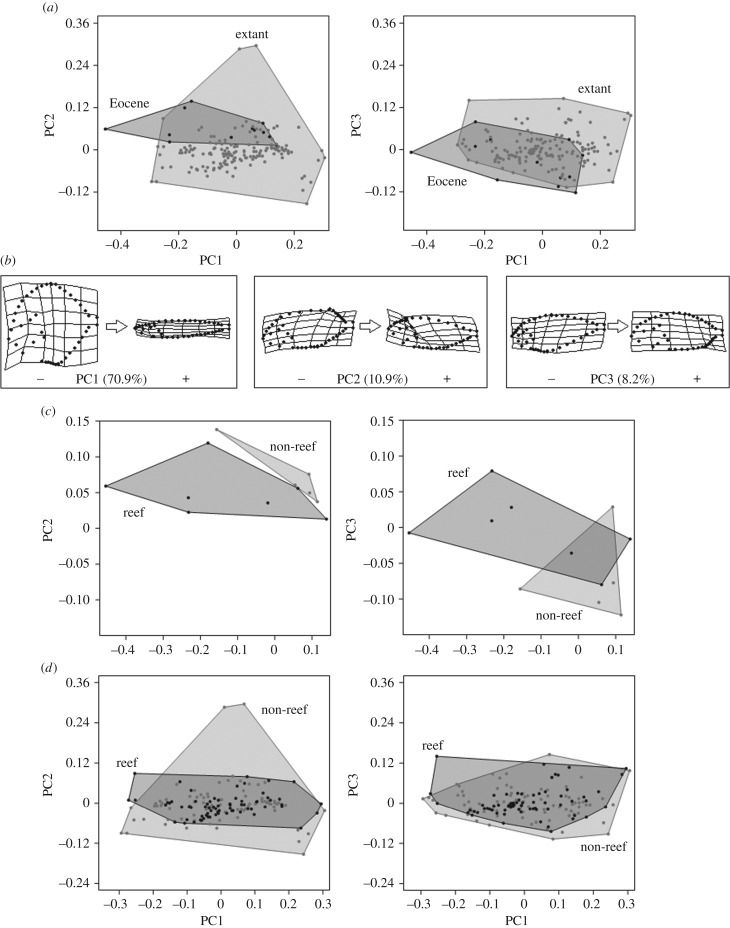
The shape variation along PCs is very similar with or without the inclusion of fossil species (figure 3; electronic supplementary material, S2). So, hereafter, we describe shape variation within the morphospace defined by PC axes including extant and Eocene species (figure 3). The principal component analysis was summarized by 90 PC axes, with the first three together accounting for about 90% of the overall shape variance (PC1 = 70.9%, PC2 = 10.9% and PC3 = 8.2% of shape variation; electronic supplementary material, table S2). PC1 mainly describes body elongation. For example, negative scores are related to deep-bodied carangoids (e.g. Selene vomer, †Ceratoichthys pinnatiformis), whereas anteroposteriorly elongated species lie on positive values (e.g. Phtheirichthys lineatus; figure 3). PC2 explains variation of the anatomical origin of the dorsal fin (posterior lobe): extreme negative scores are related to taxa with backward origin of the functional dorsal fin (e.g. Remora osteochir), whereas species with a dorsal fin extending the length of the body (e.g. Coryphaena hippurus; figure 3) have positive scores. Along axis PC3, the main shape variation is related to anal fin length: negative scores are related to taxa with long anal fin (e.g. Oligoplites saliens, Parastromateus niger), whereas positive values are associated with species with short anal fin (e.g. Seriola fasciata). All the cited taxa of the extreme scores of each PC axis lie on the periphery of morphospace and contribute highly to the morphological variance. Conversely, the centre of morphospace is occupied by taxa with a more generalized body plan, such as most members of the Carangini.
Figure 3.
Morphospaces of carangoids built on the first three PC axes explaining the greatest shape variance. (a) Morphospaces including extant and fossil species; (b) deformation grids illustrating shape variation along the first three PC axes from negative to positive scores (with percentage of the relative explained morphological variation between brackets); (c) morphospaces including Eocene species grouped by polygons enclosing reef and non-reef taxa; (d) morphospaces including extant species grouped by polygons enclosing reef and non-reef taxa.
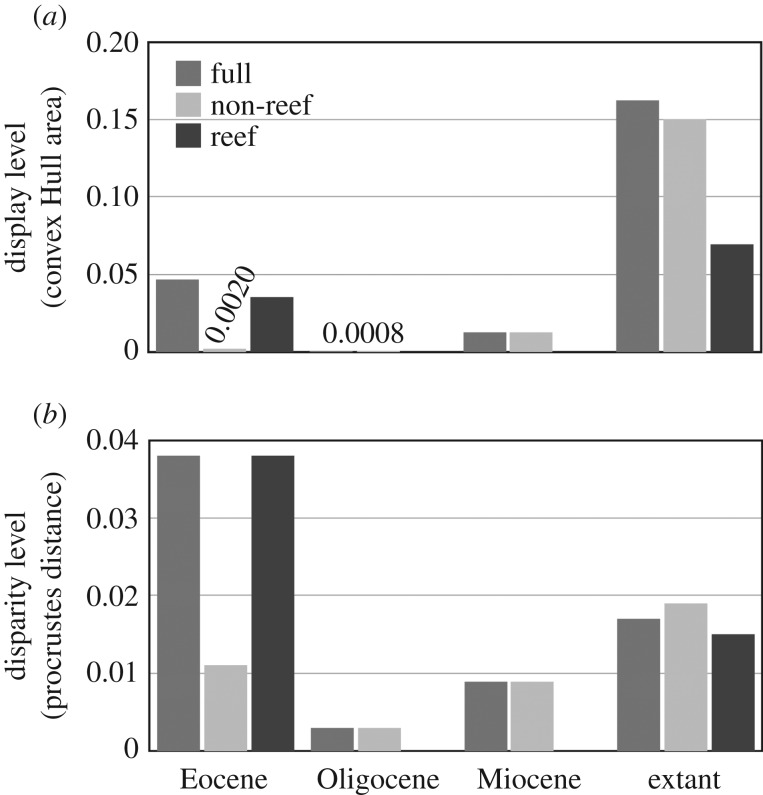
When using the convex Hulls metric, the extant taxa showed the highest level of shape disparity in the morphospace made of the first two PCs (figure 4a). However, the shape disparity level based on Procrustes variance was higher for the Eocene taxa than the extant taxa (p < 0.001; figure 4b). This high level of shape disparity observed during the Eocene was mainly explained by taxa from Bolca, which probably inhabited a reef environment (figure 4). For the extant taxa, however, species from non-reef environments showed a significantly higher level of disparity than reef group (p = 0.026; figure 4b). The disparity level of extant taxa is mainly explained by non-reef species (figure 4). The shift through time of shape disparity level between environments is mainly explained by deep-bodied carangoids living in non-reef habitats (figure 3c,d). Studied taxa from Oligocene and Miocene were living in non-reef environments (electronic supplementary material, table S1). Even if their number was limited, we have calculated the shape disparity level for the two periods (figure 4). Both metrics suggested lower level of shape disparity than the Eocene taxa (p < 0.007), but their Procrustes variance did not differ from extant taxa (p > 0.25; figure 4b).
Figure 4.
Levels of body shape disparity among periods and habitats. Disparity levels were calculated using (a) convex Hull area from the morphospace defined by the first two PCs and (b) Procrustes variance.
Habitat shifts, rather than diet, impact the morphological evolutionary rates in carangoids. Indeed, BMS_habitat was always among the best-supported models for all the studied traits (i.e. size and shape; table 2). Brownian rate parameters (σ2) were always two times higher for the lineages living in a non-reef environment than for reef-associated taxa (electronic supplementary material, table S3).
Table 2.
Results from fitting morphological diversification models. For each studied morphological trait (body size and shape), the models are ranked from best to worst, according to AICc (small-sample corrected AIC) scores and Akaike weights (wtAICc). ΔAICc scores indicate the difference between the candidate model and the best-fitting model. Refer to text for model description.
| morphological trait | model | AICc | ΔAICc | wtAICc | |
|---|---|---|---|---|---|
| body size | logTL | BMS_habitat | 58.65 | 0 | 1.00 |
| BM1 | 71.96 | 13.31 | 0 | ||
| BMS_diet | 72.26 | 13.61 | 0 | ||
| body shape | PC1 | BMS_habitat | −281.70 | 0 | 0.95 |
| BMS_diet | −275.93 | 5.77 | 0.05 | ||
| BM1 | −264.37 | 17.33 | 0 | ||
| PC2 | BMS_diet | −504.30 | 0 | 0.62 | |
| BMS_habitat | −503.30 | 1.00 | 0.38 | ||
| BM1 | −495.11 | 9.19 | 0.01 | ||
| PC3 | BMS_habitat | −495.18 | 0 | 0.97 | |
| BMS_diet | −487.91 | 7.27 | 0.03 | ||
| BM1 | −468.32 | 26.86 | 0.00 |
4. Discussion
Our data show that reef environments probably affected the carangoid diversification, especially during the early steps of their evolutionary history. Subsequently, non-reef habitats have promoted higher rates of morphological diversification and these environments currently sustain the main extant diversity of this group. Trophic shifts did not impact the rates of lineage diversification, even though the tempo of dietary shifts varies across the history of carangoids.
Several recent studies investigating the evolution of marine fishes have shown that reef association has generally triggered an increase of diversification rates in many teleost groups and carcharhinid sharks [11–14]. Except for the requiem sharks, the fish families showing the highest rates of diversification in reefs are mostly formed by species that feed on planktonic or small-to-medium benthic prey [11,12]. Here, we illustrate that non-reef environments may also have promoted increased rates of diversification in a fish clade where piscivory is common. Other species-rich groups that include large proportions of piscivorous species, such as groupers (Epinephelidae), emperors (Lethrinidae) or snappers (Lutjanidae), could have experienced higher rates of diversification in reef than in non-reef habitats, but this has not yet been tested.
Our study suggests that the ancestral carangoid fish was piscivorous, but we cannot state the origin of its habitat (figure 2). The analyses of morphospace reveal that during the Eocene the level of shape disparity was higher in reef environments than in non-reef habitats. Consequently, we hypothesize that during the Eocene carangoid fishes observed a first major radiation in reef environments, even though non-reef taxa were already present. This pattern subsequently shifted during the last 35 Ma, and the level of morphological diversity in non-reef environments is now higher for the extant species. Unfortunately, the number of Oligocene and Miocene taxa is currently limited and may thus provide a biased picture of the dynamic of disparity during these periods. While caution when interpreting these data is warranted, all the available Oligocene and Miocene taxa were associated with non-reef environments (electronic supplementary material, table S1), tentatively supporting a scenario of transition of diversity from reefs to non-reef environments.
A general summary of the carangoid morphospace (figure 3; electronic supplementary material, figure S2) shows that the dominant axis of shape variation in extant and fossil carangoid species is body elongation (PC1, figure 3), a pattern broadly observed across many fish clades [50]. The high level of shape disparity associated with extant non-reef environments is probably related to multiple invasions of non-reef habitats by distantly related lineages (figure 3). For example, the extant carangoids living in non-reef environments include powerful swimmers with deep-bodied forms (e.g. Selene spp.) as well as species with fusiform body shape with a dorsal fin extending the length of the body (e.g. Coryphaena spp.), whereas the fossil species living in non-reef environments mostly showed an anteroposteriorly elongate body (e.g. †Archaeus oblongus and †Seriola natgeosoc). The morphological diversification of non-reef carangoids is probably related to the exploitation of new ecological strategies and the filling of vacated functional roles [25,51]. Indeed, the disappearance of many large predatory fishes during the Late Cretaceous probably freed previously occupied ecological space and resources for surviving lineages [51]. The high rates of size and shape diversification for non-reef carangoids are in agreement with this scenario. Trophic resource partitioning in marine systems is often highly structured on the basis of body size [52], and we thus expect size variation to be one of the main axes of ecological diversification in zooplanktivorous and piscivorous fish species. The large size variation among piscivorous carangoids (from 50 to 200 cm of total length; electronic supplementary material, table S1) is probably related to different trophic status [53]. Body shape variation may reflect differences in swimming performance. Efficient, economical swimming suited for pelagic habitats is certainly optimized in streamlined and elongated-bodied Seriola, Trachurus, Decapterus and Coryphaena [35]. All these tuna-like species show a large anterior region separated from the caudal fin by a caudal peduncle of reduced cross-section [35]. Deep-bodied forms (e.g. Selene and Trachinotus spp.) may confer high acceleration performance, well suited for coastal environments [54]. However, differences in the morphology of efficient swimmers are not simply linked to locomotor performance, as other selective forces may have shaped their design [54]. Body forms probably represent a compromise between competing demands. Accordingly, large body depths of Selene might act as a deterrent for predators that are gape-limited [55] or might optimize the reflectance of their skin for camouflage [56]. The evolution of the symbiotic suckerfishes (i.e. Remora, Echeneis and Phtheirichthys) also had a profound impact on carangoid morphofunctional diversity. These fishes evolved peculiar body morphologies, with the first dorsal fin modified into an oval, sucker-like organ with slat-like structures that open and close to create suction [32,57] and take a firm hold against the skin of large hosts. In our morphometric data, the variation of the dorsal fins shape is mainly explained by PC2 (figure 3), and the observed increase of morphological diversity between fossil and extant groups living in non-reef environments is mainly related to this axis.
Our analyses of model fitting reveal that habitat shifts induced variation in the rates of body size and shape diversification but did not trigger an increase of speciation rates of carangoids. One possible explanation for this pattern could be that this clade experienced high levels of extinction that erased the signal of a burst of speciation associated with habitat shift. This scenario could be supported by the long stem ages that lead to a number of carangoid subclades in the timetree (electronic supplementary material, figure S1). The rich carangoid fossil record is understudied and cannot currently be brought to bear on this question. There is however evidence from other marine groups, such as cetaceans, that a high level of extinction can produce such type of pattern [19].
Piscivory has been shown to affect the pattern of diversification in some fishes [29]. Here, models of discrete trait evolution support the idea that piscivory may also impact the tempo of diversification. The evolutionary history of carangoid fishes suggests that a transition from non-piscivory to piscivory seems easier than a shift in the opposite direction. Various factors may explain this unbalanced pattern of trophic diversification. First, extrinsic factors such as ecological opportunities probably allowed easy dietary shifts to piscivory. Since the late Oligocene and Miocene, the global oceanographic conditions changed and a massive increase in the amount of food in the ocean occurred. This was mostly owing to geological and climatic factors, such as the origin of Antarctic and Arctic ice caps, which resulted in the origin of temperate climatic belt and an increase nutrient input in the ocean [58,59]. Consequently, an increased availability of food could have promoted the rise of piscivory in carangoids, but also in scombrids [60] and sparids [61], at this time. Second, intrinsic lineage characteristics, such as the general body plan of carangoids (e.g. large body size and wide gape), probably allowed them to be competitive in the capture of fast-swimming, energy-rich food such as other fishes.
5. Conclusion
To date, the promoting effect of reef environment on the diversity of fish families that include large proportions of piscivorous species has been poorly documented. Our study illustrates that some non-reef environments, such as open waters or coastal soft-bottom habitats, may also promote the tempo of diversification in some marine fish clades. We show that habitat, rather than diet, influenced the diversity of extant carangoid fishes. Reefs probably played a role during the first steps of the carangoid diversification during the Eocene, while this important group underwent a major radiation in non-reef environments during the last 35 Myr.
Supplementary Material
Acknowledgements
We thank three anonymous reviewers for their helpful comments that improved earlier versions of this paper. We also thank Dave Catania, Jon Fong, Mysi Hoang and Luiz Rocha (CAS), Rick Feeney and Christine Thacker (LACNHM), and James Maclaine and Oliver Crimmen (NHM London) for access to specimens in the collections of their institutions.
Data accessibility
Data available from the Dryad Digital Respository: http://dx.doi.org/10.5061/dryad.fs618 [62].
Authors' contributions
B.F. and F.S. conceived the study and performed research. B.F. analysed data and drafted the manuscript. F.S. photographed the museum specimens. G.M. and G.C. collected geometric and fossil data. G.M. helped with the morphometric analyses. All authors contributed to manuscript drafts and approved the final version.
Competing interests
We declare we have no competing interests.
Funding
B.F. was supported by the ‘Fonds National de la Recherche Scientifique of Belgium’ (F.R.S-FNRS). B.F. is currently a post-doctoral fellow from the Belgian Science Policy. F.S. acknowledges support from an XSEDE grant (TG-DEB140025).
References
- 1.Vermeij GJ. 1973. Adaptation, versatility, and evolution. Syst. Zool. 22, 466–477. ( 10.2307/2412953) [DOI] [Google Scholar]
- 2.Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. 2008. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution 62, 76–85. ( 10.1111/j.1558-5646.2007.00259.x) [DOI] [PubMed] [Google Scholar]
- 3.Rojas D, Vale A, Ferrero V, Navarro L. 2012. The role of frugivory in the diversification of bats in the Neotropics. J. Biogeogr 39, 1948–1960. ( 10.1111/j.1365-2699.2012.02709.x) [DOI] [Google Scholar]
- 4.Price SA, Wainwright PC, Bellwood DR, Kazancioglu E, Collar DC, Near TJ. 2010. Functional innovations and morphological diversification in parrotfish. Evolution 64, 3057–3068. ( 10.1111/j.1558-5646.2010.01036.x) [DOI] [PubMed] [Google Scholar]
- 5.Dornburg A, Sidlauskas B, Santini F, Sorenson L, Near TJ, Alfaro ME. 2011. The influence of an innovative locomotor strategy on the phenotypic diversification of triggerfish (family: Balistidae). Evolution 65, 1912–1926. ( 10.1111/j.1558-5646.2011.01275.x) [DOI] [PubMed] [Google Scholar]
- 6.Higham TE, Birn-Jeffery AV, Collins CE, Hulsey CD, Russell AP. 2015. Adaptive simplification and the evolution of gecko locomotion: morphological and biomechanical consequences of losing adhesion. Proc. Natl Acad. Sci. USA 112, 809–814. ( 10.1073/pnas.1418979112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbuckle K, Speed MP. 2015. Antipredator defenses predict diversification rates. Proc. Natl Acad. Sci. USA 112, 13 597–13 602. ( 10.1073/pnas.1509811112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 9.Losos JB. 2011. Convergence, adaptation, and constraint. Evolution 65, 1827–1840. ( 10.1111/j.1558-5646.2011.01289.x) [DOI] [PubMed] [Google Scholar]
- 10.Alfaro ME, Santini F, Brock CD. 2007. Do reefs drive diversification in marine teleosts? Evidence from the pufferfish and their allies (order tetraodontiformes). Evolution 61, 2104–2126. ( 10.1111/j.1558-5646.2007.00182.x) [DOI] [PubMed] [Google Scholar]
- 11.Price SA, Holzman R, Near TJ, Wainwright PC. 2011. Coral reefs promote the evolution of morphological diversity and ecological novelty in labrid fishes. Ecol. Lett. 14, 462–469. ( 10.1111/j.1461-0248.2011.01607.x) [DOI] [PubMed] [Google Scholar]
- 12.Cowman PF, Bellwood DR. 2011. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 24, 2543–2562. ( 10.1111/j.1420-9101.2011.02391.x) [DOI] [PubMed] [Google Scholar]
- 13.Sorenson L, Santini F, Alfaro ME. 2014. The effect of habitat on modern shark diversification. J. Evol. Biol. 27, 1536–1548. ( 10.1111/jeb.12405) [DOI] [PubMed] [Google Scholar]
- 14.Price SA, Claverie T, Near TJ, Wainwright PC. 2015. Phylogenetic insights into the history and diversification of fishes on reefs. Coral Reefs 34, 997–1009. ( 10.1007/s00338-015-1326-7) [DOI] [Google Scholar]
- 15.Bellwood DR, Goatley CHR, Bellwood O. 2016. The evolution of fishes and corals on reefs: form, function and interdependence. Biol. Rev. ( 10.1111/brv.12259) [DOI] [PubMed] [Google Scholar]
- 16.Santini F, Nguyen MTT, Sorenson L, Waltzek TB, Alfaro JWL, Eastman JM, Alfaro ME. 2013. Do habitat shifts drive diversification in teleost fishes? An example from the pufferfishes (Tetraodontidae). J. Evol. Biol. 26, 1003–1018. ( 10.1111/jeb.12112) [DOI] [PubMed] [Google Scholar]
- 17.Betancur RR, Ortí G, Stein AM, Marceniuk AP, Alexander Pyron R. 2012. Apparent signal of competition limiting diversification after ecological transitions from marine to freshwater habitats. Ecol. Lett. 15, 822–830. ( 10.1111/j.1461-0248.2012.01802.x) [DOI] [PubMed] [Google Scholar]
- 18.Kröger B, Yun-Bai Z. 2009. Pulsed cephalopod diversification during the Ordovician. Palaeogeogr. Palaeoclimatol. Palaeoecol. 273, 174–183. ( 10.1016/j.palaeo.2008.12.015) [DOI] [Google Scholar]
- 19.Slater GJ, Price SA, Santini F, Alfaro ME. 2010. Diversity versus disparity and the radiation of modern cetaceans. Proc. R. Soc. B 277, 3097–3104. ( 10.1098/rspb.2010.0408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miya M, et al. 2013. Evolutionary origin of the Scombridae (tunas and mackerels): members of a Paleogene adaptive radiation with 14 other pelagic fish families. PLoS ONE 8, e73535 ( 10.1371/journal.pone.0073535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobato FL, Barneche DR, Siqueira AC, Liedke AMR, Lindner A, Pie MR, Bellwood DR, Floeter SR. 2014. Diet and diversification in the evolution of coral reef fishes. PLoS ONE 9, e102094 ( 10.1371/journal.pone.0102094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blake RW. 2004. Fish functional design and swimming performance. J. Fish Biol. 65, 1193–1222. ( 10.1111/j.1095-8649.2004.00568.x) [DOI] [Google Scholar]
- 23.Frédérich B, Olivier D, Litsios G, Alfaro ME, Parmentier E. 2014. Trait decoupling promotes evolutionary diversification of the trophic and acoustic system of damselfishes. Proc. R. Soc. B 281, 20141047 ( 10.1098/rspb.2014.1047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marramà G, Garbelli C, Carnevale G. 2016. A morphospace for the Eocene fish assemblage of Bolca, Italy: a window into the diversification and ecological rise to dominance of modern tropical marine fishes. Boll. Soc. Pal. It. 55, 11–21. [Google Scholar]
- 25.Friedman M. 2010. Explosive morphological diversification of spiny-finned teleost fishes in the aftermath of the end-Cretaceous extinction. Proc. R. Soc. B 277, 1675–1683. ( 10.1098/rspb.2009.2177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eschmeyer WN, Fricke R, van der Laan R.2016. Catalog of fishes (accessed July 2016). See http://research.calacademy.org/research/ichthyology/catalog/fishcatmain.asp . [DOI] [PubMed]
- 27.Santini F, Carnevale G. 2015. First multilocus and densely sampled timetree of trevallies, pompanos and allies (Carangoidei, Percomorpha) suggests a Cretaceous origin and Eocene radiation of a major clade of piscivores. Mol. Phylogenet. Evol. 83, 33–39. ( 10.1016/j.ympev.2014.10.018) [DOI] [PubMed] [Google Scholar]
- 28.Froese R, Pauly D. 2015. FishBase (version 05/2015). See www.fishbase.org.
- 29.Collar DC, O'Meara BC, Wainwright PC, Near TJ. 2009. Piscivory limits diversification of feeding morphology in centrarchid fishes. Evolution 63, 1557–1573. ( 10.1111/j.1558-5646.2009.00626.x) [DOI] [PubMed] [Google Scholar]
- 30.Bannikov AF. 1990. Fossil carangids and apolectids of the USSR. Moscow, Russia: Nauka. [Google Scholar]
- 31.Blot J. 1969. Les poissons fossiles du Monte Bolca. Classés jusqu'ici dans les familles des Carangidae, Menidae, Ephippidae, Scatophagidae. Stud. Ric. Giaciam. Terz. Bolca 1, 1–525. [Google Scholar]
- 32.Friedman M, Johanson Z, Harrington RC, Near TJ, Graham MR. 2013. An early fossil remora (Echeneoidea) reveals the evolutionary assembly of the adhesion disc. Proc. R. Soc. B 280, 20131200 ( 10.1098/rspb.2013.1200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelditch ML, Swiderski DL, Sheets HD, Fink WL. 2004. Geometric morphometrics for biologists: a primer. San Diego, CA: Academic Press. [Google Scholar]
- 34.Rohlf FJ.2004. TpsDig (version 1.40), a software program for landmark data acquisition. Department of Ecology and Evolution, State University of New York at Stony Brook. See http://life.bio.sunysb.edu/morph/ .
- 35.Eloy C. 2013. On the best design for undulatory swimming. J. Fluid Mech. 717, 48–89. ( 10.1017/jfm.2012.561) [DOI] [Google Scholar]
- 36.Harper DG, Blake RW. 1990. Fast-start performance of rainbow trout Salmo gairdneri and northern pike Esox Lucius. J. Exp. Biol. 150, 321–342. [Google Scholar]
- 37.Frédérich B, Sorenson L, Santini F, Slater GJ, Alfaro ME. 2013. Iterative ecological radiation and convergence during the evolutionary history of damselfishes (Pomacentridae). Am. Nat. 181, 94–113. ( 10.1086/668599) [DOI] [PubMed] [Google Scholar]
- 38.Nelson JS. 2006. Fishes of the world. 4th edn Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 39.Huelsenbeck JP, Nielsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158. ( 10.1080/10635150390192780) [DOI] [PubMed] [Google Scholar]
- 40.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 41.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://R-project.org/. [Google Scholar]
- 42.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 43.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 44.Beaulieu JM, O'Meara B. 2016. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601. ( 10.1093/sysbio/syw022) [DOI] [PubMed] [Google Scholar]
- 45.Rabosky DL. 2010. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824. ( 10.1111/j.1558-5646.2009.00926.x) [DOI] [PubMed] [Google Scholar]
- 46.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer. [Google Scholar]
- 47.Beaulieu JM, Jhwueng D-C, Boettiger C, O'Meara BC. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66, 2369–2383. ( 10.1111/j.1558-5646.2012.01619.x) [DOI] [PubMed] [Google Scholar]
- 48.Adams DC, Otárola-Castillo E. 2013. Geomorph: an r package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399. ( 10.1111/2041-210X.12035) [DOI] [Google Scholar]
- 49.Hammer O, Harper DAT, Ryan PD. 2001. PAST: paleontological Statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9. [Google Scholar]
- 50.Claverie T, Wainwright PC. 2014. A morphospace for reef fishes: elongation is the dominant axis of body shape evolution. PLoS ONE 9, e112732 ( 10.1371/journal.pone.0112732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman M. 2009. Ecomorphological selectivity among marine teleost fishes during the end-Cretaceous extinction. Proc. Natl Acad. Sci. USA 106, 5218–5223. ( 10.1073/pnas.0808468106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross ST. 1986. Resource partitioning in fish assemblages: a review of field studies. Copeia 1986, 352–368. ( 10.2307/1444996) [DOI] [Google Scholar]
- 53.Jennings S, Pinnegar JK, Polunin NVC, Boon TW. 2001. Weak cross-species relationships between body size and trophic level belie powerful size-based trophic structuring in fish communities. J. Anim. Ecol. 70, 934–944. ( 10.1046/j.0021-8790.2001.00552.x) [DOI] [Google Scholar]
- 54.Domeneci P, Blake RW. 1997. The kinematics and performance of fish fast-start swimming. J. Exp. Biol. 200, 1165–1178. [DOI] [PubMed] [Google Scholar]
- 55.Brönmark C, Miner JG. 1992. Predator-induced phenotypical change in body morphology in crucian carp. Science 258, 1348–1350. ( 10.1126/science.258.5086.1348) [DOI] [PubMed] [Google Scholar]
- 56.Zhao S, Brady PC, Gao M, Ian Etheredge R, Kattawar GW, Cummings ME. 2015. Broadband and polarization reflectors in the lookdown, Selene vomer. J. R. Soc. Interface 12, 20141390 ( 10.1098/rsif.2014.1390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Britz R, Johnson GD. 2012. Ontogeny and homology of the skeletal elements that form the sucking disc of remoras (Teleostei, Echeneoidei, Echeneidae). J. Morphol. 273, 1353–1366. ( 10.1002/jmor.20063) [DOI] [PubMed] [Google Scholar]
- 58.Jacobs DK, Haney TA, Louie KD. 2004. Genes, diversity, and geologic process on the Pacific Coast. Annu. Rev. Earth Planet. Sci. 32, 601–652. ( 10.1146/annurev.earth.32.092203.122436) [DOI] [Google Scholar]
- 59.Grant KM, Dickens GR. 2002. Coupled productivity and carbon isotope records in the southwest Pacific Ocean during the late Miocene–early Pliocene biogenic bloom. Palaeogeogr. Palaeoclimatol. Palaeoecol. 187, 61–82. ( 10.1016/S0031-0182(02)00508-4) [DOI] [Google Scholar]
- 60.Santini F, Carnevale G, Sorenson L. 2013. First molecular scombrid timetree (Percomorpha: Scombridae) shows recent radiation of tunas following invasion of pelagic habitat. Ital. J. Zool. 80, 210–221. ( 10.1080/11250003.2013.775366) [DOI] [Google Scholar]
- 61.Santini F, Carnevale G, Sorenson L. 2014. First multi-locus timetree of seabreams and porgies (Percomorpha: Sparidae). Ital. J. Zool. 81, 55–71. ( 10.1080/11250003.2013.878960) [DOI] [Google Scholar]
- 62.Frederich B, Marramà G, Carnevale G, Santini F. 2016. Data from: Non-reef environments impact the diversification of extant jacks, remoras and allies (Carangoidei, Percomorpha). Dryad Digital Repository. 10.5061/dryad.fs618. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Respository: http://dx.doi.org/10.5061/dryad.fs618 [62].